Abstract
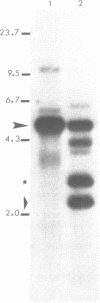
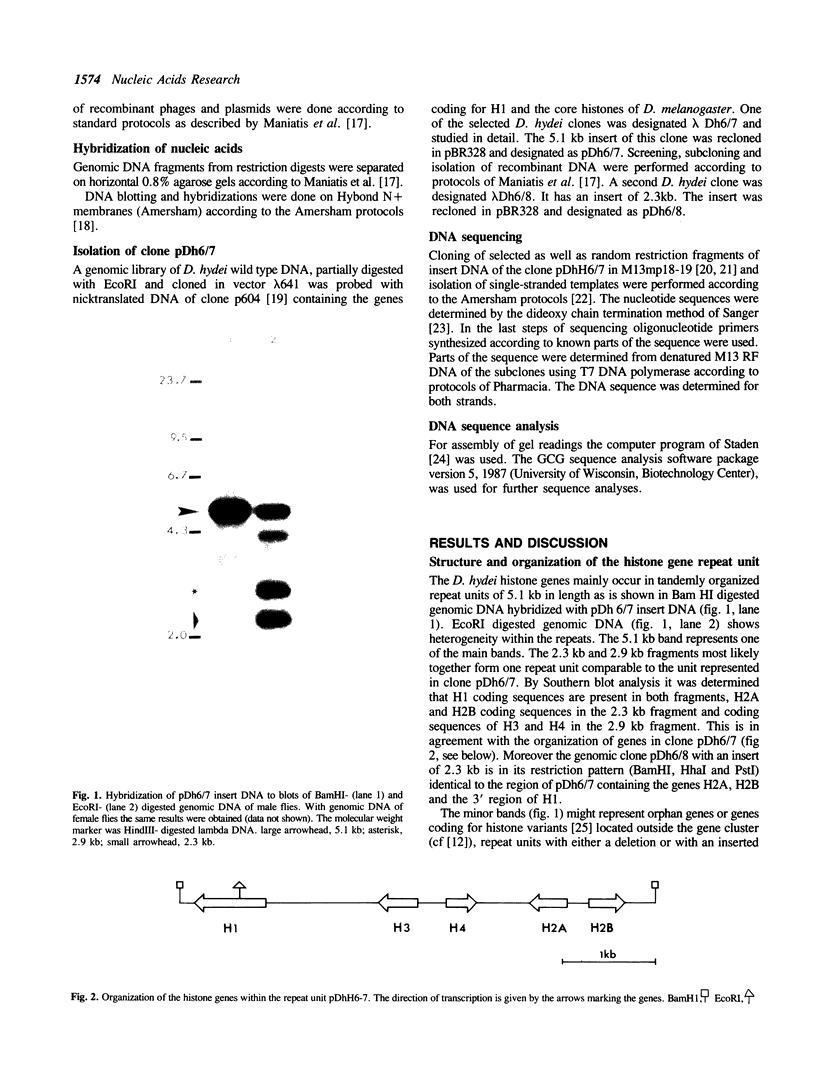
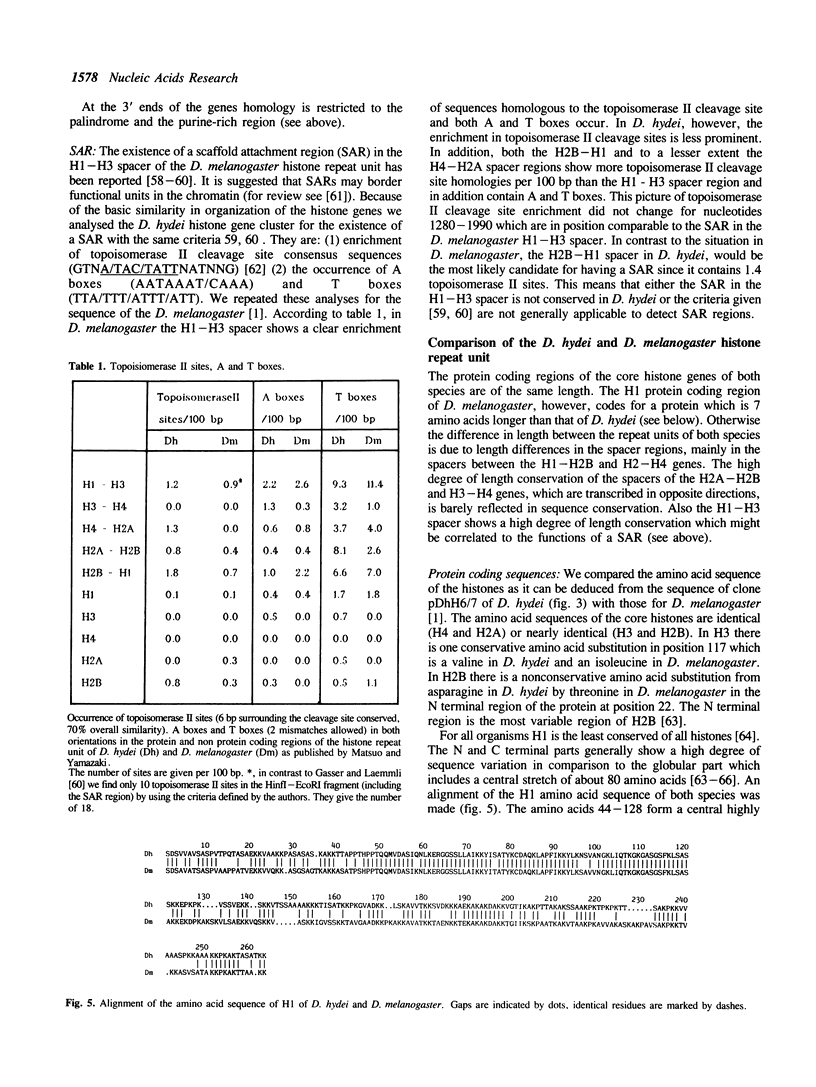
Histone genes in D. hydei are organized in tandemly repeated clusters., accomodating in total 120-140 repeat units. We cloned one of the repeat units and analysed the nucleotide sequence. The repeat unit has a size of 5.1 x 10(3) base-pairs and contains one copy of each of the genes coding for the core histones and one copy coding for the histone H1. In the promoter regions of the genes we identified the presumptive cap sites and TATA boxes. Two additional sequence elements are shared by all five Drosophila hydei histone genes in the cluster. The sequence CCCTCT/G1 is found in the region upstream of the presumptive CAP sites. The sequence element AGTGAA occurs downstream of the presumptive cap sites and is, in contrast to the promoter element, also seen in the histone genes of Drosophila melanogaster. Cell-cycle dependent regulation of transcription of the Drosophila histone genes may be different from that in other eukaryotes since sequence elements involved in the regulation of cell-cycle dependent transcription are absent. Also other regulatory elements for transcription differ from those of other genes. The highly conserved H1-specific promoter sequence AAACACA and the H2B specific promoter sequence ATTTGCAT, which are involved in the cell-cycle dependent transcription of those histone genes in eukaryotes, are missing in the Drosophila genes. However at the 3' end of the genes the palindrome and the purine-rich region, both conserved sequence elements in histone genes of eukaryotes, are present. The spacer regions show a simple sequence organization. The silent site substitution rate between the coding regions of the D. hydei and D. melanogaster histone genes is at least 1.5 times higher for Drosophila than for sea urchin histone genes.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beverley S. M., Wilson A. C. Molecular evolution in Drosophila and the higher Diptera II. A time scale for fly evolution. J Mol Evol. 1984;21(1):1–13. doi: 10.1007/BF02100622. [DOI] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Britten R. J. Rates of DNA sequence evolution differ between taxonomic groups. Science. 1986 Mar 21;231(4744):1393–1398. doi: 10.1126/science.3082006. [DOI] [PubMed] [Google Scholar]
- Bucher P., Trifonov E. N. CCAAT box revisited: bidirectionality, location and context. J Biomol Struct Dyn. 1988 Jun;5(6):1231–1236. doi: 10.1080/07391102.1988.10506466. [DOI] [PubMed] [Google Scholar]
- Busslinger M., Portmann R., Birnsteil M. L. A regulatory sequence near the 3' end of sea urchin histone genes. Nucleic Acids Res. 1979 Jul 11;6(9):2997–3008. doi: 10.1093/nar/6.9.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busslinger M., Portmann R., Irminger J. C., Birnstiel M. L. Ubiquitous and gene-specific regulatory 5' sequences in a sea urchin histone DNA clone coding for histone protein variants. Nucleic Acids Res. 1980 Mar 11;8(5):957–977. doi: 10.1093/nar/8.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busslinger M., Rusconi S., Birnstiel M. L. An unusual evolutionary behaviour of a sea urchin histone gene cluster. EMBO J. 1982;1(1):27–33. doi: 10.1002/j.1460-2075.1982.tb01119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernyshev A. I., Bashkirov V. N., Leibovitch B. A., Khesin R. B. Increase in the number of histone genes in case of their deficiency in Drosophila melanogaster. Mol Gen Genet. 1980;178(3):663–668. doi: 10.1007/BF00337876. [DOI] [PubMed] [Google Scholar]
- Childs G., Maxson R., Cohn R. H., Kedes L. Orphons: dispersed genetic elements derived from tandem repetitive genes of eucaryotes. Cell. 1981 Mar;23(3):651–663. doi: 10.1016/0092-8674(81)90428-1. [DOI] [PubMed] [Google Scholar]
- Cole R. D. A minireview of microheterogeneity in H1 histone and its possible significance. Anal Biochem. 1984 Jan;136(1):24–30. doi: 10.1016/0003-2697(84)90303-8. [DOI] [PubMed] [Google Scholar]
- Coles L. S., Wells J. R. An H1 histone gene-specific 5' element and evolution of H1 and H5 genes. Nucleic Acids Res. 1985 Jan 25;13(2):585–594. doi: 10.1093/nar/13.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cool D., Banfield D., Honda B. M., Smith M. J. Histone genes in three sea star species: cluster arrangement, transcriptional polarity, and analyses of the flanking regions of H3 and H4 genes. J Mol Evol. 1988;27(1):36–44. doi: 10.1007/BF02099728. [DOI] [PubMed] [Google Scholar]
- Cotten M., Gick O., Vasserot A., Schaffner G., Birnstiel M. L. Specific contacts between mammalian U7 snRNA and histone precursor RNA are indispensable for the in vitro 3' RNA processing reaction. EMBO J. 1988 Mar;7(3):801–808. doi: 10.1002/j.1460-2075.1988.tb02878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton S., Wells J. R. A gene-specific promoter element is required for optimal expression of the histone H1 gene in S-phase. EMBO J. 1988 Jan;7(1):49–56. doi: 10.1002/j.1460-2075.1988.tb02782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domier L. L., Rivard J. J., Sabatini L. M., Blumenfeld M. Drosophila virilis histone gene clusters lacking H1 coding segments. J Mol Evol. 1986;23(2):149–158. doi: 10.1007/BF02099909. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Control of eukaryotic messenger RNA synthesis by sequence-specific DNA-binding proteins. 1985 Aug 29-Sep 4Nature. 316(6031):774–778. doi: 10.1038/316774a0. [DOI] [PubMed] [Google Scholar]
- Elgin S. C., Weintraub H. Chromosomal proteins and chromatin structure. Annu Rev Biochem. 1975;44:725–774. doi: 10.1146/annurev.bi.44.070175.003453. [DOI] [PubMed] [Google Scholar]
- Engel J. D., Dodgson J. B. Histone genes are clustered but not tandemly repeated in the chicken genome. Proc Natl Acad Sci U S A. 1981 May;78(5):2856–2860. doi: 10.1073/pnas.78.5.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser S. M., Laemmli U. K. Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D. melanogaster. Cell. 1986 Aug 15;46(4):521–530. doi: 10.1016/0092-8674(86)90877-9. [DOI] [PubMed] [Google Scholar]
- Gasser S. M., Laemmli U. K. The organisation of chromatin loops: characterization of a scaffold attachment site. EMBO J. 1986 Mar;5(3):511–518. doi: 10.1002/j.1460-2075.1986.tb04240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves B. J., Johnson P. F., McKnight S. L. Homologous recognition of a promoter domain common to the MSV LTR and the HSV tk gene. Cell. 1986 Feb 28;44(4):565–576. doi: 10.1016/0092-8674(86)90266-7. [DOI] [PubMed] [Google Scholar]
- Heintz N., Zernik M., Roeder R. G. The structure of the human histone genes: clustered but not tandemly repeated. Cell. 1981 Jun;24(3):661–668. doi: 10.1016/0092-8674(81)90092-1. [DOI] [PubMed] [Google Scholar]
- Hentschel C. C., Birnstiel M. L. The organization and expression of histone gene families. Cell. 1981 Aug;25(2):301–313. doi: 10.1016/0092-8674(81)90048-9. [DOI] [PubMed] [Google Scholar]
- Hentschel C., Irminger J. C., Bucher P., Birnstiel M. L. Sea urchin histone mRNA termini are located in gene regions downstream from putative regulatory sequences. Nature. 1980 May 15;285(5761):147–151. doi: 10.1038/285147a0. [DOI] [PubMed] [Google Scholar]
- Isenberg I. Histones. Annu Rev Biochem. 1979;48:159–191. doi: 10.1146/annurev.bi.48.070179.001111. [DOI] [PubMed] [Google Scholar]
- Kaumeyer J. F., Weinberg E. S. Sequence, organization and expression of late embryonic H3 and H4 histone genes from the sea urchin, Strongylocentrotus purpuratus. Nucleic Acids Res. 1986 Jun 11;14(11):4557–4576. doi: 10.1093/nar/14.11.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedes L. H. Histone genes and histone messengers. Annu Rev Biochem. 1979;48:837–870. doi: 10.1146/annurev.bi.48.070179.004201. [DOI] [PubMed] [Google Scholar]
- LaBella F., Sive H. L., Roeder R. G., Heintz N. Cell-cycle regulation of a human histone H2b gene is mediated by the H2b subtype-specific consensus element. Genes Dev. 1988 Jan;2(1):32–39. doi: 10.1101/gad.2.1.32. [DOI] [PubMed] [Google Scholar]
- Lai Z. C., Childs G. Characterization of the structure and transcriptional patterns of the gene encoding the late histone subtype H1-beta of the sea urchin Strongylocentrotus purpuratus. Mol Cell Biol. 1988 Apr;8(4):1842–1844. doi: 10.1128/mcb.8.4.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. H., Tanimura M., Sharp P. M. An evaluation of the molecular clock hypothesis using mammalian DNA sequences. J Mol Evol. 1987;25(4):330–342. doi: 10.1007/BF02603118. [DOI] [PubMed] [Google Scholar]
- Lifton R. P., Goldberg M. L., Karp R. W., Hogness D. S. The organization of the histone genes in Drosophila melanogaster: functional and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):1047–1051. doi: 10.1101/sqb.1978.042.01.105. [DOI] [PubMed] [Google Scholar]
- Matsuo Y., Yamazaki T. tRNA derived insertion element in histone gene repeating unit of Drosophila melanogaster. Nucleic Acids Res. 1989 Jan 11;17(1):225–238. doi: 10.1093/nar/17.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxson R., Cohn R., Kedes L., Mohun T. Expression and organization of histone genes. Annu Rev Genet. 1983;17:239–277. doi: 10.1146/annurev.ge.17.120183.001323. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Mirkovitch J., Mirault M. E., Laemmli U. K. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell. 1984 Nov;39(1):223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- Mowry K. L., Steitz J. A. Identification of the human U7 snRNP as one of several factors involved in the 3' end maturation of histone premessenger RNA's. Science. 1987 Dec 18;238(4834):1682–1687. doi: 10.1126/science.2825355. [DOI] [PubMed] [Google Scholar]
- Murphy T. J., Blumenfeld M. Nucleotide sequence of a Drosophila melanogaster H1 histone gene. Nucleic Acids Res. 1986 Jul 11;14(13):5563–5563. doi: 10.1093/nar/14.13.5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue M. L., Kedes L. H., Weinberg E. S., Birnstiel M. L. Localization of sequences coding for histone messenger RNA in the chromosomes of Drosophila melanogaster. Chromosoma. 1977 Aug 25;63(2):135–151. doi: 10.1007/BF00292726. [DOI] [PubMed] [Google Scholar]
- Perler F., Efstratiadis A., Lomedico P., Gilbert W., Kolodner R., Dodgson J. The evolution of genes: the chicken preproinsulin gene. Cell. 1980 Jun;20(2):555–566. doi: 10.1016/0092-8674(80)90641-8. [DOI] [PubMed] [Google Scholar]
- Perry M., Thomsen G. H., Roeder R. G. Genomic organization and nucleotide sequence of two distinct histone gene clusters from Xenopus laevis. Identification of novel conserved upstream sequence elements. J Mol Biol. 1985 Oct 5;185(3):479–499. doi: 10.1016/0022-2836(85)90065-8. [DOI] [PubMed] [Google Scholar]
- Roberts S. B., Weisser K. E., Childs G. Sequence comparisons of non-allelic late histone genes and their early stage counterparts. Evidence for gene conversion within the sea urchin late stage gene family. J Mol Biol. 1984 Apr 25;174(4):647–662. doi: 10.1016/0022-2836(84)90088-3. [DOI] [PubMed] [Google Scholar]
- Saigo K., Millstein L., Thomas C. A., Jr The organization of Drosophila melanogaster histone genes. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):815–827. doi: 10.1101/sqb.1981.045.01.100. [DOI] [PubMed] [Google Scholar]
- Sander M., Hsieh T. S. Drosophila topoisomerase II double-strand DNA cleavage: analysis of DNA sequence homology at the cleavage site. Nucleic Acids Res. 1985 Feb 25;13(4):1057–1072. doi: 10.1093/nar/13.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro C., Mermod N., Andrews P. C., Tjian R. A family of human CCAAT-box-binding proteins active in transcription and DNA replication: cloning and expression of multiple cDNAs. Nature. 1988 Jul 21;334(6179):218–224. doi: 10.1038/334218a0. [DOI] [PubMed] [Google Scholar]
- Satchwell S. C., Drew H. R., Travers A. A. Sequence periodicities in chicken nucleosome core DNA. J Mol Biol. 1986 Oct 20;191(4):659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- Schaffner W., Kunz G., Daetwyler H., Telford J., Smith H. O., Birnstiel M. L. Genes and spacers of cloned sea urchin histone DNA analyzed by sequencing. Cell. 1978 Jul;14(3):655–671. doi: 10.1016/0092-8674(78)90249-0. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. On the rate of DNA sequence evolution in Drosophila. J Mol Evol. 1989 May;28(5):398–402. doi: 10.1007/BF02603075. [DOI] [PubMed] [Google Scholar]
- Sierra F., Lichtler A., Marashi F., Rickles R., Van Dyke T., Clark S., Wells J., Stein G., Stein J. Organization of human histone genes. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1795–1799. doi: 10.1073/pnas.79.6.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittman D. B., Chiu I. M., Pan C. J., Cohn R. H., Kedes L. H., Marzluff W. F. Isolation of two clusters of mouse histone genes. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4078–4082. doi: 10.1073/pnas.78.7.4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M., Hunkapiller M., Yuen D., Silvert D., Fristrom J., Davidson N. Cuticle protein genes of Drosophila: structure, organization and evolution of four clustered genes. Cell. 1982 Jul;29(3):1027–1040. doi: 10.1016/0092-8674(82)90466-4. [DOI] [PubMed] [Google Scholar]
- Soldati D., Schümperli D. Structural and functional characterization of mouse U7 small nuclear RNA active in 3' processing of histone pre-mRNA. Mol Cell Biol. 1988 Apr;8(4):1518–1524. doi: 10.1128/mcb.8.4.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Automation of the computer handling of gel reading data produced by the shotgun method of DNA sequencing. Nucleic Acids Res. 1982 Aug 11;10(15):4731–4751. doi: 10.1093/nar/10.15.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson E. C., Erba H. P., Gall J. G. Histone gene clusters of the newt notophthalmus are separated by long tracts of satellite DNA. Cell. 1981 Jun;24(3):639–647. doi: 10.1016/0092-8674(81)90090-8. [DOI] [PubMed] [Google Scholar]
- Sturm R. A., Dalton S., Wells J. R. Conservation of histone H2A/H2B intergene regions: a role for the H2B specific element in divergent transcription. Nucleic Acids Res. 1988 Sep 12;16(17):8571–8586. doi: 10.1093/nar/16.17.8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sures I., Levy S., Kedes L. H. Leader sequences of Strongylocentrotus purpuratus histone mRNAs start at a unique heptanucleotide common to all five histone genes. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1265–1269. doi: 10.1073/pnas.77.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sures I., Lowry J., Kedes L. H. The DNA sequence of sea urchin (S. purpuratus) H2A, H2B and H3 histone coding and spacer regions. Cell. 1978 Nov;15(3):1033–1044. doi: 10.1016/0092-8674(78)90287-8. [DOI] [PubMed] [Google Scholar]
- Tautz D., Renz M. Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res. 1984 May 25;12(10):4127–4138. doi: 10.1093/nar/12.10.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tönjes R., Munk K., Doenecke D. Conserved organization of an avian histone gene cluster with inverted duplications of H3 and H4 genes. J Mol Evol. 1989 Mar;28(3):200–211. doi: 10.1007/BF02102477. [DOI] [PubMed] [Google Scholar]
- Von Holt C., Strickland W. N., Brandt W. F., Strickland M. S. More histone structures. FEBS Lett. 1979 Apr 15;100(2):201–218. doi: 10.1016/0014-5793(79)80337-3. [DOI] [PubMed] [Google Scholar]
- Wells D. E. Compilation analysis of histones and histone genes. Nucleic Acids Res. 1986;14 (Suppl):r119–r149. doi: 10.1093/nar/14.suppl.r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkfein R. J., Connor W., Mezquita J., Dixon G. H. Histone H4 and H2B genes in rainbow trout (Salmo gairdnerii). J Mol Evol. 1985;22(1):1–19. doi: 10.1007/BF02105800. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- van Daal A., White E. M., Gorovsky M. A., Elgin S. C. Drosophila has a single copy of the gene encoding a highly conserved histone H2A variant of the H2A.F/Z type. Nucleic Acids Res. 1988 Aug 11;16(15):7487–7497. doi: 10.1093/nar/16.15.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]