Abstract
Despite intensive breeding efforts, potato late blight, caused by the oomycete pathogen Phytophthora infestans, remains a threat to potato production worldwide because newly evolved pathogen strains have consistently overcome major resistance genes. The potato RB gene, derived from the wild species Solanum bulbocastanum, confers resistance to most P. infestans strains through recognition of members of the pathogen effector family IPI-O. While the majority of IPI-O proteins are recognized by RB to elicit resistance (e.g. IPI-O1, IPI-O2), some family members are able to elude detection (e.g. IPI-O4). In addition, IPI-O4 blocks recognition of IPI-O1, leading to inactivation of RB-mediated programmed cell death. Here, we report results that elucidate molecular mechanisms governing resistance elicitation or suppression of RB by IPI-O. Our data indicate self-association of the RB coiled coil (CC) domain as well as a physical interaction between this domain and the effectors IPI-O4 and IPI-O1. We identified four amino acids within IPI-O that are critical for interaction with the RB CC domain and one of these amino acids, at position 129, determines hypersensitive response (HR) elicitation in planta. IPI-O1 mutant L129P fails to induce HR in presence of RB while IPI-O4 P129L gains the ability to induce an HR. Like IPI-O4, IPI-O1 L129P is also able to suppress the HR mediated by RB, indicating a critical step in the evolution of this gene family. Our results point to a model in which IPI-O effectors can affect RB function through interaction with the RB CC domain.
Author Summary
The potato late blight pathogen, Phytophthora infestans, is able to rapidly evolve to overcome resistance genes. The pathogen accomplishes this by secreting an arsenal of proteins, termed effectors, that function to modify host cells. Although hundreds of candidate effectors have been identified in P. infestans, their roles in pathogenicity or virulence remains basically unknown. Our results showed that one of these effectors functions to turn off resistance mediated by the potato gene RB. This effector accomplishes this by directly interacting with RB, which likely modifies its ability to turn on host resistance. Further molecular analysis identified two amino acids within the effector that determine interaction, which can assist in developing appropriate disease control strategies.
Introduction
Plant resistance to microbial pathogens is a complex process and includes a variety of constitutive and inducible defense mechanisms [1]. Recognition and response to microbes by plants involve multiple layers of defense. The first, basal defense, relies on the recognition of conserved microbial associated molecular patterns by host receptors [2]. However, the basal defense response can be suppressed by pathogen proteins, termed effectors, that are delivered into the apoplast or plant cell cytoplasm, resulting in effector triggered susceptibility [3]–[5]. Plants have therefore evolved a second layer of defense, called effector triggered immunity (ETI), in which host protein receptors recognize the presence of pathogen effectors and elicit responses to inhibit colonization [6]. ETI relies on resistance (R) proteins to directly or indirectly recognize the presence of specific effector molecules and activate resistance signaling.
The majority of plant R proteins have nucleotide binding (NB) and leucine-rich repeat (LRR) motifs, and can be divided into two sub-classes based on a variable N-terminal domain, which typically contains coiled-coil (CC) motifs or includes homology to the Toll-interleukin-1 receptor (TIR) [7]. Regardless of the motif present, the N-terminal domains of plant NB-LRR proteins are proposed to initiate resistance signaling or pathogen recognition [8]–[14]. The interaction and coordinated activity of these different domains is likely required for activation of plant NB-LRR proteins and the signaling needed to elicit resistance responses [15]–[17].
The mechanisms of pathogen effector perception by R proteins include both direct and indirect protein interactions. The R proteins Pi-ta [18], RRS1-R [19], N [17], L5/L6 [20], M [21], and RPP1 [22] all physically interact with their corresponding effectors. Alternatively, plant R proteins can indirectly recognize the presence of pathogen effectors by monitoring target host cellular proteins [23]. Through monitoring the integrity of host targets, R proteins detect effectors indirectly, which explains how relatively conserved plant R proteins can detect highly varied pathogen effectors [23].
Potato and tomato late blight is caused by the oomycete pathogen Phytophothora infestans (Mont.) de Bary. The late blight R gene, RB (also known as Rpi-blb1), from the wild potato species S. bulbocastanum, encodes a CC-NB-LRR protein and confers broad-spectrum, partial resistance to most strains of the pathogen due to the almost ubiquitous presence of the corresponding effector IPI-O [24]–[29]. IPI-O is a multigene effector family and the IPI-O locus can be extremely variable between pathogen strains [24], [25], [30]. IPI-O belongs to the class of Phytophthora effectors with a highly conserved N-terminal RXLR motif and a C-terminal W motif [24], [31]–[34]. IPI-O variants have been divided into three classes based on diversity of their deduced amino acid sequences [24], [25]. Class I variants (e.g. IPI-O1), which are found in the majority of P. infestans isolates, are recognized by RB, while class III variants (e.g. IPI-O4) are not [26]. In addition, P. infestans strains lacking a class I IPI-O are virulent on plants carrying RB [24], and P. infestans strains with class III variants are more aggressive on plants with RB [25]. Interestingly, IPI-O4 not only eludes detection by RB, but is also capable of inhibiting the HR elicited IPI-O1 [25].
In the present study, we investigated intra- and intermolecular interactions of the RB protein using specific R protein domains and IPI-O variants. We identified amino acids that play a key role not only in the interaction between the RB CC domain and IPI-O4, but also in elicitation or suppression of an RB-mediated HR. Our findings suggest a model in which IPI-O4 is able to affect RB function through interaction with the CC domain, possibly disrupting interactions that would otherwise lead to R protein activation.
Results
RB CC domain self-associates and physically interacts with IPI-O1 and IPI-O4
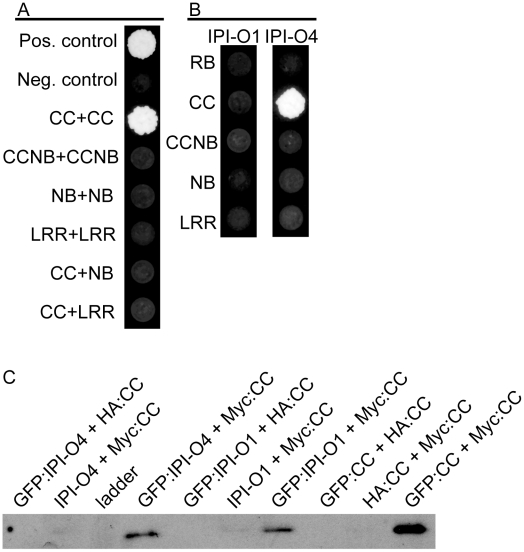
To investigate physical interactions between RB domains, pair-wise combinations of protein domains were assayed using a directed yeast two-hybrid interaction assay. Our results showed that none of the separate or fused domains of RB interacted with dissimilar domains (Figure 1A, Figure S1). Self-association of the CC domain was observed, but no self-association was detected among the NB, LRR or CCNB domains (Figure 1A). To study whether recognition of IPI-O1 by RB involves a direct R protein-effector interaction, associations between IPI-O1 or IPI-O4 with the RB CC, NB, LRR, and CCNB domains and full length RB were tested. A physical interaction was observed between the CC domain of RB and IPI-O4 (Figure 1B). However, no such interaction was found between the CC domain and IPI-O1 in yeast. No interactions between IPI-O1 or IPI-O4 with any other RB domain was observed. Also, no interaction between IPI-O1 and IPI-O4 was observed, however an interaction between IPI-O4 molecules was detected, indicating possible oligomerization of this effector (Figure S1). Protein blotting showed that all of the IPI-O and RB proteins in the pSOS expression vector were stable in yeast (Figure S2).
Figure 1. Inter- and intramolecular interactions between RB protein domains and IPI-O effectors.
A) Yeast two-hybrid interactions between domains of RB or domain self-association; B) Interactions between domains of RB and IPI-O1 or IPI-O4; C) Results of co-immunoprecipitation of RB CC domain with IPI-O fusion proteins. The indicated protein combinations were expressed in N. benthamiana leaves and total proteins were incubated with green fluorescent protein (GFP) antibody and agarose beads. Precipitated proteins were detected using Myc-tag antibody. Proteins with no fusion or with a hemagglutinin (HA) tag were used as negative controls.
We verified the yeast two-hybrid data using co-immunoprecipitation in Nicotiana benthamiana leaves. Expression of versions of the RB CC domain, IPI-O1, and IPI-O4 fused with green fluorescent protein (GFP) showed proteins of the expected sizes in leaves after detection with GFP antibodies, although some degradation products were observed (Figure S3). Compared to GFP:RBCC and GFP:IPI-O4, full-length GFP:IPI-O1 protein only accumulated to low levels indicating that full-length versions of this protein were present, but possibly unstable. No degradation products of the RB CC domain were observed when proteins were expressed as fusions with a Myc-tag (Figure S3B). Co-immunoprecipitation confirmed oligomerization of the RB CC domain and the interaction between the RB CC domain and IPI-O4 (Figure 1C). In contrast to the yeast two-hybrid results, we also detected IPI-O1 after precipitation with the RB CC domain, indicating in planta interaction between these proteins.
IPI-O1 and IPI-O4 interact with RB CC fragments from wild potato
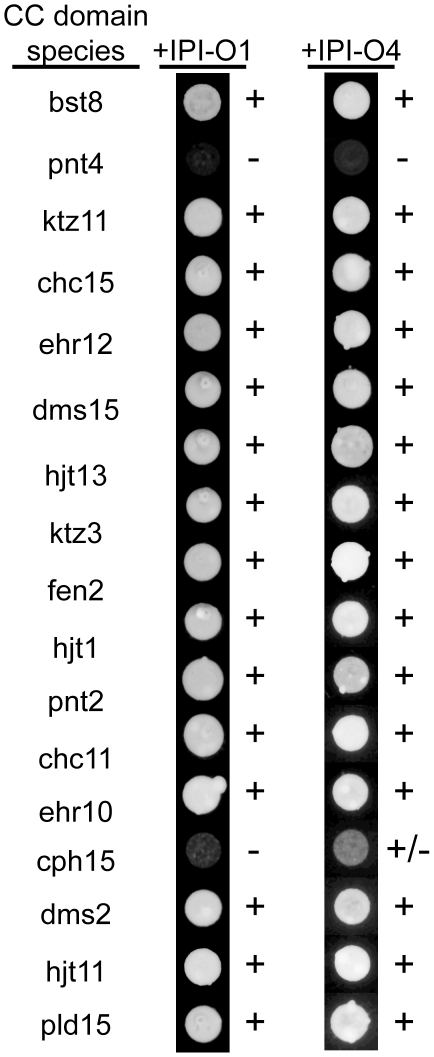
The interaction between IPI-O4 and CC domains of RB-like sequences from ten wild species of potato was tested. These ten species were not chosen based on their late blight resistance phenotype but rather because they capture a large amount of diversity within a small number of species. Seventeen different CC domains from these species, sharing deduced amino acid sequence similarities ranging between 65.9% and 98.2% (Figs. S4, S5), were chosen for interaction testing with IPI-O1 and IPI-O4. Fifteen of the 17 RB-like CC fragments interacted with both IPI-O1 and IPI-O4 in yeast (Figure 2). Only one fragment from S. cardiophyllum showed a similar interaction pattern as the S. bulbocastanum RB CC domain in yeast. It did not interact with IPI-O1, but we did observe a weak but consistent interaction with IPI-O4. This weak interaction was characterized by slower growth of the yeast during selection. One fragment from S. pinnatisectum showed no interaction with either IPI-O1 or IPI-O4. Protein blotting showed that the pnt4 and cph15 CC domains in the pSOS expression vector were stable in yeast (Figure S2).
Figure 2. Interactions between RB-like CC domains from potato species with either IPI-O1 or IPI-O4.
The three-letter abbreviation for each species is shown. Numbers after the species name represents the PCR clone number. + and − signs to the right of the yeast colonies indicate positive or negative interaction, respectively. Photos were taken after 8 days of growth on selective media. Photos represent the results of three independent yeast transformations.
Specific mutations within IPI-O4 abolish interaction with the RB CC domain
Twenty amino acids differentiate IPI-O1 from IPI-O4 [30]. In order to determine which residue(s) plays a role in the RB CC/IPI-O4 interaction, individual IPI-O1 and IPI-O4 amino acids were mutated to the corresponding residue in the other effector. Fifteen IPI-O1 mutants and 13 IPI-O4 mutants with single amino acid changes were tested for interaction with the RB CC domain. Despite extensive efforts, we could not obtain mutations at the other locations. None of the 15 IPI-O1 single amino acid mutants interacted with the CC domain (Figure 3). However, four IPI-O4 mutations, K82Y, G86V, P129L and G135S abolished or weakened the RB CC domain/IPI-O4 interaction (Figure 3). In order to determine whether variant amino acids at these four sites function together to alter the IPI-O/CC interaction phenotype, 6 IPI-O1 double amino acid mutants, and 5 IPI-O4 double amino acid mutants were tested (Figure 3). The results showed that none of the IPI-O1 mutants interacted with the CC domain. One IPI-O4 double amino acid mutant, IPI-O4 P129L/G135S, regained its interaction with the RB CC domain. Protein blotting showed that all of the IPI-O single and double mutants in the pSOS expression vector were stable in yeast (Figure S2).
Figure 3. Yeast two-hybrid interactions between the CC domain of RB and IPI-O1/4 single and double amino acid mutants.
+ and − signs to the right of the yeast colonies indicate positive or negative interaction, respectively. Pictures were taken after 8 days of growth on selective media. All the experiments were performed three times.
Amino acid 129 of IPI-O1 is critical for RB HR elicitation
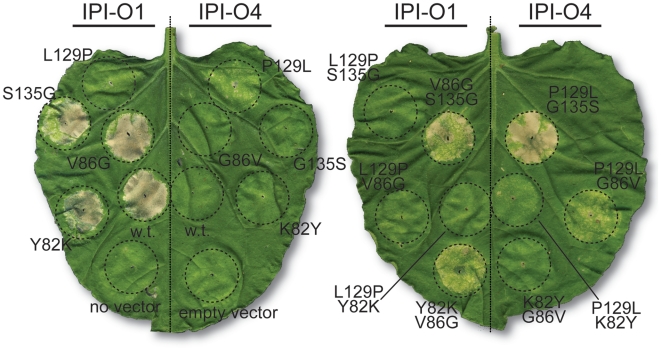
The four mutations at amino acids 82, 86, 129, and 135 of IPI-O1 and IPI-O4 were assayed for an effect of activation of HR elicitation by agroinfiltration in RB-transgenic N. benthamiana. These four amino acids were chosen because they significantly impacted the interaction between the RB CC domain and IPI-O4. The mutant IPI-O1 L129P failed to elicit the HR, while the other single amino acid mutants were still capable of inducing the HR (Figure 4). Consistent with these results, IPI-O1 double mutants containing L129P, namely IPI-O1 Y82K/L129P, V86G/L129P, and L129P/S135G, also lost the ability to elicit an RB-mediated HR. The two IPI-O1 mutants Y82K/V86G and V86G/S135G induced an HR, demonstrating that they were still recognized by RB. When present, the cell death response began approximately 48 hours after infiltration (hai). No visible differences were observed regarding timing of the onset of the HR or the cell death intensity between wild type IPI-O1 and the mutants. Protein blotting indicated that IPI-O1 mutants containing L129P were present at detectable amounts and stable in N. benthamiana leaves (Figure S6).
Figure 4. Induction of hypersensitive cell death by IPI-O1 and IPI-O4 mutants in planta.
A. tumefaciens strains expressing IPI-O, IPI-O mutants, or the indicated controls were infiltrated into leaves of RB transgenic N. benthamiana plants. Leaves were photographed at 6 days after infiltration.
The HR induction phenotype of IPI-O4 mutants also supports the conclusion that L129P is important in RB activation. All P129L-containing mutants of IPI-O4 (P129L, K82Y/P129L, G86V/P129L and P129L/G135S) gained the ability to elicit the HR in N. benthamiana with RB (Figure 4). In contrast, mutants of IPI-O4 K82Y, G86V, G135S and K82Y/G86V did not elicit the HR.
Effect of mutations on suppression of RB HR induction
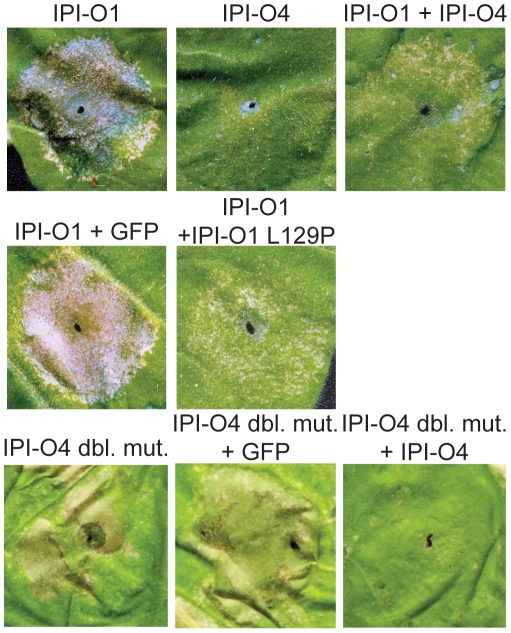
We coinfiltrated RB-transgenic N. benthamiana leaves with IPI-O1 and IPI-O1 L129P to test whether this mutant is able to suppress RB-mediated HR elicitation (Figure 5; Figure S7). Compared to IPI-O1 alone or coinfiltration of IPI-O1 with GFP, no HR or faint HR was observed in areas co-infiltrated with IPI-O1 and IPI-O1 L129P, indicating a suppression of cell death by IPI-O1 L129P. However, cell death was not suppressed when INF1 and IPI-O1 L129P were coexpressed (Figure S7), demonstrating that IPI-O1 L129P suppression of cell death is RB-specific.
Figure 5. Suppression of hypersensitive cell death of IPI-O1 by IPI-O1 L129P and IPI-O4 P129L/G135S (double mutant) by IPI-O4.
A. tumefaciens strains expressing the indicated IPI-O variants or a GFP control were infiltrated or co-infiltrated into leaves of RB transgenic N. benthamiana plants. Leaves were photographed at 6 days after infiltration.
As noted previously, the double mutant IPI-O4 P129L/G135S is able to induce the HR in the presence of RB. We tested the ability of IPI-O4 to suppress this HR response by co-infiltrating RB-transgenic N. benthamiana leaves with these two effectors (Figure 5). HR induced in RB transgenic leaves by IPI-O4 P129L/G135S alone was observed 48 hai. Similar to regions exposed to GFP or IPI-O4 alone (Figure S8), no HR or faint HR was observed in areas co-infiltrated with IPI-O4 P129L/G135S and IPI-O4, indicating a suppression of cell death in the presence of IPI-O4 (Figure 5). Coexpression of GFP with IPI-O1 or IPI-O4 P129L/G135S did not abrogate HR elicitation, nor did coexpression of IPI-O4 and INF1, demonstrating specificity for RB. We then tested the effect that single amino acid changes within IPI-O4 had on its ability to suppress the HR. Coexpression of the IPI-O4 single amino acid mutants K82Y, G86V, and G135S with IPI-O1 revealed an effect on HR suppression, although their suppressive activity appeared to be lower than IPI-O4 or the IPI-O1 L129P mutant (Figure S9). IPI-O4 P129L was not tested because it is able to elicit the HR itself.
Discussion
The interaction between R proteins and pathogen effectors determines the resistance or susceptibility phenotype of the host. P. infestans has repeatedly proven itself capable of overcoming major resistance genes. Although many resistance genes have been identified in wild potato species and have been integrated into new potato varieties, this has typically provided only temporary late blight control. Our ability to understand how these resistance genes are able to recognize pathogen effectors and elucidate the mechanisms that allow pathogens to overcome resistance will assist us in predicting the ability of P. infestans to overcome resistance and permit engineering or identifying genes that can resist suppression by specific effectors. The P. infestans effectors IPI-O1 and IPI-O4 provide opportunities to study the complex molecular mechanisms of R protein-effector interactions, since IPI-O1 elicits RB-mediated resistance while IPI-O4 functions to suppress resistance elicitation.
In this study, self-association of the RB CC domain was detected, suggesting that RB may dimerize or polymerize through this domain. Dimerization or polymerization has been reported in different types of R proteins after they are activated upon recognition of their cognate effectors. The tobacco N protein oligomerizes in the presence of the effector p50 [9]. N protein oligomerization correlates with resistance, since mutations in the TIR domain that abolish self-association also abolish resistance [9]. The CC domain of Arabidopsis R protein RPS5 also forms oligomers [11], but it is currently unknown whether interaction with the AvrPphB effector or the PBS1 accessory protein affects R protein oligomerization. The tomato Prf protein forms oligomers through self-association of the N-terminus [35]. This region of the protein is also important in interactions with important cofactors, such as Pto, Fen, Pth3, and Pth5 [35]. The barley MLA protein also self-associates through interaction of its CC domain [36], but unlike RB, MLA effector recognition specificity lies within the LRR and not the CC domain [37]. Our results indicating oligomerization of RB are based on protein-protein interaction tests in yeast and in planta using only the CC domain and, without data using the full-length protein we can only hypothesize that RB oligomerization is necessary for proper activity. We have provided additional evidence that this region of RB is important in HR elicitation through its interaction with IPI-O effectors. Since R protein oligomerization is an important function in similar resistance responses, it is not unexpected that suppression of this event would be a logical target for a pathogen effector. We observed an interaction between IPI-O4 and the CC domain of RB in yeast and in planta and between IPI-O1 and the CC domain in planta, suggesting that IPI-O may affect RB function through interaction with this region, thereby preventing CC oligomerization or blocking an interaction with other signaling components. In addition, we observed an interaction between most RB-like CC fragments from wild species of potato and IPI-O1, as well as with IPI-O4. This observation suggests that IPI-O effectors may have defeated ancient RB alleles by preventing CC oligomerization or blocking an interaction with other signaling components.
Effectors from different types of pathogens have been found to suppress host basal defense or hypersensitive cell death induced by elicitors or effectors even in the presence of cognate R proteins [38]. The Avr1 effector from Fusarium oxysporum f.sp. lycopersici (Fol) suppresses resistance mediated by the tomato I-2 and I-3 genes, but tomato plants with either I or I-1 are able to recognize Avr1 to elicit resistance [39]. The tomato protein kinase Pto confers resistance to P. syringae expressing AvrPtoB, a ubiquitin ligase [40], [41]. Despite 80% amino acid similarity to Pto, the Fen kinase is not able to elicit resistance in the presence of AvrPtoB. This is due to suppression of Fen through ubiquitination and degradation by AvrPtoB [42]. Pto phosphorylates AvrPtoB to inactivate its E3 ligase activity and mediate ETI together with Prf [43], [44]. We have previously shown that IPI-O4 is capable of inhibiting the HR elicited by IPI-O1 in N. benthamiana expressing RB [25]. In the present study, no physical interaction between IPI-O1 and IPI-O4 was detected, suggesting that IPI-O4 does not inhibit IPI-O1 recognition by interacting with IPI-O1 directly. However, we cannot rule out that conditional post-translational modifications or the environment in planta might result in a direct interaction. The ability of the IPI-O1 L129P mutant to suppress IPI-O1 elicitation of the HR in the presence of RB demonstrates a critical step in the evolution of this effector family. This finding suggests that a simple mutation at this location in IPI-O1 could alter pathogen virulence when RB is present. Our previous analysis of IPI-O variants from P. infestans isolates identified two alleles outside of the IPI-O4 class that contain a proline at amino acid 129 [25]. The first, CMPh0-07.04 derived from a Thai isolate, is unique but falls within the IPI-O3 family. The second, 68.12 from a Guatemalan isolate, is likely the result of a recombination event between IPI-O4 and another IPI-O family member. The virulence of isolate CMPh0-07 was not tested, but isolate 68 showed increased aggressiveness on plants with and without the RB gene indicating its ability to suppress resistance responses [25].
The utilization of transient effector expression in N. benthamiana has allowed us to rapidly identify specific amino acids within a pathogen effector that condition host resistance. Four amino acids in IPI-O play an important role in RB CC/IPI-O4 interaction and one of them also determines IPI-O1 recognition by RB. All four amino acids are located C-terminal to the RXLR motif, which is consistent with the fact that the RXLR motif is responsible for translocation [33], [34], while the C-terminal half conditions virulence and avirulence [24], [45]. Amino acid 129 is within the W motif and 135 is adjacent to the W motif [24], [25]. The conformational rigidity of proline and the relative flexibility of glycine suggest that they might have an important role in structural changes that alter the presentation of loops within the IPI-O4 protein. The IPI-O amino acid sequence shares no similarity to any known protein molecules and no definitive function within the host cytoplasm has been determined. The IPI-O mutants obtained in this study will help to predict the structure and properties of IPI-O, and additionally, the crystal structure of IPI-O in the presence of the RB CC domain would help to complete elucidation of the molecular mechanisms surrounding the IPI-O/RB interaction.
The two single amino acid mutants of IPI-O4, P129L and G135S, lost the ability to interact with the RB CC domain. However, the double mutant IPI-O4 P129L/G135S elicited the HR and regained the ability to interact with the CC domain, demonstrating a key link between IPI-O/CC interaction and resistance elicitation. These results also suggest that HR elicitation by IPI-O4 P129L/G135S is epistatic to interaction with the CC domain. In other words, interaction with the CC domain is not sufficient to inhibit the HR in planta if the effector molecule itself is recognized by RB and triggers the HR. The important role of CC domain interaction is further demonstrated by the fact that IPI-O4 mutants that are compromised in RB CC domain interactions are also compromised in suppressing HR induced by IPI-O1.
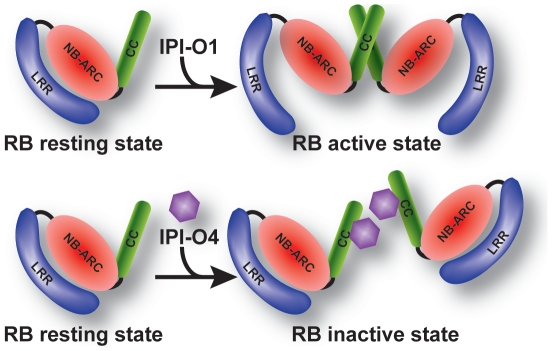
Collectively, our data, combined with data from other CC-NB-LRR proteins, is consistent with the model shown in Figure 6. The model suggests that in the absence of IPI-O, RB remains in a resting state. CCNB stabilizes this state. A conformational change occurs upon recognition of IPI-O1, which enables RB to oligomerize through the CC domain and expose a platform for signaling components or leads to an activated protein state. However, when IPI-O4 is present, this effector interacts with the CC domain and prevents CC oligomerization, or blocks an interaction with other signaling components, thus suppressing RB activation. Our data indicates that IPI-O1 also interacts with the RB CC domain in planta, but not in yeast. The IPI-O1 fusion protein is stable in yeast, suggesting that perhaps an additional accessory protein, found only in planta, is necessary for stability of the interaction with the RB CC domain. Our simplified model does not account for this. Combined with the fact that GFP:IPI-O1 fusion protein was not as abundant as GFP:IPI-O4 in N. benthamiana leaves, we hypothesize that IPI-O1 protein accumulation is affected by factors in the plant cell that interfere with its ability to suppress RB oligomerization and elicit resistance.
Figure 6. Model for RB/IPI-O interactions.
The top panel represents a resistance response. In the absence of IPI-O1, RB remains in a resting state. The presence of IPI-O1 elicits a conformational change that enables RB oligomerization through the CC domain and leads to an activated protein state. As shown in the bottom panel, when IPI-O4 is present, this effector interacts with the CC domain and prevents CC oligomerization, thus suppressing RB activation.
Our data demonstrates a co-evolutionary arms race between IPI-O and RB. The fact that several late blight susceptible potato species contain RB-like proteins with CC domains that interact with both IPI-O1 and IPI-O4, suggests that P. infestans delivery of IPI-O effectors into the plant cell could be responsible for the pathogen's ability to elude recognition and cause disease. If this is true, S. bulbocastanum RB is one of the latest in a line of R proteins that are able to recognize the presence of IPI-O1 while escaping suppression by this effector. Our ability to understand the molecular interactions that condition resistance and suppression will hopefully allow us to design or identify RB alleles that can continue to effectively control disease epidemics. A RB homolog that recognizes IPI-O4 has been found in the wild species of potato S. stoloniferum [26] demonstrating that variants of the R protein exist that are able to avoid suppression by IPI-O4. In our study, a CC homolog from S. pinnatisectum showed no interaction with either IPI-O1 or IPI-O4. We are currently pursuing research focused on answering whether an engineered RB containing the CC domain from S. pinnatisectum may also escape suppression by IPI-O4. Despite the fact that P. infestans isolates that overcome RB resistance have been found [24], [25], RB remains valuable for potato breeding due to the almost ubiquitous existence of IPI-O1 in strains from major potato growing regions. However, due to the adaptive nature P. infestans and the continual proliferation of strains of P. infestans, we expect that the benefits of using the RB gene for late blight resistance will be ephemeral. Significant efforts must continue to be made to identify and integrate novel resistance. Pyramiding of RB, RB orthologs, and modified RB recognizing different IPI-O variants is one strategy that could be used to expand the durability of this gene.
Materials and Methods
Yeast two-hybrid assays
The CytoTrap system (Agilent) was used to detect protein-protein interactions. IPI-O1, IPI-O4, full length RB [amino acids (aa) 1–970], the CC domain of RB (aa 1–165), the NB domain of RB (aa 166–521), the CC-NB domain of RB (aa 1–521) and the LRR domain of RB (aa 522–970) were amplified with primers listed in Table S1 using Platinum PCR SuperMix High Fidelity (Invitrogen). IPI-O1 (PexRD6-1) and IPI-O4 (PexRD6-3) targets were those described in Vleeshouwers et al. [26]. The PCR products were ligated into BamHI and SalI digested pSos or EcoRI and SalI digested pMyr, respectively.
Yeast protein extraction and blotting
Cdc25 yeast strains containing RB domains or IPI-O variants in pSos were grown to OD600 = 0.6 at 24°C in 10 ml SD/glucose (-L) media. Yeast cells with no pSos vector were grown in YPAD broth. Protein purification was performed according to the CytoTrap system instruction manual. Protein pellets were resuspended in 300 µl of SU buffer. 20 µl of each protein sample was heated at 65°C for 3 minutes, separated by 10% SDS-PAGE and transferred to Hybond-P (GE Healthcare). Immunoblotting was performed using a mouse monoclonal anti-Sos primary antibody (1∶250, BD Biosciences) and an anti-mouse secondary antibody produced in goat (1∶10000, Sigma). Antibody detection was carried out using an ECL Western blotting detection system (GE Healthcare) according to the manufacturer's instructions.
Cloning of RB-like CC domains from wild potato
Seeds from 10 wild species of potato, shown in Table 1, were obtained from the National Research Support Program (NRSP)-6 potato GeneBank in Sturgeon Bay, Wisconsin. Seedlings were grown under greenhouse conditions (23°C day/15°C night temperatures with 14 hours of light) and watered as needed. Total RNA was extracted from young leaves using a Total RNA Extraction Kit (Sigma) according the manufacturer's instructions. One µg of DNase-treated RNA was reverse transcribed using a First-strand Synthesis Kit (Bio-Rad). cDNA was used as a template to amplify the RB homologous CC fragments using primers RBFORBAMHI and RBCCREVSALI (Table S1). PCR conditions and cloning into pSos vectors was carried out as described above.
Table 1. Wild potato species used for amplification of RB homologous CC fragments.
| Species | Abbreviation | PI number |
| Solanum brachistotrichum | bst | 279244 |
| Solanum chacoense | chc | 275138 |
| Solanum cardiophyllum | cph | 283062 |
| Solanum demissum | dms | 161366 |
| Solanum ehrenbergii | ehr | 184762 |
| Solanum fendleri | fen | 225661 |
| Solanum hjertingii | hjt | 283103 |
| Solanum kurtzianum | ktz | 472923 |
| Solanum polyadenium | pld | 320342 |
| Solanum pinnatisectum | pnt | 186553 |
Site-directed mutagenesis
Mutants of P. infestans effectors IPI-O1 and IPI-O4 were generated by circular PCR using IPI-O1 and IpiO4 in pMyr as templates. A pair of oligonucleotide primers containing the desired nucleotide substitution (Table S1), each complementary to the opposite strands of the same target sequence, were extended during temperature cycling using PfuTurbo DNA polymerase (Agilent). After the temperature cycling, the PCR products were treated with DpnI for 2 hr at 37°C to digest the methylated parental DNA template and then transformed into E. coli. The resultant mutated plasmid constructs were verified by sequencing.
HR induction assays in N. benthamiana
The wild type and mutant IPI-O1 and IPI-O4 in pMyr were used as templates for PCR amplification to incorporate BamHI and SacI restriction enzyme sites. Primer sequences are listed in Table S1. PCR products were ligated into BamHI and SacI digested binary vector pBI121. All constructs were introduced into A. tumefaciens strain GV3101 by electroporation.
Wild type and RB-transgenic N. benthamiana plants [46] were grown in pots in a walk-in growth chamber with 28°C day/27°C night temperatures, 16 hours of light, and were watered and fertilized as needed. Four to six weeks old plants were used for agroinfiltration. Agroinfiltration was carried out as previously described [26]. Infiltrated plants were maintained in a walk-in growth chamber (22°C day/18°C night temperatures with 16 h of light). When testing the HR suppression, HR inducing constructs (INF1, IPI-O1, IPI-O4 P129L/G135S) were infiltrated at the same infiltration spot 24 hours after the infiltration of non-HR inducing constructs (GFP, IPI-O4, IPI-O1 L129P, IPI-O4 K82Y, IPI-O4 G86V, IPI-O4 G135S).
Protein extraction, blotting and co-immunoprecipitation
The wild type and mutant IPI-O1, IPI-O4 and the RB CC domain in pMyr were used as templates for PCR amplification to incorporate BamHI and XhoI restriction enzyme sites. Primer sequences are listed in Table S1. PCR products were ligated into BamHI and XhoI digested pENTR1a (Invitrogen). Gateway LR reactions (Invitrogen) were performed to introduce wild type IPI-O1 and mutants into the binary vector pEG201 [47] with an N-terminal HA tag, and introduce IPI-O1, IPI-O4, and the RB CC domain into pGWB6 and pGWB18 [48] with N-terminal GFP and Myc tags respectively. Infiltrated areas of N. benthamiana leaves were harvested 21 hours after agroinfiltration. To prepare total protein, plant material was ground in liquid nitrogen and thawed in extraction buffer containing 50 mM Tris, pH 7.5, 150 mM NaCl, 0.1% Triton X-100, and a protease inhibitor cocktail according to the manufacturer's instructions (Sigma). Samples were incubated on ice for 1 hour. The suspension was centrifuged at 4000 g at 4°C 6 times, each time for 10 min. In the protein blot assay, 100 mg of ground plant material was thawed in 100 µl of extraction buffer, and 25 µl of the supernatant was resolved on a 12% SDS-PAGE gel and transferred to Hybond-P (GE Healthcare) according to the manufacturers recommendations. Protein was detected using an anti-HA-peroxidase antibody (Roche) with an ECL Western blotting detection system (GE Healthcare) according the manufacturer's instructions. In the co-immnunoprecipitation assay, 1 gram of ground plant material was thawed in 1 ml of extraction buffer. Seven µl of anti-GFP antibody (Clontech) was added to each protein sample, followed by incubation for 1 hour with end-to-end shaking at 4°C. 50 µl of pre-equlibrated protein A agarose (Roche) was then added to each sample and incubated for 4 hours with end-to-end shaking at 4°C. The beads were spun at 4000 g at 4°C and washed with 1 ml of wash buffer (50 mM Tris (pH 7.5), 150 mM NaCl) 6 times (15 min each time) with end-to-end shaking at 4°C. After washing, the beads were eluted with 45 µl of 2× SDS loading buffer and heated at 95°C for 7 min. The eluted protein was separated on a 10% SDS-PAGE gel and transferred to PVDF membrane and detected with anti-Myc-HRP antibodies (Sigma) using an ECL Western blotting detection system (GE Healthcare).
Supporting Information
Yeast two-hybrid screening of RB domain interactions. Each panel shows three independent transformants of identical genotypes. Positive and negative controls were provided by the CytoTrap system manufacturer. RB = full-length RB; CC = RB coiled-coil domain; NB = RB nucleotide binding domain; LRR = RB leucine-rich repeat domain. All spots contained similar quantities of yeast at the time of plating. Pictures were taken after 8 days of growth on selective media.
(TIF)
Protein blotting showing stability of IPI-O and CC SOS-domain fusions in yeast. A–D) Total yeast proteins were separated on separate acrylamide gels, blotted, and probed with SOS-specific antibody. Ponceau S stained PVDF membranes are shown below the results of protein blotting and antibody detection.
(TIF)
Protein blotting of the input protein for co-immunoprecipitation. Total protein from leaf sections agroinfiltrated with the indicated constructs was extracted and separated on an acrylamide gel. Protein was blotted and detected using a GFP-specific antibody (A) or a Myc-tag specific antibody (B). Ponceau S stained PVDF membranes are shown to demonstrate equal loading.
(TIF)
Amino acid sequence identity chart showing the pairwise percent identity between RB CC domains amplified from wild potato species. Sequence names contain the three-letter abbreviation for each species. Numbers after the species name represents the PCR clone number.
(TIF)
Sequence alignment of RB CC domain deduced amino acid sequences. Letters in black boxes are identical to the consensus sequence (shown at top of each row). Sequence names contain the three-letter abbreviation for each species. Numbers after the species name represents the PCR clone number.
(TIF)
IPI-O1 mutants containing L129P are stable in RB-transgenic N. benthamiana leaves. A protein blot was performed using total protein extracts following agroinfiltration with constructs expressing indicated HA-IPI-O1 mutants. The 18-kDa protein band represents the expected size of recombinant IPI-O1 mutants. Lane1: wild type IPI-O1; Lane2: non-infiltrated N. benthamiana; Lane3: IPI-O1 L129P; Lane4: IPI-O1 Y82K/L129P; Lane5: IPI-O1 V86G/L129P; Lane6: IPI-O1 L129P/S135G.
(TIF)
IPI-O1 L129P inhibits the HR induced by IPI-O1. A. tumefaciens strains expressing IPI-O mutants or the indicated controls were infiltrated into leaves of RB transgenic N. benthamiana plants. Leaves were photographed at 6 days after infiltration.
(TIF)
IPI-O4 inhibits the HR induced by the IPI-O4 P129L/G135S double mutant. A. tumefaciens strains expressing IPI-O mutants or the indicated controls were infiltrated into leaves of RB transgenic N. benthamiana plants. Leaves were photographed at 6 days after infiltration.
(TIF)
IPI-O4 K82Y, G86V, and G135S inhibit the HR induced by IPI-O1. A. tumefaciens strains expressing IPI-O mutants or the indicated controls were infiltrated into leaves of RB transgenic N. benthamiana plants. Leaves were photographed at 6 days after infiltration. Note that the inhibitory effect of these mutants is not as strong as that of IPI-O1 L129P since some cell death was still observed in the area coinfiltrated with IPI-O1 and IPI-O4 K82Y (A), G86V (B), or G135S (C).
(TIF)
Acknowledgments
We would like to thank Gail Middleton for expert technical assistance. This article reports the results of research only. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U. S. Department of Agriculture.
Footnotes
The authors have declared that no competing interests exist.
Y.C. was supported by the UW Dept. of Plant Pathology, Z.L. by the USDA-ARS and UW Graduate School, and D.H.'s laboratory was supported by the USDA-ARS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Dangl JL, Jones JD. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- 2.Bent AF, Mackey D. Elicitors, effectors, and R genes: The new paradigm and a lifetime supply of questions. Ann Rev Phytopathol. 2007;45:399–436. doi: 10.1146/annurev.phyto.45.062806.094427. [DOI] [PubMed] [Google Scholar]
- 3.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 4.He P, Shan L, Lin N-C, Martin GB, Kemmerling B, et al. Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell. 2006;125:563–575. doi: 10.1016/j.cell.2006.02.047. [DOI] [PubMed] [Google Scholar]
- 5.Nomura K, DebRoy S, Lee YH, Pumplin N, Jones J, et al. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science. 2006;313:220–223. doi: 10.1126/science.1129523. [DOI] [PubMed] [Google Scholar]
- 6.Thomma BPHJ, Nürnberger T, Joosten MHAJ. Of PAMPs and effectors: The blurred PTI-ETI dichotomy. Plant Cell. 2011;23:4–15. doi: 10.1105/tpc.110.082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 8.Mackey D, Holt BF, Wiig A, Dangl JL. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- 9.Mestre P, Baulcombe DC. Elicitor-mediated oligomerization of the tobacco N disease resistance protein. Plant Cell. 2006;18:491–501. doi: 10.1105/tpc.105.037234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mucyn TS, Clemente A, Andriotis VM, Balmuth AL, Oldroyd GE, et al. The tomato NBARC-LRR protein Prf interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell. 2006;18:2792–2806. doi: 10.1105/tpc.106.044016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ade J, DeYoung BJ, Golstein C, Innes RW. Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci U S A. 2007;104:2531–2536. doi: 10.1073/pnas.0608779104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burch-Smith TM, Schiff M, Caplan JL, Tsao J, Czymmek K, et al. A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol. 2007;5:e68. doi: 10.1371/journal.pbio.0050068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, et al. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science. 2007;315:1098–1103. doi: 10.1126/science.1136372. [DOI] [PubMed] [Google Scholar]
- 14.Rairdan GJ, Collier SM, Sacco MA, Baldwin TT, Boettrich T, et al. The coiled-coil and nucleotide binding domains of the potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell. 2008;20:739–751. doi: 10.1105/tpc.107.056036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moffett P, Farnham G, Peart J, Baulcombe DC. Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J. 2002;21:4511–4519. doi: 10.1093/emboj/cdf453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rairdan GJ, Moffett P. Distinct domains in the ARC region of the potato resistance protein Rx mediate LRR binding and inhibition of activation. Plant Cell. 2006;18:2082–2093. doi: 10.1105/tpc.106.042747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ueda H, Yamaguchi Y, Sano H. Direct interaction between the tobacco mosaic virus helicase domain and the ATP-bound resistance protein, N factor during the hypersensitive response in tobacco plants. Plant Mol Biol. 2006;61:31–45. doi: 10.1007/s11103-005-5817-8. [DOI] [PubMed] [Google Scholar]
- 18.Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 2000;19:4004–4014. doi: 10.1093/emboj/19.15.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, et al. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci U S A. 2003;100:8024–8029. doi: 10.1073/pnas.1230660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dodds PN, Lawrence GJ, Catanzariti AM, Teh T, Wang CI, et al. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc Natl Acad Sci U S A. 2006;103:8888–8893. doi: 10.1073/pnas.0602577103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Catanzariti A-M, Dodds PN, Ve T, Kobe B, Ellis JG, et al. The AvrM effector from flax rust has a structured C-terminal domain and interacts directly with the M resistance protein. Mol Plant Microbe Interact. 2010;23:49–57. doi: 10.1094/MPMI-23-1-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krasileva KV, Dahlbeck D, Staskawicz BJ. Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell. 2010;22:2444–2458. doi: 10.1105/tpc.110.075358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van der Biezen EA, Jones JD. Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem Sci. 1998;23:454–456. doi: 10.1016/s0968-0004(98)01311-5. [DOI] [PubMed] [Google Scholar]
- 24.Champouret N, Bouwmeester K, Rietman H, van der Lee T, Maliepaard C, et al. Phytophthora infestans isolates lacking class I ipiO variants are virulent on Rpi-blb1 potato. Mol Plant Microbe Interact. 2009;22:1535–1545. doi: 10.1094/MPMI-22-12-1535. [DOI] [PubMed] [Google Scholar]
- 25.Halterman D, Chen Y, Sopee J, Berduo-Sandoval J, Sánchez-Pérez A. Competition between Phytophthora infestans effectors leads to increased aggressiveness on plants containing broad-spectrum late blight resistance. PLoS One. 2010;5:e10536. doi: 10.1371/journal.pone.0010536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vleeshouwers VG, Rietman H, Krenek P, Champouret N, Young C, et al. Effector genomics accelerates discovery and functional profiling of potato disease resistance and phytophthora infestans avirulence genes. PLoS ONE. 2008;3:e2875. doi: 10.1371/journal.pone.0002875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y, Halterman D. Phenotypic characterization of potato late blight resistance mediated by the broad-spectrum resistance gene RB. Phytopathology. 2011;101:263–270. doi: 10.1094/PHYTO-04-10-0119. [DOI] [PubMed] [Google Scholar]
- 28.Song J, Bradeen JM, Naess SK, Raasch JA, Wielgus SM, et al. Gene RB cloned from Solanum bulbocastanum confers broad spectrum resistance to potato late blight. Proc Natl Acad Sci U S A. 2003;100:9128–9133. doi: 10.1073/pnas.1533501100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Vossen E, Sikkema A, Hekkert BtL, Gros J, Stevens P, et al. An ancient R gene from the wild potato species Solanum bulbocastanum confers broad-spectrum resistance to Phytophthora infestans in cultivated potato and tomato. Plant J. 2003;36:867–882. doi: 10.1046/j.1365-313x.2003.01934.x. [DOI] [PubMed] [Google Scholar]
- 30.van West P, de Jong AJ, Judelson HS, Emons AMC, Govers F. The ipiO gene of Phytophthora infestans is highly expressed in invading hyphae during infection. Fungal Genet Biol. 1998;23:126–138. doi: 10.1006/fgbi.1998.1036. [DOI] [PubMed] [Google Scholar]
- 31.Haas BJ, Kamoun S, Zody MC, Jiang RHY, Handsaker RE, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461:393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- 32.Jiang RHY, Tripathy S, Govers F, Tyler BM. RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc Natl Acad Sci U S A. 2008;105:4874–4879. doi: 10.1073/pnas.0709303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whisson SC, Boevink PC, Moleleki L, Avrova AO, Morales JG, et al. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature. 2007;450:115–118. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]
- 34.Kale SD, Gu B, Capelluto DGS, Dou D, Feldman E, et al. External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell. 2010;142:284–295. doi: 10.1016/j.cell.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 35.Gutierrez JR, Balmuth AL, Ntoukakis V, Mucyn TS, Gimenez-Ibanez S, et al. Prf immune complexes of tomato are oligomeric and contain multiple Pto-like kinases that diversify effector recognition. Plant J. 2010;61:507–518. doi: 10.1111/j.1365-313X.2009.04078.x. [DOI] [PubMed] [Google Scholar]
- 36.Maekawa T, Cheng W, Spiridon L, Töller A, Lukasik E, et al. Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe. 2011;9:187–199. doi: 10.1016/j.chom.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 37.Shen Q, Bieri S, Zhou F, Haizel T, Shirasu K, et al. Recognition specificity and RAR1/SGT1 dependency in barley Mla disease resistance alleles to the powdery mildew fungus. Plant Cell. 2003;15:732–744. doi: 10.1105/tpc.009258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 39.Houterman PM, Cornelissen BJC, Rep M. Suppression of plant resistance gene-based immunity by a fungal effector. PLoS Pathogens. 2008;4:e1000061. doi: 10.1371/journal.ppat.1000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pedley KF, Martin GB. Molecular basis of Pto-mediated resistance to bacterial speck disease in tomato. Annu Rev Phytopathol. 2003;41:215–243. doi: 10.1146/annurev.phyto.41.121602.143032. [DOI] [PubMed] [Google Scholar]
- 41.Janjusevic R, Abramovitch RB, Martin GB, Stebbins CE. A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science. 2006;311:222–226. doi: 10.1126/science.1120131. [DOI] [PubMed] [Google Scholar]
- 42.Rosebrock TR, Zeng L, Brady JJ, Abramovitch RB, Xiao F, et al. A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature. 2007;448:370–374. doi: 10.1038/nature05966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ntoukakis V, Mucyn TS, Gimenez-Ibanez S, Chapman HC, Gutierrez JR, et al. Host inhibition of a bacterial virulence effector triggers immunity to infection. Science. 2009;324:784–787. doi: 10.1126/science.1169430. [DOI] [PubMed] [Google Scholar]
- 44.Kim YJ, Lin N-C, Martin GB. Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell. 2002;109:589–598. doi: 10.1016/s0092-8674(02)00743-2. [DOI] [PubMed] [Google Scholar]
- 45.Dou D, Kale SD, Wang X, Jiang RHY, Bruce NA, et al. RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. Plant Cell. 2008;20:1930–1947. doi: 10.1105/tpc.107.056093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Z, Halterman D. Analysis of proteins differentially accumulated during potato late blight resistance mediated by the RB resistance gene. Physiol Mol Plant Pathol. 2009;74:151–160. [Google Scholar]
- 47.Earley KW, Haag JR, Pontes O, Opper K, Juehne T, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 48.Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, et al. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng. 2007;104:34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Yeast two-hybrid screening of RB domain interactions. Each panel shows three independent transformants of identical genotypes. Positive and negative controls were provided by the CytoTrap system manufacturer. RB = full-length RB; CC = RB coiled-coil domain; NB = RB nucleotide binding domain; LRR = RB leucine-rich repeat domain. All spots contained similar quantities of yeast at the time of plating. Pictures were taken after 8 days of growth on selective media.
(TIF)
Protein blotting showing stability of IPI-O and CC SOS-domain fusions in yeast. A–D) Total yeast proteins were separated on separate acrylamide gels, blotted, and probed with SOS-specific antibody. Ponceau S stained PVDF membranes are shown below the results of protein blotting and antibody detection.
(TIF)
Protein blotting of the input protein for co-immunoprecipitation. Total protein from leaf sections agroinfiltrated with the indicated constructs was extracted and separated on an acrylamide gel. Protein was blotted and detected using a GFP-specific antibody (A) or a Myc-tag specific antibody (B). Ponceau S stained PVDF membranes are shown to demonstrate equal loading.
(TIF)
Amino acid sequence identity chart showing the pairwise percent identity between RB CC domains amplified from wild potato species. Sequence names contain the three-letter abbreviation for each species. Numbers after the species name represents the PCR clone number.
(TIF)
Sequence alignment of RB CC domain deduced amino acid sequences. Letters in black boxes are identical to the consensus sequence (shown at top of each row). Sequence names contain the three-letter abbreviation for each species. Numbers after the species name represents the PCR clone number.
(TIF)
IPI-O1 mutants containing L129P are stable in RB-transgenic N. benthamiana leaves. A protein blot was performed using total protein extracts following agroinfiltration with constructs expressing indicated HA-IPI-O1 mutants. The 18-kDa protein band represents the expected size of recombinant IPI-O1 mutants. Lane1: wild type IPI-O1; Lane2: non-infiltrated N. benthamiana; Lane3: IPI-O1 L129P; Lane4: IPI-O1 Y82K/L129P; Lane5: IPI-O1 V86G/L129P; Lane6: IPI-O1 L129P/S135G.
(TIF)
IPI-O1 L129P inhibits the HR induced by IPI-O1. A. tumefaciens strains expressing IPI-O mutants or the indicated controls were infiltrated into leaves of RB transgenic N. benthamiana plants. Leaves were photographed at 6 days after infiltration.
(TIF)
IPI-O4 inhibits the HR induced by the IPI-O4 P129L/G135S double mutant. A. tumefaciens strains expressing IPI-O mutants or the indicated controls were infiltrated into leaves of RB transgenic N. benthamiana plants. Leaves were photographed at 6 days after infiltration.
(TIF)
IPI-O4 K82Y, G86V, and G135S inhibit the HR induced by IPI-O1. A. tumefaciens strains expressing IPI-O mutants or the indicated controls were infiltrated into leaves of RB transgenic N. benthamiana plants. Leaves were photographed at 6 days after infiltration. Note that the inhibitory effect of these mutants is not as strong as that of IPI-O1 L129P since some cell death was still observed in the area coinfiltrated with IPI-O1 and IPI-O4 K82Y (A), G86V (B), or G135S (C).
(TIF)