Abstract
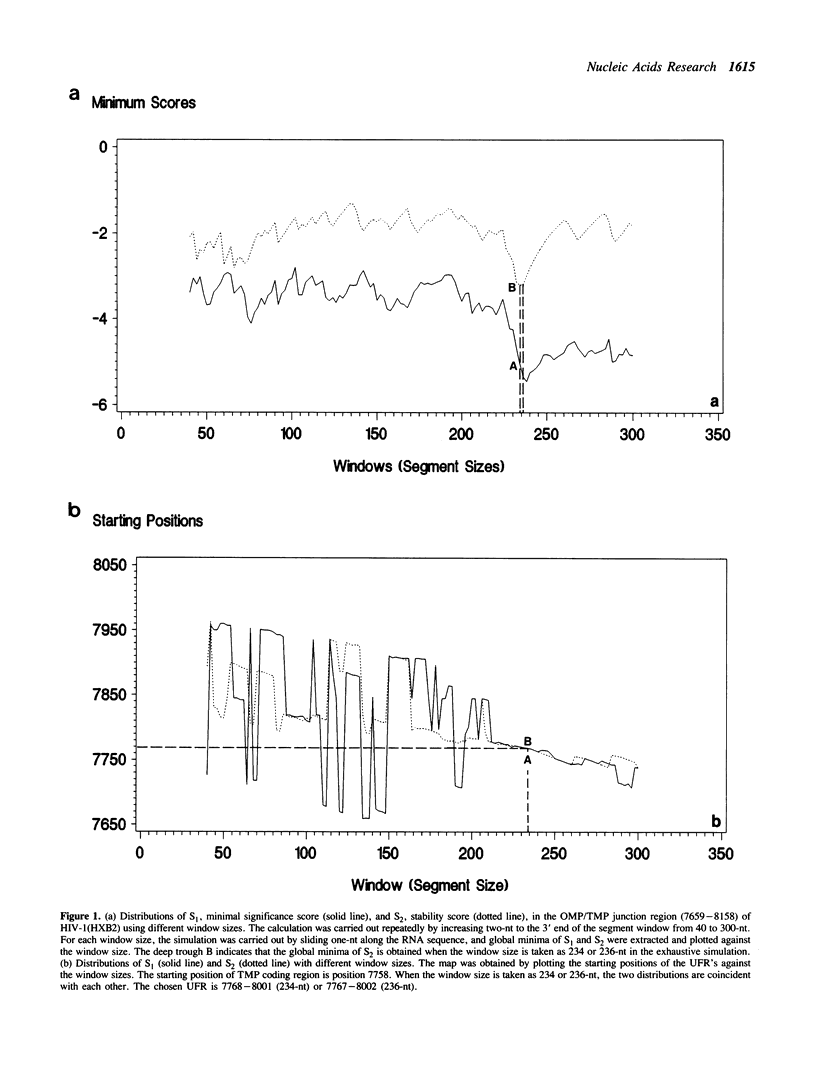
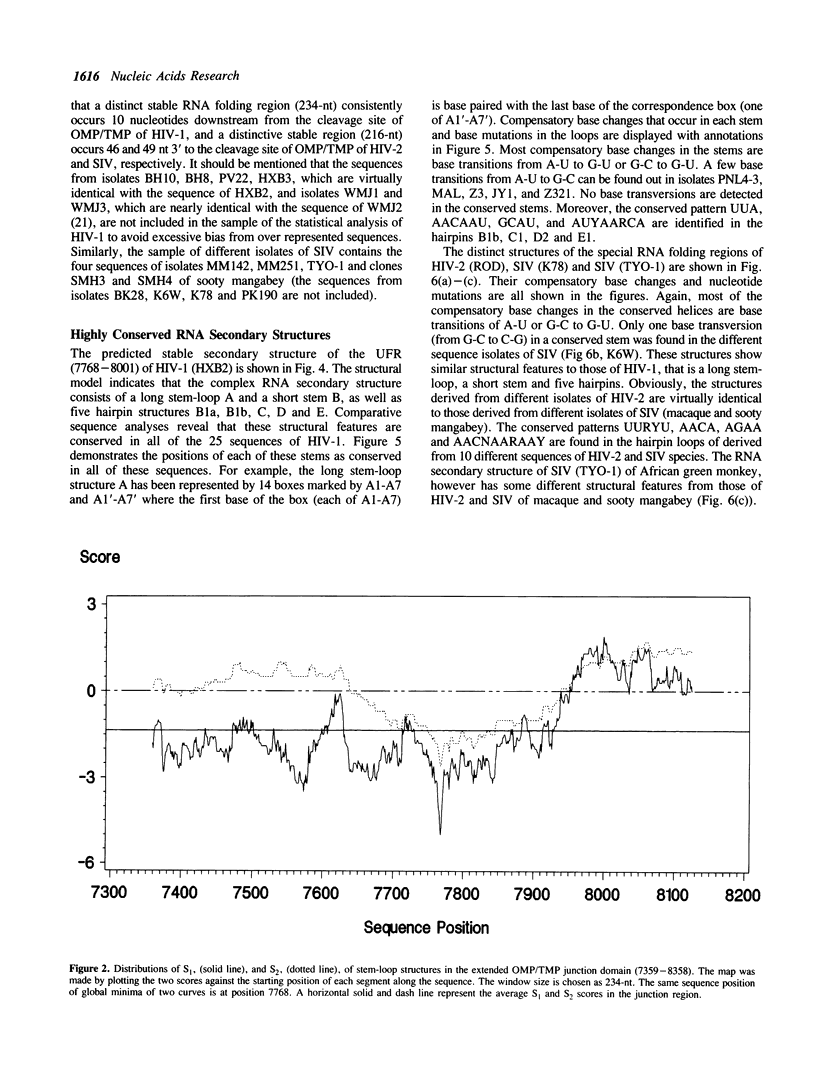
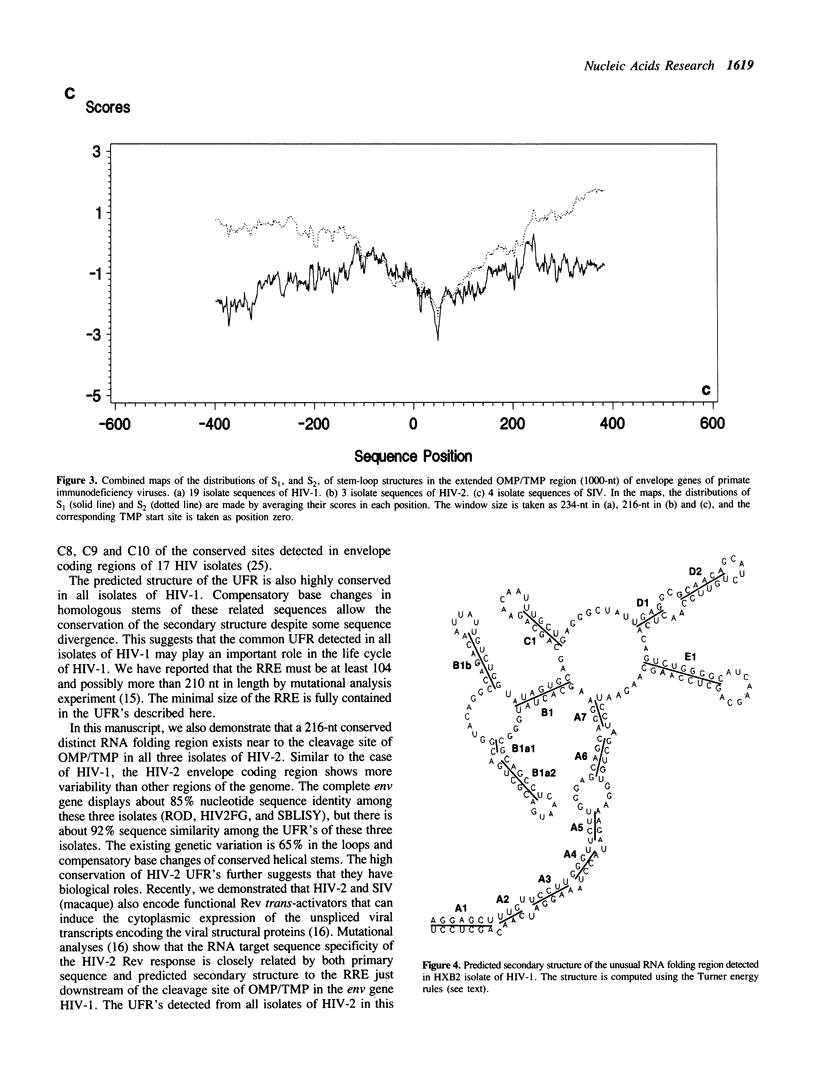
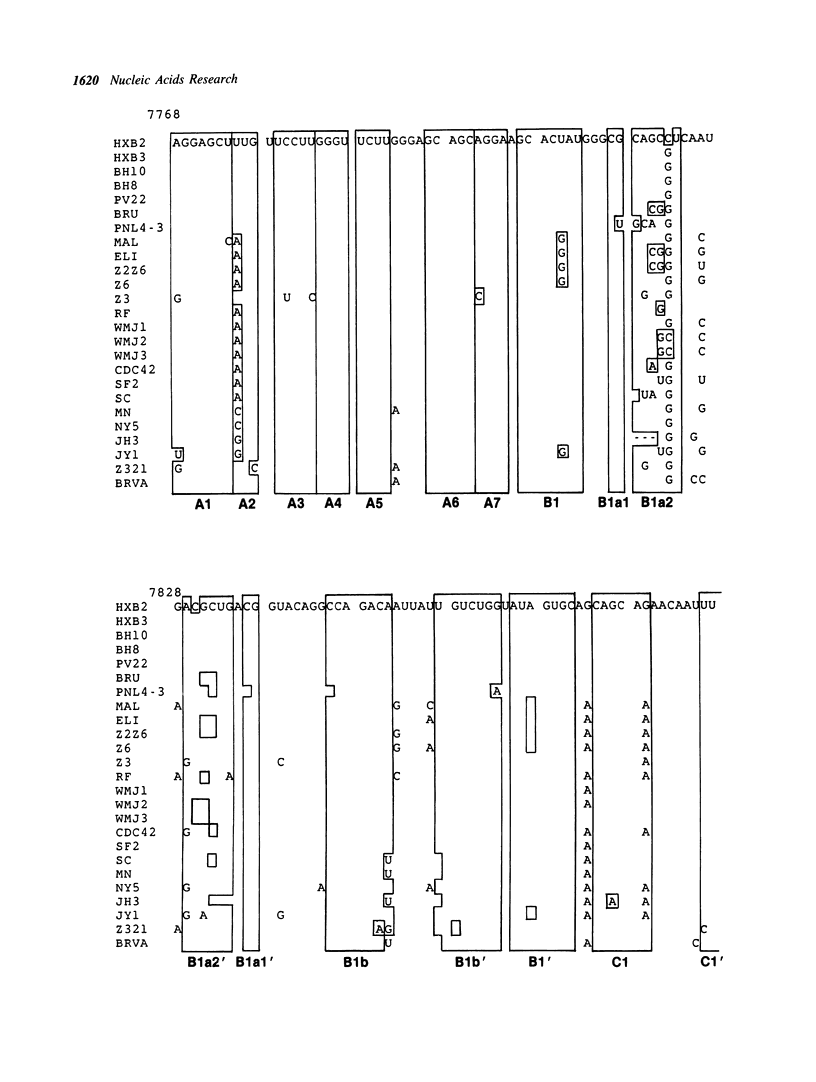
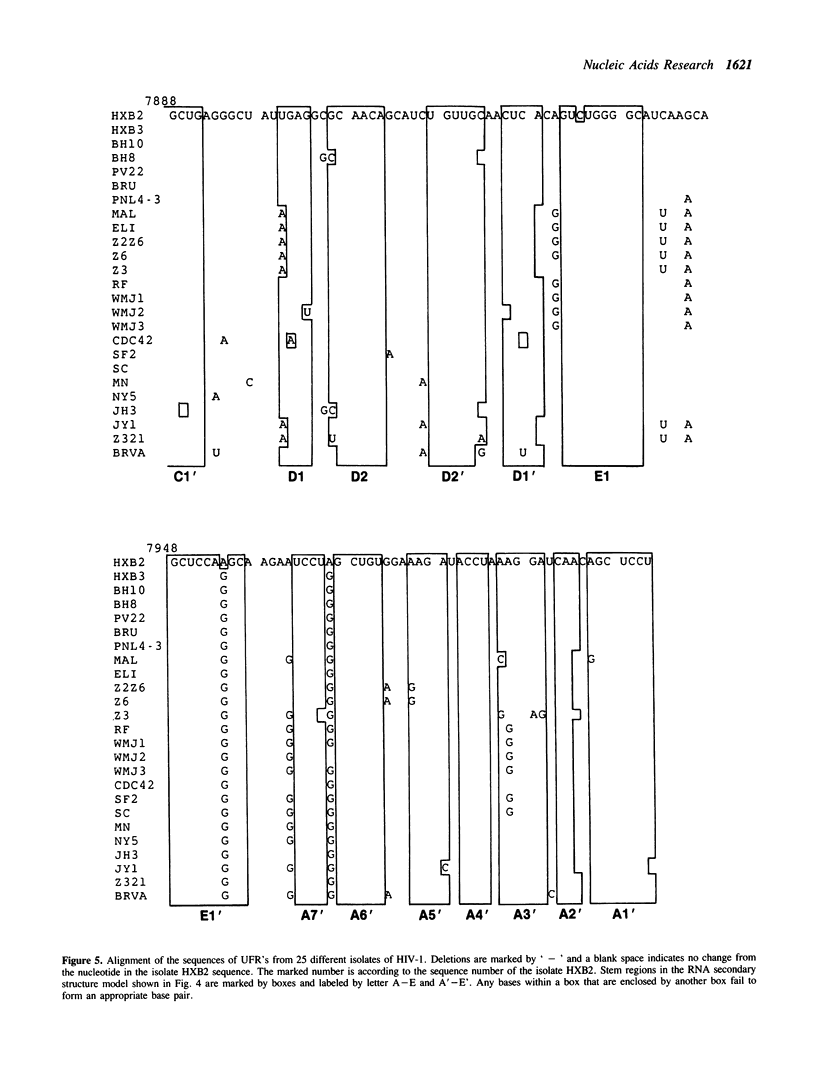
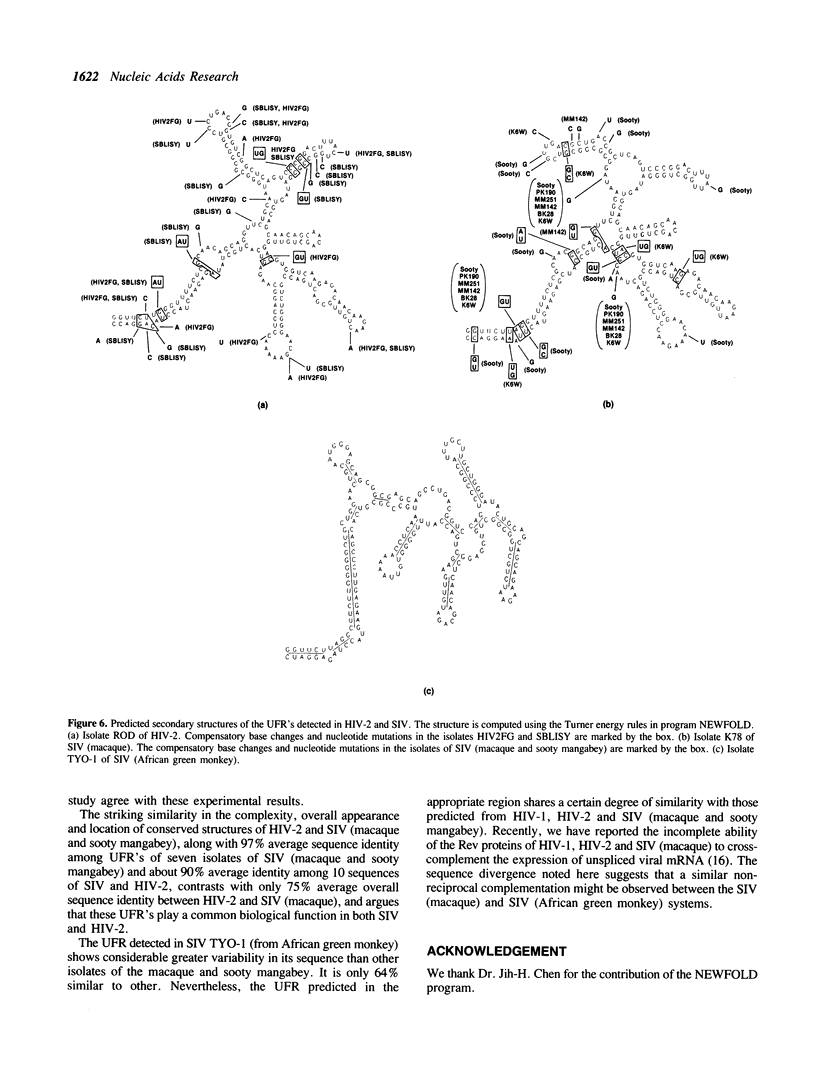
A series of unusual folding regions (UFR) immediately 3' to the cleavage site of the outer membrane protein (OMP) and transmembrane protein (TMP) were detected in the envelope gene RNA of the human immunodeficiency virus (HIV-1, HIV-2) and simian immunodeficiency virus (SIV) by an extensive Monte Carlo simulation. These RNA secondary structures were predicted to be both highly stable and statistically significant. In the calculation, twenty-five different sequence isolates of HIV-1, three isolates of HIV-2 and eight sequences of SIV were included. Although significant sequence divergence occurs in the env coding regions of these viruses, a distinct UFR of 234-nt is consistently located ten nucleotides 3' to the cleavage site of the OMP/TMP in HIV-1, and a 216-nt UFR occurs forty-six and forty-nine nucleotides downstream from the OMP/TMP cleavage site of HIV-2 and SIV, respectively. Compensatory base changes in the helical stem regions of these conserved RNA secondary structures are identified. These results support the hypothesis that these special RNA folding regions are functionally important and suggest that the role of this sequence as the Rev response element (RRE) is mediated by secondary structure as well as primary RNA sequence.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cech T. R., Tanner N. K., Tinoco I., Jr, Weir B. R., Zuker M., Perlman P. S. Secondary structure of the Tetrahymena ribosomal RNA intervening sequence: structural homology with fungal mitochondrial intervening sequences. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3903–3907. doi: 10.1073/pnas.80.13.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayton A. I., Sodroski J. G., Rosen C. A., Goh W. C., Haseltine W. A. The trans-activator gene of the human T cell lymphotropic virus type III is required for replication. Cell. 1986 Mar 28;44(6):941–947. doi: 10.1016/0092-8674(86)90017-6. [DOI] [PubMed] [Google Scholar]
- Emerman M., Guyader M., Montagnier L., Baltimore D., Muesing M. A. The specificity of the human immunodeficiency virus type 2 transactivator is different from that of human immunodeficiency virus type 1. EMBO J. 1987 Dec 1;6(12):3755–3760. doi: 10.1002/j.1460-2075.1987.tb02710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Holland E. C. HIV-1 tat trans-activation requires the loop sequence within tar. Nature. 1988 Jul 14;334(6178):165–167. doi: 10.1038/334165a0. [DOI] [PubMed] [Google Scholar]
- Fisher A. G., Feinberg M. B., Josephs S. F., Harper M. E., Marselle L. M., Reyes G., Gonda M. A., Aldovini A., Debouk C., Gallo R. C. The trans-activator gene of HTLV-III is essential for virus replication. 1986 Mar 27-Apr 2Nature. 320(6060):367–371. doi: 10.1038/320367a0. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauber J., Cullen B. R. Mutational analysis of the trans-activation-responsive region of the human immunodeficiency virus type I long terminal repeat. J Virol. 1988 Mar;62(3):673–679. doi: 10.1128/jvi.62.3.673-679.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobovits A., Smith D. H., Jakobovits E. B., Capon D. J. A discrete element 3' of human immunodeficiency virus 1 (HIV-1) and HIV-2 mRNA initiation sites mediates transcriptional activation by an HIV trans activator. Mol Cell Biol. 1988 Jun;8(6):2555–2561. doi: 10.1128/mcb.8.6.2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S. V., Chen J. H., Currey K. M., Maizel J. V., Jr A program for predicting significant RNA secondary structures. Comput Appl Biosci. 1988 Mar;4(1):153–159. doi: 10.1093/bioinformatics/4.1.153. [DOI] [PubMed] [Google Scholar]
- Le S. Y., Chen J. H., Braun M. J., Gonda M. A., Maizel J. V. Stability of RNA stem-loop structure and distribution of non-random structure in the human immunodeficiency virus (HIV-I). Nucleic Acids Res. 1988 Jun 10;16(11):5153–5168. doi: 10.1093/nar/16.11.5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S. Y., Chen J. H., Chatterjee D., Maizel J. V. Sequence divergence and open regions of RNA secondary structures in the envelope regions of the 17 human immunodeficiency virus isolates. Nucleic Acids Res. 1989 Apr 25;17(8):3275–3288. doi: 10.1093/nar/17.8.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S. Y., Maizel J. V., Jr A method for assessing the statistical significance of RNA folding. J Theor Biol. 1989 Jun 22;138(4):495–510. doi: 10.1016/s0022-5193(89)80047-5. [DOI] [PubMed] [Google Scholar]
- Malim M. H., Hauber J., Le S. Y., Maizel J. V., Cullen B. R. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989 Mar 16;338(6212):254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- Muesing M. A., Smith D. H., Capon D. J. Regulation of mRNA accumulation by a human immunodeficiency virus trans-activator protein. Cell. 1987 Feb 27;48(4):691–701. doi: 10.1016/0092-8674(87)90247-9. [DOI] [PubMed] [Google Scholar]
- Papanicolaou C., Gouy M., Ninio J. An energy model that predicts the correct folding of both the tRNA and the 5S RNA molecules. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):31–44. doi: 10.1093/nar/12.1part1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaie M. R., Benter T., Wong-Staal F. Site-directed mutagenesis of two trans-regulatory genes (tat-III,trs) of HIV-1. Science. 1988 Feb 19;239(4842):910–913. doi: 10.1126/science.3277284. [DOI] [PubMed] [Google Scholar]
- Salser W. Globin mRNA sequences: analysis of base pairing and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):985–1002. doi: 10.1101/sqb.1978.042.01.099. [DOI] [PubMed] [Google Scholar]
- Smith T. F., Srinivasan A., Schochetman G., Marcus M., Myers G. The phylogenetic history of immunodeficiency viruses. Nature. 1988 Jun 9;333(6173):573–575. doi: 10.1038/333573a0. [DOI] [PubMed] [Google Scholar]
- Stall R., McKusick L., Wiley J., Coates T. J., Ostrow D. G. Alcohol and drug use during sexual activity and compliance with safe sex guidelines for AIDS: the AIDS Behavioral Research Project. Health Educ Q. 1986 Winter;13(4):359–371. doi: 10.1177/109019818601300407. [DOI] [PubMed] [Google Scholar]
- Turner D. H., Sugimoto N., Jaeger J. A., Longfellow C. E., Freier S. M., Kierzek R. Improved parameters for prediction of RNA structure. Cold Spring Harb Symp Quant Biol. 1987;52:123–133. doi: 10.1101/sqb.1987.052.01.017. [DOI] [PubMed] [Google Scholar]
- Varmus H. Regulation of HIV and HTLV gene expression. Genes Dev. 1988 Sep;2(9):1055–1062. doi: 10.1101/gad.2.9.1055. [DOI] [PubMed] [Google Scholar]
- Viglianti G. A., Mullins J. I. Functional comparison of transactivation by simian immunodeficiency virus from rhesus macaques and human immunodeficiency virus type 1. J Virol. 1988 Dec;62(12):4523–4532. doi: 10.1128/jvi.62.12.4523-4532.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]



