Introduction
We are interested in the three-dimensional architecture of the genome and how higher order chromatin structure affects gene expression. Chromatin loops that juxtapose distal regions of the genome have been identified as important regulators of gene expression in mammalian cells (Chambeyron and Bickmore, 2004; Fraser, 2006; Marenduzzo et al., 2007; Misteli, 2007). Loops can cluster related genes into discrete transcriptional “hubs,” or insulate chromatin from neighboring domains. Chromatin loops have also been identified in yeast, but rather than position enhancer regions near cognate promoters, gene loops in yeast juxtapose promoter-terminator regions (Ansari and Hampsey, 2005; O’Sullivan et al., 2004). Here we describe the discovery of gene loops, the proteins required for looping, and the role of loops in transcription by RNA polymerase II.
Genetic suppression and the discovery of gene loops
Early genetic results from our laboratory hinted at the existence of gene loops. Two decades ago, we developed a genetic selection to identify factors that affect the accuracy of transcription start site selection by RNA polymerase II (Pol II) (Hampsey et al., 1991). The selection scheme was based on suppression of a translation initiation mutation: a point mutation at the CYC1 locus created an ATG codon (uATG) that initiated a short open reading frame downstream of the transcription start site, but upstream and out-of-frame with the ATG codon that initiates synthesis of iso-1-cytochrome c. This mutant allele (cycl-5000), fails to express iso-1-cytochrome c and consequently cells fail to grow on nonfermentable carbon sources such as lactate.
We reasoned that genes involved in Pol II start site selection might turn up in a genetic selection for suppressors of cycl-5000 by shifting the normal transcription start site downstream of the uATG, thereby restoring the normal ATG as the start codon most proximal to the 5′-end of the mRNA. To facilitate isolation of trans-acting suppressors, rather than mutations that simply eliminate the uATG or other cis-acting uATG suppressors, we constructed a plasmid that expresses the E. coli lacZgene behind the cycl-5000 promoter and transformed it into the cycl-5000 mutant. Accordingly, the cycl-5000 uATG conferred two phenotypes: failure to grow on lactate as the sole carbon source and a white colony phenotype on X-gal indicator medium. Revertants that acquired the ability to grow on lactate medium (express iso-1-cytochrome c) and turned blue (express beta-galactosidase) were likely to be the result of extragenic mutations (unlinked to cycl) that suppress the uATG. Genetic analysis of a collection of two dozen revertants defined nine different complement groups, sual – sua9 (suppression of uATG) (Pinto et al., 1992a).
The sua suppressors included two complementation groups, sua7 and sua8, that shifted Pol II transcription start site selection downstream of normal (Berroteran et al., 1994; Pinto et al., 1992b). SUA7 codes for the general transcription factor TFIIB and sua8 is allelic to RPB1, which codes for the largest subunit of Pol II. These results defined TFIIB and Rpb1 as determinants of Pol II start site selection.
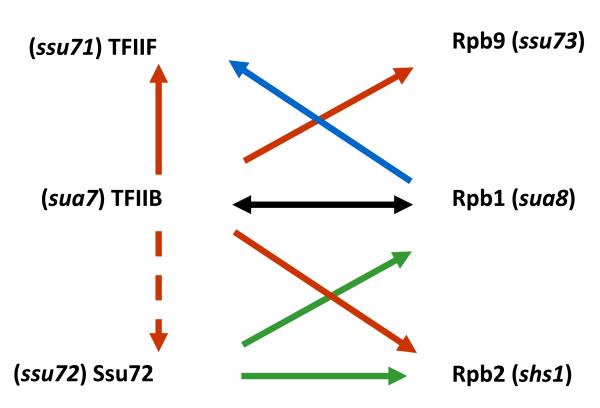
Interestingly, all sua7 and sua8 suppressors are cold-sensitive, failing to grow on rich medium at 16C. We exploited this phenotype to identify factors that functionally interact with TFIIB and Rpb1 (Sun and Hampsey, 1996). These “suppressors of suppressors” turned up four additional genes that affect start site selection, including the RPB2 and RPB9 genes, encoding two additional subunits of Pol II, TFG1, which encodes a subunit of the general transcription factor TFIIF, and a novel gene, designated SSU72 (Fig. 1).
Fig. 1.
Genetic interactions among RNA Pol II transcription factors. The sua7 and sua8 (RPB1) alleles were identified as suppressors of the cyc1-5000 uATG defect. The shs1 (RPB2), ssu71 (TFG1) and ssu73 (RBP9) genes were identified as suppressor of the sua7-1 cold-sensitive growth defect. The ssu72 allele was identified in the same strain as the ssu73 allele; unlike ssu73, ssu72 acts as an enhancer, rather than as a suppressor of sua7-1 (Sun and Hampsey, 1996). The rpb1 and rpb2 genes were also identified as suppressor of the ssu72-2 heat sensitive growth defect (Reyes-Reyes and Hampsey, 2007).
Ssu72 is a Pol II CTD phosphatase
SSU72 encodes a small protein of 206 amino acids, essential for cell viability in yeast, and conserved among all eukaryotic organisms sequenced to date. The Ssu72 sequence includes a phosphatase motif (CX5RS…DX2D); recombinant yeast and human Ssu72 proteins were subsequently shown to hydrolyze ρ-nitro-phenyl phosphate (Ganem et al., 2003; Meinhart et al., 2003). Based on its phosphatase activity and genetic interactions with Pol II, we suspected that Ssu72 might catalyze removal of phosphate from either Ser2-P or Ser5-P of the reiterated heptad repeat of the carboxy terminal domain (CTD) of Pol II. Indeed, Ssu72 is turned out to be a Pol II CTD Ser5-P phosphatase (Krishnamurthy et al., 2004). Consistent with its genetic and physical interactions with components of the initiation machinery, Ssu72 affects the initiation/elongation transition in a manner dependent upon its catalytic activity (Reyes-Reyes and Hampsey, 2007). Collectively, these results demonstrate that Ssu72 plays a key role early in the transcription cycle.
Ssu72 is an integral component of the CPF 3′-end processing complex
Interestingly, however, Ssu72 was found to be an integral component of the yeast pre-RNA 3′-end processing complex (CPF in yeast, CPSF in mammalian cells) that couples processing with transcription termination (He et al., 2003). How could a component of the CPF 3′-end processing complex be interacting with TFIIB and affect initiation? One possibility is that the transcription preinitiation complex and the 3-end processing complex are not spatially distinct, but interact to juxtapose the promoter and terminator regions, in effect creating a gene loop. Indeed, Proudfoot and colleagues had recently shown juxtaposition of the promoter-terminator regions of the yeast FMP27 gene (O’Sullivan et al., 2004).
Chromosome conformation capture
Our efforts to characterize gene loops were made feasible by a technique called “chromosome conformation capture” (3C), a method that detects and quantifies the frequency of interaction between any two genomic loci (Dekker, 2006; Dekker et al., 2002). 3C converts physical chromatin interactions into specific ligation products that are then detected and quantified by PCR. Although 3C was developed to detect long-range chromatin interactions, we adapted this technique to detect short-range interactions, including physical proximity of the ends of genes in yeast, typically spanning less than 10 kbp in yeast (Ansari and Hampsey, 2005; Singh et al., 2009).
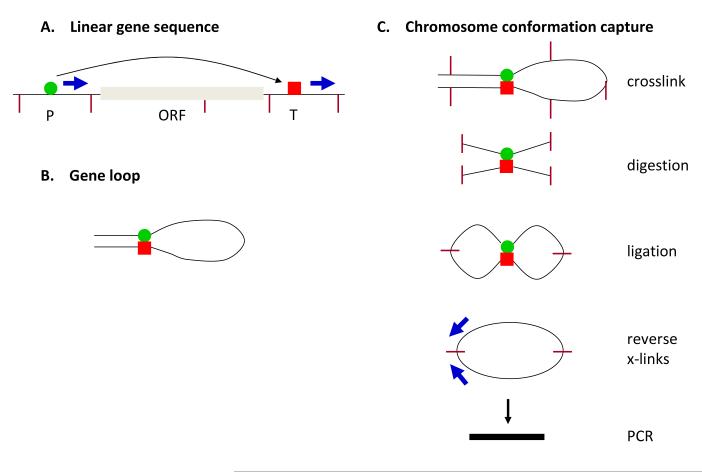
The essence of the technique is as follow. Transient chromatin interactions are stabilized in vivo by formaldehyde crosslinking. Chromatin is then isolated and digested with a restriction enzyme, chosen based on its ability to cut DNA upstream of a promoter, downstream of the terminator and at multiple sites within the ORF. Following restriction digestion, chromatin samples are ligated in dilute solution, favoring intramolecular ligation. Crosslinks are then released and products are analyzed by PCR. Accordingly, tandemly-oriented primer pairs that anneal to promoter and terminator regions of a linear gene become convergently-oriented to yield PCR products in a manner dependent upon crosslinking and ligation (Fig. 2).
Fig. 2.
Chromosome Conformation Capture (3C). (A) Linear depiction of a gene with promoter on the left, terminator on the right. (B) A transient gene loop formed by juxtaposition of the promoter and the terminator. (C) Schematic depiction of 3C methods to detect gene loops. The tandem primer pair in the linear structure (panel a) becomes a convergent primer pair following the 3C protocol. According to 3C, the abundance of the ligation product, as detected by PCR, is a measurement of the frequency with which two loci interact (Dekker et al., 2002; Dekker, 2006).
Characteristics of gene loops
We chose the BUD3 and SEN1 genes for our initial characterization of gene loops simply because they are relatively long genes (~6 kbp) and include restriction sites that make their analyses amenable to 3C. In our initial experiment we found 3C PCR products corresponding to promoter-terminator juxtaposition; control experiments confirm that these PCR products were dependent upon crosslinking, restriction digestion and ligation (Ansari and Hampsey, 2005). Mapping experiments using primer pairs that spanned the entire length of the 6.4 kbp BLM10 gene, as well as regions both upstream and downstream of the promoter and terminator, established that looping occurs specifically between restriction fragments encompassing the BLM10 promoter and terminator (Ansari and Hampsey, 2005; Singh et al., 2009).
Two experiments were done to determine whether gene loops are static structures of the genome or dynamic structures whose formation might correlate with transcription (Ansari and Hampsey, 2005). First, we asked if gene loops at BUD3 and SEN1 were diminished when transcription was shut-off. For this experiment we used an rpb1-1 mutant, which encodes an altered form of Pol II that rapidly ceases mRNA synthesis upon temperature shift from 24 C to 37 C. The looping signal for both genes was significantly diminished at 37 C, yet this temperature shift had no effect on looping in the wild type strain. Next, we replaced the normal BUD3 and SEN1 promoters with the inducible GAL1 promoter at their normal chromosomal loci. 3C analysis revealed a marked increased in the looping signal when the carbon source was shifted from glucose (repression) to galactose (induction). These results demonstrate that gene loops are dynamic structures that form in a transcription-dependent manner.
Gene loops are dependent upon components of promoter and terminator complexes
We initially suspected the existence of gene loops based on genetic interactions between TFIIB (sua7-1) and Ssu72. We therefore asked if looping is affected in TFIIB and Ssu72 mutants. Using an isogenic wild type and sua7-1 strain pair, we found that the sua7-1 mutation diminished looping at all genes tested, including BUD3, SEN1, BLM10 and HEM3 (Singh and Hampsey, 2007). Importantly, this effect was not due to impaired transcription, as the sua7-1 mutation has no effect on the transcript levels of these genes. Similar results were found by Ansari and colleagues (El Kaderi et al., 2009).
To assess the potential involvement of Ssu72 in loop formation, we used a ssu72-td degron mutant that results in rapid depletion of Ssu72 when cells are shifted to 37 C. Gene loops were readily defected at BUD3, SEN1, BLM10 and HEM3 in cells grown at the permissive temperature, but were diminished following the temperature shift (Ansari and Hampsey, 2005). Similar results were obtained using a pta1-td degron mutant. Pta1 is another component of the CPF complex and interacts directly with Ssu72 (He et al., 2003). Taken together, these results establish that the TFIIB component of the initiation complex and the Ssu72 and Pta1 components of the CPF 3′-end processing complex are required for gene looping.
TFIIB at the terminator
The TFIIB requirement for looping suggests that TFIIB associates not only with promoter DNA, but also with the terminator. To address this issue, we performed chromatin immunoprecipitation (ChIP) using antibody to TFIIB, monitoring TFIIB occupancy along the length of the PMA1 gene. As expected, TFIIB was detected at the promoter and absent along the length of the ORF. Surprisingly, however, TFIIB was also found to occupy PMA1 just downstream of its two poly(A) sites (Singh and Hampsey, 2007). In the same experiment, TATA binding protein (TBP) associated with the promoter, but not with the terminator, thereby establishing that TFIIB occupancy of the PMA1 terminator was not due to the presence of a cryptic promoter. Similar results - TFIIB occupancy of both promoter and terminator regions - was also observed at the BLM10 gene (Singh and Hampsey, 2007).
We next asked if TFIIB occupancy of the PMA1 and BLM10 terminator regions is affected in the sua7-1 mutant. Remarkably, association of TFIIB with the terminator region of both genes was eliminated in the sua7-1 mutant, with no effect on promoter occupancy (Singh and Hampsey, 2007).
We then repeated the TFIIB ChIP experiments using the ssu72-td strain grown at permissive (24 C) and restrictive (37 C) temperatures. Whereas Ssu72 depletion at 37 C was without effect on TFIIB occupancy of the PMA1 and BLM10 promoters, TFIIB no longer associated with the terminator of either gene at 37 C. These results confirm that TFIIB associates with the terminator region of these two genes and is dependent upon the Ssu72 component of the CPF 3′-end processing complex (Singh and Hampsey, 2007).
A model for gene looping
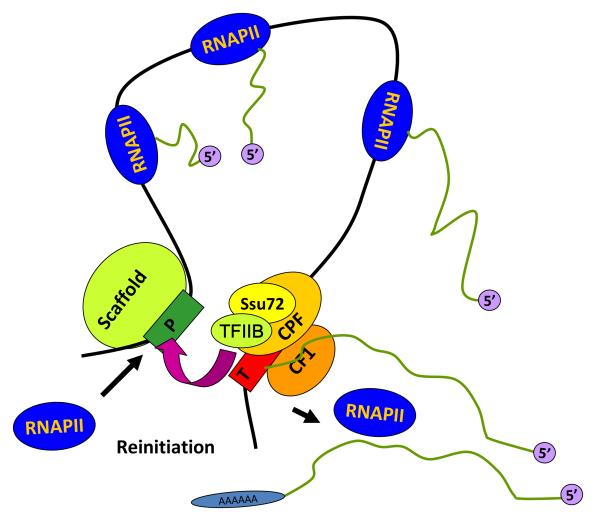
Immediately following transcription initiation, Pol II clears the promoter and TFIIB dissociates from the initiation complex. Many components of the initiation complex, however, remain intact at the promoter. This complex has been referred to as a “scaffold” that facilitates subsequent rounds of transcription (Yudkovsky et al., 2000). Accordingly, transcription reinitiation would occur by a mechanism different from de novo initiation. We suggest that TFIIB dissociates from the promoter, then reassociates with Pol II at the terminator following endonucleolytic cleavage and polyadenylation of mRNA. This terminator-Pol II-TFIIB complex then binds the scaffold, forming a loop that facilitates formation of a reinitiation complex (Singh and Hampsey, 2007). Pol II translocation from terminator to promoter might be facilitated by Ssu72-catalyzed dephosphorylation of the CTD, creating the hypophosphorylated form of Pol II required for promoter assembly. Accordingly, a pioneer round of transcription would be required for loops to form; once formed, loops could then facilitate subsequent rounds of transcription by hand-off of Pol II from terminator to promoter (Fig. 3).
Fig. 3.
A model for gene looping. We propose that a gene loop forms by juxtaposition of the promoter and terminator following a pioneer round of transcription. The loop is a transient structure, maintained by physical interaction between components of the transcription initiation and termination complexes. The gene loop might promote subsequent rounds of transcription by “hand-off” of Pol II from terminator to promoter, a process facilitated by the Ssu72 CTD phosphatase (Ansari and Hampsey, 2005).
Gene loops and transcriptional memory
Mutations that block looping have no apparent effect on transcription, yet induction of the GAL1 promoter induces looping (Ansari and Hampsey, 2005; Singh and Hampsey, 2007). These results suggest that looping might be required specifically for high levels of gene expression. To test this idea, we assayed the kinetics of GAL10 activation following a shift from glucose to galactose. Consistent with earlier results from many laboratories, we observed relatively slow activation kinetics, requiring more than 1 h to achieve maximum GAL10 mRNA accumulation (Laine et al., 2009). Activation occurred with the same kinetics in both the wild type and sua7-1 looping-defective strains.
We next assayed the kinetics of GAL10 reactivation following a cycle of galactose activation and glucose repression. In this case, activation occurred much more rapidly, requiring less than 5 min for full activation (Laine et al., 2009). This effect of slow activation and rapid reactivation following an initial cycle of activation and repression had been noted before, defined as “transcriptional memory” (Brickner et al., 2007; Kundu et al., 2007; Zacharioudakis et al., 2007). Rapid reactivation, however, required gene looping: GAL10 mRNA accumulation occurred with the same slow kinetics in the sua7-1 mutant upon activation and reactivation. Thus, transcriptional memory correlates with gene looping, suggesting that gene loops are required for memory.
Similar results were reported by Proudfoot and colleagues (Tan-Wong et al., 2009). They showed slow activation kinetics for the galactose-inducible HXK1 gene upon initial exposure to galactose, but rapid reactivation following a cycle of activation and repression. This effect was lost in the sua7-1 mutant and in an mlp1 mutant, which encodes a myelin-like protein required for translocation of active genes to the nuclear pore. Accordingly, their results also support a role for TFIIB in memory and implicate the nuclear pore in looping and memory.
Perspective
We have addressed loop formation at only a handful of yeast genes, chosen primarily based on their amenability to analysis by 3C. The technology now exists, though, to readily query looping on a genome-wide scale using a variation of 3C that incorporates biotin tags by end fill-in of restriction sites, coupled with massively parallel sequencing of paired ends (Lieberman-Aiden et al., 2009). We anticipate that Hi-C can be adapted to query looping of the entire yeast genome.
Although looping is transcription-dependent and requires specific components of initiation and 3′-end processing/termination components, the mechanism involved in formation and maintenance of loops remains to be determined. It will be especially interesting to learn whether loop formation requires the nuclear pore, or, conversely, whether gene translocation to the pore requires an intact loop. The looping requirement for transcriptional memory has so far been demonstrated only for galactose-inducible genes. Does looping facilitate memory of other classes of genes and, if so, does memory always involve the nuclear pore? Finally, does looping affect other aspects of gene expression, including, perhaps, mRNA export from nucleus to cytoplasm?
Gene loops are not unique to yeast. The HIV-1 provirus forms a loop between the 5′ long-terminal repeat and the 3′ LTR poly(A) signal and does so in a transcription-dependent manner (Perkins et al., 2008). Looping between promoter-terminator regions has also been reported for the mammalian genes encoding the immunohistological marker CD68 (O’Reilly and Greaves, 2007) and the breast cancer gene BRCA1 (Tan-Wong et al., 2008). Interestingly, different BRCA1 loops are formed in response to estrogen stimulation, and in normal versus cancer cell lines (Tan-Wong et al., 2008). These results underscore the dynamic nature of gene loops and support the premise that looping affects gene regulation.
Summary
Gene loops are dynamic structures that juxtapose promoter-terminator regions of Pol II-transcribed genes. Although first described in yeast, gene loops have now been identified in yeast and mammalian cells. Looping requires components of the transcription preinitiation complex, the pre-mRNA 3′-end processing machinery, and subunits of the nuclear pore complex. Loop formation is transcription-dependent, but neither basal nor activated transcription requires looping. Rather, looping appears to affect cellular memory of recent transcriptional activity, enabling a more rapid response to subsequent stimuli. The nuclear pore has been implicated in both memory and looping. Our working model is that loops are formed and/or maintained at the nuclear pore to facilitate hand-off of Pol II form the terminator to the promoter, thereby bypassing Pol II recruitment as the rate-limiting step in reactivation of transcription. Involvement of the nuclear pore also suggests that looping might facilitate mRNA export to the cytoplasm. The technology now exists to test these ideas.
Acknowledgments
The work in Dr. Hampsey’s laboratory is supported by NIH grants GM39484 (to M.H.) and GM68887 (to M. H. and Claire Moore (Tufts Medical School)).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ansari A, Hampsey M. A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes Dev. 2005;19:2969–2978. doi: 10.1101/gad.1362305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berroteran RW, Ware DE, Hampsey M. The sua8 suppressors of Saccharomyces cerevisiae encode replacements of conserved residues within the largest subunit of RNA polymerase II and affect transcription start site selection similarly to sua7 (TFIIB) mutations. Mol Cell Biol. 1994;14:226–237. doi: 10.1128/mcb.14.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH. H2A. Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron S, Bickmore WA. Does looping and clustering in the nucleus regulate gene expression? Curr Opin Cell Biol. 2004;16:256–262. doi: 10.1016/j.ceb.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Dekker J. The three ‘C’ s of chromosome conformation capture: controls, controls, controls. Nat Methods. 2006;3:17–21. doi: 10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- El Kaderi B, Medler S, Raghunayakula S, Ansari A. Gene looping is conferred by activator-dependent interaction of transcription initiation and termination machineries. J Biol Chem. 2009;284 doi: 10.1074/jbc.M109.007948. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser P. Transcriptional control thrown for a loop. Curr Opin Genet Dev. 2006;16:490–495. doi: 10.1016/j.gde.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Ganem C, Devaux F, Torchet C, Jacq C, Quevillon-Cheruel S, Labesse G, Facca C, Faye G. Ssu72 is a phosphatase essential for transcription termination of snoRNAs and specific mRNAs in yeast. Embo J. 2003;22:1588–1598. doi: 10.1093/emboj/cdg141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M, Na JG, Pinto I, Ware DE, Berroteran RW. Extragenic suppressors of a translation initiation defect in the cyc1 gene of Saccharomyces cerevisiae. Biochimie. 1991;73:1445–1455. doi: 10.1016/0300-9084(91)90177-3. [DOI] [PubMed] [Google Scholar]
- He X, Khan AU, Cheng H, Pappas DL, Jr., Hampsey M, Moore CL. Functional interactions between the transcription and mRNA 3′ end processing machineries mediated by Ssu72 and Sub1. Genes Dev. 2003;17:1030–1042. doi: 10.1101/gad.1075203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy S, He X, Reyes-Reyes M, Moore C, Hampsey M. Ssu72 Is an RNA polymerase II CTD phosphatase. Mol Cell. 2004;14:387–394. doi: 10.1016/s1097-2765(04)00235-7. [DOI] [PubMed] [Google Scholar]
- Kundu S, Horn PJ, Peterson CL. SWI/SNF is required for transcriptional memory at the yeast GAL gene cluster. Genes Dev. 2007;21:997–1004. doi: 10.1101/gad.1506607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine JP, Singh BN, Krishnamurthy S, Hampsey M. A physiological role for gene loops in yeast. Genes Dev. 2009;23:2604–2609. doi: 10.1101/gad.1823609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marenduzzo D, Faro-Trindade I, Cook PR. What are the molecular ties that maintain genomic loops? Trends Genet. 2007;23:126–133. doi: 10.1016/j.tig.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Meinhart A, Silberzahn T, Cramer P. The mRNA transcription/processing factor ssu72 is a potential tyrosine phosphatase. J Biol Chem. 2003;278:15917–15921. doi: 10.1074/jbc.M301643200. [DOI] [PubMed] [Google Scholar]
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- O’Reilly D, Greaves DR. Cell-type-specific expression of the human CD68 gene is associated with changes in Pol II phosphorylation and short-range intrachromosomal gene looping. Genomics. 2007;90:407–415. doi: 10.1016/j.ygeno.2007.04.010. [DOI] [PubMed] [Google Scholar]
- O’Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J, Proudfoot NJ. Gene loops juxtapose promoters and terminators in yeast. Nat Genet. 2004;36:1014–1018. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- Pappas DL, Jr., Hampsey M. Functional interaction between Ssu72 and the Rpb2 subunit of RNA Polymerase II in Saccharomyces cerevisiae. Molecular & Cellular Biology. 2000;20:8343–8351. doi: 10.1128/mcb.20.22.8343-8351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins KJ, Lusic M, Mitar I, Giacca M, Proudfoot NJ. Transcription-Dependent Gene Looping of the HIV-1 Provirus Is Dictated by Recognition of Pre-mRNA Processing Signals. Mol Cell. 2008;29:56–68. doi: 10.1016/j.molcel.2007.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto I, Na JG, Sherman F, Hampsey M. cis- and trans-acting suppressors of a translation initiation defect at the cyc1 locus of Saccharomyces cerevisiae. Genetics. 1992a;132:97–112. doi: 10.1093/genetics/132.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto I, Ware DE, Hampsey M. The yeast SUA7 gene encodes a homolog of human transcription factor TFIIB and is required for normal start site selection in vivo. Cell. 1992b;68:977–988. doi: 10.1016/0092-8674(92)90040-j. [DOI] [PubMed] [Google Scholar]
- Reyes-Reyes M, Hampsey M. Role for the Ssu72 C-terminal domain phosphatase in RNA polymerase II transcription elongation. Molecular & Cellular Biology. 2007;27:926–936. doi: 10.1128/MCB.01361-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh BN, Ansari A, Hampsey M. Detection of gene loops by 3C in yeast. Methods. 2009;48:361–367. doi: 10.1016/j.ymeth.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh BN, Hampsey M. A Transcription-Independent Role for TFIIB in Gene Looping. Mol Cell. 2007;27:806–816. doi: 10.1016/j.molcel.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Sun Z-W, Hampsey M. Synthetic enhancement of a TFIIB defect by a mutation in SSU72, an essential gene encoding a novel protein that affects transcription start site selection in vivo. Mol Cell Biol. 1996;16:1557–1566. doi: 10.1128/mcb.16.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan-Wong SM, French JD, Proudfoot NJ, Brown MA. Dynamic interactions between the promoter and terminator regions of the mammalian BRCA1 gene. Proc Natl Acad Sci U S A. 2008;105:5160–5165. doi: 10.1073/pnas.0801048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 2009;23:2610–2624. doi: 10.1101/gad.1823209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudkovsky N, Ranish JA, Hahn S. A transcription reinitiation intermediate that is stabilized by activator. Nature. 2000;408:225–229. doi: 10.1038/35041603. [DOI] [PubMed] [Google Scholar]
- Zacharioudakis I, Gligoris T, Tzamarias D. A yeast catabolic enzyme controls transcriptional memory. Curr Biol. 2007;17:2041–2046. doi: 10.1016/j.cub.2007.10.044. [DOI] [PubMed] [Google Scholar]