Abstract
Background
The development of cervical cancer and its high-grade precursor lesions (Cervical Intraepithelial Neoplasia grade 2/3 [CIN2/3]) result from a persistent infection with high-risk human papillomavirus (hrHPV) types and the accumulation of (epi)genetic host cell aberrations. Epidemiological studies have demonstrated variable CIN2/3 and cancer risks between different hrHPV types. Recent genomic profiling studies revealed substantial heterogeneity in the chromosomal aberrations detected in morphologically indistinguishable CIN2/3 suggestive of varying cancer risk. The current study aimed to investigate whether CIN2/3 with different hrHPV types vary with respect to their chromosomal profiles, both in terms of the number of aberrations and chromosomal loci affected.
Methods
Chromosomal profiles were determined of 43 p16INK4a-immunopositive CIN2/3 of women with long-term hrHPV infection (≥ 5 years). Sixteen lesions harboured HPV16, 3 HPV18, 14 HPV31, 1 HPV33, 4 HPV45, 1 HPV51, 2 HPV52 and 2 HPV58.
Results
Unsupervised hierarchical clustering analysis of the chromosomal profiles revealed two major clusters, characterised by either few or multiple chromosomal aberrations, respectively. A majority of 87.5% of lesions with HPV16 were in the cluster with relatively few aberrations, whereas no such unbalanced distribution was seen for lesions harbouring other hrHPV types. Analysis of the two most prevalent types (HPV16 and HPV31) in this data set revealed a three-fold increase in the number of losses in lesions with HPV31 compared to HPV16-positive lesions. In particular, losses at chromosomes 2q, 4p, 4q, 6p, 6q, 8q & 17p and gain at 1p & 1q were significantly more frequent in HPV31-positive lesions (FDR < 0.2).
Conclusions
Chromosomal aberrations in CIN2/3 are at least in part related to the hrHPV type present. The relatively low number of chromosomal aberrations observed in HPV16-positive CIN2/3 suggests that the development of these lesions is less dependent on genetic insult than those caused by other types like HPV31.
Keywords: Array CGH, Cervical cancer, Chromosomal aberrations, High-grade cervical intraepithelial neoplasia, HPV
Background
Persistent infection with mucosal high-risk human papillomaviruses (hrHPVs) has been causally related to the development of cervical cancer [1]. HPV types can be grouped into genera (α, β, γ, μ, η) with types belonging to the same genus generally sharing common characteristics, such as tissue tropism and oncogenic potential [2]. The hrHPV types encompass the α5, α6, α7, α9 and α11 species of the α-genus. There are around 13 hrHPV types, of which types 16 (α9 species) and 18 (α7 species) are the cause of approximately 70% of all cervical cancers [3]. Though infection with hrHPV is common, the majority of infections are cleared by the immune system. Only in some instances pre-cancerous lesions arise, so-called cervical intraepithelial neoplasia (CIN) [4]. Not all CIN represent direct precursor stages of cervical cancer. Low-grade CIN (CIN1) mostly reflect productive hrHPV infections, in which active viral replication and virion production are strongly related to the differentiation programme of the infected epithelium [5]. High-grade CIN (CIN2/3) usually harbour transforming infections, characterised by deregulated expression of viral oncogenes E6 and E7 in proliferating cells [5]. These lesions have the potential of malignant progression towards invasive carcinoma, largely due to the inactivation of tumour suppressors p53 and pRb by viral oncoproteins. This results in the accumulation of specific (epi)genetic changes in the host cell genome that may further drive the progression to a malignant phenotype [1,6]. One of the features of a transforming infection is over-expression of p16INK4a due to deregulated E7 expression, making p16INK4a a suitable marker to distinguish cervical pre-cancer from productive viral infections [7-10].
CIN2/3 are believed to represent a heterogeneous disease. While these may be rapidly induced within 2-3 years following hrHPV infection, progression to invasive cervical carcinoma may still take another 10-30 years [5,11,12]. The heterogeneous nature is substantiated by the fact that some features common to cervical carcinomas are only found in a subset of CIN2/3. These include for instance up-regulated hTERT, VEGF, c-fms and COX-2 expression, but also methylation-mediated silencing of (candidate) tumour suppressor genes such as CADM1, MAL, CALCA, RARβ2, TFPI2, SPARC, CCNA and hsa-miR-124 [13-20]. Our previous studies demonstrated that heterogeneity also exists at the chromosomal level [21]. Comparative genomic hybridisation microarray (arrayCGH) analysis of p16INK4a-positive CIN2/3 demonstrated two subsets, one showing few chromosomal aberrations and the second subset containing multiple aberrations akin to those found in cervical carcinomas [21]. We found that this heterogeneity is, at least in part, related to varying duration of lesion existence, as was approximated by the duration of preceding hrHPV infection [6]. CIN3 with a long-term HPV infection (≥ 5 years) had a significantly higher number of chromosomal aberrations compared to CIN3 with a short-term HPV infection (< 5 years).
There is now compelling evidence that different hrHPV types confer variable risks of CIN2/3 and cervical cancer. This likely reflects various viral properties that become manifest at different stages during cervical cancer development. Firstly, the risk of developing CIN2/3 is strongly coupled to type-specific differences in viral persistence, most likely reflecting different efficiencies in evading the immune system [22,23]. Secondly, carcinogenic properties seem to vary between different hrHPV types, as reflected by their diverse prevalence in CIN2/3 versus cervical cancer [24-31]. Particularly HPV types 16 and 18 predominate in cervical cancers, whereas other types, like 31 and 33 are relatively more prevalent in CIN2/3 [24-31]. To what extent these differences in transforming properties are reflected by type-specific differences in chromosomal aberrations is still unknown.
The aim of the current study was to investigate whether CIN2/3 containing different hrHPV types also differ with respect to their chromosomal signatures, both at the numerical level (i.e. the number of aberrations) and structural level (i.e. the specific loci affected). Since our previous study showed that the difference in number of aberrations was correlated to duration of preceding hrHPV infection, we only included cases with more than 5 years preceding hrHPV infection in order to minimise bias due to duration of existence. In addition to comparison of HPV16-positive lesions to those harbouring all other high-risk types (HPVnon16) and HPV31-positive cases, also, CIN2/3 with HPV types from the α9 species (HPVα9, types 16, 31, 33, 52, 58) were compared to those with α5&α7 species combined (HPVα5&α7, types 18, 45, 51). The latter branches are located in close proximity to each other in the phylogenetic tree of HPV types and are clearly separated from the α9 species [32].
Methods
Tissue specimens
Formalin-fixed paraffin embedded CIN2/3 biopsies (4 CIN2, 29 CIN3) of 43 women who participated in the population based screening study Amsterdam (POBASCAM) were used in this study [33]. A subset (n = 24) of these lesions has also been described previously [6]. Lesions were detected in the second screening round of the POBASCAM trial in women who at baseline, 5 years earlier, had an hrHPV-positive smear with normal cytology. The lesions contained the same hrHPV type present in the baseline smear, as determined using the GP5+/6+ RLB system [34]. Sixteen lesions harboured HPV16 (α9), 3 HPV18 (α7), 14 HPV31 (α9), 1 HPV33 (α9), 4 HPV45 (α7), 1 HPV51 (α5), 2 HPV52 (α9) and 2 HPV58 (α9). With exception of two cases the lesions only harboured one hrHPV type. In case of lesion number HPV16.12, HPV16 was the type detected at both baseline and in the biopsy. However, the biopsy contained HPV51 as well. In case of lesion number HPV18.1, HPV18 was detected at both baseline and in the biopsy, the latter contained HPV16 as well. Given the history of HPV type persistence, these lesions were classified as being caused by HPV16 and HPV18, respectively. Immunohistochemical staining for p16INK4a was performed using p16INK4a Ab-4, Clone 16P04 (Lab Vision Corporation, Neomarkers, Fremont, California, USA). All lesions showed diffuse staining for p16INK4a in at least the lower 2/3 of the epithelium, indicating the presence of a transforming infection. Histological review of lesions was performed by two experienced pathologists (dr. F.J.van Kemenade and dr. M.C.G Bleeker). The average age of the women was 41, ranging from 34-56 years. There was no significant difference in the average age of the women diagnosed with lesions containing HPV16 compared to those containing HPV31 or other types of hrHPV. This study was approved by the Institutional Review Board of the VU University Medical Center.
ArrayCGH of microdissected tissues
Microdissection of dysplastic areas of the biopsies, DNA extraction, amplification, labeling and across arrayCGH hybridisation to the 2×105 K arrayCGH platform (Agilent Technologies, Palo Alto, USA) as well as hierarchical clustering was performed as described previously [6]. ArrayCGH analysis of a subset (n = 24) of lesions included in the present study was also performed in this previous study. Array data is available from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/projects/geo/) through series accession number GSE31241.
Statistical analysis
Cluster assignment of lesions with HPV16 and those with types other than HPV16 (HPVnon16) was compared using the two-tailed Fisher exact-test. The non-parametric Mann-Whitney-U test was used for comparisons of the proportion of altered features (percentage of oligonucleotides deviating from the normal state) between lesions with HPV16 versus those with HPVnon16 or HPV31, as well as comparisons between lesions with hrHPV types from the α9 species (HPVα9) versus those from the α5 and α7 species combined (HPVα5&α7). The χ2-test (CGHtest_version_1.1) was used to determine whether there was an association between chromosomal aberration patterns of CIN2/3 and hrHPV type [6,35]. The test procedure includes a permutation-based false discovery rate (FDR) correction for multiple testing. Chromosomal profiles with < 1.5% altered probes in total were excluded from this test in order to only investigate regions that were differentially affected. It should be noted that the lesions excluded from this analysis were equally distributed over the HPV types present. Differences were considered significant if the false discovery rate (FDR) was < 0.2. Pathway analysis was performed on all genes located within the significantly different chromosomal regions using Ingenuity Pathway Analysis version 8.7, build 99759; content version 3203, build ing_merak (Ingenuity Systems, Redwood City, California).
Results
Clustering analysis displays the heterogeneity of CIN2/3
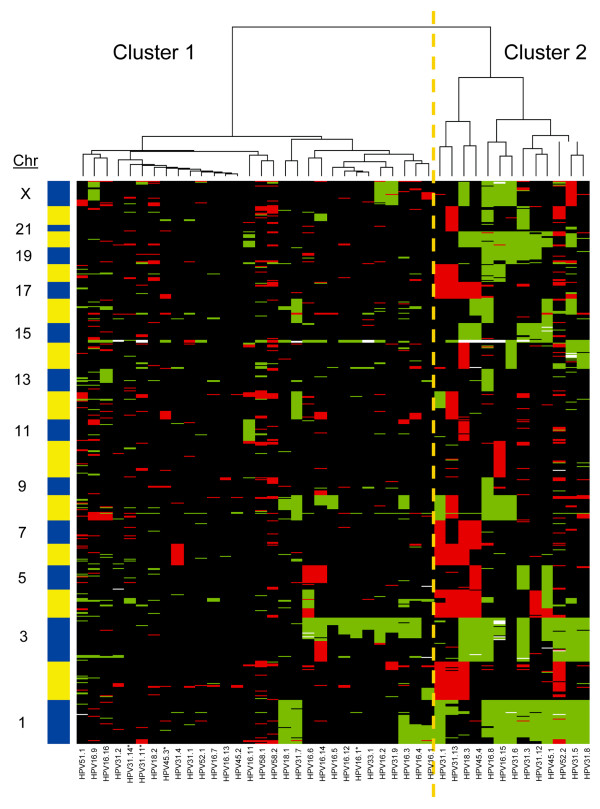
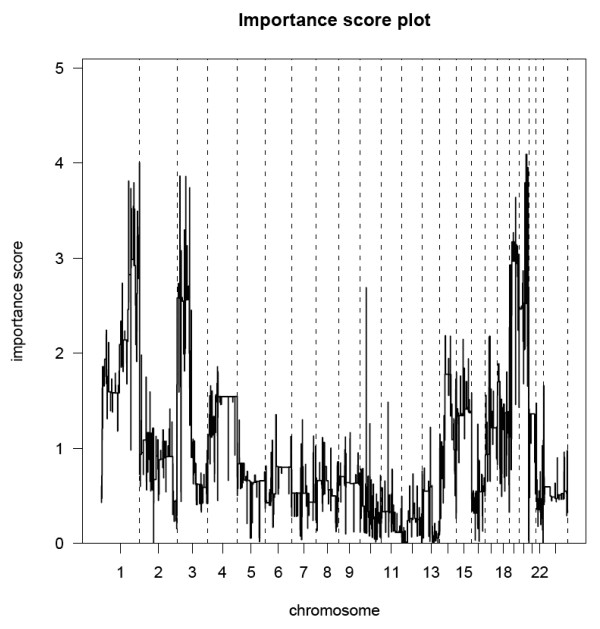
Chromosomal profiles of 43 p16INK4a-positive, microdissected CIN2/3 were determined using high-resolution arrayCGH. Unsupervised hierarchical clustering analysis was performed to compare these profiles in an unbiased manner as a first investigation into a possible association of aberration pattern with hrHPV type. Two main clusters were identified (Figure 1). Cluster 1 contained 30 samples with few aberrations, including 14 cases with HPV16, 2 with HPV18, 7 with HPV31, 1 with HPV33, 2 with HPV45, 1 with HPV51, 1 with HPV52 and 2 with HPV58. Cluster 2 contained 13 samples, including 2 cases with HPV16, 1 with HPV18, 7 with HPV31, 2 with HPV45 and 1 with HPV52. The difference in distribution of lesions with HPV16 compared to HPVnon16 over clusters 1 and 2 was borderline significant (p = 0.086), with 87.5% of lesions with HPV16 belonging to cluster 1. Lesions with HPV31 were equally distributed over the clusters. Comparison of the distribution of other hrHPV types individually was not possible due to the small numbers of each of these individual types. The major differences in chromosomal aberrations occurring in lesions of clusters 1 and 2 were determined by the maximum pair-wise symmetrised Kullback-Leibler divergence. In Figure 2 the importance scores are shown per chromosomal region, with a higher score indicating a larger contribution of the related aberration to the clustering results. This revealed gains of chromosomal regions on 1q, 3p and 20q to be most discriminatory between the clusters. Yet, within cluster 1 a subgroup of 13 lesions could be discerned that are characterised by gains of chromosome 3q and/or 1. Interestingly, the majority of these lesions (n = 9) harboured HPV16.
Figure 1.
Results of WECCA clustering on all the CIN2/3. Cluster 1 contains CIN2/3 with few chromosomal aberrations; the majority of the samples in the right subcluster primarily show a gain of 3q. Samples in cluster 2 have more chromosomal aberrations, including gained regions on chromosomes 1, 3q and 20 and more chromosomal losses. Samples with * are CIN2, the others CIN3.
Figure 2.
Importance score plot between lesions in cluster 1 versus lesions in cluster 2. For each chromosomal region the maximum pair wise symmetrised Kullback-Leibler divergence was determined. Gains of chromosomal regions 1q, 3p and 20q are revealed as the most striking differences of the lesions the two clusters.
CIN2/3 with HPV16 have fewer and different chromosomal aberrations than those with other hrHPV types
The average percentage of all the microarray oligonucleotides that deviated from the normal state (a proxy for the number of aberrations) occurring in the CIN2/3 was determined and compared between lesions with different hrHPV types (Table 1). The percentage of aberrant oligonucleotides for lesions with HPV16 was 11.4%, for lesions with HPVnon16 16.1% and those with HPV31 18.3%. The percentage of aberrant oligonucleotides in lesions with hrHPV types from the α9 species (HPVα9) was 13.6% and 17.8% for lesions with hrHPV types from the α5 and α7 species combined (HPVα5&α7). Whereas the percentage of total aberrations, or losses and gains separately, was not significantly different amongst these groups, lesions with HPVnon16 and HPV31 specifically displayed an approximately three-fold increase in the percentage of losses compared to lesions with HPV16. Comparison of lesions with HPVα9 versus HPVα5&α7 revealed a two-fold increase in the number of losses in lesions with HPVα5&α7.
Table 1.
The percentage of aberrant probes in lesions with different hrHPV types
| Losses% | Gains% | Total% | ||||
|---|---|---|---|---|---|---|
| average | range | average | range | average | range | |
| HPV16 (n = 16) | 2.19% | 0.005-9.82% | 9.18% | 0.028-43.33% | 11.37% | 0.19-43.91% |
| HPVnon16 (n = 27) | 6.65% | 0.015-37.75% | 9.49% | 0.015-37.75% | 16.14% | 0.12-54.04% |
| HPV31 (n = 14) | 6.67% | 0.015-37.75% | 11.64% | 0.081-31.40% | 18.31% | 0.20-46.46% |
| HPVα9 (n = 35) | 4.20% | 0.005-37.75% | 9.38% | 0.028-43.33% | 13.58% | 0.19-46.46% |
| HPVα5+α7 (n = 8) | 8.45% | 0.093-36.60% | 9.36% | 0.025-30.18% | 17.80% | 0.12-54.04% |
Shown are the average percentage and the range of deviating array oligonucleotides. An increased amount of aberrations can be seen in the lesions with types other than HPV16
To determine whether there was a difference in affected regions in CIN2/3 with different hrHPV types as well as different species, the frequency of gains and losses per oligonucleotide was plotted (Figure 3). Aberrations occurring in ≥ 20% of the CIN2/3 cases with HPV16 included gained regions on chromosomes 1p, 3q, 8q and X. For CIN2/3 with HPVnon16 these included gained regions on 1, 3, 15q, 20 and Xq and lost regions on 2q, 4p, 7q, 12q and 17p, and, for CIN2/3 with HPV31 gained regions on 1, 3, 8q, 14q, 19p, 20 and Xq and lost regions on 2q, 4, 6p, 7q, 8q, 12q and 17. For CIN2/3 with HPVα9, aberrations in ≥ 20% of the cases involved gains on chromosomes 1, 3q, 8q and 20 and losses on 17q. For CIN2/3 with HPVα5&α7 these were gains on 1, 3, 4q, 5q, 15q, 16q and 20 and lost regions on 2, 4, 6q, 7, 14q, 17 and Xp.
Figure 3.
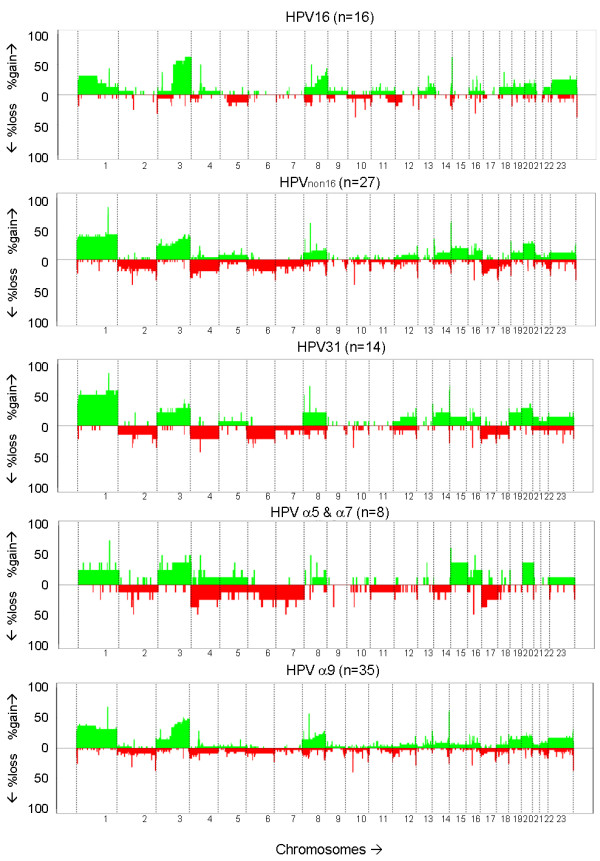
Frequency plots of the chromosomal aberrations. Lesions with HPV16 display fewer chromosomal losses than lesions with other hrHPV types or HPV31 in particular.
We next determined whether specific chromosomal regions were differentially affected between lesions harbouring the various hrHPV types. Comparison of lesions with HPV16 (n = 14) to HPVnon16 (n = 20) did not reveal regions that were significantly different. However, comparison of lesions with HPV16 (n = 14) to those with HPV31 (n = 10) indicated both lost and gained chromosomal regions that were significantly more affected in lesions harbouring HPV31 (Table 2). These included lost regions on chromosomes 2q, 4p, 4q, 6p, 6q, 8q & 17p and gained regions on 1p & 1q. Comparison of lesions with HPVα9 (n = 28) to HPVα5&α7 (n = 6) did not reveal regions that were significantly different.
Table 2.
Altered chromosomal regions showing significant differences between lesions with HPV31 compared to those with HPV16 (FDR < 0.2).
| Number of CIN2/3 cases loss vs. no loss | ||||||
|---|---|---|---|---|---|---|
| Region | Cytoband | HPV16 loss | HPV31 loss | FDR | HPV16 gain | HPV31 gain |
| chr2:222405165-227437895 | 2q36.1-q36.3 | 0 | 3 | 0.123 | 0 | 0 |
| chr2:229728860-238929377 | 2q36.3-q37.3 | 0 | 3 | 0.123 | 0 | 0 |
| chr4:49276564-62563622 | 4p11-q13.1 | 0 | 3 | 0.132 | 1 | 0 |
| chr4:71160319-78914719 | 4q13.3-q21.1 | 0 | 3 | 0.132 | 1 | 0 |
| chr4:79281456-84030000 | 4q21.21-q21.22 | 0 | 3 | 0.132 | 1 | 0 |
| chr4:88363994-132792857 | 4q21.3-q28.3 | 0 | 3 | 0.132 | 1 | 0 |
| chr4:133037098-163174205 | 4q28.3-q32.2 | 0 | 3 | 0.132 | 1 | 0 |
| chr4:163304315-191027815 | 4q32.2-q35.2 | 0 | 3 | 0.132 | 1 | 0 |
| chr6:431607-3540777 | 6p25.3-p25.2 | 0 | 3 | 0.121 | 1 | 0 |
| chr6:7902624-26085970 | 6p24.3-p22.2 | 0 | 3 | 0.121 | 0 | 0 |
| chr6:27956910-31362796 | 6p22.1-p21.33 | 0 | 3 | 0.121 | 0 | 0 |
| chr6:32718990-36975126 | 6p21.32-p21.2 | 0 | 3 | 0.121 | 0 | 0 |
| chr6:42755363-48175246 | 6p21.1-p12.3 | 0 | 3 | 0.121 | 0 | 0 |
| chr6:48648135-106496239 | 6p12.3-q21 | 0 | 3 | 0.121 | 0 | 0 |
| chr6:107106382-169915340 | 6q21-q27 | 0 | 3 | 0.121 | 0 | 0 |
| chr8:142600766-145818157 | 8q24.3 | 0 | 3 | 0.127 | 5 | 3 |
| chr17:3564934-7259932 | 17p13.2-p13.1 | 0 | 3 | 0.122 | 0 | 0 |
| chr17:7328200-10781444 | 17p13.1-p12 | 0 | 3 | 0.122 | 0 | 0 |
| chr17:12531028-22078641 | 17p12-p11.2 | 0 | 3 | 0.122 | 0 | 0 |
| Number of CIN2/3 cases gain vs. no gain |
||||||
| Region | Cytoband | HPV16gain | HPV31gain | FDR | HPV16loss | HPV31loss |
| chr1:107415557-114310216 | 1p13.3-p13.2 | 3 | 7 | 0.163 | 0 | 0 |
| chr1:114666834-146812099 | 1p13.2-q21.2 | 3 | 7 | 0.163 | 0 | 0 |
| chr1:148081768-151210846 | 1q21.2-q21.3 | 3 | 7 | 0.163 | 0 | 0 |
| chr1:151380037-167624107 | 1q21.3-q24.3 | 3 | 7 | 0.163 | 0 | 0 |
| chr1:168079451-172935386 | 1q24.3-q25.2 | 3 | 7 | 0.163 | 0 | 0 |
| chr1:172950731-177727312 | 1q25.2-q25.3 | 2 | 7 | 0.063 | 0 | 0 |
| chr1:177742624-194855793 | 1q25.3-q31.3 | 2 | 8 | 0.029 | 0 | 0 |
| chr1:195212008-208655399 | 1q31.3-q32.3 | 2 | 8 | 0.029 | 0 | 0 |
| chr1:212426726-217520338 | 1q41 | 2 | 8 | 0.029 | 0 | 0 |
| chr1:218155880-223642833 | 1q41-q42.13 | 2 | 8 | 0.029 | 0 | 0 |
| chr1:223683098-230121747 | 1q42.13-q42.2 | 2 | 7 | 0.063 | 0 | 0 |
| chr1:231048153-234780716 | 1q42.3-q43 | 2 | 7 | 0.063 | 0 | 0 |
| chr1:236021319-242608344 | 1q43-q44 | 2 | 8 | 0.029 | 0 | 0 |
| chr1:242848259-246751472 | 1q44 | 2 | 8 | 0.029 | 0 | 0 |
All lesions included in the analysis had > 1.5% aberrations
All known genes within the affected chromosomal regions differing between lesions with HPV16 compared to HPV31 were subjected to Ingenuity Pathway Analysis. The top 3 canonical pathways allocated to these genes involve the antigen presentation pathway, allograft rejection signaling and cytotoxic T-lymphocyte-mediated apoptosis of target cells. The majority of the genes in these pathways overlap and involve major histocompatibility complex molecules located at chromosome 6p, which was found to be more frequently lost in HPV31-positive lesions (see Additional file 1: Table S1).
Discussion
Chromosomal profiles of CIN2/3 infected with different hrHPV types were compared in order to determine the potential contribution of different hrHPV types to the heterogeneity in chromosomal profiles as observed previously in CIN2/3 [21].
Unsupervised hierarchical clustering analysis revealed considerable heterogeneity between CIN2/3 with respect to their chromosomal aberrations, with one subset having relatively few aberrations (cluster 1) and the other with an increased number of aberrations (cluster 2), similar to our observations in earlier studies [21]. It should be noted that, even though all lesions were associated with long-term hrHPV infections (≥ 5 years), there was still a subset of lesions with few chromosomal aberrations. This is in line with our previous study comparing lesions with < 5 years versus ≥ 5 years preceding hrHPV infection. In addition to the majority of lesions with short-term infection (< 5 years), also a subset of lesions with long-term HPV infection (≥ 5 year) showed rather few chromosomal aberrations as well. Interestingly, the majority of lesions with HPV16 (87.5%) were in the relatively quiet cluster (cluster 1) and had few aberrations. Within this cluster a subgroup could be recognised that consisted primarily of lesions with HPV16 that had gain of 3q and/or 1. The fact that CIN3 with a short-term HPV16 infection, likely reflecting the fast-progressing HPV16 lesions, also showed few chromosomal aberrations [6], indicates that the overall detection of fewer aberrations in HPV16 positive lesions is not dependent on duration of HPV16 infection. It should, however, be noted that only women over 30 years of age were included in the study since they were derived from the POBASCAM trial.
The detection of fewer copy number aberrations in HPV16-positive lesions was furthermore corroborated by hrHPV-typing analysis of an independent set of CIN2/3 previously analysed by arrayCGH [21], which also demonstrated that the majority of CIN2/3 with HPV16 (81.8%) clustered together and showed relatively few aberrations (data not shown).
Related to the clustering results, the average percentage of chromosomal losses in lesions with HPV31 was three-fold higher compared to the HPV16-positive lesions, i.e. 6.7% versus 2.2% (Table 1). No conclusion on the distribution over the two clusters of the remaining lesions harbouring other hrHPV types could be drawn due to their low numbers. However, lesions harbouring α5&α7 types had two-fold more losses compared to α9 positive lesions, i.e. 8.5% versus 4.2% (Table 1).
The observation that lesions with HPV16 tended to contain fewer aberrations than those with different types may indicate that HPV16 causes faster progression than other types and/or may not need as many chromosomal aberrations for progression to CIN2/3. The idea that lesions with HPV16 may progress faster seems to be supported by data from Vinokurova et al. [36], who reported a significant difference in the age of cervical cancer patients harbouring different hrHPV types. While the age at CIN3 diagnosis was not significantly different for HPV16-positive women compared to HPV31-positive women, women with an HPV16-positive carcinoma had a median age of 43 years, whereas those with HPV31-containing carcinomas were on average 64 years of age (p < 0.01). Also, other studies reported hrHPV type-related differences in that women with HPV16-and HPV18-positive carcinomas were younger than those infected with other types [26,31,37-39].
Comparison of lesions with HPV16 to HPV31 revealed significant differences in the affected chromosomal regions. In particular, losses at chromosomes 2q, 4p, 4q, 6p, 6q, 8q & 17p and gain of 1p & 1q were significantly more frequent in lesions with HPV31. Ingenuity Pathway Analysis indicated that genes located within significantly more frequently altered chromosomal regions in HPV31-positive lesions were particularly involved in pathways related to the immune response, most of which are related to loss of the locus encoding the major histocompatibility complex molecules (human leukocyte antigen, HLA) at chromosome 6p.
While not significant, lesions with HPV16 had the highest incidence of 3q gain, which was almost two-fold higher compared to HPVnon16 or HPV31. Gain of 3q is one of the most consistent chromosomal aberrations in cervical carcinoma [40-46] and has been suggested to predict progression of CIN [47]. Genes located at 3q, as was recently shown for PIK3CA [48], may have an important role in malignant transformation. It is tempting to speculate that HPV16-positive lesions with this particular aberration may not require many additive copy number aberrations for progression.
It should be noted that by arrayCGH analysis only copy number aberrations can be detected. Hence, it cannot be excluded that lesions with HPV16 have other, potentially more subtle genomic or (epi)genetic aberrations, such as mutations, loss of heterozygosity or DNA methylation. The fact that HPV16E6E7 have convincingly been demonstrated to induce genetic instability in in vitro model systems (reviewed by Korzenievski [49]), would argue for the presence of a genetic instable environment in HPV16-positive CIN3. We anticipate that the aberrations detected with arrayCGH result from their selective growth advantage becoming evident upon selection pressure during multiple cell divisions. This is substantiated by a recent paper by Bester et al. [50] showing an immediate induction of replication induced DNA damage upon HPV16E6E7 expression, whereas loss of heterozygosity and copy number variations only became evident after 100-250 population doublings.
Immune evasion is important for the persistence of HPV and may be achieved by various mechanisms, including HLA loci being affected by chromosomal loss, loss of heterozygosity, viral integration or mutations, which may result in functional loss [51-58]. Indeed expression of HLA class I has been reported to decrease progressively with cervical lesion grade, [59,60]. Immune evasion may also occur via direct interaction between viral and host proteins. For HPV16, for instance, direct interaction of E5 with the hydrophobic domain of HLA class I heavy chain was found to prevent its transport to the cell surface [61], whereas HPV16 E7 can trigger down-regulation of HLA class I gene transcription [62]. However, neither the mechanisms nor efficiency of HLA class I down-regulation has been investigated for HPV31 and as such it is not feasible to perform comparisons to HPV16. It may be that the respective viral proteins of other hrHPV types, such as HPV31, are less capable of interfering with antigen presentation, so that persistence in transforming infections becomes more dependent on (epi)genetic aberrations within the HLA locus. Interestingly, lesions with HPV31 also had significantly more losses at the location of the p53 gene on chromosome 17. It is still questionable to what extend p53 activity is affected in these cases. It may be that the E6 protein of HPV16 is more efficient in inactivation of p53 at the posttranslational level than E6 of HPV31 and that for this type additional events are needed to inactivate p53. Indeed, the study of Ku et al. reported that in cervical carcinomas the majority of cases with p53 mutations and LOH at 17p occurred in carcinomas harbouring types other than HPV16 [63]. However, in vitro experiments do not provide evidence of differences in degradation efficiency of the E6 proteins of these different hrHPV types [64].
To the best of our knowledge, there are no other studies in which the relation between specific hrHPV types and chromosomal aberrations in precursor lesions has been examined. So far, only a study on cervical carcinomas demonstrated the presence of type-dependent chromosomal aberrations [65]. Carcinomas without detectable HPV or containing HPV18 in a single or multiple infection showed a higher incidence of gains at 20q compared to cases containing HPV16 or other types [65]. Comparison of the frequency of 20q gain in CIN2/3 with HPV16 versus HPV18 was not possible in our data set due to the low numbers of lesions harbouring HPV18. Comparison of the frequency of 20q gain in HPVα9-versus HPVα5&α7-positive lesions, however, indicated an increased frequency in lesions harbouring types from the α5 and α7 species (i.e. 25.8% vs. 36.7%). Gain of 20q was also more frequent in lesions with HPVnon16 or HPV31 in particular, compared to those with HPV16 (29.6%, 35.7% vs. 25.0% respectively).
Conclusions
Present data indicate that the chromosomal signatures in CIN2/3 are at least in part related to the hrHPV type present. Lesions with HPV16 displayed fewer chromosomal aberrations than lesions with other hrHPV types. Specific differences, when comparing lesions with HPV16 to HPV31, included losses at chromosome 6p encoding major histocompatibility genes. One could speculate that these type-specific aberrations in lesions with HPVnon16 types facilitate viral persistence and high-grade CIN development.
Competing interests
C.J.L.M. Meijer, P.J.F. Snijders and R.D.M. Steenbergen have relationships with Self-screen B.V., a spin-off company of VU university medical center, The Netherlands. All other authors declared no conflict of interest.
Authors' contributions
MB performed experiments and drafted the manuscript. SMW, CJLMM, PJFS and RDMS participated in the design of the study, interpretation of data and revised the manuscript. WNvW and MAvdW contributed to the array data analysis. BY provided the infrastructure and know-how for the microarray experiments. All authors have read and approved the final manuscript.
Online supporting information
Array data is available from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/projects/geo/) through series accession number GSE31241.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Table S1. Genes within the top 3 canonical pathways.
Contributor Information
Mariska Bierkens, Email: mariska_bierkens@hotmail.com.
Saskia M Wilting, Email: s.wilting@vumc.nl.
Wessel N van Wieringen, Email: w.vanwieringen@vumc.nl.
Mark A van de Wiel, Email: mark.vdwiel@vumc.nl.
Bauke Ylstra, Email: B.Ylstra@vumc.nl.
Chris JLM Meijer, Email: cjlm.meijer@vumc.nl.
Peter JF Snijders, Email: PJF.Snijders@vumc.nl.
Renske DM Steenbergen, Email: r.steenbergen@vumc.nl.
Acknowledgements
Financial Support
VUmc Institute for Cancer and Immunology-VUmc Cancer Center Amsterdam.
We are grateful to dr. F.J. van Kemenade and dr. M.C.G. Bleeker for the pathological revision of tissue specimens.
References
- zur Hausen H. Papillomaviruses causing cancer: evasion from host-cell control in early events in carcinogenesis. J Natl Cancer Inst. 2000;92:690–8. doi: 10.1093/jnci/92.9.690. [DOI] [PubMed] [Google Scholar]
- Burk RD, Chen Z, Van DK. Human papillomaviruses: genetic basis of carcinogenicity. Public Health Genomics. 2009;12:281–90. doi: 10.1159/000214919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Part B: Biological Agents. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans; 2009. A review of human carcinogens; p. 100B. [Google Scholar]
- Baseman JG, Koutsky LA. The epidemiology of human papillomavirus infections. J Clin Virol. 2005;32(Suppl 1):S16–S24. doi: 10.1016/j.jcv.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Snijders PJ, Steenbergen RD, Heideman DA, Meijer CJ. HPV-mediated cervical carcinogenesis: concepts and clinical implications. J Pathol. 2006;208:152–64. doi: 10.1002/path.1866. [DOI] [PubMed] [Google Scholar]
- Bierkens M, Wilting SM, Van Wieringen WN, van Kemenade FJ, Bleeker MC, Jordanova ES, Chromosomal profiles of high-grade cervical intraepithelial neoplasia relate to duration of preceding high-risk human papillomavirus infection. Int J Cancer. 2011. doi: 10.1002/ijc.26496. [DOI] [PubMed]
- Laughlin-Drubin ME, Crum CP, Munger K. Human papillomavirus E7 oncoprotein induces KDM6A and KDM6B histone demethylase expression and causes epigenetic reprogramming. Proc Natl Acad Sci USA. 2011;108:2130–5. doi: 10.1073/pnas.1009933108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert AP, Anschau F, Schmitt VM. p16INK4A expression in cervical premalignant and malignant lesions. Exp Mol Pathol. 2006;80:192–6. doi: 10.1016/j.yexmp.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Queiroz C, Silva TC, Alves VA, Villa LL, Costa MC, Travassos AG. et al. P16(INK4a) expression as a potential prognostic marker in cervical pre-neoplastic and neoplastic lesions. Pathol Res Pract. 2006;202:77–83. doi: 10.1016/j.prp.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Wentzensen N, von Knebel DM. Biomarkers in cervical cancer screening. Dis Markers. 2007;23:315–30. doi: 10.1155/2007/678793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winer RL, Kiviat NB, Hughes JP, Adam DE, Lee SK, Kuypers JM. et al. Development and duration of human papillomavirus lesions, after initial infection. J Infect Dis. 2005;191:731–8. doi: 10.1086/427557. [DOI] [PubMed] [Google Scholar]
- McCredie MR, Sharples KJ, Paul C, Baranyai J, Medley G, Jones RW. et al. Natural history of cervical neoplasia and risk of invasive cancer in women with cervical intraepithelial neoplasia 3: a retrospective cohort study. Lancet Oncol. 2008;9:425–34. doi: 10.1016/S1470-2045(08)70103-7. [DOI] [PubMed] [Google Scholar]
- Wisman GB, De JS, Meersma GJ, Helder MN, Hollema H, de Vries EG. et al. Telomerase in (pre)neoplastic cervical disease. Hum Pathol. 2000;31:1304–12. doi: 10.1053/hupa.2000.19299. [DOI] [PubMed] [Google Scholar]
- Wilting SM, van Boerdonk RA, Henken FE, Meijer CJ, Diosdado B, Meijer GA. et al. Methylation-mediated silencing and tumour suppressive function of hsa-miR-124 in cervical cancer. Mol Cancer. 2010;9:167. doi: 10.1186/1476-4598-9-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenbergen RD, Kramer D, Braakhuis BJ, Stern PL, Verheijen RH, Meijer CJ. et al. TSLC1 gene silencing in cervical cancer cell lines and cervical neoplasia. J Natl Cancer Inst. 2004;96:294–305. doi: 10.1093/jnci/djh031. [DOI] [PubMed] [Google Scholar]
- Snijders PJ, van Duin M, Walboomers JM, Steenbergen RD, Risse EK, Helmerhorst TJ. et al. Telomerase activity exclusively in cervical carcinomas and a subset of cervical intraepithelial neoplasia grade III lesions: strong association with elevated messenger RNA levels of its catalytic subunit and high-risk human papillomavirus DNA. Cancer Res. 1998;58:3812–8. [PubMed] [Google Scholar]
- Overmeer RM, Henken FE, Snijders PJ, Claassen-Kramer D, Berkhof J, Helmerhorst TJ. et al. Association between dense CADM1 promoter methylation and reduced protein expression in high-grade CIN and cervical SCC. J Pathol. 2008;215:388–97. doi: 10.1002/path.2367. [DOI] [PubMed] [Google Scholar]
- Overmeer RM, Henken FE, Bierkens M, Wilting SM, Timmerman I, Meijer CJ. et al. Repression of MAL tumour suppressor activity by promoter methylation during cervical carcinogenesis. J Pathol. 2009;219:327–36. doi: 10.1002/path.2598. [DOI] [PubMed] [Google Scholar]
- Yang N, Nijhuis ER, Volders HH, Eijsink JJ, Lendvai A, Zhang B. et al. Gene promoter methylation patterns throughout the process of cervical carcinogenesis. Cell Oncol. 2010;32:131–43. doi: 10.3233/CLO-2009-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammes LS, Tekmal RR, Naud P, Edelweiss MI, Kirma N, Valente PT. et al. Up-regulation of VEGF, c-fms and COX-2 expression correlates with severity of cervical cancer precursor (CIN) lesions and invasive disease. Gynecol Oncol. 2008;110:445–51. doi: 10.1016/j.ygyno.2008.04.038. [DOI] [PubMed] [Google Scholar]
- Wilting SM, Steenbergen RD, Tijssen M, Van Wieringen WN, Helmerhorst TJ, van Kemenade FJ. et al. Chromosomal signatures of a subset of high-grade premalignant cervical lesions closely resemble invasive carcinomas. Cancer Res. 2009;69:647–55. doi: 10.1158/0008-5472.CAN-08-2478. [DOI] [PubMed] [Google Scholar]
- Berkhof J, Bulkmans NW, Bleeker MC, Bulk S, Snijders PJ, Voorhorst FJ. et al. Human papillomavirus type-specific 18-month risk of high-grade cervical intraepithelial neoplasia in women with a normal or borderline/mildly dyskaryotic smear. Cancer Epidemiol Biomarkers Prev. 2006;15:1268–73. doi: 10.1158/1055-9965.EPI-05-0764. [DOI] [PubMed] [Google Scholar]
- Bulkmans NW, Berkhof J, Bulk S, Bleeker MC, van Kemenade FJ, Rozendaal L. et al. High-risk HPV type-specific clearance rates in cervical screening. Br J Cancer. 2007;96:1419–24. doi: 10.1038/sj.bjc.6603653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford GM, Smith JS, Aguado T, Franceschi S. Comparison of HPV type distribution in high-grade cervical lesions and cervical cancer: a meta-analysis. Br J Cancer. 2003;89:101–5. doi: 10.1038/sj.bjc.6601024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz AT, Reid R, Jenson AB, Greenberg MD, Lancaster W, Kurman RJ. Human papillomavirus infection of the cervix: relative risk associations of 15 common anogenital types. Obstet Gynecol. 1992;79:328–37. doi: 10.1097/00006250-199203000-00002. [DOI] [PubMed] [Google Scholar]
- Wheeler CM, Hunt WC, Joste NE, Key CR, Quint WG, Castle PE. Human papillomavirus genotype distributions: implications for vaccination and cancer screening in the United States. J Natl Cancer Inst. 2009;101:475–87. doi: 10.1093/jnci/djn510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson K, Taddeo FJ, Benediktsdottir KR, Olafsdottir K, Sigvaldason H, Oddsson K. et al. HPV genotypes in CIN 2-3 lesions and cervical cancer: a population-based study. Int J Cancer. 2007;121:2682–7. doi: 10.1002/ijc.23034. [DOI] [PubMed] [Google Scholar]
- Zuna RE, Allen RA, Moore WE, Mattu R, Dunn ST. Comparison of human papillomavirus genotypes in high-grade squamous intraepithelial lesions and invasive cervical carcinoma: evidence for differences in biologic potential of precursor lesions. Mod Pathol. 2004;17:1314–22. doi: 10.1038/modpathol.3800223. [DOI] [PubMed] [Google Scholar]
- Insinga RP, Liaw KL, Johnson LG, Madeleine MM. A systematic review of the prevalence and attribution of human papillomavirus types among cervical, vaginal, and vulvar precancers and cancers in the United States. Cancer Epidemiol Biomarkers Prev. 2008;17:1611–22. doi: 10.1158/1055-9965.EPI-07-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutlee F, Ratnam S, Ramanakumar AV, Insinga RR, Bentley J, Escott N. et al. Distribution of human papillomavirus genotypes in cervical intraepithelial neoplasia and invasive cervical cancer in Canada. J Med Virol. 2011;83:1034–41. doi: 10.1002/jmv.22081. [DOI] [PubMed] [Google Scholar]
- Carozzi FM, Tornesello ML, Burroni E, Loquercio G, Carillo G, Angeloni C. et al. Prevalence of human papillomavirus types in high-grade cervical intraepithelial neoplasia and cancer in Italy. Cancer Epidemiol Biomarkers Prev. 2010;19:2389–400. doi: 10.1158/1055-9965.EPI-10-0131. [DOI] [PubMed] [Google Scholar]
- de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur HH. Classification of papillomaviruses. Virology. 2004;324:17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Bulkmans NW, Berkhof J, Rozendaal L, van Kemenade FJ, Boeke AJ, Bulk S. et al. Human papillomavirus DNA testing for the detection of cervical intraepithelial neoplasia grade 3 and cancer: 5-year follow-up of a randomised controlled implementation trial. Lancet. 2007;370:1764–72. doi: 10.1016/S0140-6736(07)61450-0. [DOI] [PubMed] [Google Scholar]
- van den Brule AJ, Pol R, Fransen-Daalmeijer N, Schouls LM, Meijer CJ, Snijders PJ. GP5+/6+ PCR followed by reverse line blot analysis enables rapid and high-throughput identification of human papillomavirus genotypes. J Clin Microbiol. 2002;40:779–87. doi: 10.1128/JCM.40.3.779-787.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wiel MA, Picard F, Van Wieringen WN, Ylstra B. Preprocessing and downstream analysis of microarray DNA copy number profiles. Brief Bioinform. 2011;12:10–21. doi: 10.1093/bib/bbq004. [DOI] [PubMed] [Google Scholar]
- Vinokurova S, Wentzensen N, Kraus I, Klaes R, Driesch C, Melsheimer P. et al. Type-dependent integration frequency of human papillomavirus genomes in cervical lesions. Cancer Res. 2008;68:307–13. doi: 10.1158/0008-5472.CAN-07-2754. [DOI] [PubMed] [Google Scholar]
- de Sanjose S, Quint WG, Alemany L, Geraets DT, Klaustermeier JE, Lloveras B. et al. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol. 2010;11:1048–56. doi: 10.1016/S1470-2045(10)70230-8. [DOI] [PubMed] [Google Scholar]
- Sideri M, Cristoforoni P, Casadio C, Boveri S, Igidbashian S, Schmitt M. et al. Distribution of human papillomavirus genotypes in invasive cervical cancer in Italy: a representative, single institution case series. Vaccine. 2009;27(Suppl 1):A30–A33. doi: 10.1016/j.vaccine.2008.12.028. [DOI] [PubMed] [Google Scholar]
- Sideri M, Igidbashian S, Boveri S, Radice D, Casadio C, Spolti N. et al. Age distribution of HPV genotypes in cervical intraepithelial neoplasia. Gynecol Oncol. 2011;121:510–3. doi: 10.1016/j.ygyno.2011.02.018. [DOI] [PubMed] [Google Scholar]
- Heselmeyer K, Macville M, Schrock E, Blegen H, Hellstrom AC, Shah K. et al. Advanced-stage cervical carcinomas are defined by a recurrent pattern of chromosomal aberrations revealing high genetic instability and a consistent gain of chromosome arm 3q. Genes Chromosomes Cancer. 1997;19:233–40. doi: 10.1002/(SICI)1098-2264(199708)19:4<233::AID-GCC5>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Allen DG, White DJ, Hutchins AM, Scurry JP, Tabrizi SN, Garland SM. et al. Progressive genetic aberrations detected by comparative genomic hybridization in squamous cell cervical cancer. Br J Cancer. 2000;83:1659–63. doi: 10.1054/bjoc.2000.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff M, Rose H, Petersen BL, Maahr J, Gerdes T, Lundsteen C. et al. Comparative genomic hybridization reveals a recurrent pattern of chromosomal aberrations in severe dysplasia/carcinoma in situ of the cervix and in advanced-stage cervical carcinoma. Genes Chromosomes Cancer. 1999;24:144–50. doi: 10.1002/(SICI)1098-2264(199902)24:2<144::AID-GCC7>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Rao PH, rias-Pulido H, Lu XY, Harris CP, Vargas H, Zhang FF. et al. Chromosomal amplifications, 3q gain and deletions of 2q33-q37 are the frequent genetic changes in cervical carcinoma. BMC Cancer. 2004;4:5. doi: 10.1186/1471-2407-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilting SM, Snijders PJ, Meijer GA, Ylstra B, van den Ijssel PR, Snijders AM. et al. Increased gene copy numbers at chromosome 20q are frequent in both squamous cell carcinomas and adenocarcinomas of the cervix. J Pathol. 2006;209:220–30. doi: 10.1002/path.1966. [DOI] [PubMed] [Google Scholar]
- Matthews CP, Shera KA, McDougall JK. Genomic changes and HPV type in cervical carcinoma. Proc Soc Exp Biol Med. 2000;223:316–21. doi: 10.1046/j.1525-1373.2000.22345.x. [DOI] [PubMed] [Google Scholar]
- Hidalgo A, Baudis M, Petersen I, Arreola H, Pina P, Vazquez-Ortiz G. et al. Microarray comparative genomic hybridization detection of chromosomal imbalances in uterine cervix carcinoma. BMC Cancer. 2005;5:77. doi: 10.1186/1471-2407-5-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heselmeyer-Haddad K, Sommerfeld K, White NM, Chaudhri N, Morrison LE, Palanisamy N. et al. Genomic amplification of the human telomerase gene (TERC) in pap smears predicts the development of cervical cancer. Am J Pathol. 2005;166:1229–38. doi: 10.1016/S0002-9440(10)62341-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henken FE, Banerjee NS, Snijders PJ, Meijer CJ, De-Castro AJ, Rosl F. et al. PIK3CA-mediated PI3-kinase signalling is essential for HPV-induced transformation in vitr. Mol Cancer. 2011;10:71. doi: 10.1186/1476-4598-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzeniewski N, Spardy N, Duensing A, Duensing S. Genomic instability and cancer: lessons learned from human papillomaviruses. Cancer Lett. 2011;305:113–22. doi: 10.1016/j.canlet.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M. et al. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145:435–46. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullokandov MR, Kholodilov NG, Atkin NB, Burk RD, Johnson AB, Klinger HP. Genomic alterations in cervical carcinoma: losses of chromosome heterozygosity and human papilloma virus tumor status. Cancer Res. 1996;56:197–205. [PubMed] [Google Scholar]
- Mitra AB, Murty VV, Li RG, Pratap M, Luthra UK, Chaganti RS. Allelotype analysis of cervical carcinoma. Cancer Res. 1994;54:4481–7. [PubMed] [Google Scholar]
- Kersemaekers AM, van d Kenter GG, Fleuren GJ. Genetic alterations during the progression of squamous cell carcinomas of the uterine cervix. Genes Chromosomes Cancer. 1999;26:346–54. doi: 10.1002/(SICI)1098-2264(199912)26:4<346::AID-GCC9>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Rader JS, Gerhard DS, O'Sullivan MJ, Li Y, Li L, Liapis H. et al. Cervical intraepithelial neoplasia III shows frequent allelic loss in 3p and 6p. Genes Chromosomes Cancer. 1998;22:57–65. doi: 10.1002/(SICI)1098-2264(199805)22:1<57::AID-GCC8>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Chatterjee A, Pulido HA, Koul S, Beleno N, Perilla A, Posso H. et al. Mapping the sites of putative tumor suppressor genes at 6p25 and 6p21.3 in cervical carcinoma: occurrence of allelic deletions in precancerous lesions. Cancer Res. 2001;61:2119–23. [PubMed] [Google Scholar]
- Vermeulen CF, Jordanova ES, Zomerdijk-Nooijen YA, Ter Haar NT, Peters AA, Fleuren GJ. Frequent HLA class I loss is an early event in cervical carcinogenesis. Hum Immunol. 2005;66:1167–73. doi: 10.1016/j.humimm.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Sheu BC, Chiou SH, Chang WC, Chow SN, Lin HH, Chen RJ. et al. Integration of high-risk human papillomavirus DNA correlates with HLA genotype aberration and reduced HLA class I molecule expression in human cervical carcinoma. Clin Immunol. 2005;115:295–301. doi: 10.1016/j.clim.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Koopman LA, van der Slik AR, Giphart MJ, Fleuren GJ. Human leukocyte antigen class I gene mutations in cervical cancer. J Natl Cancer Inst. 1999;91:1669–77. doi: 10.1093/jnci/91.19.1669. [DOI] [PubMed] [Google Scholar]
- Wang JF, Wang CX, Wang LS, Zhang J, Yang XJ, Liu M. et al. Association of human papillomavirus type 16 E7 and HLA class I antigen expression in cervical premalignant and malignant lesions. Int J Biol Markers. 2007;22:124–31. doi: 10.1177/172460080702200206. [DOI] [PubMed] [Google Scholar]
- Cromme FV, Meijer CJ, Snijders PJ, Uyterlinde A, Kenemans P, Helmerhorst T. et al. Analysis of MHC class I and II expression in relation to presence of HPV genotypes in premalignant and malignant cervical lesions. Br J Cancer. 1993;67:1372–80. doi: 10.1038/bjc.1993.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi GH, Haghshenas M, Marchetti B, Campo MS. E5 protein of human papillomavirus 16 downregulates HLA class I and interacts with the heavy chain via its first hydrophobic domain. Int J Cancer. 2006;119:2105–12. doi: 10.1002/ijc.22089. [DOI] [PubMed] [Google Scholar]
- Li H, Zhan T, Li C, Liu M, Wang QK. Repression of MHC class I transcription by HPV16E7 through interaction with a putative RXRbeta motif and NF-kappaB cytoplasmic sequestration. Biochem Biophys Res Commun. 2009;388:383–8. doi: 10.1016/j.bbrc.2009.08.019. [DOI] [PubMed] [Google Scholar]
- Ku WH, Liu IL, Yen MS, Chang Chien CC, Yue CT, Ma YY. et al. Genomic deletion and p53 inactivation in cervical carcinoma. Int J Cancer. 1997;72:270–6. doi: 10.1002/(SICI)1097-0215(19970717)72:2<270::AID-IJC12>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Fu L, Van DK, Chen Z, Ristriani T, Masson M, Trave G. et al. Degradation of p53 by human Alphapapillomavirus E6 proteins shows a stronger correlation with phylogeny than oncogenicity. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0012816. doi:10.1371/journal.pone.0012816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotto L, Narayan G, Nandula SV, rias-Pulido H, Subramaniyam S, Schneider A. et al. Identification of copy number gain and overexpressed genes on chromosome arm 20q by an integrative genomic approach in cervical cancer: potential role in progression. Genes Chromosomes Cancer. 2008;47:755–65. doi: 10.1002/gcc.20577. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Genes within the top 3 canonical pathways.