Abstract
Background/Aims
Transforming growth factor beta1 (TGF-β1) is a key cytokine in the production of extracellular matrix. A genetic polymorphism at codon 10 of the TGF-β1 gene is associated with liver fibrosis. We investigated the effect of genetic polymorphisms at codon 10 on the development of alcoholic liver cirrhosis (ALC).
Methods
In total, 119 controls and 182 patients with ALC, were enrolled in the study. Clinical and laboratory data including total lifetime alcohol intake were collected at enrollment. The genotype at codon 10 was determined for each patient by single-strand conformation polymorphism.
Results
There were three types of genetic polymorphism at codon 10: homozygous proline (P/P), heterozygous proline/leucine (P/L), and homozygous leucine (L/L). Among the controls, the proportions of P/P, P/L, and L/L were 26.1%, 44.5%, and 29.4%, respectively in the ALC group, these proportions were 23.1%, 43.4%, and 33.5%, respectively. The genotype distribution did not differ between the controls and the ALC group. In the ALC group, age, total lifetime alcohol intake, and distribution of Child-Pugh class did not differ with the genotype. Of the male patients with ALC (n=164), the proportions of P/P, P/L, and L/L were 20.1%, 44.5%, and 35.4%, respectively the genotype distribution did not differ between the male controls and the male ALC patients.
Conclusions
The genotype at codon 10 in TGF-β1 does not appear to influence the development of ALC. Further study is needed to investigate other genetic factors that influence the development of ALC in patients with chronic alcohol intake.
Keywords: Transforming growth factor beta1, Alcoholic liver cirrhosis, Genetic polymorphism
INTRODUCTION
According to the Korea national health and nutrition examination survey conducted by the Korean Center of Disease Control in 2008, the monthly alcohol consumption rate (rate of alcohol consumption more than once a month) of Korean adults older than 19 years is 58.7%, which is slightly lower than 61.5% in U.S.A. When it is divided to the male and the female, the monthly alcohol consumption rate of the male was 74.8%, which is higher than 68.2% in U.S.A. The heavy alcohol consumption rate (more than 80 g of alcohol drinking on one occasion or drinking more than two times a week) was 20.2%.1,2
Until now, in Korea, studies on liver damage by alcohol consumption have not been conducted in nationwide. According to recent studies, 7-31% of chronic liver diseases are alcoholic liver diseases.3,4 Nevertheless, because drinking rates are substantial in chronic hepatitis B and hepatitis C patients, more patients than expected may be affected by alcohol.
It is well known that alcohol consumption caused inflammation in the liver, and furthermore, it induces liver fibrosis. Alcohol induces liver fibrosis by the following mechanism. First, acetaldehyde which is the metabolite of alcohol increases the synthesis of collagen by directly acting on hepatic stellate cells, alternately, it increases the synthesis of collagen by accelerating the synthesis of transforming growth factor (TGF)-β1 in hepatic stellate cells. Second, reactive oxygen species produced in liver cells by alcohol metabolism induces the synthesis of collagen in hepatic stellate cells. Third, hepatocyte apoptic bodies induced by alcohol are phagocytized by Kupffer cells and hepatic stellate cells, which activates stellate cells and increases TGF-β1.5,6
However, not all drinkers develop liver fibrosis or cirrhosis, and even in heavy drinkers, only approximately 15-20% cases develop liver fibrosis or cirrhosis.5 The risk factors for cirrhosis are drinking habits, the duration of drinking, the content of alcohol, the activity of enzymes involved in alcohol metabolism, gender, estrogen, age, the presence or absence of associated liver diseases, etc.5-7 Among them, as the host factors for the development of alcoholic liver fibrosis, the accumulation of acetaldehyde, acceleration of oxidative stress, and genetic factors involved in the production of inflammatory cytokines have been suggested.5
As described above, TGF-β1 is involved in the mechanism of liver fibrosis induced by alcohol.8,9 The genetic polymorphism of TGF-β1 can effect on the level of its production.10 Therefore, this genetic polymorphism may explain the role of host factors for the progression to alcoholic liver cirrhosis (ALC) in chronic alcohol drinkers. TGF-β1 gene is located in chromosome 19q13. Total 7 regions of genetic polymorphisms are present: -988, -800 and -509 of the promoter region, the nontranslated region +72, codon 10 and 25 of the signal sequence region, and codon 629 of the gene coding region.11-13 Nonetheless, in Koreans and other Asians, the genetic polymorphism was detected only in -509 and codon 10.14-16
Among the polymorphic regions of TGF-β1 gene, codon 10 and 25 of the signal sequence region are involved in pulmonary and liver fibrosis, and they are the regions investigated most extensively.10,12-14,16 However, there is no genetic polymorphism in codon 25 in Korean.14-16 There is 3 genotypes at codon 10; nucleotide sequence CCG/CCG that is proline homozygote (P/P), CCG/CTG that is proline/leucine heterozygote (P/L), and CTG/CTG that is leucine homozygote (L/L).10,17-19
Therefore, we investigated whether the genotype at codon 10 in TGF-β1 gene is associated with the development of ALC.
MATERIALS AND METHODS
Patients
This study was conducted from December 1999 to June 2001 on 119 subjects who were negative for both hepatitis B surface antigen (HBsAg, RIA, General Biologicals Corp, Hsin Chu, Taiwan) and anti-hepatitis C virus (anti-HCV, RIA, General Biologicals Corp) with normal liver function (Control group), and from December 2000 to December 2002 on 182 ALC patients with the significant drinking history and negative for both HBsAg and anti-HCV (Cirrhosis group). The definition of cirrhosis was cases satisfying 2 of the followings: (1) gastric or esophageal varix in the upper gastrointestinal endoscopy, (2) regenerated or cirrhotic liver surface, and (3) splenomegaly in abdominal ultrasonography or computed tomography. The definition of the significant drinking history was that in the male, daily alcohol consumption is more than 80 g, more than 5 times per week, and more than 10 years of drinking. In the female, daily alcohol consumption is more than 40 g, drink more than 5 times per week, and longer than 10 years of drinking.20,21
Collection of clinical data and Estimation of alcohol consumption
The amount of alcohol consumption was measured by individual history taking. The content of inquiry included the average amount of alcohol consumed daily, the frequency of drinking per week, and the duration of drinking. Considering the inaccuracy of the calculation of the amount of alcohol consumption, the total amount of alcohol consumption was categorized as follows: less than 500 kg, 500-999 kg, 1,000 -1,499 kg, and more than 1,500 kg. All clinical data were collected at the time of enrollment. The study protocol was approved by the institutional review board of Gil Hospital, Incheon, Korea.
Examination of the genotype at the codon 10 in TGF-β1 gene
The whole blood sample was collected from all controls and patients. The buffy coat was separated and stored frozen at -80℃. Genomic DNAs were extracted from peripheral blood leukocytes using proteinase K and phenol/chloroform. Polymerase chain reaction (PCR) was performed by using a primer set (sense 5'-TGT TCG CGC TCT CGG CAG T-3', anti-sense 5'-TCA CCA GCT CCA TGT CGA TA-3') that amplified the DNA fragment including codon 10. PCR mixture consisted of MgCl2 1.5 mM/L, KCl 50 mM/L, Tris-HCl 10 mM/L, dNTP 200 M/L, primer 0.5 µM/L, Taq polymerase 1U (Bioneer, Daejeon, Korea), and genomic DNA 50 ng. After initial denaturation at 94℃ for 5 minutes, thermocycling consisted of denaturation at 94℃ for 30 seconds, annealing at 60℃ for 30 seconds, extension at 72℃ for 30 seconds for 35 cycles, and followed by a final extension at 72℃ for 5 minutes.
For the single stranded conformational polymorphism (SSCP) analysis, 2 µL of the PCR product were mixed with 8 µL of denaturing solution containing formamide, 250 mM NaOH, 25 mM EDTA, and 0.05% bromophenol blue. Then the DNA samples were denatured at 94℃ for 5 minutes, dipped in ice and loaded onto a 13.5% polyacrylamide (acrylamide to bisacrylamide ratio of 29:1) gel. Single-stranded DNA migrated through the gel at 40 W power for 6 hours in a 4℃ cold room and visualized by silver staining. The genotypes were determined according to the genotype of SSCP used by Yu et al.22
Statistical analysis
Continuous variables were presented as a mean±standard deviation and compared by the Student's t-test. Categorical variables were presented as the frequency (percentage), and compared by the χ2-test. To compare the continuous variables according to genotypes, ANOVA was used. The χ2-test was used to compare the genotype at the codon 10 between the control group and the cirrhosis group.
A P-value less than 0.05 was considered statistically significant. For statistical analysis, the SPSS version 12.0 (SPSS, Chicago, IL, USA) was used.
RESULTS
Clinical characteristics of the control group and the cirrhosis group
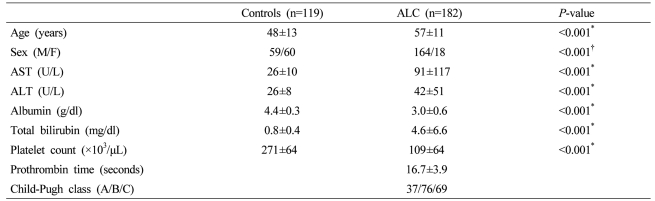
The mean age of the control group was 48±13 years and the mean age of the cirrhosis group was 57±11 years. The ratio of the male and the female of the control group was 59/60 and the ratio of the male and the female of the cirrhosis group was 164/18. The two groups showed significant differences in clinical characteristics (Table 1). Twenty four patients in the cirrhosis group had hepatocellular carcinoma (HCC).
Table 1.
Clinical characteristics of the controls and the patients with alcoholic liver cirrhosis (ALC)
Data represent mean±SD or number.
*Student's t-test, †χ2-test.
ALC, alcoholic liver cirrhosis; ALT, alanine aminotransferase; AST, asparatate aminotransferase; M, male; F, female.
The genetic polymorphism at the codon 10 in TGF-β1 gene
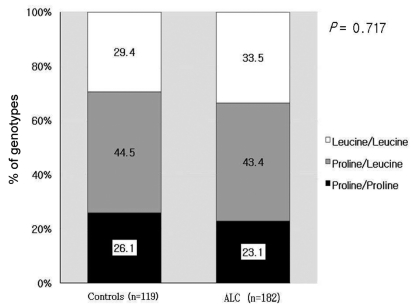
In the control group, the proportions of P/P, P/L, and L/L genotypes were 26.1%, 44.5%, and 29.4%, respectively. In the cirrhosis group, the proportions of P/P, P/L, and L/L genotypes were 23.1%, 43.4%, and 33.5%, respectively. There was no significant difference between the two groups (Fig. 1). Because 24 patients with HCC were included, the distribution of genotype at codon 10 in the cirrhosis group excluding patients with HCC was examined. The proportions of P/P, P/L, and L/L genotypes were 23%, 43%, and 34%, respectively. There was no significant difference between the control group and the cirrhosis group excluding HCC. Since cirrhosis was diagnosed clinically, subgroup analysis was performed in overt cirrhotic patients with Child-Pugh class B and C. The proportions of P/P, P/L, and L/L genotypes were 24.1%, 44.1%, and 31.7%, respectively. There was no significant difference between the control group and the overt cirrhotic patients.
Figure 1.
Comparison of genetic polymorphisms at codon 10 between controls and patients with alcoholic liver cirrhosis (ALC). The genotype distribution at codon 10 did not differ between controls and ALC patients.
Clinical characteristics of each genotype in the cirrhosis group
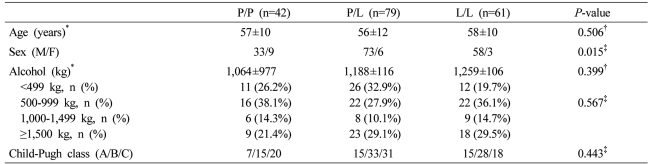
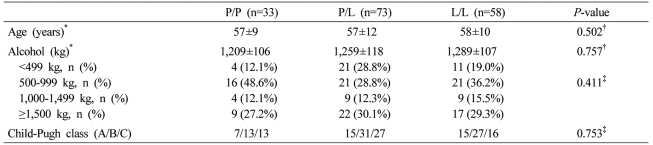
Age and Child-Pugh class of each genotype in the cirrhosis group were not different. In regard to the amount of alcohol consumption, not only the total amount but also the distribution of the subdivided amount of alcohol consumption was not different. Nevertheless, the P/P genotype was 33 males and 9 females, and in comparison with the P/L genotype (male/female: 73/6) and the L/L genotype (male/female: 58/3), females were more abundant (P=0.015, Table 2).
Table 2.
Clinical characteristics of the ALC group according to the genetic polymorphism at codon 10
*Mean±SD, †ANOVA, ‡χ2-test.
M, male; F, female; P/P, proline homozygous genotype; P/L, proline/leucine heterozygous genotype; L/L, leucine homozygous genotype.
The distribution of the genotype at codon 10 in the male cirrhosis group and clinical characteristics of each genotype
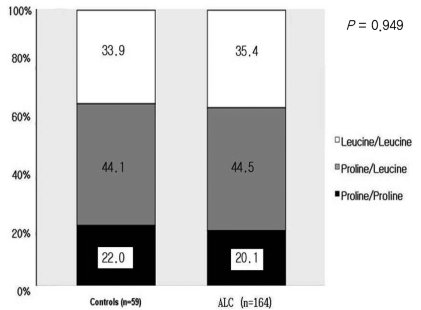
In the male cirrhosis group (n=164), the proportions of P/P, P/L, and L/L genotypes were 20.1%, 44.5%, and 35.4%, respectively. There was no significant difference between the control group and the male cirrhosis group (Fig. 2). In 59 males of the control group, the proportions of P/P, P/L, and L/L genotypes were 22.0%, 44.1%, and 33.9%, respectively, which was not different from the male cirrhosis group. When clinical characteristics according to each genotype were compared in the male cirrhosis group, age, the amount of alcohol consumption, and the distribution of Child-Pugh class were not different (Table 3).
Figure 2.
Comparison of genetic polymorphism at codon 10 between male controls and male patients with ALC. The genotype distribution at codon 10 did not differ between controls and ALC patients.
Table 3.
Clinical characteristics of the male ALC patients according to the genetic polymorphism at codon 10
*Mean±SD, †ANOVA, ‡χ2-test.
P/P, proline homozygous genotype; P/L, proline/leucine heterozygous genotype; L/L, leucine homozygous genotype.
DISCUSSION
Whether the genotype at codon 10 in TGF-β1 gene is associated with elevated TGF-β1 synthesis and fibrosis is controversial. In the L/L genotype, the serum concentration of TGF-β1 is higher and pulmonary fibrosis is more accelerated than other genotypes.10 In liver fibrosis induced by hepatitis B virus (HBV), the L/L genotype accelerate cirrhosis in comparison with other genotypes.22 Nevertheless, contradictory to this, it has been also reported that in the P/P genotype, the serum concentration of TGF-β1 is higher than other genotypes,23-26 and liver fibrosis progresses rapidly in the genotype containing proline.13,26 On the other hand, it has been also reported that the genotype at codon 10 is not associated with liver fibrosis.9,12,27,28 It is thought that the reason of such results contradictory to each other may be the effect of the sample size, races, and the difference of the cause of liver diseases such as HBV, hepatitis C virus (HCV), and alcohol.
In this study, the distribution of the genotype at codon 10 in the cirrhosis group was not different from the control group. This implies that the genetic polymorphism at codon 10 does not exert great effects on the development of ALC as a host factor. In the study reported by Oliver et al9 and Bathgate et al29 the development of ALC was not associated with the genetic polymorphism at the codon 10, which is consistent with the results of this study.
Liver damage induced by alcohol is different according to gender. In the female, liver damage induced by alcohol appears earlier than the male, and females are more vulnerable to alcohol toxicity.7 Because females excrete less alcohol dehydrogenase than males and thus the alcohol metabolism rate in the stomach is lower, the rate of the removal of alcohol from body is lower and thus the concentration of serum alcohol is high, which worsens liver damage.7,30,31 In addition, the female hormone estrogen sensitizes Kupffer cells to endotoxin, alcohol enhances the expression of endotoxin receptor resulting in the acceleration of the synthesis of tumor necrosis factor (TNF), interleukin (IL)-1, IL-6 and other proinflammatory cytokines, and induces inflammation in the liver.7 In addition, estrogen exerts effects on the expression of cytochrom P450 2E1 (CYP2E1) which is one of enzymes of alcohol metabolism.7 Therefore, males were selected from the cirrhosis group, and the genetic polymorphism at codon 10 in the cirrhosis group was compared with that in the control group. However, even in such analysis, there is no difference between the two groups.
This may be due to other host factors, in other words, other genetic polymorphism mediating effects on the activation of alcohol dehydrogenase, acetaldehyde dehydrogenase, and cytochrome P-450 system involved in alcohol metabolism.5,6,30 In addition, the genetic polymorphism of TNF-α or other cytokines gene may play more important roles.5-7,12,28 Additional studies on them are required in the future.
Another factor exerting effects on the development of ALC is the total amount of alcohol consumption. Generally, alcoholic liver diseases are developed in individuals consumed more than 80 g alcohol daily for longer than 10 years.20 In females, individuals consumed smaller amounts (more than 40 g alcohol) daily for longer than 10 years develop alcoholic liver diseases.21 Therefore, in this study, the significant alcohol consumption was defined as more than 80 g in the male and more than 40 g in the female. In addition, since the amount of alcohol consumption was assessed by history taking only, considering the inaccuracy of the measurement of the amount of alcohol consumption, the amount was subdivided. If the amount of alcohol consumption according to the each genotype is not comparable, in other words, if the amount of alcohol consumption is high in particular genotypes, the analysis of the effect at the codon 10 genotype on the development of ALC may not be correct. Therefore, the amount of alcohol consumption according to each genotype was analyzed, and it was not different. This implies that the analysis was performed in the equal condition in regard to the amount of alcohol consumption. In such condition, the genotype at codon 10 could not exert effects on the development of ALC.
It has been reported that TGF-β1 is also associated with the development of HCC.14,19,32 The L/L genotype at codon 10 is associated with the development of HCC in comparison with other genotypes.22,32 In this study, 24 patients with HCC who have underlying cirrhosis were included. It may influence on the analysis of the genotypes at codon 10 for the development of cirrhosis. Therefore, the subgroup analysis of the genetic polymorphism at codon 10 was performed on 158 ALC patients excluding HCC. Nonetheless, the result of 158 ALC patients was not different from that of the control group (data was not shown).
This study has several limitations. First, there is an inaccuracy in estimating the amount of alcohol consumption. It was determined by interviewing with patients or family members. This method may measure the amount of alcohol consumption inaccurately due to the bias of information or the bias of memory.33 Although this study adjusted the bias of the amount of alcohol consumption by categorizing alcohol dose, there will be the possibility of erroneous information still. Second, cirrhosis was diagnosed by endoscopy or imaging methods. The liver biopsy is the most accurate tool for the diagnosis of liver cirrhosis. However, it takes only a small portion of liver, and thus it dose not represent the pathology of the entire liver. Furthermore, it may be fatal in decompensated cirrhotic patients. To compensate clinical diagnosis of cirrhosis, the subgroup analysis was done for comparing the genetic polymorphism at codon 10 between patients with advanced cirrhosis (Child-Pugh class B or C) and the control group. Nevertheless, the analysis also did not show differences. Third, if drinkers with significant alcohol consumption but without liver cirrhosis were recruited as the control group, more accurate effects of the genotype at codon 10 on the development of cirrhosis could be examined. However, those drinkers could not be recruited. Fourth, the number of the control group was not large, and thus they may not represent the genotype at codon 10 in the entire Koreans. However, the distribution of the genotype in the healthy controls in Asia including Korea was not greatly different from the distribution of the genotype in this study.14-16
In conclusion, the polymorphism at the codon 10 in TGF-β1 gene has no effect on the development of ALC. Further studies comparing with the habitual drinking group without alcoholic cirrhosis are required. In addition, studies on other genetic factors associated with the development of ALC are also needed.
Abbreviations
- ALC
alcoholic liver cirrhosis
- ALT
alanine aminotransferase
- anti-HCV
anti-hepatitis C virus
- AST
aspartate aminotransferase
- HBsAg
hepatitis B surface antigen
- IL-1
interleukin-1
- LC
liver cirrhosis
- SSCP
single stranded conformational polymorphism
- TGF-β1
transforming growth factor beta1
- TNF-α
tumor necrosis factor alpha
- F
female
- HCC
hepatocelluar carcinoma
- M
male
- PCR
polymerase chain reaction
References
- 1.Ministry of Health and Welfare; Korea Centers for Disease Control and Prevention. 2008 National Health Statistics-The Fourth Korean National Health and Nutrition Examination Survey (KNHANES IV-2) (2008) Seoul: Korea Centers for Disease Control and Prevention; 2009. pp. 22–23. [Google Scholar]
- 2.U. S. Department of Health and Human Services; Centers for Disease Control and Prevention; National Center for Health Statistics. Health, United States, 2009: with special feature on medical technology. Maryland: National Center for Health Statistics (US); 2010. pp. 286–288. [Google Scholar]
- 3.Lee CH The Korean Socienty of Gastroenterology, editors. Hepatitis. 2nd ed. Seoul: Koonja; 2005. Perspectives of chronic liver disease in Korea; pp. 1–2. [Google Scholar]
- 4.Kim KA. Current status of liver diseases in Korea: toxic and alcoholic liver diseases. Korean J Hepatol. 2009;15(Suppl 6):S29–S33. doi: 10.3350/kjhep.2009.15.S6.S29. [DOI] [PubMed] [Google Scholar]
- 5.Purohit V, Brenner DA. Mechanisms of alcohol-induced hepatic fibrosis: a summary of the Ron Thurman Symposium. Hepatology. 2006;43:872–878. doi: 10.1002/hep.21107. [DOI] [PubMed] [Google Scholar]
- 6.Walsh K, Alexander G. Alcoholic liver disease. Postgrad Med J. 2000;76:280–286. doi: 10.1136/pmj.76.895.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Müller C. Liver, alcohol and gender. Wien Med Wochenschr. 2006;156:523–526. doi: 10.1007/s10354-006-0348-8. [DOI] [PubMed] [Google Scholar]
- 8.Branton MH, Kopp JB. TGF-beta and fibrosis. Microbes Infect. 1999;1:1349–1365. doi: 10.1016/s1286-4579(99)00250-6. [DOI] [PubMed] [Google Scholar]
- 9.Oliver J, Agúndez JA, Morales S, Fernández-Arquero M, Fernández-Gutierrez B, de la Concha EG, et al. Polymorphisms in the transforming growth factor-beta gene (TGF-beta) and the risk of advanced alcoholic liver disease. Liver Int. 2005;25:935–939. doi: 10.1111/j.1478-3231.2005.01150.x. [DOI] [PubMed] [Google Scholar]
- 10.Awad MR, El-Gamel A, Hasleton P, Turner DM, Sinnott PJ, Hutchinson IV. Genotypic variation in the transforming growth factor-beta1 gene: association with transforming growth factor-beta1 production, fibrotic lung disease, and graft fibrosis after lung transplantation. Transplantation. 1998;66:1014–1020. doi: 10.1097/00007890-199810270-00009. [DOI] [PubMed] [Google Scholar]
- 11.Cambien F, Ricard S, Troesch A, Mallet C, Générénaz L, Evans A, et al. Polymorphisms of the transforming growth factor-beta 1 gene in relation to myocardial infarction and blood pressure. The Etude Cas-Témoin de l'Infarctus du Myocarde (ECTIM) Study. Hypertension. 1996;28:881–887. doi: 10.1161/01.hyp.28.5.881. [DOI] [PubMed] [Google Scholar]
- 12.Powell EE, Edwards-Smith CJ, Hay JL, Clouston AD, Crawford DH, Shorthouse C, et al. Host genetic factors influence disease progression in chronic hepatitis C. Hepatology. 2000;31:828–833. doi: 10.1053/he.2000.6253. [DOI] [PubMed] [Google Scholar]
- 13.Gewaltig J, Mangasser-Stephan K, Gartung C, Biesterfeld S, Gressner AM. Association of polymorphisms of the transforming growth factor-beta1 gene with the rate of progression of HCV-induced liver fibrosis. Clin Chim Acta. 2002;316:83–94. doi: 10.1016/s0009-8981(01)00738-0. [DOI] [PubMed] [Google Scholar]
- 14.Kwon OS, Song SH, Ju KT, Chung MG, Park DK, Kim SS, et al. Polymorphism in codons 10 and 25 of the transforming growth factor-beta1 gene in Korean population and in patients with liver cirrhosis and hepatocellular carcinoma. Korean J Gastroenterol. 2003;42:212–219. [PubMed] [Google Scholar]
- 15.Zhang Y, Liu B, Jin M, Ni Q, Liang X, Ma X, et al. Genetic polymorphisms of transforming growth factor-beta1 and its receptors and colorectal cancer susceptibility: a population-based case-control study in China. Cancer Lett. 2009;275:102–108. doi: 10.1016/j.canlet.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 16.Kikuchi K, Tanaka A, Matsushita M, Kitazawa E, Hosoya N, Kawashima Y, et al. Genetic polymorphisms of transforming growth factor beta-1 promoter and primary biliary cirrhosis in Japanese patients. Ann N Y Acad Sci. 2007;1110:15–22. doi: 10.1196/annals.1423.003. [DOI] [PubMed] [Google Scholar]
- 17.Blobe GC, Schiemann WP, Lodish HF. Role of transforming growth factor beta in human disease. N Engl J Med. 2000;342:1350–1358. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- 18.Jakowlew SB, Mead JE, Danielpour D, Wu J, Roberts AB, Fausto N. Transforming growth factor-beta (TGF-beta) isoforms in rat liver regeneration: messenger RNA expression and activation of latent TGF-beta. Cell Regul. 1991;2:535–548. doi: 10.1091/mbc.2.7.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bissell DM, Roulot D, George J. Transforming growth factor beta and the liver. Hepatology. 2001;34:859–867. doi: 10.1053/jhep.2001.28457. [DOI] [PubMed] [Google Scholar]
- 20.McCullough AJ. Alcoholic liver disease. In: Schiff ER, Sorrell MF, Maddrey WC, editors. Schiff's Diseases of the Liver. 8th ed. Vol 2. Philadelphia: Lippincott-Raven; 1999. pp. 941–972. [Google Scholar]
- 21.Norton R, Batey R, Dwyer T, MacMahon S. Alcohol consumption and the risk of alcohol related cirrhosis in women. Br Med J (Clin Res Ed) 1987;295:80–82. doi: 10.1136/bmj.295.6590.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu SK, Kwon OS, Jung HS, Bae KS, Kwon KA, Kim YK, et al. Influence of transforming growth factor-beta1 gene polymorphism at codon 10 on the development of cirrhosis in chronic hepatitis B virus carriers. J Korean Med Sci. 2010;25:564–569. doi: 10.3346/jkms.2010.25.4.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamada Y, Miyauchi A, Goto J, Takagi Y, Okuizumi H, Kanematsu M, et al. Association of a polymorphism of the transforming growth factor-beta1 gene with genetic susceptibility to osteoporosis in postmenopausal Japanese women. J Bone Miner Res. 1998;13:1569–1576. doi: 10.1359/jbmr.1998.13.10.1569. [DOI] [PubMed] [Google Scholar]
- 24.Yamada Y, Okuizumi H, Miyauchi A, Takagi Y, Ikeda K, Harada A. Association of transforming growth factor beta1 genotype with spinal osteophytosis in Japanese women. Arthritis Rheum. 2000;43:452–460. doi: 10.1002/1529-0131(200002)43:2<452::AID-ANR28>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 25.Yokota M, Ichihara S, Lin TL, Nakashima N, Yamada Y. Association of a T29→C polymorphism of the transforming growth factor-beta1 gene with genetic susceptibility to myocardial infarction in Japanese. Circulation. 2000;101:2783–2787. doi: 10.1161/01.cir.101.24.2783. [DOI] [PubMed] [Google Scholar]
- 26.Wang H, Mengsteab S, Tag CG, Gao CF, Hellerbrand C, Lammert F, et al. Transforming growth factor-beta1 gene polymorphisms are associated with progression of liver fibrosis in Caucasians with chronic hepatitis C infection. World J Gastroenterol. 2005;11:1929–1936. doi: 10.3748/wjg.v11.i13.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki S, Tanaka Y, Orito E, Sugauchi F, Hasegawa I, Sakurai M, et al. Transforming growth factor-beta-1 genetic polymorphism in Japanese patients with chronic hepatitis C virus infection. J Gastroenterol Hepatol. 2003;18:1139–1143. doi: 10.1046/j.1440-1746.2003.03161.x. [DOI] [PubMed] [Google Scholar]
- 28.Migita K, Miyazoe S, Maeda Y, Daikoku M, Abiru S, Ueki T, et al. Cytokine gene polymorphisms in Japanese patients with hepatitis B virus infection--association between TGF-beta1 polymorphisms and hepatocellular carcinoma. J Hepatol. 2005;42:505–510. doi: 10.1016/j.jhep.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 29.Bathgate AJ, Pravica V, Perrey C, Hayes PC, Hutchinson IV. Polymorphisms in tumour necrosis factor alpha, interleukin-10 and transforming growth factor beta1 genes and end-stage liver disease. Eur J Gastroenterol Hepatol. 2000;12:1329–1333. doi: 10.1097/00042737-200012120-00011. [DOI] [PubMed] [Google Scholar]
- 30.Baraona E, Abittan CS, Dohmen K, Moretti M, Pozzato G, Chayes ZW, et al. Gender differences in pharmacokinetics of alcohol. Alcohol Clin Exp Res. 2001;25:502–507. [PubMed] [Google Scholar]
- 31.Seitz HK, Egerer G, Simanowski UA, Waldherr R, Eckey R, Agarwal DP, et al. Human gastric alcohol dehydrogenase activity: effect of age, sex, and alcoholism. Gut. 1993;34:1433–1437. doi: 10.1136/gut.34.10.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim YJ, Lee HS, Im JP, Min BH, Kim HD, Jeong JB, et al. Association of transforming growth factor-beta1 gene polymorphisms with a hepatocellular carcinoma risk in patients with chronic hepatitis B virus infection. Exp Mol Med. 2003;35:196–202. doi: 10.1038/emm.2003.27. [DOI] [PubMed] [Google Scholar]
- 33.Midanik L. The validity of self-reported alcohol consumption and alcohol problems: a literature review. Br J Addict. 1982;77:357–382. doi: 10.1111/j.1360-0443.1982.tb02469.x. [DOI] [PubMed] [Google Scholar]