Abstract
It is commonly agreed that there is an association of chronic inflammation with tumorigenesis. COX-2, a key regulator of inflammation-producing prostaglandins, promotes cell proliferation and growth; thus, overexpression of COX-2 is often found in tumor tissues. Therefore, a better understanding of the regulatory mechanism(s) of COX-2 could lead to novel targeted cancer therapies. In this study, we investigated the mechanism of microRNA-101 (miR-101)-regulated COX-2 expression and the therapeutic potential of exogenous miR-101 for COX-2-associated cancer. A stably expressing exogenous miR-101 prostate cancer cell line (BPH1CmiR101) was generated by using lentiviral transduction as a tool for in vitro and in vivo studies. We found that miR-101 inhibited COX-2 posttranscriptional expression by directly binding to the 3′-untranslated region (3′-UTR) of COX-2 mRNA. The regulatory function of miR-101 was also confirmed by using antisense DNA. As a result, exogenous miR-101 is able to effectively suppress the growth of cultured prostate cancer cells and prostate tumor xenografts. The average tumor weight was significantly lower in the BPH1CmiR101 group (0.22 g) than the BPH1Cvec group (0.46 g). Expression levels of the cell growth regulators, such as cyclin proteins, PCNA (proliferating cell nuclear antigen), EGFR (epidermal growth factor receptor), were also studied. In conclusion, COX-2 is a direct target in miR-101 regulation of posttranscription. Exogenous miR-101 suppresses the proliferation and growth of prostate cancer cells in vitro and in vivo. These data suggest that exogenous miR-101 may provide a new cancer therapy by directly inhibiting COX-2 expression.
Introduction
Prostate cancer is currently the second leading cause of cancer death for males in United States. Chronic inflammation has been revealed to contribute to the development of malignant tumors (1-4), that showed aberrant overexpression of COX-2. COX-2 is a key rate-limiting enzyme that catalyzes the biosynthesis of prostaglandins from arachidonic acid. COX-2 is an attractive target for regulating cancer cell growth and it has been found in many types of cancers including prostate cancer (5, 6). Due to the cardiovascular toxicity of selective COX-2 inhibitors in long-term usage (7), such as celecoxib, it is appealing to investigate new endogenous and therapeutic targets to regulate COX-2 expression for prevention and treatment of COX-2-associated cancer.
MicroRNAs (miRNA) are a class of approximately 22 nucleotide-long, endogenously expressed, highly conserved noncoding RNAs with important regulatory functions in cell proliferation, apoptosis, and metastasis. miRNAs act through imperfect base pairing with the 3′-untranslated region (3′-UTR) of the target genes or bind their target mRNA through perfect base pairing (8, 9). Computational and experimental outcomes predict that miRNAs regulate at least 30% of protein-coding genes (10), and approximately 50% of miRNAs genes are located in cancer-related genomic regions (11). miRNAs are thought to regulate cancer development as tumor suppressors or oncogenes (12). Therefore, miRNAs may be ideal targets of cancer prediction and treatment.
Recent studies showed that tumor-suppressive miRNAs, such as miR-101, miR-126*, miR-146a, miR-330, miR-34 cluster, and the miR-200 family, are usually downregulated in prostate cancer compared with normal tissue (13). Loss of miR-101 was found to be concordant with overexpression of EZH2, a histone methytransferase. EZH2 expression can be regulated to inhibit prostate cancer growth by means of miR-101 expression (14, 15). Moreover, it is worth mentioning that miR-101 has ability to target COX-2 in endometrial serous adenocarcinomas (16), colon cancer (17, 18), and gastric cancer (19). However, the role of miR-101 in COX-2–associated tumor growth in vivo is unclear.
In this study, we aim to identify the mechanism of miR-101–regulated COX-2 expression and explore the therapeutic potential of miR-101 in prostate cancer by using a stably expressed exogenous miR-101 prostate cancer cell line tested in vitro and in vivo.
Material and Methods
Cell lines and culture
A benign prostatic hyperplasia cell line (BPH1) and a tumorigenic cell line BPHCAFTD (BPH1 transformed with carcinoma-associated fibroblasts) were kindly provided by Simon Hayward, Vanderbilt University, Nashville, TN; a transformed SV40 human prostatic epithelial cell line (PNT-1) was kindly provided by Bernard-kwabi Addo, Howard University; an androgen receptor-positive prostate cancer cell line (LNCap) and an androgen-negative prostate cancer cell line (PC3) were purchased from ATCC (American Type Culture Collection). All cells were grown in RPMI 1640 or other suitable media with 10% FBS.
EGFP-miR-101 expression vector
We modified the commercial pLVX-Tight-Puro vector (Clontech) with an expression cassette containing PCMV promoter, EGFP (enhanced green fluorescent protein), miRNA linker, and pre-miR-101 by replacing the Ptight promoter. The miRNA linker contained a multiple cloning site (MCS). The pre-miR-101 double-strand sequence was designed on the basis of the miRBase:Sequences 12.0 Databases. An EGFP control vector was also constructed using same expression system without the miR-101 gene. The constructed vectors were verified by DNA sequencing. Lentiviral particles containing EGFP-miR-101 vectors or EGFP control vectors were produced by using the Lentiphos HT Packaging System (Clontech) following the manufacturer’s instructions.
Stable expression miRNA-101 cell line
BPH1CAFTD cellline was selected as a candidate cell lines for this purpose because the cell line shows a high background level of COX-2 protein. Briefly, BPH1CAFTD cells were preseeded in 6-well plate overnight and then infected with 200 μL of lentivirus containing EGFP-miR-101 vectors or EGFP control vectors plus 4 μg/mL polybrene (Sigma), rocked the plate every 15 minutes for 2 hours followed by the adding 2 mL of RPMI 1640 media with 10% FBS in each well. After 48 hours, the infected cells were selected with fresh medium containing 5 ng/mL puromycin for 4 to 5 passages.Two stably expressing cell lines EGFP-miR-101 (BPH1CmiR101) or EGFP alone (BPH1Cvec) were generated, and the infected cells were easily viewed under the fluorescence microscope.
mRNA 3′-UTR of COX-2 luciferase and miR-101–targeting gene test
The miR-101–targeted gene was evaluated by using a luciferase reporter assay in 293T cells (ATCC). The 3′-UTR-Luci (3′-UTR of COX-2 luciferase) vector was constructed using the phCMV-FSR luciferase reporter vector (Genlantis) with a fragment of mRNA 3′-UTR of COX-2, which carries a putative miR-101 complementary site (NM_000963; 3′-UTR: 1,735–1,741). 293T cells were preseeded in a 24-well plate and they were 70% confluent at the time of transfection. The 3′-UTR-Luci vector and EGFP-miR-101 vector were transfected into 293T cells either alone or together using calcium phosphate method. Luciferase activity was detected after 48 hours of transfection using bioluminescence imaging with a Xenogren IVIS instrument (Caliper Life Sciences) on the basis of the manufacturer’s protocol.
miRNA expression assay
Expression of miRNAs was analyzed by using quantitative RT-PCR (20). Total RNA was extracted from cells using RNeasy Mini Kit (Qiagen) following the manufacturer’s protocol. MiR-101 forward primer (5′-TAC-AGTACTGTGA-TAACTGAA-3′) was synthesized at Sigma according to miRBase:Sequences 12.0. Total RNA was polyadenylated by Escherichia coli poly(A) polymerase, and then first-strand cDNA synthesis and quantitative PCR were conducted according to High-Specificity miRNA quantitative RT-PCR detection kit (Stratagene). Three controls were conducted in the test as follows: (i) a control test without the DNA template, (ii) a control test without poly(A) polymerase to monitor reagent contamination or false amplification, and (iii) an endogenous control test to normalize variations in the amount of cDNA template across samples. The expression of miRNAs relative to U6 RNA was determined using the ΔCt method. The average ΔCt of each group was calculated by the following formula: ΔCt = Ct_miRNA gene – Ct_u6 gene. ΔΔCt was calculated by ΔΔCt = ΔCt = ΔCt_vector – ΔCt_miR-101. The fold change for miRNA expression level was calculated using 2−ΔΔCt.
Colony formation assay
This method was used for evaluating cell proliferation and growth (21). BPH1Cvec and BPH1CmiR101 cells were seeded at a density of 500 cells per well in BD Falcon 6-well tissue culture plates. The colonies that formed after nearly 9 days were stained with 0.1% trypan blue in 50% ethanol. Any colony containing more than 50 cells was considered to represent a viable clonogenic cell. At least 2 independent experiments were conducted with triplicate test samples.
MTT assay
The MTT method was used to estimate cell viability (21). The cells were plated at an initial density of 5,000 cells per well in flat-bottom, 96-well cell culture plate and allowed to grow for 48 hours. MTT solution (Sigma) was added to each well followed by a 4-hour incubation at 37°C. After removing the media, dimethyl sulfoxide was added to each well for solubilizing the formazan formed. After 30 minutes at room temperature, the plates were scanned spectrophotometrically with a microplate reader (Bio-Rad) set at 560 nm for measuring the absorbance.
Cell-cycle analysis
Briefly, cells were collected and fixed with cold 80% fresh ethanol and stored at 4°C for 24 hours. After removing the ethanol, the cells were incubated with 1 mg/mL RNase A in PBS for 30 minutes at room temperature, and then the cells were incubated for an additional 30 minutes in the dark, adding 0.5 mL of 50 mg/mL propidium iodide. The cell cycle was analyzed by Becton-Dickinson FACScan.
Human tumor xenografts in athymic (nude) mice
Four-week-old, male, BALB/c athymic nude mice (nu/nu) were obtained from Harlan Sprague Dawley, Inc. Mice were housed in temperature-controlled rooms (74 ± 2°F) with a 12-hour alternating light–dark cycle. The mice were separated into 2 groups BPH1CmiR101 or BPH1Cvec group and each group contained 5 mice. Approximately, 2 × 106 cells were injected subcutaneously into the left and right lower backs of mice, respectively. The mice were housed for 35 days postinjection. The mice were terminated by euthanizing and the tumor tissues were removed by surgical excision. The tumor tissues were processed for RNA and protein extraction and formalin fixed for histologic studies. Tumor volume and body weight were measured once a week during the experimental period. Guidelines for the humane treatment of animals were followed as approved by the Howard University Animal Care and Use Committee.
Immunohistochemistry
The tests followed the instruction of the LSAB2 System-HRP Kit (DakoCytomation). The tissue sections were processed, deparaffinized, and treated with hydrogen peroxide. The sections were then incubated with primary antibodies of COX-2, EGFR (epidermal growth factor receptor), and PCNA (proliferating cell nuclear antigen), respectively. Following which, the sections were developed with streptavidin reagent and DAB (3,3′-diaminobenzi-dine) substrate.
Statistical analysis
Quantitative data are presented as the mean ± SD. Student’s t test was used to determine statistical significance. Differences were considered significant at P < 0.01 and P < 0.05.
Results
Expression levels of COX-2 protein and miR-101 in human tumorigenic and nontumorigenic prostate cell lines
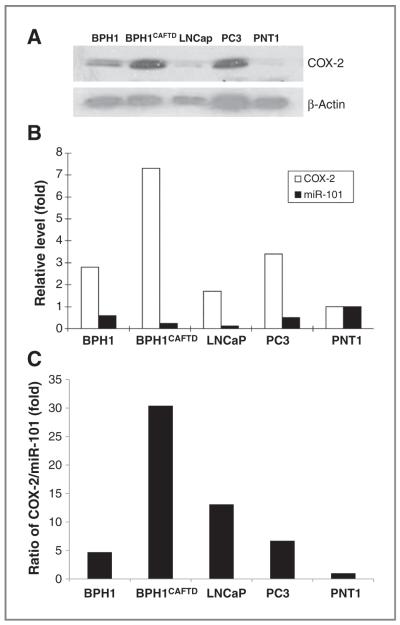
COX-2 an enzyme involved in the inflammatory response of tissues is often found in tumor cells, but not in normal cells. We evaluated COX-2 expression levels among 5 prostate cell lines, including the immortalized human prostatic PNT1 cell line, the BPH1 cell line, and the tumorigenic LNCaP, BPH1CAFTD, and PC3 cell lines by Western blot analysis (Fig. 1). The BPH1CAFTD cell line had the highest level of COX-2 among the cell lines tested. The expression levels of miR-101 were also investigated by a quantitative RT-PCR. The miR-101 levels showed an inverse correlation with COX-2 protein expression in all 5 cell lines (Fig. 1B and C). The ratio of COX-2 protein to miR-101 was considered as the miR-101 contribution to regulating COX-2 expression. The BPH1CAFTD cell line had the highest ratio. On the basis of this result, BPH1CAFTD was chosen as a candidate cell line to stably transfect with miR-101 for further investigation of miR-101 function in the regulation of COX-2 expression under cell culture and tumor xenograft conditions.
Figure 1.
The expression levels of COX-2 and miR-101 in prostate cell lines. A, comparison of COX-2 expression in various prostate cell lines. COX-2 protein levels in BPH1, androgen receptor–positive prostate tumorigenic cell lines (BPH1CAFTD and LNCap), androgen receptor–negative prostate tumorigenic cell line (PC3), and in nontumorigenic human prostatic cell line (PNT1). B, the expression levels of COX-2 and miR-101 in prostate cell lines. COX-2 levels were analyzed by Western blotting and semiquantified on the basis of COX-2/β-actin relative intensity. Bio-Rad Quantity One software was used for densitometric analysis of the Western blots. MiR-101 levels were analyzed by quantitative RT-PCR. The fold changes of each cell line were calculated by comparing with PNT1. C, the ratio of COX-2 to miR-101 in prostate cells.
Stably enforced expression of miRNA-101 in BPH1CAFTD-cultured cells and xenografts
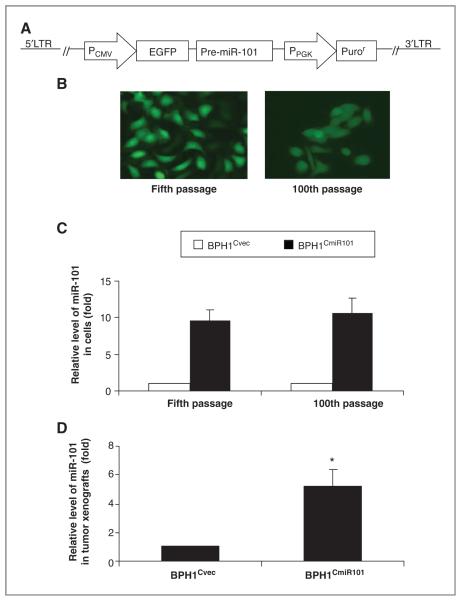
To investigate the role of miR-101 in COX-2-associated prostate cancer development in vivo, we constructed a prostate cancer cell line with stable expression of miR-101 using a lentiviral delivery system, which contained an expression cassette of the PCMV promoter, EGFP, and miR-101 precursor (Fig. 2A). miR-101–pseudotyped lentiviral particles were generated by using the Lenti-X HT Packaging System in 293T cells. BPH1CAFTD cells were transduced with lentiviral particles containing EGFP-miR-101 vectors (BPH1CmiR101) or EGFP vector (BPH1Cvec), respectively. The lentiviral-delivered EGFP-miR-101 expression was monitored under fluorescence microscope on the basis of EGFP expression (Fig. 2B) and also quantified by using real-time RT-PCR (Fig. 2C and D). After 100 passages in culture, the BPH1CmiR101 cells presented green fluorescence as bright as the fifth-passage BPH1CmiR101 cells (Fig. 2B), as well as the miR-101 expression levels (Fig. 2C). In addition, the miR-101 level was 10 times higher in BPH1CmiR101 than BPH1Cvec (Fig. 2C). A similar outcome was obtained from BPH1CmiR101 tumor xenografts that high levels of miR-101 were detected in BPH1CmiR101 tumor xenografts. The level of miR-101 was 5.2-fold higher in BPH1CmiR101 than BPH1Cvec tumor tissues (Fig. 2D).
Figure 2.
Stable expression of exogenous miR-101 in prostate cancer cells and xenografts. A, schematic representation of the design of a lentiviral-mediated delivery of a miR-101 vector, which contains an expression cassette of PCMV promoter, EGFP, and miR-101 precursor, and a selective cassette of PPGK promoter and Puror. B, EGFP expression levels of BPH1CmiR101 cells at the fifth and 100th passages were captured by fluorescence microscope at 400 magnification. C, the expression levels of miR-101 in cultured BPH1Cvec and BPH1CmiR101 cells and in BPH1Cvec and BPH1CmiR101 tumor xenografts (D) were revealed by quantitative RT-PCR. The P value was compared with vector control BPH1Cvec cells (*, P < 0.05) and the results represented the mean ± SD of 2 independent tests. LTR, long terminal repeats.
miR-101 directly inhibits COX-2 expression during posttranslation
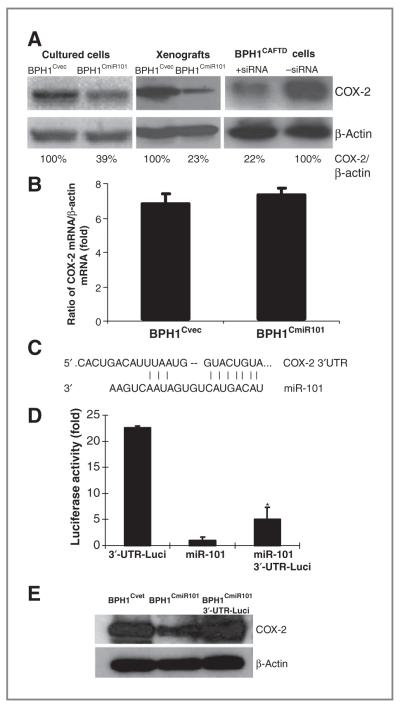
COX-2 as a direct target for miR-101 regulation was determined by using a luciferase reporter assay. The effect of miR-101 on inhibition of COX-2 expression was analyzed in a set of cell lines, including a pair of miR-101–transfected BPH1CmiR101 cells and vector control BPH1Cvec cells, a pair of BPH1CmiR101 and BPH1Cvec xenografts, and parental BPH1CAFTD cells and COX-2 siRNA-transfected BPH1CAFTD cells as positive controls by Western blot analysis (Fig. 3A). COX-2 protein level was significantly reduced in cultured BPH1CmiR101 cells and its xenografts compared with BPH1Cvec and BPH1CAFTD controls. The COX-2 protein was attenuated to 61% in cultured cells and 77% in xenografts by miR-101 or 78% by COX-2 siRNA, respectively (Fig. 3A). However, the COX-2 mRNA levels were not significantly different in both BPH1Cvec and BPH1CmiR101 cell lines by quantitative RT-PCR (Fig. 3B). The results indicate that miR-101 does not affect COX-2 mRNA transcription.
Figure 3.
miRNA-101 directly inhibits COX-2 expression at the posttranscriptional level. A, COX-2 protein levels in pairs of BPH1Cvec/BPH1CmiR101 and BPH1CAFTD+siRNA/BPH1CAFTD cell lines were evaluated by Western blotting and semiquantified on the basis of COX-2/β-actin relative intensity. The percentage indicates the relative intensity of COX-2. B, COX-2 mRNA levels of BPH1Cvec and BPH1CmiR101 cells were determined by quantitative RT-PCR and the results represented the mean ± SD of 2 independent experiments with triplicates. C, a RNA sequence map to show the 3′-UTR of COX-2 mRNA carries a complementary site (NM_000963 3′-UTR: 1,735–1,741) for the seed region of miRNA-101. D, luciferase report assay was applied to determine the miR-101 binding target. The luciferase activity was detected after transfection of 3′-UTR-Luci vector or EGFP-miR-101 vector alone, or combination of the two vectors into 293T cells. The P value was compared with vector control BPH1Cvec cells (*, P < 0.05) and the results represented the mean ± SD of 2 independent experiments with triplicates. E, the COX-2 protein levels were analyzed by Western blotting and the amount of protein was normalized by comparing the intensity of the β-actin band.
The TargetScan human 5.0 and PicTar Program predicted that COX-2 is one of the putative targets for miR-101. We utilized a luciferase reporter assay to further confirm the miR-101–binding site on the 3′-UTR region of COX-2 mRNA. Figure 3C depicts a sequence map in which the 3′-UTR of the COX-2 mRNA fragment carries a complementary site (NM_000963; 3′-UTR: 1,735–1,741) for the seed region of miRNA-101. Luciferase activity was detected 48 hours after transfection of 3′-UTR-Luci vector or EGFP-miR-101 vector alone, or both of the vectors in 293T cells. The luciferase activity was decreased 4.5-fold in 293T cells cotransfected with 3′-UTR-Luci vector and EGFP-miR-101 vector compared with 3′-UTR-Luci vector alone (Fig. 3D). These data suggest that miR-101 suppresses COX-2 protein expression by directly binding to the 3′-UTR of COX-2 mRNA. Interestingly, the inhibitory function of miR-101 on COX-2 expression can be reversed by introduction of the 3′-UTR fragment of COX-2 (Fig. 3E). COX-2 protein levels were significantly lower in BPH1CmiR101 cells than in BPH1Cvec cells; however, the protein level of COX-2 in BPH1CmiR101 cells was significantly recovered by introducing 3′-UTR of COX-2 fragments (Fig. 3E). In conclusion, miR-101 negatively regulates COX-2 protein expression by specifically binding to the 3′-UTR of the COX-2 mRNA region.
MiR-101 suppresses the proliferation of prostate cancer cells in vitro and in vivo
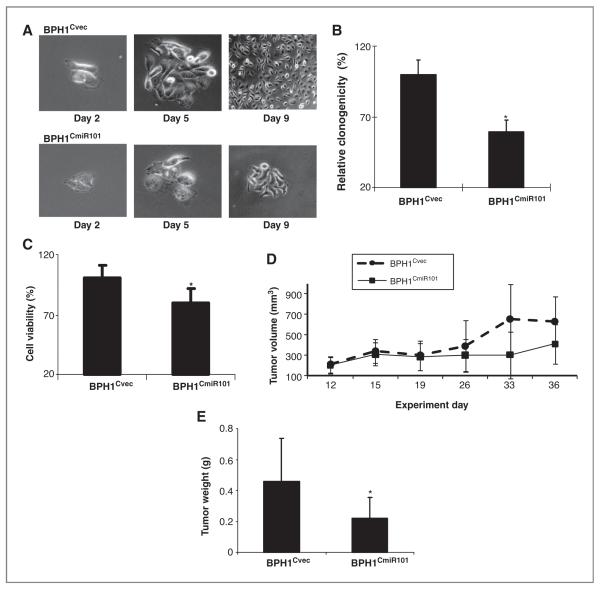
The effect of miR-101 on cell proliferation was evaluated using the colony formation assay (Fig. 4A and B) and cell viability was estimated using the MTT assay (Fig. 4C). BPH1Cvec and BPH1CmiR101 cells were separately seeded in 6-well plates and the cell growth was daily monitored under the microscope for less than 12 days. The culture was stopped when the colonies were clearly visible by the naked eye. As expected, BPH1CmiR101 cells displayed a significant proliferation delay, leading to the formation of fewer and smaller colonies even on day 9, in comparison to BPH1Cvec cells which grew more vigorously (Fig. 4A and B). The colony-forming ability of BPH1CmiR101 was 41% less than that of BPH1Cvec cells (Fig. 4B). The MTT assay showed that enforced expression of miR-101 in BPH1CmiR101 cells led to 21% decrease in cell proliferation relative to 100% proliferation observed in the case of BPH1Cvec (Fig. 4C).
Figure 4.
Exogenous miR-101 suppresses proliferation and growth of prostate cancer cells in vitro and in vivo. Cell proliferation and cell viability of cultured BPH1CmiR101 and BPH1Cvec cell lines were compared under light microscope (A), colony formation assay (B), and MTT assay (C). The results from colony formation and MTT assays represented the mean ± SD of 2 independent experiments with triplicates. D, comparison of tumor xenograft growth between BPH1CmiR101 and BPH1Cvec groups. The day of cell inoculation was the experimental start day and all mice were sacrificed on day 36. The growth of solid tumor xenografts was monitored every other day and measured using vernier calipers. E, The tumor mass (g) was measured on the final experiment day immediately after the tumor tissue was removed from the mouse by surgical excision. The average tumor mass is indicated as a bold bar in each group. The P value was compared with vector control BPH1Cvec group, *, P < 0.05.
The miR-101 function was further tested in a BPH1CmiR101 tumor xenograft animal model. Five weeks after subcutaneous inoculation of BPH1Cvec or BPH1CmiR101 cells into BALB/c athymic nude mice, each cell inoculation developed into a solid tumor xenograft. The growth curve of tumor xenografts showed that enforced expression of miR-101 slowed tumor growth than control BPH1Cvec cells (Fig. 4D) since day 26. The experimental mice were euthanized on the day 36. The average tumor weight in the BPH1Cvec group was significantly higher (0.46 g) than the BPH1CmiR101 group (0.22 g; Fig. 4E). These findings indicated that exogenous miR-101 is able to inhibit COX-2–associated cancer cell growth.
Exogenous miR-101 suppressed cancer cell proliferation
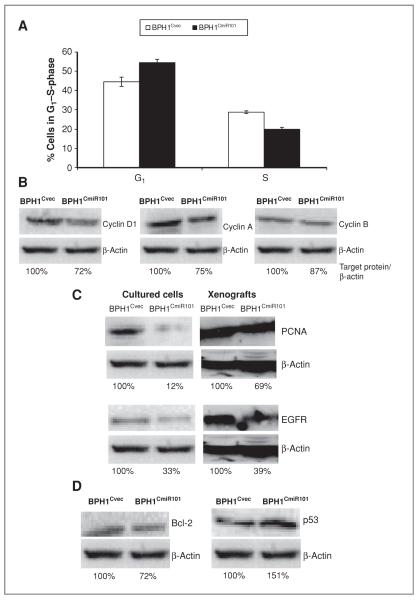
To better understand the role of miR-101 in repressing cell proliferation, we analyzed a set of cell-cycle markers in BPH1CmiR101 and BPH1Cvec cells. Figure 5A showed that BPH1CmiR101 exhibited a greater tendency for G1/S arrest (defined as an increased G1 peak with a decreased S peak) than BPH1Cvec cells. All levels of cell-cycle–related proteins, such as cyclin A, cyclin B, cyclin D1, were lower in BPH1CmiR101 cells than BPH1Cvec cells determined by Western blot analysis. The level of cyclin D1, which is important for the G1 to S-phase transition, was decreased by 28% in BPH1CmiR101 cells, and cyclin A and cyclin B1 were also 25% and 13% lower in BPH1CmiR101 cells than in BPH1Cvec cells (Fig. 5B).
Figure 5.
Effects of exogenous miR-101 on inducing G1/S arrest and altering cell growth–associated proteins. A, cell-cycle profiles of BPH1CmiR101 and BPH1Cvec cell lines. The cell distribution of G1 and S-phases in each group was analyzed by flow cytometry. B–D, the levels of cell growth–associated proteins between BPH1CmiR101 and BPH1Cvec cell lines and xenografts were analyzed by Western blotting and semiquantified on the basis of targeted protein/β-actin relative intensity. The percentage indicates relative intensity of specific protein.
Cell proliferation is regulated by multiple proteins. We analyzed additional cell proliferation markers, such as PCNA, EGFR, antiapoptotic protein Bcl-2, and proapoptotic protein p53. The levels of PCNA and EGFR were 88% lower and 67% lower in cultured BPH1CmiR101 cells than in BPH1Cvec cells, respectively (Fig. 5C). In BPH1CmiR101 xenografts, the PCNA level was 31% lower and EGFR was 61% lower than in BPH1Cvec xenografts (Fig. 5C). The Bcl-2 level was 28% downregulated, whereas p53 was 51% upregulated in BPH1CmiR101 cells than in BPH1Cvec cells (Fig. 5D).
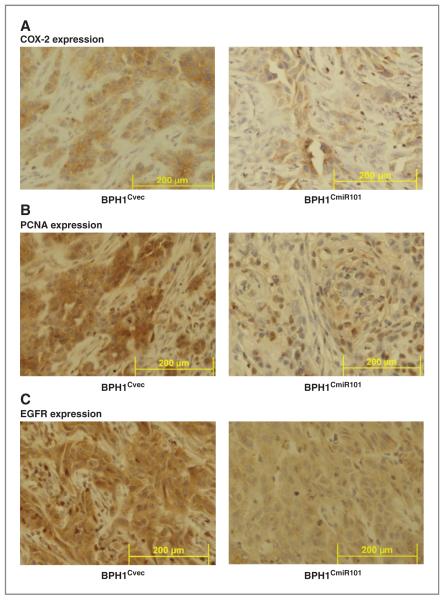
Immunohistochemical staining of the tumor xenografts of BPH1CmiR101 and BPH1Cvec further confirmed the Western blot results. We found that exogenous miR-101 resulted not only in decreased COX-2 expression but also reduced PCNA and EGFR expression in the BPH1CmiR101 xenografts compared with BPH1Cvec xenografts (Fig. 6A–C). The consistent expression pattern of cell proliferation and apoptotic markers in cultured cells and in tumor xenografts supported that exogenous miR-101 is able to inhibit prostate cancer growth by directly and indirectly modulating cell-cycle regulators to suppress cell proliferation.
Figure 6.
Expression of COX-2, PCNA, and EGFR in BPH1CmiR101 and BPH1Cvec tumor xenografts. The expression levels of COX-2 (A), PCNA (B), and EGFR (C) in the BPH1CmiR101 tumor xenografts were compared with the corresponding levels in BPH1Cvec by immunohistochemical staining method. As described under Materials and Methods, the tumor xenografts were removed at the end of the experiment, fixed in formalin, and then stained with specific monoclonal antibody. Magnification × 400.
Discussion
MicroRNAs are predicted to regulate more than 30% of all gene expression and may account for some of the aberrant gene expression in cancer cells. There is strong evidence that miR-101 plays a role as a tumor suppressor in various cancers. For instance, miR-101 inhibits Mcl-1 protein (an antiapoptotic gene of Bcl-2 family; ref. 22), EZH2 (a histone methyltransferase; refs. 14, 15), and DNA-PKcs (an essential factor for nonhomologous end-joining repair), and ATM (ataxia telangiectasia mutated; an important checkpoint regulator for promoting homologous recombination repair; ref. 23) in cancer. Although several studies have reported that miR-101 can inhibit cancer cell growth via blocking COX-2 expression, the research is still hovering at cellular level. Therefore, we are committed to such a work around exogenous miR-101 in inhibiting prostate cancer cell growth through modulation of COX-2 pathway in vivo.
There are 2 precursors of miR-101 pre-miR-101-1 located at chromosome 1p31.3 and pre-miR-101-2 at chromosome 9p24.2. Both of the loci are lost in 37.5% of organ-confined prostate cancer cells and 66.7% of metastatic disease cells (18). This event seems to be common to a broad range of cancer types, according to public domain CGH data sets. In contrast, overexpression of miR-101 has been found to suppress cell proliferation and impair the invasive potential in prostate cancer (18), bladder carcinoma (15), and colon cancer as a tumor suppressor (17). We found that exogenous miR-101 significantly inhibited the cell proliferation (Fig. 4A–C) and tumor growth (Fig. 4D and E) in COX-2–associated prostate cancer either in cultured BPH1CmiR101 cells or in BPH1CmiR101 xenografts by directly suppressing COX-2 protein expression, resulting in suppression of COX-2–associated cell growth factors (Fig. 3).
Numerous evidence has revealed that COX-2 is an inducible enzyme that is elevated and catalyzes arachidonic acid to prostaglandins during inflammatory and tumorigenic (22). Thus, COX-2 is a critical target that has implications for both prevention and treatment of prostate cancer (24). The prediction based on PicTar and TargetScan DNA analysis software suggest that COX-2 could be one direct target for miR-101 because miR-101 has a seed region, which is able to bind to the COX-2 mRNA 3′-UTR (Fig. 3C). The observations from this present study support an inverse correlation between miR-101 and COX-2 expression in prostate cell lines (Fig. 1). This hypothesis was confirmed by following approaches: First, we successfully established an enforced EGFP-miR-101 expression prostate cell line (BPH1CmiR101) by using stable gene transfection technology (Fig. 2). In addition, the cell was uniquely cotransfected with the EGFP gene sharing the same PCMV promoter with the miR-101 gene. Therefore, we can easily monitor miR-101 expression in the cell under a fluorescent microscope (Fig. 2B). The miR-101 levels were detected to be more than 10 times higher in the miR-101–transfected cells (BPH1CmiR101) than in the vector control cells (BPH1Cvec; Fig. 2C and D), and the miR-101 level in BPH1CmiR101 cells was maintained at the same level after 100 passages (Fig. 2B and C). As a powerful tool, this enforced EGFP-miR-101 expression cell line allowed us to study the role of miR-101 in regulation of COX-2 in cultured cells and tumor xenografts.
Second, we examined the regulatory target of miR-101 using a luciferase report assay (Fig. 3). We constructed a luciferase reporter vector (3′-UTR-Luci) containing the 3′-UTR of COX-2 fragment, which carried a putative miR-101 complementary region (NM_000963; 3′-UTR: 1,735–1,741). Theoretically, luciferase expression can be blocked and a decreased luciferase activity can be observed when the miR-101 gene binds to the complementary region of 3′-UTR of COX-2 in this system. We found a significantly lower level of luciferase activity when the 293T cells were cotransfected with EGFP-miR-101 vector and 3′-UTR-Luci vector (Fig. 3D) indicating a direct interaction between miR-101 and COX-2 mRNA. In contrast, the level of COX-2 protein was able to be restored in BPH1CmiR101 after transfecting the 3′-UTR of COX-2 fragment (Fig. 3E). However, depending on the extent of sequence complementarity between a miRNA and its target gene, miRNA-guided posttranscriptional regulation of gene expression follows 2 distinct mechanisms target mRNA cleavage and translational repression.
Third, we evaluated whether the miR-101 is able to alter the mRNA expression of COX-2. We found that there was no statistical significance on the levels of COX-2 mRNA between enforced expression of miR-101 BPH1CmiR101 cells and vector control BPH1Cvec cells (Fig. 3B). Above all, the observations from our study clearly show that miR-101 regulates COX-2 by directly binding to 3′-UTR of COX-2 mRNA and causing translational repression.
Our previous studies reported that the COX2/PGE2 pathway plays a key role in regulating cancer development along with other signaling pathways (25). Interestingly, in this study, we found that exogenous miR-101 significantly inhibits prostate cancer cell growth both in vitro and in vivo (Fig. 4). Thus, we further investigated the therapeutic potential of exogenous miR-101 for COX-2–associated prostate cancer and the mechanism(s) by which miR-101 modulated COX-2/PGE2/EGFR pathways. We found exogenous miR-101 not only can reduce COX-2 protein expression but can also concurrently decrease the EGFR level in cultured BPH1CmiR101 cells and xenograft tissues. EGFR exists on the cell surface and is activated by binding of ligand. Activation of EGFR in turn initiates its down-stream signal transduction cascades leading to DNA synthesis and cell proliferation. Abnormally high expression of EGFR has been observed in many types of cancers including prostate cancer. The high expression of COX-2 leads to an EGFR-stimulated cell proliferation (26) and COX-2 products, such as PGE2, activate EGFR signaling pathways to promote cancer growth (27, 28). Hence, COX-2/PGE2/EGFR arrest has been revealed to be a potential mechanism of miR-101 inhibition of cancer cell proliferation.
In addition, flow cytometric analysis showed that miR-101 caused cell-cycle arrest at the G1/S border (Fig. 5A and B). There are a series of regulatory proteins involved in cell proliferation and cell fate. Cyclin D1, which is important for G1 to S-phase transition, increases in response to extracellular signals from a growth factor (e.g., EGFR). The cyclin D–CDK4 (cyclin-dependent kinase) complex activates E2F and results in transcription of other cell-cycle–related genes such as cyclin A, cyclin E, and PCNA. Cyclin A and cyclin E push the cell from G1 to S-phase transition, and cyclin B–cdc2 complex initiates the G2 to M phase transition. PCNA, a member of DNA-sliding clamp family, interacts with multiple partners involved in DNA repair, DNA replication, chromatin remodeling, and cell-cycle regulation. PCNA is essentially required to bind to cyclin D–CDK4, cyclin E–CDK2, and cyclin A–CDK2 complexes for progression from G1 to S-phase in the cell cycle (29). We found that the levels of cyclin D1, cyclin A, and cyclin B were notably decreased in the prostate cancer cells with exogenous miR-101 (Fig. 5B). The decrease in PCNA expression was similarly found in BPH1CmiR101 contributing to the G1/S arrest (Fig. 5C). In addition, the p53 protein is characterized as an essential mediator of cell-cycle arrest that can halt progression of the cell cycle in G1 by blocking the activity of CDK2 and senses DNA damage to apoptosis induction (30, 31). p53 showed an increased (151%) level in BPH1CmiR101 than in BPH1Cvec, whereas the expression of the antiapoptotic protein Bcl-2 which negatively correlated with p53 displayed a decrease (28%) in BPH1CmiR101 compared with BPH1Cvec. This result indicated that miR-101 decreasing cyclin D1, cyclin A, PCNA along with Bcl-2 led to the block in the G1 to S-phase transition.
In conclusion, the role of miR-101 in inhibiting COX-2 protein expression was studied in a stably enforced expression of miR-101 cell line grown in vitro and in vivo. MiR-101 is able to inhibit COX-2 protein expression through directly binding to 3′-UTR region of COX-2 mRNA. Therefore, exogenous miR-101 has a high potential for treating COX-2–associated cancers through directly and indirectly modulating the COX-2/PGE2/EGFR pathways.
Acknowledgments
The authors thank Dan Zhang for her excellent technical assistance, Rajagopalan Sridhar for valuable discussion during the manuscript preparation, Simon Hayward for the prostate cell lines (BPH1 and BPH1CAFTD), and Bernard-kwabi Addo for PNT1 cell line.
Grant Support
This work was supported in part by funding P20CA118770 and U54CA91431 from National Cancer Institute, NIH.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Davies G, Martin LA, Sacks N, Dowsett M. Cyclooxygenase-2 (COX-2), aromatase and breast cancer: a possible role for COX-2 inhibitors in breast cancer chemoprevention. Ann Oncol. 2002;13:669–78. doi: 10.1093/annonc/mdf125. [DOI] [PubMed] [Google Scholar]
- 2.Marrogi A, Pass HI, Khan M, Metheny-Barlow LJ, Harris CC, Gerwin BI. Human mesothelioma samples overexpress both cyclooxygen-ase-2 (COX-2) and inducible nitric oxide synthase (NOS2): in vitro antiproliferative effects of a COX-2 inhibitor. Cancer Res. 2000;60:3696–700. [PubMed] [Google Scholar]
- 3.Sun Y, Tang XM, Half E, Kuo MT, Sinicrope FA. Cyclooxygenase-2 overexpression reduces apoptotic susceptibility by inhibiting the cytochrome c-dependent apoptotic pathway in human colon cancer cells. Cancer Res. 2002;62:6323–8. [PubMed] [Google Scholar]
- 4.Taketo MM. COX-2 and colon cancer. Inflamm Res. 1998;47(Suppl 2):S112–6. doi: 10.1007/s000110050295. [DOI] [PubMed] [Google Scholar]
- 5.Sugar LM. Inflammation and prostate cancer. Can J Urol. 2006;13(Suppl 1):46–7. [PubMed] [Google Scholar]
- 6.Sooriakumaran P, Langley SE, Laing RW, Coley HM. COX-2 inhibition: a possible role in the management of prostate cancer? J Chemother. 2007;19:21–32. doi: 10.1179/joc.2007.19.1.21. [DOI] [PubMed] [Google Scholar]
- 7.Solomon SD, McMurray JJ, Pfeffer MA, Wittes J, Fowler R, Finn P, et al. Cardiovascular risk associated with celecoxib in a clinical trial for colorectal adenoma prevention. N Engl J Med. 2005;352:1071–80. doi: 10.1056/NEJMoa050405. [DOI] [PubMed] [Google Scholar]
- 8.Garzon R, Pichiorri F, Palumbo T, Iuliano R, Cimmino A, Aqeilan R, et al. MicroRNA fingerprints during human megakaryocytopoiesis. Proc Natl Acad Sci U S A. 2006;103:5078–83. doi: 10.1073/pnas.0600587103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 11.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vrba L, Jensen TJ, Garbe JC, Heimark RL, Cress AE, Dickinson S, et al. Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PLoS One. 2010;5:e8697. doi: 10.1371/journal.pone.0008697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pang Y, Young CY, Yuan H. MicroRNAs and prostate cancer. Acta Biochim Biophys Sin. 2010;42:363–9. doi: 10.1093/abbs/gmq038. [DOI] [PubMed] [Google Scholar]
- 14.Cao P, Deng Z, Wan M, Huang W, Cramer SD, Xu J, et al. MicroRNA-101 negatively regulates Ezh2 and its expression is modulated by androgen receptor and HIF-1alpha/HIF-1beta. Mol Cancer. 2010;9:108. doi: 10.1186/1476-4598-9-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman JM, Liang G, Liu CC, Wolff EM, Tsai YC, Ye W, et al. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009;69:2623–9. doi: 10.1158/0008-5472.CAN-08-3114. [DOI] [PubMed] [Google Scholar]
- 16.Hiroki E, Akahira J, Suzuki F, Nagase S, Ito K, Suzuki T, et al. Changes in microRNA expression levels correlate with clinicopathological features and prognoses in endometrial serous adenocarcinomas. Cancer Sci. 2010;101:241–9. doi: 10.1111/j.1349-7006.2009.01385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strillacci A, Griffoni C, Sansone P, Paterini P, Piazzi G, Lazzarini G, et al. MiR-101 downregulation is involved in cyclooxygenase-2 overexpression in human colon cancer cells. Exp Cell Res. 2009;315:1439–47. doi: 10.1016/j.yexcr.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 18.Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–9. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang HJ, Ruan HJ, He XJ, Ma YY, Jiang XT, Xia YJ, et al. MicroRNA-101 is down-regulated in gastric cancer and involved in cell migration and invasion. Eur J Cancer. 2010;46:2295–303. doi: 10.1016/j.ejca.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 20.Zhao Y, Hao Y, Ji H, Fang Y, Guo Y, Sha W, et al. Combination effects of salvianolic acid B with low-dose celecoxib on inhibition of head and neck squamous cell carcinoma growth in vitro and in vivo. Cancer Prev Res. 2010;3:787–96. doi: 10.1158/1940-6207.CAPR-09-0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hao Y, Xie T, Korotcov A, Zhou Y, Pang X, Shan L, et al. Salvianolic acid B inhibits growth of head and neck squamous cell carcinoma in vitro and in vivo via cyclooxygenase-2 and apoptotic pathways. Int J Cancer. 2009;124:2200–9. doi: 10.1002/ijc.24160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palapattu GS, Sutcliffe S, Bastian PJ, Platz EA, De Marzo AM, Isaacs WB, et al. Prostate carcinogenesis and inflammation: emerging insights. Carcinogenesis. 2005;26:1170–81. doi: 10.1093/carcin/bgh317. [DOI] [PubMed] [Google Scholar]
- 23.Yan D, Ng WL, Zhang X, Wang P, Zhang Z, Mo YY, et al. Targeting DNA-PKcs and ATM with miR-101 sensitizes tumors to radiation. PLoS One. 2010;5:e11397. doi: 10.1371/journal.pone.0011397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenhough A, Smartt HJ, Moore AE, Roberts HR, Williams AC, Paraskeva C, et al. The COX-2/PGE2 pathway: key roles in the hallmarks of cancer and adaptation to the tumour microenvironment. Carcinogenesis. 2009;30:377–86. doi: 10.1093/carcin/bgp014. [DOI] [PubMed] [Google Scholar]
- 25.Zhao Y, Hao Y, Ji H, Fang Y, Guo Y, Sha W, et al. Combination effects of salvianolic acid B with low-dose celecoxib on inhibition of head and neck squamous cell carcinoma growth in vitro and in vivo. Cancer Prev Res. 2010;3:787–96. doi: 10.1158/1940-6207.CAPR-09-0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ceccarelli C, Piazzi G, Paterini P, Pantaleo MA, Taffurelli M, Santini D, et al. Concurrent EGFr and Cox-2 expression in colorectal cancer: proliferation impact and tumour spreading. Ann Oncol. 2005;16(Suppl 4):iv74–9. doi: 10.1093/annonc/mdi912. [DOI] [PubMed] [Google Scholar]
- 27.Thomas SM, Bhola NE, Zhang Q, Contrucci SC, Wentzel AL, Freilino ML, et al. Cross-talk between G protein-coupled receptor and epidermal growth factor receptor signaling pathways contributes to growth and invasion of head and neck squamous cell carcinoma. Cancer Res. 2006;66:11831–9. doi: 10.1158/0008-5472.CAN-06-2876. [DOI] [PubMed] [Google Scholar]
- 28.Buchanan FG, Wang D, Bargiacchi F, DuBois RN. Prostaglandin E2 regulates cell migration via the intracellular activation of the epidermal growth factor receptor. J Biol Chem. 2003;278:35451–7. doi: 10.1074/jbc.M302474200. [DOI] [PubMed] [Google Scholar]
- 29.Maga G, Hubscher U. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J Cell Sci. 2003;116:3051–60. doi: 10.1242/jcs.00653. [DOI] [PubMed] [Google Scholar]
- 30.Bartek J, Lukas J. Mammalian G1- and S-phase checkpoints in response to DNA damage. Curr Opin Cell Biol. 2001;13:738–47. doi: 10.1016/s0955-0674(00)00280-5. [DOI] [PubMed] [Google Scholar]
- 31.Hemann MT, Lowe SW. The p53-Bcl-2 connection. Cell Death Differ. 2006;13:1256–9. doi: 10.1038/sj.cdd.4401962. [DOI] [PMC free article] [PubMed] [Google Scholar]