Abstract
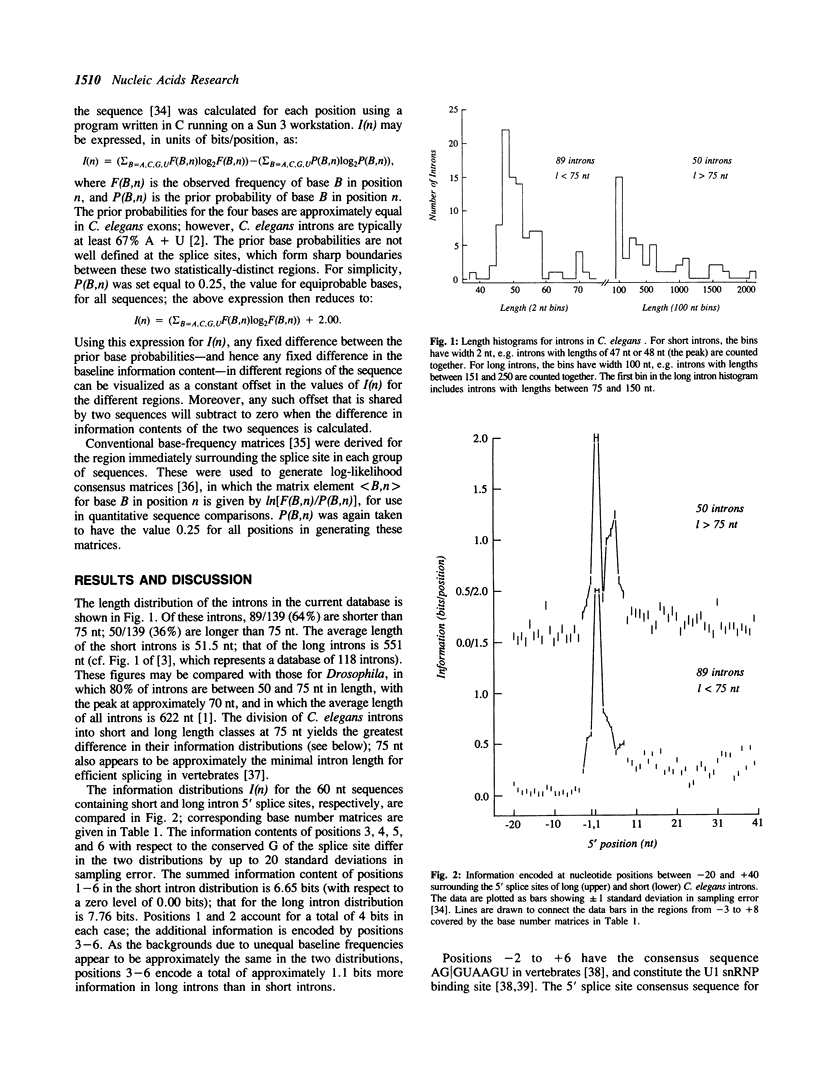
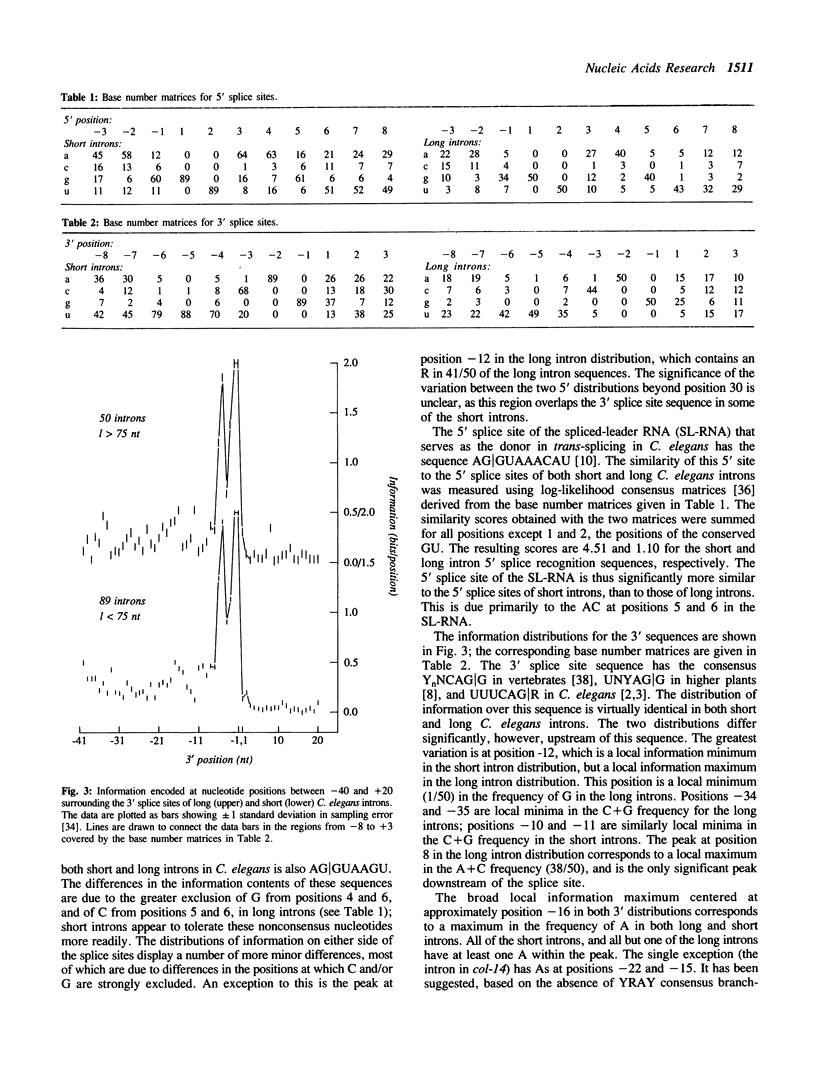
A database of sequences of 139 introns from the nematode Caenorhabditis elegans was analyzed using the information measure of Schneider et al. (1986) J. Mol. Biol. 128: 415-431. Statistically significant information is encoded by at least the first 30 nt and last 20 nt of C. elegans introns. Both the quantity and the distribution of information in the 5' splice site sequences differs between the typical short (length less than 75 nt) and rarer long (length greater than 75 nt) introns, with the 5 sites of long introns containing approximately one bit more information. 3' splice site sequences of long and short C. elegans introns differ significantly in the region between -20 and -10 nt.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barstead R. J., Waterston R. H. The basal component of the nematode dense-body is vinculin. J Biol Chem. 1989 Jun 15;264(17):10177–10185. [PubMed] [Google Scholar]
- Bektesh S. L., Hirsh D. I. C. elegans mRNAs acquire a spliced leader through a trans-splicing mechanism. Nucleic Acids Res. 1988 Jun 24;16(12):5692–5692. doi: 10.1093/nar/16.12.5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benian G. M., Kiff J. E., Neckelmann N., Moerman D. G., Waterston R. H. Sequence of an unusually large protein implicated in regulation of myosin activity in C. elegans. Nature. 1989 Nov 2;342(6245):45–50. doi: 10.1038/342045a0. [DOI] [PubMed] [Google Scholar]
- Berg O. G., von Hippel P. H. Selection of DNA binding sites by regulatory proteins. Statistical-mechanical theory and application to operators and promoters. J Mol Biol. 1987 Feb 20;193(4):723–750. doi: 10.1016/0022-2836(87)90354-8. [DOI] [PubMed] [Google Scholar]
- Blumenthal T., Thomas J. Cis and trans mRNA splicing in C. elegans. Trends Genet. 1988 Nov;4(11):305–308. doi: 10.1016/0168-9525(88)90107-2. [DOI] [PubMed] [Google Scholar]
- Brown J. W. A catalogue of splice junction and putative branch point sequences from plant introns. Nucleic Acids Res. 1986 Dec 22;14(24):9549–9559. doi: 10.1093/nar/14.24.9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox G. N., Fields C., Kramer J. M., Rosenzweig B., Hirsh D. Sequence comparisons of developmentally regulated collagen genes of Caenorhabditis elegans. Gene. 1989;76(2):331–344. doi: 10.1016/0378-1119(89)90173-x. [DOI] [PubMed] [Google Scholar]
- Cummins C., Anderson P. Regulatory myosin light-chain genes of Caenorhabditis elegans. Mol Cell Biol. 1988 Dec;8(12):5339–5349. doi: 10.1128/mcb.8.12.5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibb N. J., Maruyama I. N., Krause M., Karn J. Sequence analysis of the complete Caenorhabditis elegans myosin heavy chain gene family. J Mol Biol. 1989 Feb 5;205(3):603–613. doi: 10.1016/0022-2836(89)90229-5. [DOI] [PubMed] [Google Scholar]
- Finney M., Ruvkun G., Horvitz H. R. The C. elegans cell lineage and differentiation gene unc-86 encodes a protein with a homeodomain and extended similarity to transcription factors. Cell. 1988 Dec 2;55(5):757–769. doi: 10.1016/0092-8674(88)90132-8. [DOI] [PubMed] [Google Scholar]
- Goodall G. J., Filipowicz W. The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell. 1989 Aug 11;58(3):473–483. doi: 10.1016/0092-8674(89)90428-5. [DOI] [PubMed] [Google Scholar]
- Hawkins J. D. A survey on intron and exon lengths. Nucleic Acids Res. 1988 Nov 11;16(21):9893–9908. doi: 10.1093/nar/16.21.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. Y., Barrios L. A., Vonkhorporn P., Honda S., Albertson D. G., Hecht R. M. Genomic organization of the glyceraldehyde-3-phosphate dehydrogenase gene family of Caenorhabditis elegans. J Mol Biol. 1989 Apr 5;206(3):411–424. doi: 10.1016/0022-2836(89)90490-7. [DOI] [PubMed] [Google Scholar]
- Jacob M., Gallinaro H. The 5' splice site: phylogenetic evolution and variable geometry of association with U1RNA. Nucleic Acids Res. 1989 Mar 25;17(6):2159–2180. doi: 10.1093/nar/17.6.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa H., Gengyo K., McLachlan A. D., Brenner S., Karn J. Paramyosin gene (unc-15) of Caenorhabditis elegans. Molecular cloning, nucleotide sequence and models for thick filament structure. J Mol Biol. 1989 May 20;207(2):311–333. doi: 10.1016/0022-2836(89)90257-x. [DOI] [PubMed] [Google Scholar]
- Karn J., Brenner S., Barnett L. Protein structural domains in the Caenorhabditis elegans unc-54 myosin heavy chain gene are not separated by introns. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4253–4257. doi: 10.1073/pnas.80.14.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay R. J., Russnak R. H., Jones D., Mathias C., Candido E. P. Expression of intron-containing C. elegans heat shock genes in mouse cells demonstrates divergence of 3' splice site recognition sequences between nematodes and vertebrates, and an inhibitory effect of heat shock on the mammalian splicing apparatus. Nucleic Acids Res. 1987 May 11;15(9):3723–3741. doi: 10.1093/nar/15.9.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer J. M., Cox G. N., Hirsh D. Comparisons of the complete sequences of two collagen genes from Caenorhabditis elegans. Cell. 1982 Sep;30(2):599–606. doi: 10.1016/0092-8674(82)90256-2. [DOI] [PubMed] [Google Scholar]
- Kramer J. M., Johnson J. J., Edgar R. S., Basch C., Roberts S. The sqt-1 gene of C. elegans encodes a collagen critical for organismal morphogenesis. Cell. 1988 Nov 18;55(4):555–565. doi: 10.1016/0092-8674(88)90214-0. [DOI] [PubMed] [Google Scholar]
- Krause M., Hirsh D. A trans-spliced leader sequence on actin mRNA in C. elegans. Cell. 1987 Jun 19;49(6):753–761. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M., Wild M., Rosenzweig B., Hirsh D. Wild-type and mutant actin genes in Caenorhabditis elegans. J Mol Biol. 1989 Aug 5;208(3):381–392. doi: 10.1016/0022-2836(89)90503-2. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Reed R. The role of small nuclear ribonucleoprotein particles in pre-mRNA splicing. Nature. 1987 Feb 19;325(6106):673–678. doi: 10.1038/325673a0. [DOI] [PubMed] [Google Scholar]
- Russnak R. H., Candido E. P. Locus encoding a family of small heat shock genes in Caenorhabditis elegans: two genes duplicated to form a 3.8-kilobase inverted repeat. Mol Cell Biol. 1985 Jun;5(6):1268–1278. doi: 10.1128/mcb.5.6.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvato M., Sulston J., Albertson D., Brenner S. A novel calmodulin-like gene from the nematode Caenorhabditis elegans. J Mol Biol. 1986 Aug 5;190(3):281–289. doi: 10.1016/0022-2836(86)90002-1. [DOI] [PubMed] [Google Scholar]
- Savage C., Hamelin M., Culotti J. G., Coulson A., Albertson D. G., Chalfie M. mec-7 is a beta-tubulin gene required for the production of 15-protofilament microtubules in Caenorhabditis elegans. Genes Dev. 1989 Jun;3(6):870–881. doi: 10.1101/gad.3.6.870. [DOI] [PubMed] [Google Scholar]
- Schneider T. D., Stormo G. D., Gold L., Ehrenfeucht A. Information content of binding sites on nucleotide sequences. J Mol Biol. 1986 Apr 5;188(3):415–431. doi: 10.1016/0022-2836(86)90165-8. [DOI] [PubMed] [Google Scholar]
- Sharp P. A. Splicing of messenger RNA precursors. Science. 1987 Feb 13;235(4790):766–771. doi: 10.1126/science.3544217. [DOI] [PubMed] [Google Scholar]
- Spieth J., Denison K., Zucker E., Blumenthal T. The nucleotide sequence of a nematode vitellogenin gene. Nucleic Acids Res. 1985 Oct 11;13(19):7129–7138. doi: 10.1093/nar/13.19.7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G. D. Computer methods for analyzing sequence recognition of nucleic acids. Annu Rev Biophys Biophys Chem. 1988;17:241–263. doi: 10.1146/annurev.bb.17.060188.001325. [DOI] [PubMed] [Google Scholar]
- Thomas J. D., Conrad R. C., Blumenthal T. The C. elegans trans-spliced leader RNA is bound to Sm and has a trimethylguanosine cap. Cell. 1988 Aug 12;54(4):533–539. doi: 10.1016/0092-8674(88)90075-x. [DOI] [PubMed] [Google Scholar]
- Way J. C., Chalfie M. mec-3, a homeobox-containing gene that specifies differentiation of the touch receptor neurons in C. elegans. Cell. 1988 Jul 1;54(1):5–16. doi: 10.1016/0092-8674(88)90174-2. [DOI] [PubMed] [Google Scholar]
- Wiebauer K., Herrero J. J., Filipowicz W. Nuclear pre-mRNA processing in plants: distinct modes of 3'-splice-site selection in plants and animals. Mol Cell Biol. 1988 May;8(5):2042–2051. doi: 10.1128/mcb.8.5.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieringa B., Hofer E., Weissmann C. A minimal intron length but no specific internal sequence is required for splicing the large rabbit beta-globin intron. Cell. 1984 Jul;37(3):915–925. doi: 10.1016/0092-8674(84)90426-4. [DOI] [PubMed] [Google Scholar]
- Yarbrough P. O., Hayden M. A., Dunn L. A., Vermersch P. S., Klass M. R., Hecht R. M. The glyceraldehyde-3-phosphate dehydrogenase gene family in the nematode, Caenorhabditis elegans: isolation and characterization of one of the genes. Biochim Biophys Acta. 1987 Jan 28;908(1):21–33. doi: 10.1016/0167-4781(87)90018-2. [DOI] [PubMed] [Google Scholar]
- Yochem J., Greenwald I. glp-1 and lin-12, genes implicated in distinct cell-cell interactions in C. elegans, encode similar transmembrane proteins. Cell. 1989 Aug 11;58(3):553–563. doi: 10.1016/0092-8674(89)90436-4. [DOI] [PubMed] [Google Scholar]
- Yochem J., Weston K., Greenwald I. The Caenorhabditis elegans lin-12 gene encodes a transmembrane protein with overall similarity to Drosophila Notch. Nature. 1988 Oct 6;335(6190):547–550. doi: 10.1038/335547a0. [DOI] [PubMed] [Google Scholar]
- von Mende N., Bird D. M., Albert P. S., Riddle D. L. dpy-13: a nematode collagen gene that affects body shape. Cell. 1988 Nov 18;55(4):567–576. doi: 10.1016/0092-8674(88)90215-2. [DOI] [PubMed] [Google Scholar]



