Abstract
Background
Increased biotic and abiotic plant stresses due to climate change together with an expected global human population of over 9 billion by 2050 intensifies the demand for agricultural production on marginal lands. Soil salinity is one of the major abiotic stresses responsible for reduced crop productivity worldwide and the salinization of arable land has dramatically increased over the last few decades. Consequently, as land becomes less amenable for conventional agriculture, plants grown on marginal soils will be exposed to higher levels of soil salinity. Forage grasses are a critical component of feed used in livestock production worldwide, with many of these same species of grasses being utilized for lawns, erosion prevention, and recreation. Consequently, it is important to develop a better understanding of salt tolerance in forage and related grass species.
Findings
A gene encoding a ZnF protein was identified during the analysis of a salt-stress suppression subtractive hybridization (SSH) expression library from the forage grass species Festuca arundinacea. The expression pattern of FaZnF was compared to that of the well characterized gene for delta 1-pyrroline-5-carboxylate synthetase (P5CS), a key enzyme in proline biosynthesis, which was also identified in the salt-stress SSH library. The FaZnF and P5CS genes were both up-regulated in response to salt and drought stresses suggesting a role in dehydration stress. FaZnF was also up-regulated in response to heat and wounding, suggesting that it might have a more general function in multiple abiotic stress responses. Additionally, potential downstream targets of FaZnF (a MAPK [Mitogen-Activated Protein Kinase], GST [Glutathione-S-Transferase] and lipoxygenase L2) were found to be up-regulated in calli overexpressing FaZnF when compared to control cell lines.
Conclusions
This work provides evidence that FaZnF is an AN1/A20 zinc finger protein that is involved in the regulation of at least two pathways initiated by the salt stress response, thus furthering our understanding of the mechanisms of cellular action during a stress that is applicable to commercial crops worldwide.
Introduction
With an expected population of over 9 billion by 2050 and added abiotic and biotic plant stresses due to climate change, there is an increased demand for agricultural production on marginal lands. Additionally, the salinization of arable land has dramatically increased over the last few decades [1,2]. Soil salinity is one of the major abiotic stresses responsible for reduced crop productivity worldwide [3]. As land becomes less amenable for conventional agriculture, plants grown on marginal soils will be exposed to higher levels of soil salinity. Forage grasses are a critical component of feed used in livestock production worldwide, with many of these same species of grasses being utilized for lawns, erosion prevention, and recreation. Consequently, it is important to develop a better understanding of salt tolerance in forage and related grass species.
Since plants are sessile, they have developed mechanisms that enable them to sense stressful environmental conditions and to elicit complex interactions between signaling molecules and pathways to adapt to various stresses. In the genomic era, new methods for looking at plant abiotic stress responses have evolved. The availability of the genome sequence and other resources such as microarrays, knockout mutants, and ease of transformation of the model dicot, Arabidopsis, greatly advanced our understanding of how plants respond to stress, including knowledge on the various components utilized in the signaling and response pathways. More recently, the availability of sequencing in other model species as well as some crop species facilitated the use of microarrays to analyze genes that are up- or down-regulated in response to specific stresses [4,5]. As sequencing became less cost prohibitive, high-resolution transcript profiling has been used to identify stress related genes and pathways in both monocot and dicot species [5]. In response to salt or dehydration stress, small molecules such as ABA and calcium are utilized by plants to induce various signalling cascades. These pathways use various proteins such as phospholipases, kinases, calmodulin, calcium-binding proteins and transcription factors to activate genes and pathways necessary for water-related stress tolerance (for reviews see: [2,6-11]) and are often targets for genetic modification to improve salt/drought tolerance in plants [12,13].
One particular class of proteins which are involved in plant response to abiotic stresses are zinc finger proteins. Zinc finger proteins were first characterized by a motif present in a protein (TFIIIA) that contained zinc and had the ability to bind DNA [14]. Members of the zinc finger transcription factors are characterized by the number and arrangement of cysteine (C) and histidine (H) within the zinc finger (C2H2, C3H, C2C2, C3HC4, C2HC5) in combination with other hydrophobic amino acids essential for stabilizing the zinc finger [14,15]. The first plant-specific zinc finger, identified in petunia, belonged to the C2H2 type zinc finger family and contained a plant specific "QALGGH" domain [16]. In Arabidopsis, data mining revealed 171 genes coding for C2H2-type zinc finger proteins, of which 77 contained the plant specific "QALGGH" domain [17]. Similarly, 189 C2H2 zinc finger proteins were data mined in rice, of which 26 were up-regulated in response to abiotic stress (drought, and/or salt and/or cold), while 21 genes were down-regulated during abiotic stress [18]. Several Arabidopsis C2H2 zinc-finger proteins function as transcription repressors during drought, cold and high salinity stress conditions [19]. Many C2H2-type zinc finger proteins have been shown to have a role in stress responses in plants [16,19-25], and many of the genes for these proteins, when overexpressed in Arabidopsis or tobacco, were shown to improve stress tolerance.
Another family of zinc finger proteins is characterized by the presence of A20 and AN1 zinc finger domains. The A20 domain was first identified in the A20 protein which was induced by a tumor-necrosis-factor in human endothelial cells [26]. The AN1 domain was initially identified as a putative zinc finger domain in an ubiquitin-like protein (AN1) from Xenopus laevis [27]. The first plant A20/AN1 zinc finger protein (OSISAP1) identified and characterized from rice, was shown to be induced by multiple stresses including cold, desiccation, salt, submergence, heavy metals, and injury, as well as ABA (Abscisic Acid). When this gene was overexpressed in tobacco, it conferred tolerance to multiple stresses (cold, dehydration and salt) during germination and early seedling growth stages [28]. Subsequently Rice Stress Associated Proteins (OsSAP 1-18) containing A20 and/or AN1 zinc fingers were identified by in silico analysis and compared to Arabidopsis A20/AN1 SAPs (AtSAP 1-10) [29]. There are 11 Rice SAPs (OsSAP1-11) that contain single A20 and AN1 zinc finger domains at the N and C termini, respectively. OsSAP12 contains two A20 zinc finger domains at the N-terminus and an AN1 zinc finger at the C terminus. There are several OsSAP proteins that only contain one (OsSAP13-15) or two (OsSAP16-17) AN1 zinc finger domains, while OsSAP18 only contains the A20 zinc finger domain. Arabidopsis has 10 SAPs (AtSAP1-10) which contain both the A20 and AN1 zinc finger domains, 3 SAPs (AtSAP11-13) with two AN1 zinc-finger domains and one SAP (AtSAP14) with a single AN1 zinc finger [29].
Expression analysis under different abiotic stressful conditions revealed that many of these genes were induced in response to salt and dehydration stress. Overexpression of several of these genes or homologs from other species has been shown to increase tolerance to one or more of various abiotic stresses (salt, drought, cold, and/or heat) [28,30-33], but in one case overexpression increased tolerance to cold, but increased sensitivity to salt and drought [34]. Overexpression of AtSAP10 confers tolerance to heavy metals (nickel, manganese, zinc) and high temperature stress [35]. This zinc finger family of proteins shows potential for increasing or stabilizing crop production on marginal soils and during increasing abiotic stress conditions due to climate change.
In this paper, we describe the characterization of a gene encoding an A20/AN1 zinc-finger protein that was identified during the analysis of a salt-stress suppression subtractive hybridization (SSH) expression library in F. arundinacea, a moderately salt tolerant glycophyte http://www.salinitymanagement.org/Salinity%20Management%20Guide/cp/cp_7_table-1.html[36]. The expression pattern of FaZnF was compared to the well characterized dehydration stress tolerance gene, delta 1-pyrroline-5-carboxylate synthetase (P5CS). Expression levels of both genes were analyzed in response to different levels of salinity and to seven other forms of abiotic stress. F. arundinacea calli, which were transformed to over-express FaZnF, were used to identify potential downstream targets of FaZnF. In this report we present evidence that FaZnF plays a role in dehydration stress responses and is also responsive to heat and wounding stress in tall fescue.
Materials and methods
Plant materials
F. arundinacea (tall fescue) seeds were planted in 4-inch square pots (volume approximately 750 mL) in SB40 Sunshine Growing Mix (Sun Gro Horticulture, Canada). Plants were fertilized weekly using Technigro 20-18-20 all-purpose fertilizer (Sun Gro Horticulture, Canada). Plants were grown in a Conviron E15 (Conviron, Winnipeg, Canada) growth chamber under an 8-hr photoperiod at 21°C day and 18°C night. Plants were grown for 6 weeks and pots containing 12-16 plants were then used for various stress treatments described below. At the designated times, the aerial portions of 6-7 plants were collected, immediately frozen in liquid nitrogen and stored at -80°C for future analysis.
Stress treatments
Salt Stress
Plants were subjected to salt stress by treating the soil with 500 mL of 500 mM NaCl. Plants showed signs of mild wilting in the leaf blades after 1 hr. The aerial portions of 6-7 plants were collected 12 and 24 hr after salt treatment, immediately frozen in liquid nitrogen and stored at -80°C.
Salt concentration analysis
Plants were grown in soil for 6 weeks. Each pot containing 10-14 plants was treated with 500 mL of one of the following: 0 mM, 100 mM, 200 mM 300 mM, 400 mM or 500 mM NaCl salt solution. Plants treated with 0-300 mM NaCl did not show any visible signs of wilting over the 24 hr period, but plants treated with 400 and 500 mM NaCl showed signs of mild wilting but recovered within the 24-hr treatment period. The aerial portions of 6-7 plants were collected at 0, 12 and 24 hr, immediately frozen in liquid nitrogen and stored at -80°C.
Osmotic stress
Plants were subjected to osmotic stress by treating the soil with 500 mL of 12.87% polyethylene glycol 6000 solution (PEG). Plants showed mild wilting of leaf blades after 1 hr of treatment. The aerial portions of 6-7 plants were collected after 12 and 24 hr of stress treatment.
Wilt (Drought)
To induce drought stress, watering was stopped and pots were allowed to dry overnight. After 24 hr some very mild wilting was observed. Tissue was collected at 24 and 48 hr after watering was stopped.
UV Stress
Plants were laid on their side and irradiated for 5 min. Two hand-held 254-nm UV short-wave devices (Model UVG-11, Ultra-violet Products Inc, USA) were held 12-15 cm above the stems and leaves. Tissue limpness was observed after irradiation and samples from 6-8 plants were collected 1 and 8 hr post treatment.
Heat stress
Plants were subjected to 40°C in a Conviron E15 Growth Chamber to simulate heat stress. The plants were well watered and placed in a shallow pan of water to maintain adequate hydration during heat stress treatments. Tissue from 6-8 plants was collected after 2 and 8 hr of heat stress.
Wounding
Plants were mechanically wounded by closing a hemostat perpendicularly across the leaves and stems 3-5 times. Aerial plant tissue was collected 12 hr after wounding.
Cold stress
Plants were subjected to 4°C for 24 hr. Tissue was collected from plants after 24 hr in the cold.
Control tissue
Control tissue was collected from plants that were untreated and watered normally. All tissues were collected, immediately frozen in liquid nitrogen and stored at -80°C.
Cloning of FaZnF gene and construction of Agrobacterium vector, pVec8.Ubi-ZnF
The 5' end of FaZnF was isolated previously with a partial clone from a tall fescue salt-stress Subtractive Suppression Hybridization (SSH) library. This library was constructed and analyzed as described in an earlier paper except tall fescue tissue was used instead of L. temulentum [37]. In the analysis of the tall fescue SSH library, we confirmed salt induced expression of selected genes by Northern analysis (Dombrowski and Baldwin, unpublished results). Genes were selected based on homology to genes described in other systems that were shown to be involved in salt stress. Northern analysis confirmed that FaZnF was up-regulated by salt stress.
RACE (rapid amplification of cDNA ends) was used to isolate the 3' end of the gene. Total RNA was isolated from leaf/crown tissues of F. arundinacea plants which had been subjected to a 20-hr salt stress treatment, using TRIzol reagent (Life Technologies, Gaithersburg, MD) following the manufacturer's instructions. RNA was reverse transcribed to cDNA using SuperScript III Reverse Transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol with the modified oligo(dT) primer (5'-GGCCACGCGTCGACTAGTACT17-3'). The resulting cDNA was used for PCR amplification in a 25 μl reaction containing 1× HotStar Taq Master Mix (Qiagen, Chatsworth, CA), 1 μl cDNA, and 0.4 μM of each primer (5'-GCCCCCAAAGGCCCAAGCAGGTG-3' and 5'-GGCCACGCGTCGACTAGTAC-3'). The PCR cycle was as follows: an initial denaturation at 94°C for 14 min; 7 cycles of 94°C for 1 min, 72°C for 2.25 min; 35 cycles of 94°C for 1 min, 67°C for 45 sec, 72°C for 1 min; with a final extension at 72°C for 10 min. The initial PCR was followed with a nested PCR reaction using primers (5'-CAACTGCCGGTGCGGGAACCTGTACCTC and 5'-GGCCACGCGTCGACTAGTAC-3') and the same cycling conditions as used for the primary PCR. A 510-base pair product was gel purified, ligated into the p-GEM-T Easy vector (Promega, Madison, WI), cloned and sequenced. A BLAST search confirmed high homology to the ZnF genes from other species.
Primers were designed using the 5' sequence information from the initial salt-stress clone and the sequence from 3' RACE to obtain the complete FaZnF gene. An initial PCR was performed using primers (5'-GGGCAGGTCAGAATTGCTCG-3' and 5'-CGATTACTAGTTACTATTACCGGTTGCG-3') at a concentration of 0.6 μM each with 1× HotStarTaq Master Mix (Qiagen) and 1 μl of cDNA. Reaction conditions were as follows: an initial denaturation at 94°C for 14 min followed by 35 cycles of 94°C for 1 min, 52°C for 1.5 min, 72°C for 1.5 min; with a final extension at 72°C for 10 min. This PCR reaction was followed by nested PCR using the primers (5'-ATGGATCCCGCCGGAGAG-3'; BamH I site underlined) and (5'-ATGGTACCACAGATTACAGAGTGC-3'; Kpn I site underlined) at a concentration of 0.6 μM each with 1× HotStarTaq Master Mix (Qiagen) and 0.3 μl of the primary PCR product. Cycling parameters were as in the primary reaction with the exception that the annealing temperature was 59°C. An 848-bp product was gel purified, ligated into the p-GEM-T Easy vector (Promega), cloned and sequenced. A BLAST search confirmed high homology to other ZnF genes. The purified FaZnF clone was digested with Kpn I and BamH I and ligated into the BamH I/Kpn I digested pVec8.Ubi vector using T4 DNA ligase in 1× T4 ligase buffer (New England Biolabs, Ipswich, MA). The pVec8.Ubi vector was obtained from CSIRO (Commonwealth Scientific and Industrial Research Organisation; Australia) [38,39]. The final clone was sequenced to verify complete insertion into the vector.
RACE was also used to isolate the 5' and 3' regions of the P5CS partial clones identified by PCR-based SSH library analysis [37]. Total RNA from leaf/crown tissues of L. temulentum plants subjected to a 12-hr salt stress treatment was isolated using TRIzol reagent (Life Technologies, Gaithersburg, MD) following manufacturer's instructions. RNA was prepared for 5'RACE, and first-strand cDNA synthesis was performed with the GeneRacer™ Oligo dT Primer included with the SuperScript™ III RT module following the manufacturer's instructions (Invitrogen, Carlsbad, CA). For 5' RACE, the cDNA was amplified with a gene-specific primer (5'-TGCCTCTCGGAATAACAAGGTCAATCA-3') and the kit 5' primer. For 3' RACE, the cDNA was amplified with the kit 3' RACE primer and a gene specific primer (5'-TCGGCTGACATGGATATGGCAAAACG-3'). The PCR reaction conditions were as follows: 5 cycles of 98°C for 10 sec, 70°C for 5 sec (decreasing 2°C/cycle), 72°C for 1 min 30 sec; followed by 25 cycles of 98°C for 10 sec, 60°C for 5 sec, 72°C for 1 min 30 sec; followed by a final extension at 72°C for 10 min. PCR products were purified and sequenced. Based on these sequences, primers were designed to amplify the coding sequence with an extension homologous to the vector (underlined) at each end (5'-CGACTCTAGAGGATCCATGGGCAGGGGAGGCATCGGA-3' and 5'-CGGTACCCGGGGATCCGAATCCTCTACCTGCAATCAATG-3') to facilitate In-fusion PCR Cloning (Clonetech, Ca) for future experiments.
Phylogenetic analysis of FaZnF
The FaZnF cDNA and protein sequences were subjected to BLAST searches at NCBI (National Center for Biotechnology Information; http://blast.ncbi.nlm.nih.gov/) against the nr (nonredundant) nucleotide and protein databases to provide annotation information and ortholog sequences from other species [40,41]. Closely related DNA and protein sequences, the top alignment from each genera, were used for phylogenetic analysis and tree construction at the "Phylogeny.fr" website [42]. Additionally, the Stress Associated Protein (SAP) sequences from rice and Arabidopsis and the most closely related Brachypodium sequences containing at least one of each of the A20/AN1 domains were used for a phylogenetic analysis.
Probe construction
Primer3 software [43] was used to design primers for P5CS probe synthesis (5'-CATCAAGACCCTCTGTCTTG-3' and 5'-GTATATTCTGGGATAATGACAG-3') based on the LtP5CS sequence. RNA from salt stressed tall fescue tissue was used to produce cDNA, as described above, which was used for PCR to produce an ~1.2 kb DIG-labeled P5CS probe using the PCR DIG Probe Synthesis Kit (Roche-Applied Science, IN) following the manufacturer's instructions. The PCR reaction conditions were as follows: an initial denaturation at 94°C for 4 min; followed by 30 cycles of 94°C for 30 sec, 55°C for 30 sec, 72°C for 1.0 min; with a final extension at 72°C for 10 min. Probe was used at 2 μl/mL of hybridization solution (Note: FaP5CS and LtP5CS sequences have 96% homology in 1.1 kb of this region; data not shown).
Primer3 software [43] was used to design primers for ZnF probe synthesis (5'-CGAGGGCTTTCTCGTATCAGTA-3' and 5'-GAGTGCTAGCTAAATGCGAAGC-3') to produce an ~ 800 bp DIG-labeled ZnF probe using DIG labeled dUTP (Roche-Applied Science, IN) for PCR following the manufacturer's recommendations. ExTaq Polymerase (Takara, WI) was used with the following PCR reaction conditions: an initial denaturation at 95°C for 4 min; followed by 35 cycles of 94°C for 30 sec, 60°C for 30 sec, 72°C for 1.0 min; with a final extension at 72°C for 10 min. Probe was used at 2 μl/mL of hybridization solution.
RNA gel blot analysis of genes
Genes coding for P5CS and FaZnf were subjected to further analysis by RNA blot analysis. Harvested tissue from F. arundinacea plants that had been subjected to various stresses, was ground to a powder in liquid nitrogen using a precooled mortar and pestle. Total RNA extractions from these ground tissues were performed using TRIzol reagent (Life Technologies, Gaithersburg, MD) following the manufacturer's instructions. Ten μg of total RNA isolated from selected/treated F. arundinacea plant tissue was electrophoretically separated on 1.2% denaturing formaldehyde agarose gels and blotted onto Hybond N+ nylon membranes [44]. Regions of selected genes were amplified by PCR using gene-specific primers and DIG labeled dUTP. The membranes were hybridized overnight with a DIG-labeled probe in a solution containing 15% SDS, 0.25 M NaPO4 pH 7.2, 1 mM EDTA, 0.5% blocking solution (Blocking solution: 10% Boehringer-Mannheim blocking reagent, 100 mM maleic acid pH 8.0, and 1 M NaCl). Filters were washed in a solution containing 1% SDS, 20 mM NaPO4 pH 7.2, and 1 mM EDTA at 60°C for three 30-min washes. The blots were then blocked and incubated with anti-digoxigenin-alkaline phosphatase antibodies and washed following the manufacturer's instructions (Roche; IN). The chemiluminescent substrate, CDP-Star (Roche; IN), was applied to the blots and light emission was detected on X-ray film.
Transformation of suspension cells with FaZnF
Generation of tall fescue suspension cultured cell lines
Tall fescue (TF) suspension cultured cells were derived from calli induced when juvenile plants or seeds were cultured on Callus Induction Media D (CIM D: MS salts with 5 mg/L 2,4-D, 30 g sucrose, 8 g Phytagar and 110 mg/L Nitsch & Nitsch Vitamins). After 4 weeks of incubation, callus tissues generated from diced seed or explant material were visually selected and propagated by sub-culturing on the same medium every two weeks. After 2-3 months, the friable callus tissue that developed was transferred to 30 mL of liquid suspension induction medium (SIM: 4.3 g MS salts, 10 mg 2,4-D and 30 g sucrose) [45] in a 125-mL Erlenmeyer flask and placed on a rotary shaker (195 rpm) in the dark at RT. After 1-2 months, the established suspension cultured cell lines were transferred into maintenance medium (MM: MS basal medium containing/L; 5 mg 2,4-D, 30 g sucrose, 1 mg thiamine, 100 mg myo-inositol and 1 mM EDTA). All cell lines used in this study are over seven years old. TF suspension cell lines were maintained in 40 mL of medium in 125-mL Erlenmeyer flasks on orbital shakers (195 rpm) in the dark. Cells (10 mL) were sub-cultured every 7 days into 30 mL of fresh sterile medium.
Preparation of Agrobacterium
The Agrobacterium vector pVec8.Ubi-FaZnF was introduced into Agrobacterium AGL-1 using the freeze thaw method [46]. Agrobacterium AGL-1 containing pVec8.Ubi-FaZnF was cultured on YEP agar plates containing 30 μg/mL rifampicin, 80 μg/mL carbenicillin and 100 μg/mL spectinomycin and incubated at 28°C to obtain individual colonies. An individual colony was used for transformation studies following Gelvin's protocol [47] with a slight modification. Briefly AGL-1 containing pVec8.Ubi-FaZnF was grown overnight in YEP medium containing rifampicin at 30 μg/mL, carbenicillin at 80 μg/mL, and spectinomycin at 100 μg/mL. The next day, 0.5-1 mL of the Agrobacterium overnight culture was added to 50 mL of AB sucrose minimal medium containing rifampicin, carbenicillin and spectinomycin at the same concentration. After growing overnight at 28°C, the bacteria were centrifuged and resuspended in 50 mL induction medium containing 100 μM acetosyringone. The bacterial cultures were placed in two 50-mL conical Falcon tubes on a rocker shaker overnight at room temperature. Transformation was performed using Agrobacterium in the induction medium.
Agrobacterium mediated transformation of suspension cell cultures
Suspension cell cultures eight days after subculturing were used for transformation experiments. Cells were allowed to settle and then approximately 4 mL of cells was transferred from the flask to a sterile disposable 100 × 20 mm Petri dish (Fisher Cat. No. 0875711Z) for transformation. Excess media was removed from the plates, and 1 mL of Agrobacterium was added to the cells and allowed to sit for approximately 2 hr. Transformation efficiency was improved when 10 μl of 1 M NaOH and 50 μL of cysteine (Sigma C7880; St. Louis, MO.; Stock 20 mg/mL; final concentration 0.5 mg/mL) were added as two droplets to a clear area of each original plate. An additional 1 mL of Agrobacterium culture was first mixed with the NaOH and cysteine solutions and then mixed with the suspension cells. The plates were then wrapped with Nescofilm (Karlan Reseach Products; Cottonwood, AZ) and placed in the dark at RT for three days. Following co-cultivation, 3 mL of MM medium was added to the plate and pipetted over the cells to loosen the cells and bacteria that were adhering to the plate. The medium was removed from the plate and the cells were washed with 2 mL of MM medium containing 400 mg/L timentin (GlaxoSmithKline; Research Triangle Park, NC) and 40 mg/L L-cysteine. This medium was removed and replaced with 10 mL of the same medium. Cultures were grown at room temperature in the dark on an Innova platform shaker at 80 rpm. Three days later, hygromycin (Cat. #10687-010; Invitrogen; Carlsbad, CA) was added to a final concentration of 40 mg/L to select for transformants. The culture medium was removed each week and replaced with fresh media. After approximately 4-6 weeks, the transformed cells grew into larger clusters which were removed and placed on solid MM2 media containing 400 mg/L timentin, 40 mg/L hygromycin, 40 mg/L cysteine and 3 g/L gelrite (Research Products International; Mt. Prospect, IL). After approximately 2-4 weeks, clusters that grew were spread thinly on new plates and were allowed to grow for 2 weeks to maximize the possibility that the selected clusters were derived from a single transformation event. A single homogenous culture representing each original cluster was selected and grown up as an individual line. Five independent lines overexpressing FaZnF and two non-transformed control lines were used for the Reverse Transcription quantitative PCR experiments (RT qPCR).
RNA extraction and cDNA synthesis
Two non-transformed control calli (obtained from the same original suspension cell culture used for transformation) and five FaZnF overexpressing calli were harvested for RNA extraction. Briefly, 0.5 g of calli was submerged in Trizol and homogenized for 30 seconds (Ultra-Turrax® T25 at a setting of 4). Samples were then frozen at -80°C, quickly thawed at 37°C, and centrifuged for 5 min at ~12500 × g to remove cellular debris. Continuation of RNA extraction followed the manufacturer's protocol for Trizol (Invitrogen, Carlsbad, Ca). After RNA purification, samples were treated with Turbo DNase following the manufacturer's instructions (Ambion, Austin, TX), and cDNA was synthesized using 5 μg of RNA from each sample with the SUPERSCRIPT III RT kit (Invitrogen, Carlsbad, CA) primed with random hexamers.
RT qPCR analysis
Based on contig sequences for each potential stress-associated target gene (MYC, lipoxygenase L2, Gst24, and eIF1) and sequences of reference genes GAPDH and UBC for F. arundinacea from the TIGR database, specific qPCR primers were designed (Table 1). Two reference genes were used in this study to reduce the possibility that the selected genes might themselves be changing with overexpression of FAZnF and to increase confidence in the fold changes of other genes relative to reference genes. Sequences of all contigs/genes used in this paper are reported in Supplementary Figure 1. Roche LightCycler Probe Design Software 2.0 was used to design primers with an average melting temperature of 62°C that, when used for PCR, would produce a product between 75 and 125 bp. Quantitative PCR was performed in 20 μl reactions in 96-well plates with BioRad iQSyber Supermix using a BioRad iQ5 Real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA). Serial dilution standard curves from pooled cDNA samples were utilized to test the efficiency and specificity of each primer (Table 1). The efficiency calculation is based on the slope of the serially diluted points: E = -1 + 10(-1/slope), and the specificity of each primer was analyzed by its single point dissociation curve. The close range of efficiencies between the targets and controls allowed for a ΔΔCT analysis using GAPDH or UBC as calibrators [48]. Calculation of relative mRNA followed the equation: 2(-ΔCtSample - ΔCtCalibrator) as described by Livak and Schmittgen [49]. For each 20 μl reaction, 0.5 μl of sample cDNA was used. Analysis was performed from five independently transformed biological replicates of overexpressing calli and repeated on three independent qPCR plates. Reactions were performed with the following PCR cycle: Initial denaturation for 3 min at 95°C; 50 cycles of 15 sec at 95°C, 1 minute at 59°C; followed by a dissociation curve. Error was calculated as Standard Error of the Mean (SEM).
Table 1.
Gene target information.
| Name | Top Reference Alignment | Arabidopsis Homolog | Position | Forward primer Reverse primer 5'-3' |
% efficiency |
|---|---|---|---|---|---|
| eIF1 | ref|XP_478516.1| translational initiation factor eIF1 Oryza sativa |
AT1G20010 | 97 | GAAGAACGTCTCAAATTTCCTCG CAGTTGCTCAGAAACCATGAATC | 99 |
| GST24 | ref|XP_463733.1| glutathione S-transferase GST 24 | Oryza sativa |
AT1G10370 | 80 | AGAAGATCTCACCAACAAGAGC TCGCCGTGGAGGAGAAC | 95 |
| Lipoxy-genase L2 | ref|XP_469655.1| lipoxygenase L-2; Oryza sativa |
AT1G72520 | 361 | CACGAGCCTGCCATTGATTA TGTGGTTGTTCTTGACGATGA | 98 |
| MAPK1 | ref|XP_470659.1| Putative MAP kinase 1 Oryza sativa |
AT3G45640 | 148 | CCACGGAGAATTTGATAAAGGAAATAC TCCATCAGATTATTCGCTCAAATCAAG | 95 |
| FaZnF | TIGR Transcript Assembly TA567_4606 Putative Zn finger Festuca arundinacea |
n/a | 632 | TGTGCTACCTCACCGTCA TCAGGATGCCCAACAACTAGA | 96 |
| GAPDH | TIGR Transcript Assembly TA626_4606 Glycerol aldehyde dehydrogenase Festuca arundinacea |
n/a | 951 | ATGGGTTATGTTGAGGAGGATT TTGACGAAGTTGTCGTTCAGAG | 99 |
| UBC | TIGR Transcript Assembly DT703874 Ubiquitin conjugating enzyme Festuca arundinacea |
n/a | 239 | CGGCGGCTTCAACTACA CTCGCCAGCATAGAGTG | 102 |
Quantitative PCR primers for each gene and the relative position of the amplicon (center) in the Tall Fescue genes/contigs are indicated. Amplicon length was designed to be between 75-125 bp. Gene target references are also noted along with the PCR efficiency of each primer set. Where applicable, the corresponding Arabidopsis gene reference number of genes found to be up-regulated in Arabidopsis plants overexpressing OsSAP11 [32] are included.
Figure 1.
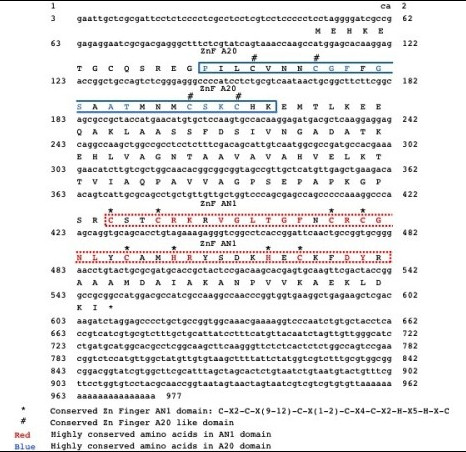
Festuca arundinacea zinc finger protein. cDNA with translated protein shown above with functional regions of the zinc finger protein designated. Asterisks (*) indicate the conserved AN1 zinc finger domain (C-X2-C-X(9-12)-C-X(1-2)-C-X4-C-X2-H-X5-H-X-C); number symbols (#) indicate a conserved A20-like zinc finger domain. The solid red outlined box indicates the highly conserved amino acids in the AN1 domain and the blue dotted box represents the highly conserved amino acids in the A20 domain.
Results and discussion
Cloning and sequence analysis of the ZnF protein gene from F. arundinacea
Suppression subtractive hybridization (SSH) was used to identify differentially expressed genes related to salt stress in F. arundinacea. Analysis of the SSH library (SSH) revealed a partial clone coding for a ZnF protein. The preliminary screening of selected genes by RNA blot analysis confirmed that FaZnF was up-regulated by salt stress. BLAST search results indicated high homology to a transcription factor. Since transcription factors have the potential to regulate plant responses to multiple stresses and given the high homology of FaZnF to stress associated proteins across species, we focused our attention on this gene/protein as a possible transcription factor capable of directing the salt stress response in tall fescue.
The whole gene sequence was obtained using 3' RACE (See Figure 1; Genbank Accession JN790818, to be released in April). Sequence analysis showed a full-length cDNA of 977 nucleotides, containing an open reading frame (ORF) of 501 nucleotides (ATG, 108-110; TAG, 609-611). To determine the function of this gene, this cDNA sequence was subjected to a BLAST search [50] against the nr nucleotide collection at the NCBI database and phylogenetic trees were constructed. The first gene listed on the BLAST results to FaZnF was from Triticum aestivum accession AK330210. The second gene on the list was a gene identified in osmotically stressed maize seedlings (Accession DQ244548) (See Phylogenetic Tree, Figure 2). Also included in the phylogenetic tree is "TA567_4606_Festuca" which was the top hit when FaZnF was subject to a BLAST search against the TIGR Plant Transcript Assemblies [51]. The identified ORF encoded a protein of 165 amino acids, with a potential molecular mass of 17.6 kDa and a pI of 8.6 (Figure 1). Using a BLAST protein similarity search, an additional phylogenetic tree was constructed [40]. The protein was most closely related to a predicted protein from Hordeum vulgare subsp vulgare (Accession number BAK03538.1) and the OsSAP8 from rice (Os06g0612800) (See Figure 3). Finally, to identify the most closely related stress associated zinc fingers containing at least one of each of the A20/AN1 domains from rice (OsSAPs) and Arabidopsis (AtSAPs) as well as the most closely related Brachypodium A20/AN1 genes (Brachypodium distachyon Acc ##; Bradi##), a phylogenetic tree was constructed. From this phylogenetic analysis, FaZnF was shown to be most closely related to Brachypodium Bradi1g36050.1, OsSAP 8 and 4, and AtSAP2 (See Figure 4) [42]. A gene closely related to OsSAP8 that was isolated from indica rice (OsiSAP8) has been characterized extensively and was also induced in response to multiple abiotic stresses including salt, drought, temperature, desiccation, submergence, wounding, heavy metals and ABA [52]. An OsiSAP8/GFP fusion protein was shown to be localized to the cytoplasm and the A20 and AN1 zinc finger domains were shown to interact using the yeast two-hybrid system suggesting that this zinc finger protein may function via protein-protein interactions [52].
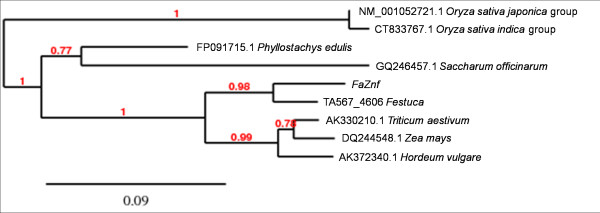
Figure 2.
Phylogenetic tree of FaZnF and related genes. The top BLAST hits from each genera obtained when FaZnF was used for BLAST analysis at NCBI [49]. Also included is "TA567_4606_Festuca" which was the top BLAST hit when TIGR Plant Transcript Assembly http://plantta.jcvi.org/search.shtml was queried with FaZnF [50]. This transcript assembly contained ESTs from a heat stressed cDNA subtraction library from tall fescue [80] and was most closely related to FaZnF. The scale bar represents .09 nucleotide substitutions per site, or 9 nucleotide differences per 100 nucleotides.
Figure 3.
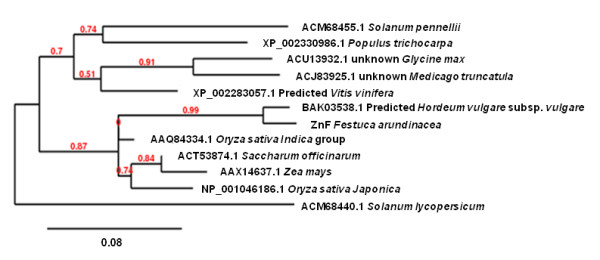
Phylogenetic tree of FaZnF and related proteins. The top BLAST hits from different genera (only the top hit from each genera is included) obtained when FaZnF was used for a protein BLAST analysis at NCBI [50]. The protein most closely related to FaZnF is a predicted protein from Hordeum vulgare which was identified in a subtraction cDNA library from seedling shoot and root treated with ABA. The scale bar represents .08 amino acid substitutions per site, or 8 amino acid differences per 100 amino acids.
Figure 4.
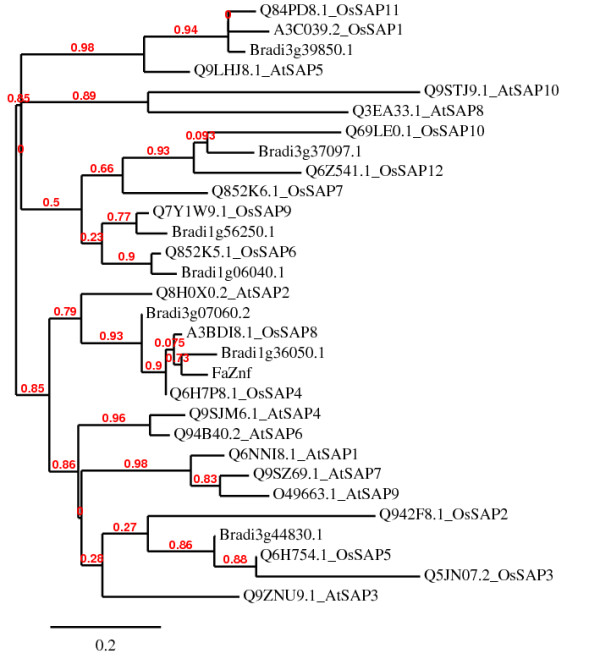
Alignment of Stress Associated Proteins. Alignment of Stress Associated Proteins (SAP) with both AN1/A20 domains from rice and Arabidopsis and most closely AN1/A20 domain-containing proteins from Brachypodium. FaZnF is most closely related to Brachypodium Bradi1g36050.1 and OsSAP8. The scale bar represent 0.2 amino acid substitutions per site, indicating 2 amino acid differences per 10 amino acids.
Response of FaZnF to salt stress
Since the FaZnF gene was initially isolated from salt stressed F. arundinacea plants, we wanted to investigate the kinetics of salt induced expression of this gene compared to the well characterized dehydration stress tolerance gene encoding a key enzyme in proline biosynthesis, delta 1-pyrroline-5-carboxylate synthetase (P5CS) [53]. Proline is an important osmoprotectant produced in plants in response to water related stresses [54-57] and recently was shown to be increased in tall fescue cultivars under drought stress conditions [58]. The P5CS gene has been shown previously to be involved in the biosynthesis of compatible solutes in response to salt stress in other plant species. Northern analysis of these two transcripts (FaZnF and P5CS) was performed in plants exposed to 500 mM NaCl over a 24 hr period. As shown in Figure 5A, FaZnF displayed a low level constitutive expression prior to salt stress (0 hr) and remained relatively constant until it started increasing at 4 hr post-stress induction and continued to increase through 16 hr and then decreased slightly from the 16-hr level to the 24-hr level. P5CS was not detectable prior to salt induction and started to increase slightly at 2 hr post-induction, gradually increasing to a maximum at 16 hr post-induction and slightly decreasing from that level at 24 hr.
Figure 5.
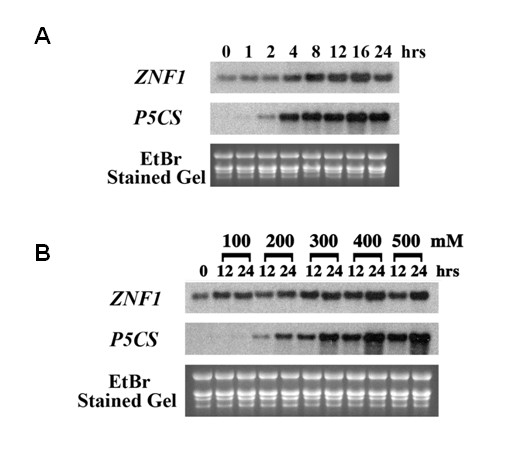
Northern Blot analysis of FaZnF and P5CS expression. (A) Northern Blot analysis of FaZnF and P5CS expression in response to salt stress for various time periods (0-24 hr). Total RNA was extracted from the aerial portions of plants that had been treated with 500 mM NaCl for the indicated number of hours. (B) Analysis of transcript levels of FaZnF and P5CS from plants treated with different levels of NaCl (0-500 mM) for 12 or 24 hr. Total RNA was extracted from the aerial portions of the plants that had been treated with different levels of NaCl (100-500 mM) for 12 or 24 hr, or with water (0 hr control). Ten μg of total RNA from each treatment was separated on a denaturing (formaldehyde) agarose gel. Equal loading was confirmed by visualization of the rRNA bands with ethidium bromide before (gel) and after transfer to the nylon membrane (blot). The RNA blots were hybridized with DIG-labeled DNA probes for ZNF1 and P5CS.
To investigate the sensitivity of FaZnF and P5CS to different levels of salt stress, eight-week-old mature plants were treated with increasing concentrations of NaCl for 12 and 24 hr, and gene expression was assessed by Northern blot analysis. As shown in Figure 5B, FaZnF was expressed at a low level in the absence of salt (0), increased slightly at 12 and 24 hr with both 100 and 200 mM salt, but was increased greatly at 300, 400 and 500 mM NaCl, indicating that FaZnF has a role in the salt stress response in tall fescue. P5CS started to show induction at 12 and 24 hr with 200 mM salt and increased steadily as the salt concentration increased to 500 mM.
The effect of different abiotic stresses on gene expression
The expression patterns of P5CS and FaZnF genes were examined in response to various abiotic stress conditions. In Figure 6, as shown in the previous figures, both genes were activated by treatment with 500 mM NaCl for 12 and 24 hr. PEG treatments resulted in a slight elevation of the FaZnF gene at 12 hr, which was back to control levels at 24 hr. FaZnF gene was induced by heat (moderate increase at 8 hr), wilting (slight increase at 24 hr, greatly increased at 48 hr) and wounding similar to OsiSAP8, but was not induced by cold or UV whereas OsiSAP8 was cold induced [52]. OsSAP4 was also shown to be induced by salt and dehydration [29]. Similar to most SAP genes that have been studied in plants, FaZnF was induced by multiple abiotic stresses. P5CS showed a slight induction 8 hr post UV treatment, during wilting (greatly increased at 48 hr), and was slightly increased after cold stress.
Figure 6.
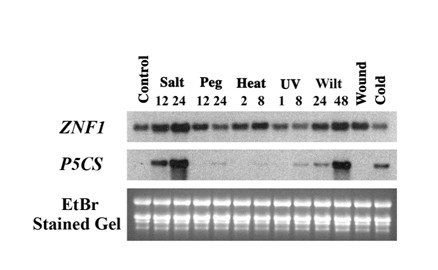
Gene expression analysis of FaZnF and P5CS after exposure to different abiotic stresses. Total RNA was extracted from the aerial portions of control plants (untreated and watered normally), from plants that had been subjected to different stresses for the indicated time periods, and from plants at specific time points after exposure to indicated stress (Salt: 500 mM NaCl; PEG: 12.87% PEG 6000; Heat: 40°C; UV: time after 5 min exposure to short-wave ultraviolet light; Wilting (Drought): no water for 24 or 48 hr; Wounding: 12 hr after mechanically wounded 3-5 times perpendicularly across the leaves; Cold: 4°C for 24 hr. Ten μg of total RNA from each treatment was separated on a denaturing (formaldehyde) agarose gel. Equal loading was confirmed by visualization of the rRNA bands with ethidium bromide before (gel) and after transfer to the nylon membrane (blot). The RNA blots were hybridized with DIG-labeled DNA probes for ZNF1 and P5CS genes.
Identification of possible targets of FaZnF
By identifying conserved domains within FaZnF, potential targets can be predicted using published target lists of orthologous proteins. To identify possible targets of FaZnf, SMART analysis was utilized to predict conserved protein domains http://smart.embl-heidelberg.de/. From this analysis, two specific zinc finger domains were identified including motif A20 (SMART accession SM00259) and zinc finger motif AN1 (SMART accession SM00154) with e-values of 1.53e-09 and 5.15e-16, respectively (Figure 1).
Some SAP proteins in Arabidopsis and rice have been localized to the nucleus [32,35] suggesting possible roles as transcription factors. Given the considerable amount of literature on A20/AN1 stress associated proteins (SAPs) in the model organism Arabidopsis, including microarrays documenting transcriptional changes in transgenic Arabidopsis overexpressing rice OsSAP11 or a protein that interacts with OsSAP11 (OsRLCK253) [32], we were able to compare a known Arabidposis A20/AN1 target list to the list of genes identified in the tall fescue salt SSH library. Of the 447 contig sequences from our salt SSH library, 38 sequences closely matched the Arabidopsis A20/AN1 data set through top reference alignment similarities. Confirmation of homology was then performed by tBLASTx analysis to Arabidopsis (SDSC workbench). Four representative contig sequences, each representative of a specific arm of the salt stress response, were identified and analyzed to determine if overexpression of FaZnF influences transcriptional activation; MAPK, GST24, lipoxygenase L-2 and eIF1.
MAPK gene expression is elevated in calli overexpressing FaZnF
Signal transduction through MAPK activation during salt stress is essential for activation of the high osmolarity glycerol pathway (HOG) [59] which in turn enables survival during high osmotic stress [60]. Though MAPK is post-transcriptionally activated through a cascade of phosphorylation events, transcriptional induction also occurs during salt stress in Arabidopsis [61]. Consistent with these observations, our salt subtraction library also indicates that MAPK transcription is induced with salt stress in F. arundinacea. However, the general mechanism of transcriptional activation of the MAPK gene during salt stress is not clear. To address whether FaZnF can influence the transcription of MAPK, RNA was isolated from control calli and calli overexpressing FaZnF and changes in transcription were quantified by RT qPCR. Collectively, these overexpressing calli showed an increase in FaZnF expression by greater than 12-fold compared to housekeeping genes GAPDH or UBC (Figure 7). Interestingly, expression of MAPK was also increased almost 3-fold in overexpressing FaZnF calli, thus suggesting the possibility that the A20/AN1 domain containing FaZnF enzyme might regulate expression of MAPK, directly or indirectly.
Figure 7.
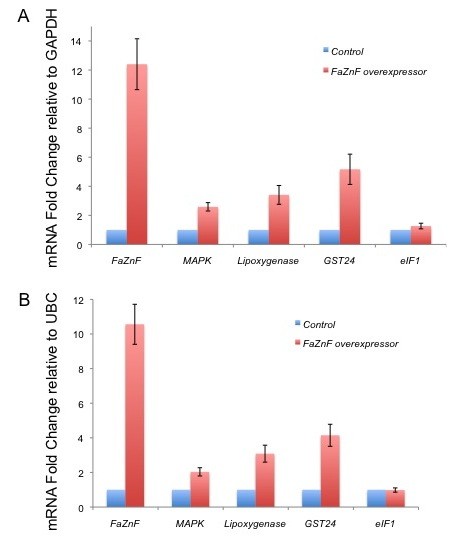
Overexpression of FaZnF increases transcription of selected salt stress genes. A) RT qPCR analysis of Festuca arundinacea calli using ΔΔCt quantification with GAPDH as the housekeeping gene. Values are normalized to control sample values (non-overexpressing calli) in order to represent relative expression changes. Error bars represent the SEM (Standard Error of the Mean). B) RT qPCR analysis of Festuca arundinacea calli using Ct quantification with UBC as the housekeeping gene. Values are normalized to control sample values (non-overexpressing calli) in order to represent relative expression changes. Error bars represent the SEM. Note that both housekeeping gene normalizations maintained the same trends in fold induction.
FaZnF influences transcription of oxidative stress pathway genes
Abiotic stressors such as salt stress increase the production of Reactive Oxygen Species (ROS), thus activating the oxidative stress pathway. Enzymes such as glutathione S-transferases are transcriptionally up-regulated to scavenge the elevated ROS to protect the organism [62]. Our salt subtraction library identified one major GST, GsT24 as being up-regulated with salt stress. Considering that overexpression of the highly homologous A20/AN1 SAP1 enzyme influenced transcription levels of many GSTs, experiments were performed to investigate whether FaZnF also influenced transcription of GsT24. Again, to determine transcriptional induction changes in mRNA, the expression of GsT24 was monitored in F. arundinacea control calli and FaZnF overexpressing calli by RT qPCR. Similar to MAPK expression, levels of GsT24 expression increased more than 5-fold in calli overexpressing FaZnF compared to control calli (Figure 7), strongly suggesting that this A20/AN1 protein influences the transcription of GsT24.
While combating oxidative stress, plants will utilize ROS as signaling molecules to either maximize the detoxification response or to accelerate cell death [62,63]. One way to intensify the production of ROS and potentially the oxidative stress response is through the up-regulation of lipoxygenases [64,65] which catalyze polyunsaturated fatty acid dioxygenation or pyridine nucleotide oxidation [66]. Many members of the lipoxygenase family are transcriptionally up-regulated with salt stress [67]. Similarly, lipoxygenase is up-regulated with salt stress in our salt SSH library, and overexpression of FaZnF results in a 3-fold increase in lipoxygenase L-2 transcript levels compared to housekeeping genes as determined by RT qPCR (Figure 7).
To ensure the most efficient use of cellular resources during salt or oxidative stress, translation is restricted [68,69]. Translation efficiency can be reduced by post-translational modification of the translation apparatus or through transcriptional regulation of key translation factors. However, similar to the heat shock stress response, the salt stress response can also be accompanied by salt stress recovery, where the cellular machinery anticipates increases in gene expression by increasing the transcription of translation factors [70]. Interestingly, overexpression of translation initiation factor eIF1A has been reported to increase salt tolerance in multiple organisms [71], and eIF1 was identified in our salt subtraction library as being up-regulated with salt stress. However, transcriptional analysis to determine whether overexpression of the A20/AN1 FaZnF results in increased eIF1 transcription did not show enhanced induction compared to housekeeping genes GAPDH or UBC (Figure 7). This result suggests that additional signals during salt stress might be required for activation of this translation initiation gene.
Conclusions
In summary, one of the biggest challenges to agricultural yield worldwide is increased soil salinity [3,72]. Plants responding to increased salinity have at least two survival response strategies: activation of the MAPK pathway to deal with subsequent osmotic stress, and activation of the oxidative stress pathway to deal with large increases in reactive oxygen species (ROS). For many plants, including forage crops such as F. arundinacea, increased soil salinity induces osmotic and oxidative stress responses [73]. However, the cellular mechanisms driving these stress responses in grasses have not been fully delineated. Here we describe a highly conserved, from humans to plants, Stress Associated Protein (SAP) that contains the unique combination of dual zinc finger motifs, A20/AN1, in the forage grass F. arundinacea. This protein could act both as a transcription factor in the nucleus and as a mediator of stability and function in the mitochondria and cytoplasm, similar to the mammalian stress factors p53 and XBP1 [74-77]. This work provides evidence that FaZnF is involved in the regulation of at least two pathways initiated by the salt stress response. It is not yet known if the FaZnF protein acts through ubiquitin-related mechanisms like AtSAP5 [30] and animal ZNF216 proteins [78], or perhaps through interactions with protein kinases like OsSAP11/1 [32] and ZNF216 [79], or through interactions with other SAP/nonSAP proteins or transcription factors, or as a transcription factor itself.
Future studies are necessary to determine the cellular localization and mechanism of function for FaZnF, though it is quite possible that FaZnF has many functions as an abiotic stress factor, as do many stress related proteins. It will also be of additional interest to determine if overexpression of FaZnF will lead to greater abiotic stress tolerance in grasses.
Abbreviations
ABA: abscisic acid; BLAST: basic local alignment search tool; CIM: callus induction medium; 2,4-D: 2,4-Dichlorophenoxyacetic acid; DNase: deoxyribonuclease; EDTA: ethylenediaminetetraacetic acid: FAO: Food and Agriculture Organization; FaZnF: Festuca arundinacea zinc finger protein; GST: glutathione-S-transferase; MAPK: mitogen-activated protein kinase; NaCl: sodium chloride; NaPO4: combination of NaH2PO4 and Na2HPO4; P5CS: 1-pyrroline-5-carboxylate synthetase; PCR: polymerase chain reaction; PEG: polyethylene glycol; RACE: random amplification of cDNA ends; ROS: reactive oxygen species; RT qPCR: reverse transcription quantitative PCR; SAP: stress associated protein; SDS: Sodium dodecyl sulfate; SSH: suppression subtraction hybridization; UV: ultraviolet.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
JED, JCB, and RCM conceived of the study and participated in its design. RCM was responsible for northern analysis and callus transformation. KGC designed and performed the RT qPCR analysis. RCM, KGC and JEB helped to draft the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Sequence of all genes/contigs used in this study are listed. Protein sequence of the open reading frame of FaZnF is also included. Note that primers used for quantitative RT-PCR are highlighted in yellow. Contigs refer to the contig number of clones from the Tall Fescue salt SSH library that were sequenced.
Contributor Information
Ruth C Martin, Email: Ruth.Martin@ars.usda.gov.
Kira Glover-Cutter, Email: Kira.Glover-Cutter@ars.usda.gov.
James C Baldwin, Email: James.Baldwin@wpafb.af.mil.
James E Dombrowski, Email: Jim.Dombrowski@ars.usda.gov.
Acknowledgements
Special thanks is extended to Dr. Ming-Bo Wang and Dr. Richard Brettell (CSIRO, Canberra Australia) for providing the pVec8 plasmid, to Thomas Lockwood (USDA-ARS, Corvallis OR) for his invaluable inputs and work on the tissue culture, transformation experiments and Northern Analysis, and to Vicky Hollenbeck for cloning the FaZnF gene and construction of the pVec FaZnF vector. Experimental methods performed in this research complied with current laws and regulations of the U.S.A. The use of trade, firm, or corporation names in this publication is for the information and convenience of the reader. Such use does not constitute an official endorsement or approval by the United States Department of Agriculture or the Agricultural Research Service of any product or service to the exclusion of others that may be suitable.
Relevance to ARS National Programs. NP205: Rangeland, Pasture and Forages: Component "Plant Resources". NP302: Plant Biological and Molecular Processes: Component "Biological Processes that Determine Plant Productivity and Quality".
References
- Wang W, Vinocur B, Altman A. Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta. 2003;218:1–14. doi: 10.1007/s00425-003-1105-5. [DOI] [PubMed] [Google Scholar]
- Munns R. Genes and salt tolerance: bringing them together. New Phytol. 2005;167:645–663. doi: 10.1111/j.1469-8137.2005.01487.x. [DOI] [PubMed] [Google Scholar]
- Food and Agriculture Organization, Rome. World agriculture: towards 2015/2030. Summary report. 2002. http://www.fao.org/docrep/004/y3557e/y3557e00.htm
- Deyholos MK. Making the most of drought and salinity transcriptomics. Plant Cell Environ. 2010;33:648–654. doi: 10.1111/j.1365-3040.2009.02092.x. [DOI] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K. Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J. 2010;61:1041–1052. doi: 10.1111/j.1365-313X.2010.04124.x. [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Schumaker K, Zhu JK. Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J Exp Bot. 2004;55:225–236. doi: 10.1093/jxb/erh005. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- Bartels D, Sunkar R. Drought and salt tolerance in plants. Crit Rev Plant Sci. 2005;24:23–58. doi: 10.1080/07352680590910410. [DOI] [Google Scholar]
- Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- Pardo JM. Biotechnology of water and salinity stress tolerance. Curr Opin Biotechnol. 2010;21:185–196. doi: 10.1016/j.copbio.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Ito Y, Yamaguchi-Shinozaki K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009;149:88–95. doi: 10.1104/pp.108.129791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain SS, Kayani MA, Amjad M. Transcription factors as tools to engineer enhanced drought stress tolerance in plants. Biotechnol Prog. 2011;27:297–306. doi: 10.1002/btpr.514. [DOI] [PubMed] [Google Scholar]
- Peleg Z, Apse MP, Blumwald E. Engineering salinity and water-stress tolerance in crop plants. Adv Bot Res. 2011;57:405–443. [Google Scholar]
- Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu Rev Biochem. 2010;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- Sugano S, Kaminaka H, Rybka Z, Catala R, Salinas J, Matsui K, Ohme-Takagi M, Takatsuji H. Stress-responsive zinc finger gene ZPT2-3 plays a role in drought tolerance in petunia. Plant J. 2003;36:830–841. doi: 10.1046/j.1365-313X.2003.01924.x. [DOI] [PubMed] [Google Scholar]
- Englbrecht CC, Schoof H, Böhm S. Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genomics. 2004;5:39. doi: 10.1186/1471-2164-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal P, Arora R, Ray S, Singh A, Singh V, Takatsuji H, Kapoor S, Tyagi A. Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol Biol. 2007;65:467–485. doi: 10.1007/s11103-007-9199-y. [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Maruyama K, Sakuma Y, Meshi T, Iwabuchi M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol. 2004;136:2734–2746. doi: 10.1104/pp.104.046599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Yang X, Wang M-M, Tang H-J, Ding L-Y, Shen Y, Zhang H-S. A novel rice C2H2-type zinc finger protein lacking DLN-box/EAR-motif plays a role in salt tolerance. Biochim Biophys Acta Gene Struct Expression. 2007;1769:220–227. doi: 10.1016/j.bbaexp.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Pan L, Yang Q, Chi X, Chen M, He Y, Yu S. AhZFP1, a cDNA encoding C2H2-type zinc finger protein, induced by salt stress in peanut (Arachis hypogaea L.) Bioinformatics and Biomedical Engineering (iCBBE), 2010 4th International Conference on June 18-20. 2010. pp. 1–7.
- An Y, Wang Y, Lou L, Zheng T, Qu G-Z. A novel zinc-finger-like gene from Tamarix hispida is involved in salt and osmotic tolerance. J Plant Res. 2011;124:689–697. doi: 10.1007/s10265-011-0403-4. [DOI] [PubMed] [Google Scholar]
- Gourcilleau D, Lenne C, Armenise C, Moulia B, Julien JL, Bronner G, Leblanc-Fournier N. Phylogenetic study of plant Q-type C2H2 zinc finger proteins and expression analysis of poplar genes in response to osmotic, cold and mechanical stresses. DNA Res. 2011;18:77–92. doi: 10.1093/dnares/dsr001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S-J, Guo S-Q, Yang X, Bao Y-M, Tang H-J, Sun H, Huang J, Zhang H-S. Functional analysis of a novel Cys2/His2-type zinc finger protein involved in salt tolerance in rice. J Exp Bot. 2010;61:2807–2818. doi: 10.1093/jxb/erq120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Wang X, Chen J. Zinc finger protein 1 (ThZF1) from salt cress Thellungiella halophila is a Cys-2/His-2-type transcription factor involved in drought and salt stress. Plant Cell Rep. 2007;26:497–506. doi: 10.1007/s00299-006-0248-9. [DOI] [PubMed] [Google Scholar]
- Opipari AW, Boguski MS, Dixit VM. The A20 cDNA induced by tumor necrosis factor alpha encodes a novel type of zinc finger protein. J Biol Chem. 1990;265:14705–14708. [PubMed] [Google Scholar]
- Linnen JM, Bailey CP, Weeks DL. Two related localized mRNAs from Xenopus laevis encode ubiquitin-like fusion proteins. Gene. 1993;128:181–188. doi: 10.1016/0378-1119(93)90561-G. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay A, Vij S, Tyagi AK. Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc Natl Acad Sci USA. 2004;101:6309–6314. doi: 10.1073/pnas.0401572101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vij S, Tyagi AK. Genome-wide analysis of the stress associated protein (SAP) gene family containing A20/AN1 zinc-finger(s) in rice and their phylogenetic relationship with Arabidopsis. Mol Genet Genomics. 2006;276:565–575. doi: 10.1007/s00438-006-0165-1. [DOI] [PubMed] [Google Scholar]
- Kang M, Fokar M, Abdelmageed H, Allen R. Arabidopsis SAP5 functions as a positive regulator of stress responses and exhibits E3 ubiquitin ligase activity. Plant Mol Biol. 2011;75:451–466. doi: 10.1007/s11103-011-9748-2. [DOI] [PubMed] [Google Scholar]
- Ben Saad R, Zouari N, Ben Ramdhan W, Azaza J, Meynard D, Guiderdoni E, Hassairi A. Improved drought and salt stress tolerance in transgenic tobacco overexpressing a novel A20/AN1 zinc-finger gene isolated from the halophyte grass Aeluropus littoralis. Plant Mol Biol. 2010;72:171–190. doi: 10.1007/s11103-009-9560-4. [DOI] [PubMed] [Google Scholar]
- Giri J, Vij S, Dansana PK, Tyagi AK. Rice A20/AN1 zinc-finger containing stress-associated proteins (SAP1/11) and a receptor-like cytoplasmic kinase (OsRLCK253) interact via A20 zinc-finger and confer abiotic stress tolerance in transgenic Arabidopsis plants. New Phytol. 2011;191:721–732. doi: 10.1111/j.1469-8137.2011.03740.x. [DOI] [PubMed] [Google Scholar]
- Huang J, Wang M-M, Jiang Y, Bao Y-M, Huang X, Sun H, Xu D-Q, Lan H-X, Zhang H-S. Expression analysis of rice A20/AN1-type zinc finger genes and characterization of ZFP177 that contributes to temperature stress tolerance. Gene. 2008;420:135–144. doi: 10.1016/j.gene.2008.05.019. [DOI] [PubMed] [Google Scholar]
- Xuan N, Jin Y, Zhang H, Xie Y, Liu Y, Wang G. A putative maize zinc-finger protein gene, ZmAN13, participates in abiotic stress response. Plant Cell Tiss Org Cult. 2011;107:1–12. doi: 10.1007/s11240-011-9950-6. [DOI] [Google Scholar]
- Dixit AR, Dhankher OP. A novel stress-associated protein 'AtSAP10' from Arabidopsis thaliana confers tolerance to nickel, manganese, zinc, and high temperature stress. PLoS ONE. 2011;6:e20921. doi: 10.1371/journal.pone.0020921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels JG, Rink R, Jensen K. Stress tolerance and biotic interactions determine plant zonation patterns in estuarine marshes during seedling emergence and early establishment. J Ecol. 2011;99:277–287. doi: 10.1111/j.1365-2745.2010.01745.x. [DOI] [Google Scholar]
- Baldwin JC, Dombrowski JE. Evaluation of Lolium temulentum as a model grass species for the study of salinity stress by PCR-based subtractive suppression hybridization analysis. Plant Sci. 2006;171:459–469. doi: 10.1016/j.plantsci.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Wang M-B, Matthews PR, Upadhyaya MN, Waterhouse PM. Improved vectors for Agrobacterium tumefaciens-mediated plant transformation. Acta Hortic. 1998;461:401–407. [Google Scholar]
- Wang MB, Upadhyaya MN, Brettell RIS, Waterhouse PM. Intron mediated improvement of a selectable marker gene for plant transformation using Agrobacterium tumefaciens. J Genet Breeding. 1997;51:325–334. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M. et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M. et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. In: Bioinformatics Methods and Protocols: Methods in Molecular Biology. Krawetz S, Misener S, editor. Totowa, NJ: Humana Press; 2000. Primer3 on the WWW for general users and for biologist programmers; pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russel DW, editor. Molecular Cloning, A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Press; 2001. [Google Scholar]
- Dalton SJ. Plant regeneration from cell suspension protoplasts of Festuca arundinacea Schreb, Lolium perenne L and Lolium multiflorum Lam. Plant Cell Tiss Org Cult. 1988;12:137–140. doi: 10.1007/BF00040075. [DOI] [Google Scholar]
- Hofgen R, Willmitzer L. Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 1988;16:9877. doi: 10.1093/nar/16.20.9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agro Induction Protocol.doc. http://plant-tc.cfans.umn.edu/listserv/2002/log0206/msg00166.html
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL. et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- Childs KL, Hamilton JP, Zhu W, Ly E, Cheung F, Wu H, Rabinowicz PD, Town CD, Buell CR, Chan AP. The TIGR plant transcript assemblies database. Nucleic Acids Res. 2007;35:D846–851. doi: 10.1093/nar/gkl785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanneganti V, Gupta AK. Overexpression of OsiSAP8, a member of stress associated protein (SAP) gene family of rice confers tolerance to salt, drought and cold stress in transgenic tobacco and rice. Plant Mol Biol. 2008;66:445–462. doi: 10.1007/s11103-007-9284-2. [DOI] [PubMed] [Google Scholar]
- Hu C-AA, Delauney AJ, Verma PS. A bifunctional enzyme (Δl-pyrroline-5-carboxylate synthetase) catalyzes the first two steps in proline biosynthesis in plants. Proc Natl Acad Sci USA. 1992;89:9354–9358. doi: 10.1073/pnas.89.19.9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delauney AJ, Verma DPS. Proline biosynthesis and osmoregulation in plants. Plant J. 1993;4:215–223. doi: 10.1046/j.1365-313X.1993.04020215.x. [DOI] [Google Scholar]
- Yoshiba Y, Kiyosue T, Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K. Regulation of levels of proline as an osmolyte in plants under water stress. Plant Cell Physiol. 1997;38:1095–1102. doi: 10.1093/oxfordjournals.pcp.a029093. [DOI] [PubMed] [Google Scholar]
- Kishor PBK, Sangam S, Amrutha RN, Laxmi PS, Naidu KR, Rao KRSS, Rao S, Reddy KJ, Theriappan P, Sreenivasulu N. Regulation of proline biosynthesis, degradation, uptake and transport in higher plants: Its implications in plant growth and abiotic stress tolerance. Curr Sci. 2005;88:424–438. [Google Scholar]
- Szabados L, Savouré A. Proline: a multifunctional amino acid. Trends Plant Sci. 2010;15:89–97. doi: 10.1016/j.tplants.2009.11.009. [DOI] [PubMed] [Google Scholar]
- Man D, Bao Y-X, Han L-B. Drought tolerance associated with proline and hormone metabolism in two tall fescue cultivars. HortScience. 2011;46:1027–1032. [Google Scholar]
- Covic L, Silva NF, Lew RR. Functional characterization of ARAKIN (ATMEKK1): a possible mediator in an osmotic stress response pathway in higher plants. Biochim Biophys Acta Gene Regul Mech. 1999;1451:242–254. doi: 10.1016/s0167-4889(99)00096-8. [DOI] [PubMed] [Google Scholar]
- Westfall PJ, Patterson JC, Chen RE, Thorner J. Stress resistance and signal fidelity independent of nuclear MAPK function. Proc Natl Acad Sci USA. 2008;105:12212–12217. doi: 10.1073/pnas.0805797105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Irie K, Hirayama T, Hayashida N, Yamaguchi-Shinozaki K, Matsumoto K, Shinozaki K. A gene encoding a mitogen-activated protein kinase kinase kinase is induced simultaneously with genes for a mitogen-activated protein kinase and an S6 ribosomal protein kinase by touch, cold, and water stress in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1996;93:765–769. doi: 10.1073/pnas.93.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke A, Forzani C, Hirt H. Forum Review: Reactive oxygen species signaling in plants. Antioxid Redox Signal. 2006;8:1757–1764. doi: 10.1089/ars.2006.8.1757. [DOI] [PubMed] [Google Scholar]
- Van Breusegem F, Dat JF. Reactive oxygen species in plant cell death. Plant Physiol. 2006;141:384–390. doi: 10.1104/pp.106.078295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiteller G. The relationship between changes in the cell wall, lipid peroxidation, proliferation, senescence and cell death. Physiol Plant. 2003;119:5–18. doi: 10.1034/j.1399-3054.2003.00097.x. [DOI] [Google Scholar]
- Roy P, Roy SK, Mitra A, Kulkarni AP. Superoxide generation by lipoxygenase in the presence of NADH and NADPH. Biochim Biophys Acta Lipids Lipid Met. 1994;1214:171–179. doi: 10.1016/0005-2760(94)90041-8. [DOI] [PubMed] [Google Scholar]
- Needleman P, Truk J, Jakschik BA, Morrison AR, Lefkowith JB. Arachidonic Acid Metabolism. Annu Rev Biochem. 1986;55:69–102. doi: 10.1146/annurev.bi.55.070186.000441. [DOI] [PubMed] [Google Scholar]
- Kreps JA. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002;130:2129–2141. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenton D. Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J Biol Chem. 2006;281:29011–29021. doi: 10.1074/jbc.M601545200. [DOI] [PubMed] [Google Scholar]
- Rausell A, Kanhonou R, Yenush L, Serrano R, Ros R. The translation initiation factor eIF1A is an important determinant in the tolerance to NaCl stress in yeast and plants. Plant J. 2003;34:257–267. doi: 10.1046/j.1365-313X.2003.01719.x. [DOI] [PubMed] [Google Scholar]
- Gu R, Fonseca S, Puskas LG, Hackler LJ, Zvara A, Dudits D, Pais MS. Transcript identification and profiling during salt stress and recovery of Populus euphratica. Tree Physiol. 2004;24:265–276. doi: 10.1093/treephys/24.3.265. [DOI] [PubMed] [Google Scholar]
- Diédhiou CJ, Popova OV, Dietz KJ, Golldack D. The SUI-homologous translation initiation factor eIF-1 is involved in regulation of ion homeostasis in rice. Plant Biol. 2008;10:298–309. doi: 10.1111/j.1438-8677.2008.00037.x. [DOI] [PubMed] [Google Scholar]
- Zhu J-K. Plant salt tolerance. Trends Plant Sci. 2001;6:66–71. doi: 10.1016/S1360-1385(00)01838-0. [DOI] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu J-K, Bohnert HJ. Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:463–499. doi: 10.1146/annurev.arplant.51.1.463. [DOI] [PubMed] [Google Scholar]
- Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM. p53 has a direct apoptogenic role at the mitochondria. Mol Cell. 2003;11:577–590. doi: 10.1016/S1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- Morselli E, Galluzzi L, Kroemer G. Mechanisms of p53-mediated mitochondrial membrane permeabilization. Cell Res. 2008;18:708–710. doi: 10.1038/cr.2008.77. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Lee J, Reno CM, Sun C, Park SW, Chung J, Fisher SJ, White MF, Biddinger SB, Ozcan U. Regulation of glucose homeostasis through a XBP-1-FoxO1 interaction. Nat Med. 2011;17:356–365. doi: 10.1038/nm.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Uemura A, Mori K. pXBP1(U), a negative regulator of the unfolded protein response activator pXBP1(S), targets ATF6 but not ATF4 in proteasome-mediated degradation. Cell Struct Funct. 2009;34:1–10. doi: 10.1247/csf.06028. [DOI] [PubMed] [Google Scholar]
- Hishiya A, Lemura S, Natsume T, Takayama S, Ikeda K, Watanabe K. A novel ubiquitin-binding protein ZNF216 functioning in muscle atrophy. EMBO J. 2006;25:554–564. doi: 10.1038/sj.emboj.7600945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Teng L, Li L, Liu T, Chen D, Xu LG, Zhai Z, Shu HB. ZnF216 is an A20-like and IκB kinase γ-interacting inhibitor of NFκB activation. J Biol Chem. 2004;279:16847–16853. doi: 10.1074/jbc.M309491200. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Mian MA, Chekhovskiy K, So S, Kupfer D, Lai H, Roe BA. Differential gene expression in Festuca under heat stress conditions. J Exp Bot. 2005;56:897–907. doi: 10.1093/jxb/eri082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence of all genes/contigs used in this study are listed. Protein sequence of the open reading frame of FaZnF is also included. Note that primers used for quantitative RT-PCR are highlighted in yellow. Contigs refer to the contig number of clones from the Tall Fescue salt SSH library that were sequenced.