Abstract
Objective:
To investigate the role of SMN1 and SMN2 copy number variation and point mutations in amyotrophic lateral sclerosis (ALS) pathogenesis in a large population.
Methods:
We conducted a genetic association study including 847 patients with ALS and 984 controls. We used multiplexed ligation-dependent probe amplification (MLPA) assays to determine SMN1 and SMN2 copy numbers and examined effects on disease susceptibility and disease course. Furthermore, we sequenced SMN genes to determine if SMN mutations were more prevalent in patients with ALS. A meta-analysis was performed with results from previous studies.
Results:
SMN1 duplications were associated with ALS susceptibility (odds ratio [OR] 2.07, 95% confidence interval [CI] 1.34–3.20, p = 0.001). A meta-analysis with previous data including 3,469 individuals showed a similar effect: OR 1.85, 95% CI 1.18–2.90, p = 0.008). SMN1 deletions and SMN2 copy number status were not associated with ALS. SMN1 or SMN2 copy number variants had no effect on survival or the age at onset of the disease. We found no enrichment of SMN point mutations in patients with ALS.
Conclusions:
Our data provide firm evidence for a role of common SMN1 duplications in ALS, and raise new questions regarding the disease mechanisms involved. Neurology® 2012;78:776–780
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease that selectively affects motor neurons in the spinal cord and motor cortex, leading to progressive paralysis and invariably death. The majority of cases have no familial history of the disease and are said to be sporadic. The pathogenesis of sporadic ALS is thought to be an interplay of genetic and environmental risk factors contributing to increased disease susceptibility. One gene that has been claimed to modulate susceptibility and disease course in ALS is the survival motor neuron (SMN) gene.1 It is present in 2 copies: SMN1 and a centromeric copy SMN2, with about 10% of the biological activity of SMN1.2 Homozygous deletion of SMN1 causes spinal muscular atrophy (SMA), a congenital motor neuron disease, and higher SMN2 copy numbers are associated with milder SMA phenotypes.3 In a minority of cases, SMA is caused by point mutations in SMN1, rather than by homozygous deletion.4
Because of the phenotypic similarities between SMA and ALS, the role of SMN in ALS pathogenesis has been the subject of various studies, several of them reporting significant effects of SMN1 and SMN2 copy numbers on disease susceptibility or on disease duration.1,5–8 The largest study performed so far showed that abnormal (i.e., 1 and 3) SMN1 copy numbers were associated with ALS.8 This investigation did, however, incorporate samples from a prior study, and the results were inconsistent with other reports.5,9 Furthermore, the reported effects of SMN copy numbers on disease duration have been inconsistent. The role of SMN mutations in ALS has never been investigated.
We performed a large-scale association study to determine the effect of SMN1 and SMN2 copy numbers on disease susceptibility and on disease course. We genotyped a genetically homogeneous population that had not been included in previous reports. In addition, we carried out a comprehensive mutation screen to examine the role of SMN coding sequence mutations in ALS.
METHODS
Patients with ALS and healthy volunteers participating in this study were recruited in the outpatient clinic for motor neuron diseases of the Utrecht University Medical Center, or were part of a population-based study on ALS in the Netherlands. This population has been described in detail elsewhere.10 Patients and controls participating in previous studies on SMN were excluded from the copy number analyses. Patients with ALS had no family history of the disease and all fulfilled the 1994 El Escorial criteria for probable or definite ALS.11 All participants gave written informed consent. Genomic DNA of patients with ALS and controls was isolated in the same laboratory, using a salting-out procedure. In total we included 847 patients with ALS (57% male) and 984 controls (52% male) in the copy number analyses and 975 patients with ALS (60% male) and 1,044 controls (53% male) in the mutation screen (table 1).
Table 1.
Patient characteristics
Abbreviation: ALS = amyotrophic lateral sclerosis.
Multiplexed ligation-dependent probe amplification (MLPA) assays were run using standard protocols (www.mlpa.com). We used the SALSA P060 MLPA kit (MRC Holland, the Netherlands), containing 2 probes specifically targeted to SMN1, 2 probes targeted to SMN2, and control probes targeted to other chromosomal loci for normalization and assay quality control. A total of 50–100 ng of genomic DNA was used in each MLPA assay. Data normalization and analysis were performed with GeneMarker software (SoftGenetics, State College, PA) using standard parameters.
To determine the reproducibility of our MLPA assay, we ran 90 samples twice, in separate reactions, and calculated the copy numbers for both replicates of each sample as described below. For the SMN1 probes, the percentage of agreement was 99% (1 of 90 samples had different copy numbers between the 2 replicates), and 98% for SMN2 (2 out of 90 samples showed different copy numbers between replicates).
For mutation screening, we used PCR and sequencing protocols described elsewhere.12 In short, we designed 2 nested primer pairs for each amplicon, amplified exonic sequences and intron-exon boundaries, and sequenced the amplicons using di-deoxy sequencing. Sequencing was done on ABI 3,730 capillary sequencers with Big Dye Terminator v3.1 chemistry (Applied Biosystems, Foster City, CA). Sequence data were imported in PolyPhred software,13 and sequences were visually inspected for heterozygous sites. All putative mutations were confirmed with an independent PCR and sequencing reaction. Functional impact of identified mutations was predicted using PolyPhen software (http://genetics.bwh.harvard.edu/pph/). Primer sequences are available upon request. These primers are not specific to SMN1 or SMN2, but amplify sequences from both genes. Identified mutations cannot, therefore, be mapped specifically to 1 of the 2 genes. We chose this method because approaches to specifically sequence either SMN1 or SMN2 would be extremely laborious, and would only be justified in the case of a suspected association. Two SMA patients with known SMN1 mutations (in the presence of normal SMN2 copy numbers) were used as positive controls. The software called both mutations, thus demonstrating that our method reliably detects mutant alleles at least in a 1:3 ratio.
All statistical procedures were carried out in R 2.10.1 statistical environment (http://www.r-project.org). Because the quantitative measurement of copy number data is prone to systematic bias leading to false-positive associations,14 we used 2 different methods to test SMN1 and SMN2 copy number state for association with ALS susceptibility. First, we determined SMN1 and SMN2 copy number states for each individual using Gaussian mixture modeling with the CNVtools software package in R.14 The mean signal of the 2 probes for each gene was used as the input signal. Gaussian distributions were fitted on the signal intensity distributions and individuals were assigned to copy number states based on the highest a posteriori probability. For SMN1 a 3-component model was used (corresponding to 1, 2 and 3 copies) and for SMN2 a 5-component model was used (corresponding to 0, 1, 2, 3, and 4 copies). These copy number states were then used in a multivariate logistic regression model including SMN1 and SMN2 copy number state and with age at onset and gender as covariates. Secondly, we used a likelihood ratio association test employing CNVtools, using a linear trend model. This method was specifically designed to handle intensity data from quantitative measurements, and allows for differential bias due to possible differences in data quality between cases and controls, causing spurious associations.14 Cox regression was used to test for effect of SMN1 and SMN2 copy number on survival, using age at onset, gender, and site of onset as covariates. For the effect on age at onset, we used Cox regression with gender and site of onset as covariates. For the combined analysis of the different studies, we used the random-effects meta-analysis (DerSimonian-Laird) in the rmeta package in R. We used the Woolf test to test for significant heterogeneity between different studies. In order to test for difference in frequency of SMN mutations between patients and controls, the Fisher exact test (2-sided) was applied.
RESULTS
We included 847 patients with ALS and 984 controls in the copy number analyses (table 1). First we tested both SMN1 and SMN2 copy number states for association with ALS susceptibility using logistic regression. SMN1 duplications (i.e., 3 copies) were significantly associated with ALS (odds ratio [OR] = 2.07, 95% confidence interval [CI] = 1.34–3.20, p = 0.001) (table 2). There was no effect of SMN1 deletions (i.e., 1 SMN1 copy) on disease susceptibility (OR = 0.83, 95% CI = 0.42–1.65, p = 0.60), and we found no effect of SMN2 copy number states on disease susceptibility. The removal of SMN2 copy number states from the model that tested SMN1 and vice versa did not change the results (not shown).
Table 2.
SMN1 and SMN2 copy number association analysis
Abbreviations: ALS = amyotrophic lateral sclerosis; CI = confidence interval; OR = odds ratio; p = logistic regression p value.
We then used a likelihood ratio test for association to investigate the significance of SMN copy numbers on ALS susceptibility. Using this approach, we obtained similar results for both genes: p = 0.001 for SMN1, p = 0.99 for SMN2, thus corroborating the results obtained from the logistic regression model.
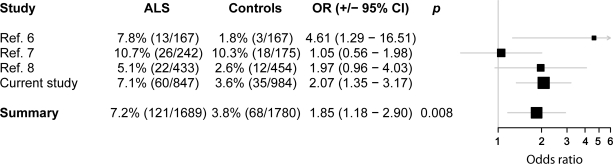
When combining data from the current study with previously published data, SMN1 duplications were significantly associated with ALS (OR = 1.85, 95% CI = 1.18–2.90, p = 0.008) (figure). There was no significant heterogeneity between different studies (p = 0.17, Woolf test). The effect of SMN1 and SMN2 deletions was not significant (SMN1: OR = 2.23, 95% CI = 0.93–5.32, p = 0.07; SMN2: OR = 1.41, 95% CI = 0.87–2.27, p = 0.16).
Figure. Combined analysis of SMN1 duplications.
The table shows per study the frequency and absolute numbers and risk estimate of SMN1 duplications. ALS = amyotrophic lateral sclerosis; CI = confidence interval; OR = odds ratio; p = logistic.
Then the effect of SMN copy number on phenotypic characteristics was tested. Complete clinical data were available for 814 patients with ALS. We found no effect of either deletions or duplications of SMN1 or SMN2 on disease duration or age at onset (p values >0.1, not shown).
Sequence data were available for 975 patients with ALS and 1,044 controls. In our mutation screen we identified 15 heterozygous sequence variants, excluding known single nucleotide polymorphisms (dbSNP build 129) (table e-1 on the Neurology® Web site at www.neurology.org). There was no enrichment of SMN mutations in patients with ALS: p = 0.30, 2-tailed Fisher exact test. Two variants, G26D and P198L, will result in amino acid changes. A G26D mutation was identified in 1 patient with ALS; this variant is predicted to be “possibly damaging” by PolyPhen. A P198L mutation was found in a healthy control, and is predicted to be “probably damaging.” Both individuals had “normal” (i.e., 2) copy number of both SMN genes.
DISCUSSION
We found a significant effect of SMN1 duplications on ALS susceptibility, which can be considered a major risk factor for sporadic ALS. The effect size of SMN1 duplications, obtained from the combined analysis of almost 3,500 individuals, is one of the highest, compared to other established risk factors for ALS. The large sample size and the robust techniques used for data acquisition and analysis provide confidence in the solidity of the data.
Our results are in line with previous reports that showed that abnormal SMN1 copy numbers are associated with an increased susceptibility to ALS. In fact, all 3 studies measuring SMN1 duplications in the context of ALS reported higher frequency in patients with ALS, compared to controls (see figure); in 2 of the 3 studies this difference was statistically significant.6–8 This is reflected by a significant result in the combined analysis, without evidence for heterogeneity between the different studies. With these data, we conclude there is now firm evidence for an association between SMN1 duplications and ALS susceptibility.
In the current study we find no evidence for an effect of SMN1 deletions on disease susceptibility. Additionally, the results of the combined analysis weaken previous reported associations of SMN1 deletions and ALS. The same holds true for the effect of SMN2 copy number variants, which was not confirmed in our study. One possible explanation for the nonreplication of previous reported results is the fact that studies using quantitative PCR-derived data, such as in copy number studies, are inherently sensitive to different sources of bias.14 For example, DNA isolation and handling can introduce differential bias between cases and controls, leading to spurious associations.15 For this reason we used MLPA, a technique that allows simultaneous quantification of multiple probes with one primer pair.16 This reduces the chances of spurious results due to different PCR reaction properties of target and normalization primers, as might be obtained with standard quantitative PCR assays. Furthermore, we used a recently developed statistical framework, providing a robust means of association testing of copy number data. The fact that we obtained similar results using different association tests adds to the validity of our data.
In addition to the copy number analyses, we undertook a mutation screen to examine the role of SMN point mutations in ALS pathogenesis, prompted by the fact that a minority of SMA cases are caused by subtle point mutations rather than by gross deletions of SMN1. The role of SMN mutations in ALS has not been studied before. We find no evidence for a role of point mutations in ALS pathogenesis. Although our approach does not discriminate between mutations in SMN1 and SMN2, and therefore a differential clustering of SMN1 mutations in patients with ALS and SMN2 mutations in controls cannot be excluded, the lack of enrichment of mutations in patients with ALS does make it very unlikely that these mutations contribute significantly to ALS pathogenesis.
The mechanism of SMN1 duplications on disease susceptibility remains elusive. Given the initial hypothesis that low SMN protein levels increase risk for ALS, in analogy to SMA, this is counterintuitive. An explanation would be that SMN1 duplications actually produce lower amounts of SMN protein. A total of 5%–15% of copy number variants are negatively correlated with gene expression17 but it is not known if such a relation exists for SMN. If a lower amount of SMN protein mediates ALS risk, one would also expect an overrepresentation of SMN1 deletions in ALS. An alternative and intriguing explanation would be that SMN duplications produce higher SMN protein levels that are toxic to motor neurons, but to our knowledge there is as yet no experimental evidence to support this theory. Another explanation is that SMN1 duplications confer a risk factor independent of SMN protein, e.g., because of duplication of other local genomic regions, which we cannot exclude. Our MLPA assay did not include probes that are targeted at SMN-flanking genes. Therefore, this question remains to be answered and could be subject to further research in the future.
Our data provide firm evidence that SMN1 copy number variants are involved in ALS pathogenesis. Further research is needed to explain the increased risk of SMN1 duplications, rather than deletions. Given the large effect size, as compared to other established risk factors for sporadic ALS, SMN duplications are an important risk factor for ALS and further functional studies would be highly justified.
Supplementary Material
ACKNOWLEDGMENT
The authors thank all patients and healthy volunteers participating in this study.
GLOSSARY
- ALS
amyotrophic lateral sclerosis
- CI
confidence interval
- MLPA
multiplexed ligation-dependent probe amplification
- OR
odds ratio
- SMA
spinal muscular atrophy
Footnotes
Editorial, page 770.
Supplemental data at www.neurology.org
AUTHOR CONTRIBUTIONS
Dr. Blauw: drafting/revising the manuscript, study concept or design, analysis or interpretation of data, acquisition of data, statistical analysis, study supervision. Dr. Barnes: analysis or interpretation of data, statistical analysis. Dr. Van Vught: analysis or interpretation of data, contribution of vital reagents/tools/patients, acquisition of data. Dr. Van Rheenen: analysis or interpretation of data, acquisition of data. M. Verheul: study concept or design, acquisition of data. Dr. Cuppen: study concept or design, analysis or interpretation of data, acquisition of data. Dr. Veldink: drafting/revising the manuscript, study concept or design, analysis or interpretation of data, acquisition of data, statistical analysis, study supervision, obtaining funding. Dr. Van Den Berg: drafting/revising the manuscript, study concept or design, analysis or interpretation of data, acquisition of data, study supervision, obtaining funding.
DISCLOSURE
Dr. Blauw, Dr. Barnes, Dr. Van Vught, Dr. Van Rheenen, M. Verheul, Dr. Cuppen, and Dr. Veldink report no disclosures. Dr. van den Berg received travel grants and consultancy fees from Baxter; serves on scientific advisory boards for ARISLA (the Italian ALS Association), Prinses Beatrix Fonds, Theirry Latran Foundation, and Biogen Idec; serves as a consultant for and has received funding for travel from Baxter International Inc.; serves on the editorial board of Amyotrophic Lateral Sclerosis; and receives research support from the Prinses Beatrix Fonds, Netherlands ALS Foundation, VSB Fonds, Adessium Foundation, and the European Union.
REFERENCES
- 1. Corcia P, Camu W, Praline J, Gordon P, Vourch P, Andres C. The importance of the SMN genes in the genetics of sporadic ALS. Amyotroph Lateral Scler 2009; 10: 436– 440 . [DOI] [PubMed] [Google Scholar]
- 2. Lorson CL, Rindt H, Shababi M. Spinal muscular atrophy: mechanisms and therapeutic strategies. Hum Mol Genet 2010; 19: R111– 118 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gavrilov DK, Shi X, Das K, Gilliam TC, Wang CH. Differential SMN2 expression associated with SMA severity. Nat Genet 1998; 20: 230– 231 . [DOI] [PubMed] [Google Scholar]
- 4. Rochette CF, Surh LC, Ray PN, et al. Molecular diagnosis of non-deletion SMA patients using quantitative PCR of SMN exon 7. Neurogenetics 1997; 1: 141– 147 . [DOI] [PubMed] [Google Scholar]
- 5. Veldink JH, van den Berg LH, Cobben JM, et al. Homozygous deletion of the survival motor neuron 2 gene is a prognostic factor in sporadic ALS. Neurology 2001; 56: 749– 752 . [DOI] [PubMed] [Google Scholar]
- 6. Corcia P, Mayeux-Portas V, Khoris J, et al. Abnormal SMN1 gene copy number is a susceptibility factor for amyotrophic lateral sclerosis. Ann Neurol 2002; 51: 243– 246 . [DOI] [PubMed] [Google Scholar]
- 7. Veldink JH, Kalmijn S, Van der Hout AH, et al. SMN genotypes producing less SMN protein increase susceptibility to and severity of sporadic ALS. Neurology 2005; 65: 820– 825 . [DOI] [PubMed] [Google Scholar]
- 8. Corcia P, Camu W, Halimi J-M, et al. SMN1 gene, but not SMN2, is a risk factor for sporadic ALS. Neurology 2006; 67: 1147– 1150 . [DOI] [PubMed] [Google Scholar]
- 9. Veldink JH, Kalmijn S, Van der Hout AH, et al. SMN genotypes producing less SMN protein increase susceptibility to and severity of sporadic ALS. Neurology 2005; 65: 820– 825 . [DOI] [PubMed] [Google Scholar]
- 10. van Es MA, van Vught PW, Blauw HM, et al. Genetic variation in DPP6 is associated with susceptibility to amyotrophic lateral sclerosis. Nat Genet 2008; 40: 29– 31 . [DOI] [PubMed] [Google Scholar]
- 11. Brooks BR. El Escorial World Federation of Neurology criteria for the diagnosis of amyotrophic lateral sclerosis: Subcommittee on Motor Neuron Diseases/Amyotrophic Lateral Sclerosis of the World Federation of Neurology Research Group on Neuromuscular Diseases and the El Escorial “Clinical limits of amyotrophic lateral sclerosis” workshop contributors. J Neurol Sci 1994; 124 (suppl): 96–107 . [DOI] [PubMed] [Google Scholar]
- 12. van Boxtel R, Toonen PW, Verheul M, et al. Improved generation of rat gene knockouts by target-selected mutagenesis in mismatch repair-deficient animals. BMC Genomics 2008; 9: 460 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nickerson DA, Tobe VO, Taylor SL. PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res 1997; 25: 2745– 2751 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barnes C, Plagnol V, Fitzgerald T, et al. A robust statistical method for case-control association testing with copy number variation. Nat Genet 2008; 40: 1245– 1252 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Craddock N, Hurles ME, Cardin N, et al. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature 2010; 464: 713– 720 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res 2002; 30: e57 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stranger BE, Forrest MS, Dunning M, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 2007; 315: 848– 853 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.