Summary
Aggregatibacter actinomycetemcomitans establishes a tenacious biofilm that is important for periodontal disease. The tad locus encodes the components for the secretion and biogenesis of Flp pili, which are necessary for the biofilm to form. TadZ is required, but its function has been elusive. We show that tadZ genes belong to the parA/minD superfamily of genes and that TadZ from A. actinomycetemcomitans (AaTadZ) forms a polar focus in the cell independent of any other tad locus protein. Mutations indicate that regions in AaTadZ are required for polar localization and biofilm formation. We show that AaTadZ dimerizes and that all TadZ proteins are predicted to have a Walker-like A box. However, they all lack the conserved lysine at position 6 (K6) present in the canonical Walker-like A box. When the alanine residue (A6) in the atypical Walker-like A box of AaTadZ was converted to lysine, the mutant protein remained able to dimerize and localize, but it was unable to allow the formation of a biofilm. Another essential biofilm protein, the ATPase (AaTadA), also localizes to a pole. However, its correct localization depends on the presence of AaTadZ. We suggest that the TadZ proteins mediate polar localization of the Tad secretion apparatus.
Keywords: type IV pili, adherence, biofilm, Actinobacillus, tad, Walker-like A box
Introduction
The gram-negative coccobacillus Aggregatibacter actinomycetemcomitans is the causative agent of Localized Aggressive Periodontitis (LAP) and several non-oral diseases, such as infective endocarditis and brain abscesses (Slots, 1999, Zambon, 1985, Das et al., 1997, Stepanovic et al., 2005, Henderson et al., 2003). A. actinomycetemcomitans adheres tenaciously and nonspecifically to plastic, glass, and hydroxyapatite (Henderson et al., 2002, Kachlany et al., 2001a). Clinical isolates are characterized by having the following properties: rough colony morphology, tenacious adherence to the wall of the culture vessel, autoaggregation, and the presence of bundled Flp pili (Flp fibrils) associated with the cells (Fine et al., 2006, Inouye et al., 1990, Kachlany et al., 2000, Kachlany et al., 2001a, Kachlany et al., 2001b, Preus et al., 1988, Rosan et al., 1988). The Flp pili of A. actinomycetemcomitans support colonization of the oral cavity and are required for oral pathogenesis in a rat model (Schreiner et al., 2003).
The genetic basis of tenacious biofilm formation is the 14-gene tad (tight adherence) locus (Kachlany et al., 2000, Kachlany et al., 2001a, Kachlany et al., 2001b, Perez et al., 2006, Planet et al., 2003, Tomich et al., 2007). Phylogenetic analysis indicated that the tad locus has a history of horizontal transfer. Consequently, it is sometimes referred to as the “Widespread Colonization Island” (WCI) (Planet et al., 2003).
The role of the tadZ gene product in A. actinomycetemcomitans (AaTadZ) has been elusive. No tadZ homologs are present in other known secretion systems. The AaTadZ protein has no apparent transmembrane domain and is predicted to be soluble. Previous studies have shown that the product of cpaE, a tadZ homolog in Caulobacter crescentus, localizes to the old pole of the swarmer cell (Viollier et al., 2002b, Viollier et al., 2002a) and that pilus biogenesis occurs at that pole in the swarmer cell (Skerker & Shapiro, 2000, Viollier et al., 2002b). Localization of CpaC, the putative secretin, is dependent on CpaE (Viollier et al., 2002b).
We found a consistent similarity of the products of tadZ homologs to the products of parA and minD. ParA is involved in DNA segregation in bacteria, and MinD is required for proper cell division (de Boer et al., 1991 Leonard et al., 2005b, Rothfield et al., 2005, Thanbichler, Gerdes et al.). Both proteins localize to distinct regions in the cell and can mediate the localization of other proteins (Li et al., 2004, Marston & Errington, 1999, Quisel et al., 1999, Hu & Lutkenhaus, 1999, Lutkenhaus, 2002, Raskin & de Boer, 1999). To determine the relationship of tadZ and its close relatives to other members of the superfamily that includes parA and minD, we undertook a large phylogenetic analysis using 4,074 related genes.
Sequence analyses of TadZ-like proteins showed that they all harbor a close relative of the Walker-like A box. The Walker-like A box is a variant of the Walker A box. The one in the TadZ proteins is designated the atypical Walker-like A box. The amino acids of the Walker A box and its variant, the Walker-like A box, are known to be involved in binding and hydrolysis of ATP (Abrahams et al., 1994, Story & Steitz, 1992, Karkaria et al., 1990, Walker et al., 1982, Koonin, 1993, de Boer et al., 1991). Proteins harboring Walker-like A motifs are involved in variety of biological functions. These include ParA (DNA segregation), MinD (spatial regulation of cell division), FleN (flagellar regulation), ArsA (arsenite resistance), BcsQ/YhjQ (localization of cellulose biosynthesis apparatus) (Le Quere & Ghigo, 2009), and NifH (electron transfer) (Zhou et al., 2000, Leonard et al., 2005a, Hayashi et al., 2001, Lutkenhaus & Sundaramoorthy, 2003, Schindelin et al., 1997, Cordell & Lowe, 2001, Motallebi-Veshareh et al., 1990). Inactivation of the Walker-like A box in ParA or MinD is known to affect DNA plasmid partitioning (ParA) (Davis et al., 1996, Ebersbach & Gerdes, 2004, Ebersbach & Gerdes, 2001, Fung et al., 2001, Lemonnier et al., 2000, Leonard et al., 2005a, Libante et al., 2001) or spatial organization of cell division in bacteria (MinD) (Hayashi et al., 2001, Hu et al., 2002, Zhou & Lutkenhaus, 2004). Extensive mutational analyses of the ParA and MinD Walker-like A boxes have shown that the A box residues are important for function. The conservation of the atypical Walker-like A box in the predicted products of tadZ homologs led us to believe that it has an important role in the function of the proteins.
In this study we present our findings on the localization and functional characterization of AaTadZ. We provide evidence for polar localization, and we identify domains that are critical for membrane association, localization, and biofilm formation. We show that the atypical Walker-like A box of AaTadZ is important for biofilm formation, but not for dimerization or localization. Although AaTadZ localizes without any other tad locus protein, we found that the polar position of AaTadA, an ATPase (Bhattacharjee et al., 2001), is stabilized by the presence of AaTadZ. We propose that AaTadZ is needed to localize the entire Tad secretion apparatus to a pole.
Results
tadZ is related to genes of the parA/minD superfamily
Systematic searches of public databases revealed that TadZ-like amino acid sequences showed consistent, but low level, similarity (37–62%) to the products of the parA/minD superfamily (see SI). Similarity was sometimes confined to the Walker-like A box region of TadZ, but the region of similarity often extended beyond it. BLAST alignments using default parameters (low complexity filter off) ranged in size from 16 to 279 amino acids. In addition to the Walker-like A box, TadZ-like proteins also have a predicted amphipathic helical domain similar to those predicted to be in ParA- and MinD-related proteins (Fig. 1). An amphipathic helix in MinD is thought to be a membrane-associating domain (Zhou & Lutkenhaus, 2003, Hu & Lutkenhaus, 2003, Szeto et al., 2002). This could indicate that TadZ is peripherally associated with the inner membrane. Taken together these data indicated to us that tadZ genes were likely to be at least distant homologs of the parA and minD superfamily of genes.
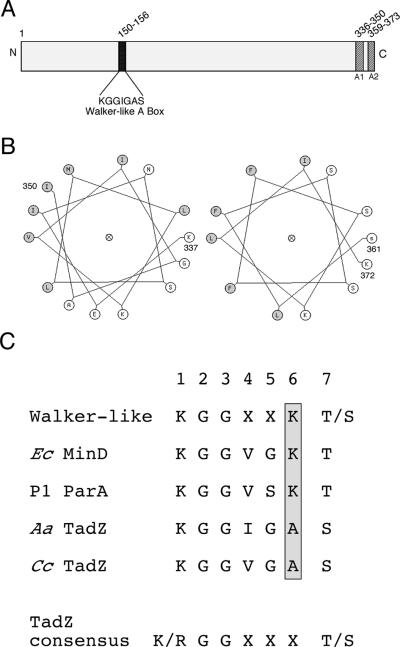
Figure 1. The AaTadZ protein.
Shown here is a diagram of the AaTadZ protein, which comprises 373 amino acids. (A) Amino acid positions and domain boundaries (darkened segments) are indicated by the numbers. A1 refers to the predicted amphipathic helix 1 and A2, to the predicted amphipathic helix 2. (B) Helical wheel representations of putative membrane-targeting domains 1 (left) and 2 (right). Secondary structure prediction programs identified two putative amphipathic helices at amino acid residues 336–349 and 359–373, both of which are located in the C-terminal region. Residues 337–350 (left) and 361–372 (right) were arranged on helical wheels and were predicted to be amphipathic. Nonpolar residues are shaded. (C) Sequences of Walker-like A box motifs, including sequences of Walker-like A box from E. coli (Ec) MinD, P1 ParA, A. actinomycetemcomitans (Aa) TadZ, and C. crescentus (Cc) TadZ. The amino acid at position 6 in the Walker-like A boxes is shaded.
Other genes whose products are involved in polar molecular structures and have been shown to be similar to those of the parA/minD superfamily are flhG/fleN of the flagellar secretion system in Vibro alginolyticus and Pseudomonas spp. (Kusumoto et al., 2006) and bcsQ/yhjQ of the cellulose synthetase complex in Escherichia coli (Le Quere & Ghigo, 2009). To test the relationship of tadZ genes to these and other parA/minD superfamily members, we undertook a phylogenetic analysis.
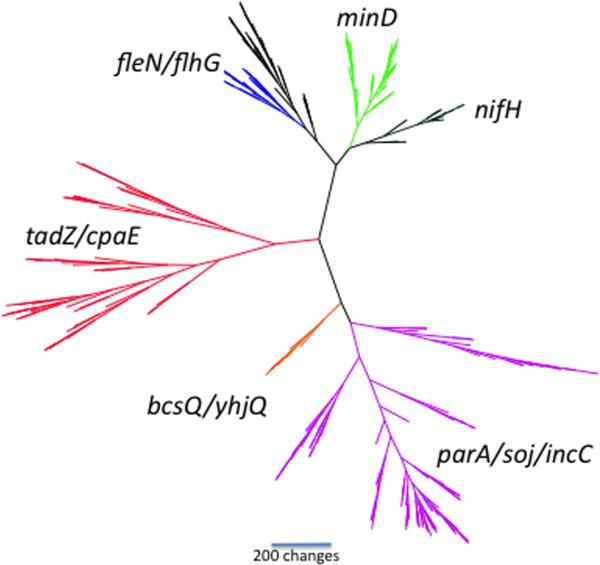
The large number of genes found in our search (n=4,074) to identify members of the parA/minD superfamily introduced serious analytical difficulties. To reduce the size of the dataset, we used a sampling technique aimed at representing as great a diversity of genes as possible. Figure 2 shows a representative tree. To test whether or not our sampled datasets were representative of the overall 4,074-gene dataset, we replicated our gene sampling technique ten times, obtaining a newly selected set of 309 genes from the superfamily each time. The trees generated from each sample had some topological differences, as would be expected when different taxa are sampled; but overall they were very similar. In particular, six major families of genes could be distinguished in all trees (Fig. 2): (1) minD, (2) parA/soj/incC, (3) nifH, (4) flhG, (5) bcsQ/yhjQ, and (6) tadZ/cpaE. The tadZ/cpaE sequences formed a cluster in 8 out of 10 sampled trees; in three of these clusters, the bcsQ/yhjQ clade was included in the tadZ/cpaE clade. The Gram-positive tadZ homologs, including ErtadZ from Eubacterium rectale (the accompanying paper, Xu et al., 2012) almost always grouped with the other putative tadZ genes (9 out of 10 trees), indicating a close relationship. These findings show that the tadZ/cpaE clade is a distinct family within the parA/minD superfamily (Fig. 2).
Figure 2. A representative phylogeny of the parA/minD superfamily.
The tree is unrooted and shows the relative positions of the major families in this superfamily. Six major families of genes are highlighted: minD (green), parA/soj/incC (purple), nifH (black), fleN/flhG (blue; the black branch is a clade of as yet unidentified genes), bcsQ/yhjQ (orange), and tadZ/cpaE (red). All datasets and tree files are available as supplementary information.
Our results also indicate that the bcsQ/yhjQ clade may be closely related to the genes constituting the tadZ/cpaE clade for the following reasons. (1) The genes are usually found immediately upstream of a tadA homolog. (2) The predicted polypeptide products of the bcsQ/yhjQ clade have the unique atypical Walker-like A box (see the next section). (3) Proteins of the bcsQ/yhjQ clade have a two-domain structure, with a predicted aminoterminal receiver domain (ARD) and an ATP-binding domain (AAD) that contains the atypical Walker-like A box motif (see the accompanying paper on ErTadZ for details).
The atypical Walker-like A box of TadZ proteins
We found that predicted TadZ proteins possess a variant of the Walker-like A box. The canonical Walker-like A box signals that the protein binds and hydrolyzes ATP. However, the TadZ protein of A. actinomycetemcomitans (AaTadZ) lacks the lysine in the 6 position (K6) in the Walker-like A box numbering system shown in Fig. 1C. That lysine is conserved in the Walker-like A box. In AaTadZ it is replaced with an alanine residue (Fig. 1C). In addition to AatadZ, all tadZ homologs examined (>300) are predicted to encode proteins that lack the conserved K6 residue of the Walker-like A box in proteins of the parA/minD superfamily (Hayashi et al., 2001, Leonard et al., 2005a, Atmakuri et al., 2007, Lutkenhaus & Sundaramoorthy, 2003). The “atypical” Walker-like A box for all predicted TadZ proteins is K/RGGXXX(T/S) (Fig. 1C). Although it was not part of our study, one predicted polypeptide is a possible exception. The predicted TadZ-TadA fusion protein encoded by the genome of Bdellovibrio bacteriovorus HD100 does not have the atypical Walker-like A box of TadZ proteins (Rendulic, S. et al., 2004).
Localization of TadZ in A. actinomycetemcomitans cells
The similarity of AaTadZ to products of the parA/minD superfamily and the studies with CpaE, the product of a tadZ homolog from C. crescentus, indicated that AaTadZ may localize to a specific site in the cell. We fused the coding region for EGFP to the 3'-end of AatadZ to determine the localization of AaTadZ by fluorescence microscopy. The tadZ-egfp fusion on a plasmid was mobilized into wild-type and tadZ mutant strains of A. actinomycetemcomitans by conjugation, and the fusion product was expressed from the IPTG-inducible tac promoter. In the presence of low IPTG (10 μM), the fusion protein was expressed and functional, as determined by its ability to partially complement the adherence defect in the tadZ mutant (Fig. S1).
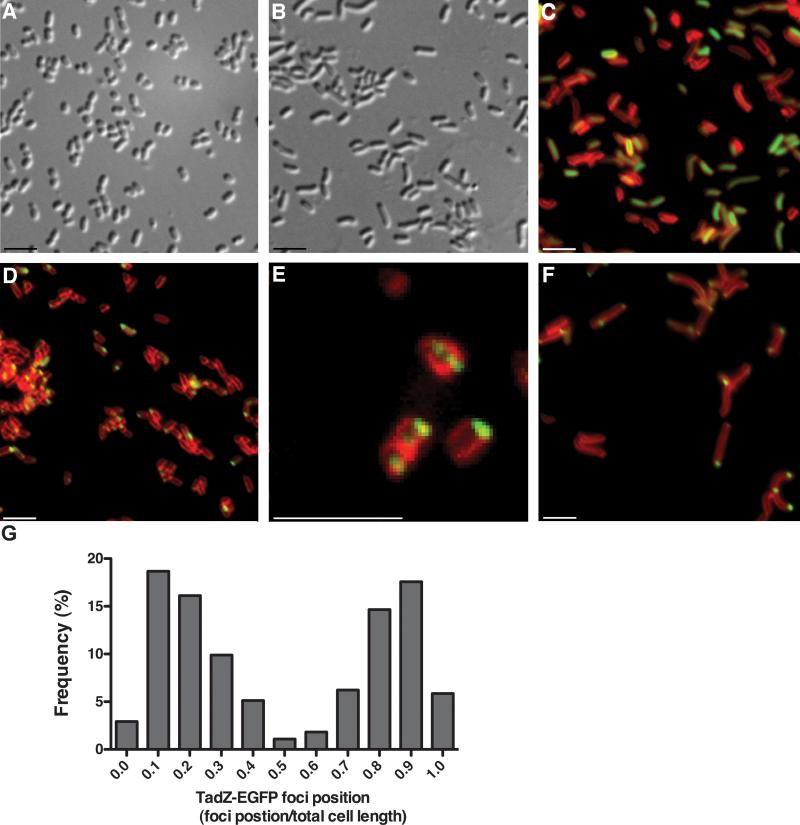
Individual cells of A. actinomycetemcomitans are significantly smaller than E. coli cells (Fig. 3A and data not shown). Therefore, the membrane stain TMA-DPH was important for localization (Fig 3C–3F). For visualization of GFP and the fusion protein, it was necessary to induce the fusion gene with 10 μM IPTG for 2–3 hours (1–1.5 cell divisions). GFP showed diffuse, cytoplasmic fluorescence, as expected (Fig. 3C). In contrast, the AaTadZ-EGFP fusion protein showed a polar focus (Fig. 3D–F). Nearly all cells with a focus showed a single focus at a pole (Fig. 3D). The apparent membrane-proximal location of fluorescent foci from AaTadZ-EGFP is consistent with evidence from a membrane-fractionation study indicating that AaTadZ associates peripherally with the inner membrane (Fig. 6).
Figure 3. AaTadZ-EGFP localization in cells of A. actinomycetemcomitans.
Images A and B were taken using differential interference contrast microscopy. (A) A. actinomycetemcomitans tadZ mutant cells and (B, C, and F) piperacillin-treated tadZ mutant showing elongated cells. C–F show fluorescence images of the cells of tadZ mutant strains treated with TMA-DPH. (C) The tadZ mutant strain expressing GFP (Aa3136); (D–F) the tadZ mutant strain expressing AaTadZ-EGFP (Aa3242). Cells were prepared and collected for fluorescence microscopy as described in Materials and Methods. Size bars are ~1.5 μm. (G) Histogram of line-scan analysis of fluorescent images that show AaTadZ-EGFP focus localization. The histogram gives focus position as a fraction of total cell length. Each bar represents a bin center. All bin centers are cell fractional lengths of 0.10. Only cells with AaTadZ-EGFP foci are included in the analysis. All foci in multi-focus cells were mapped, but cells with multiple foci were less than 5% of the total number of cells mapped. Focus position was determined by scanning the total length of the cell along the major axis. The maximum intensity of fluorescence was taken as the focus position. Because the ends of the cell are indistinguishable and the cells were arrayed randomly, there is no specificity to the end chosen to begin scanning. Consequently, the data approximate a mirror image. For example, no difference should be assigned to the foci in bin 0.1 and in bin 0.9
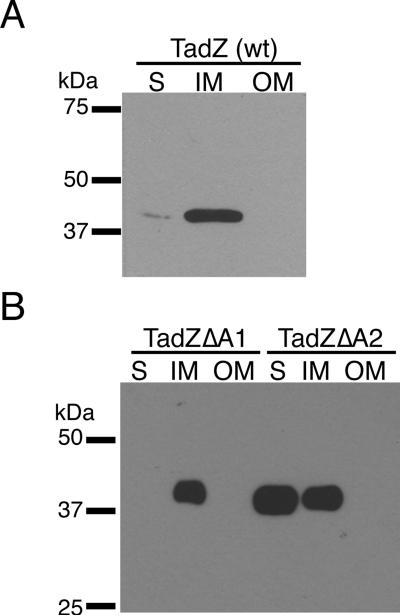
Figure 6. Subcellular localization of AaTadZ and the ΔA1 and ΔA2 mutants.
Subcellular fractions of CU1000N (wild type, wt) (A) and of Aa3341 and Aa3346 (B), which are tadZ mutants producing AaTadZΔA1 and AaTadZΔA2 from pBP346 and pBP352, respectively, were prepared by differential detergent solubilization and ultracentrifugation, as described in materials and methods. Immunoblots were probed with α-TadZ antisera. Soluble (S), inner membrane (IM), and outer membrane (OM) fractions are indicated.
We found that 49.7% of cells with a focus had at least one polar fluorescent focus. We did not detect a GFP signal in a significant portion of the population, and less than 2% of the cells had diffuse cytoplasmic localization of TadZ-EGFP. Rarely, we observed two foci at opposite poles or an additional focus at a septum at the mid-cell division site (Fig. 3E and 3G). We consider the septum to be the site for the formation of a nascent pole for the daughter cell. Overall our results indicate that AaTadZ prefers non-septum-associated poles.
Autoaggregation is a property of Tad+ A. actinomycetemcomitans (Fig. S1). Individual cells within clumps can be seen to have polar AaTadZ-EGFP foci; but, to avoid an enumeration error, we omitted clumps of cells from our quantitation. However, a smaller percentage of the free cells showed foci; and thus our calculations probably underestimate the percentage of cells that displayed TadZ-EGFP foci. Higher expression with 100 μM IPTG did not help to increase the number of cells with foci. It merely resulted in cells showing diffuse cytoplasmic localization of AaTadZ-EGFP (data not shown).
To determine better if AaTadZ is at a pole, we examined AaTadZ-EGFP in cells that were made larger than wild-type cells through elongation induced by piperacillin, which inhibits cell division in E. coli (Pogliano et al., 1997, Botta & Park, 1981) (Fig. 3B, 3C, and 3F). Addition of a subinhibitory concentration of piperacillin (2 μg ml−1) to the growth medium does not disrupt biofilm formation by A. actinomycetemcomitans, but the cells are elongated (Fig. 3). TadZ-EGFP was clearly observed to be at the poles in piperacillin-treated cells (Fig. 3F). Many of the elongated cells arising from piperacillin treatment showed foci at both ends of the cell (Fig. 3F) and, in some instances, near or at the mid-cell (Fig. 3F). In addition, we did line-scan analysis on fluorescent images of A. actinomycetemcomitans tadZ mutant cells (not treated with piperacillin) expressing AaTadZ. Frequency distribution of focus position confirmed that AaTadZ-EGFP localization was enriched at the poles. A small percentage of foci were non-polar (Fig. 3G).
AaTadZ polar localization is not dependent on other tad genes
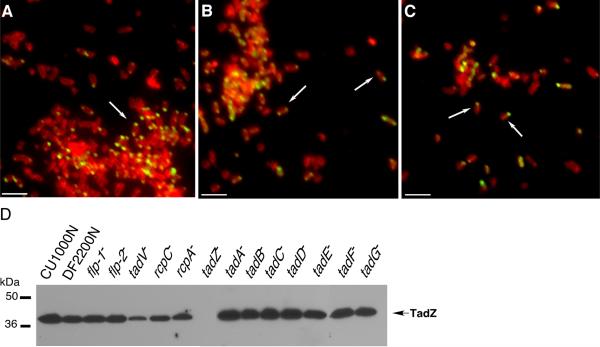
Three experiments showed that the formation of foci by AaTadZ and polar localization are independent of any other tad locus protein. (1) AaTadZ-EGFP foci are formed in all tad mutant strains. We probed whole-cell extracts of A. actinomycetemcomitans with TadZ-specific polyclonal antiserum. A band of approximately 41.6 kDa was recognized by the TadZ antibody in samples prepared from a wild-type strain (CU1000) and the isogenic tad mutants [except the strain mutated in tadZ (Fig. 4D)]. (AaTadZ accumulation was slightly reduced in mutants defective in tadV, rcpC, and rcpA.) We observed foci of AaTadZ-EGFP localized to the poles in all the mutants (Fig. 4 and S2). (2) We did localization studies in the Tad− mutant strain CU1060, a promoter mutant of CU1000 that does not produce detectable levels of any the tad-locus proteins (Kram et al., 2008). CU1060 allows AaTadZ-EGFP to form foci at the poles (data not shown). We also noted an overall decrease in AaTadZ-EGFP foci in tadZ+ cells, whereas robust polar localization was observed in tadZ− cells (Fig. 4 and S2), indicating that the wild-type AaTadZ protein can displace AaTadZ-EGFP from the poles. (3) We expressed AaTadZ-EGFP in E. coli, a heterologous species that does not harbor a tad locus. As expected, we observed diffuse cytoplasmic fluorescence for GFP (Fig. S3). In contrast, AaTadZ-EGFP was usually seen as a single focus at a pole. Increased expression of AaTadZ-EGFP by adding IPTG caused the formation of multiple, randomly distributed foci (Fig. S3). From the three results, we conclude that localization of AaTadZ does not depend on another tad-locus protein.
Figure 4. AaTadZ-EGFP polar foci and AaTadZ expression in tad gene mutants of A. actinomycetemcomitans.
Shown are representatives of fluorescence images of tad mutant strains treated with TMA-DPH and expressing AaTadZ-EGFP: (A) Aa3313 (flp-1∷IS903fkan); (B) Aa3318 (tadA∷IS903fkan), and (C) Aa3322 (tadE∷IS903fkan). Arrows show cells with foci. Cells were prepared and collected for fluorescence microscopy as described in Materials and Methods. Size bars are ~1.5 μm. (D) AaTadZ expression in wild-type A. actinomycetemcomitans strains CU1000N, DF2200N, and nonpolar tad mutants. Strains were grown for approximately 24 hr in liquid medium with Cm. Whole-cell extracts were prepared and analyzed by α-AaTadZ immunoblotting. The predicted molecular mass for AaTadZ is ~41.6 kDa.
Regions with putative amphipathic helices are required for localization and biofilm formation in AaTadZ
Using secondary structure prediction programs, we identified two putative helices, one (A1) located within amino acid residues 336–350 and the other (A2) within residues 359–373 (Fig. 1). Arrangement of these amino acids on a helical wheel revealed that residues 337–350 and 361–372 could form amphipathic helices (Fig. 1B). For both putative helices, one face comprises nonpolar hydrophobic residues, while the opposing face includes charged and uncharged polar residues (Fig. 1B). Because these regions are in the carboxy-terminus, as are the amphipathic helices in MinD, we hypothesized that the putative AaTadZ helices might perform a similar function (Hu & Lutkenhaus, 2003).
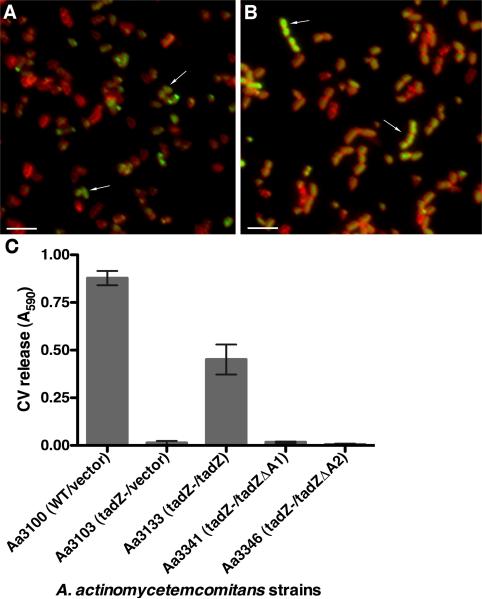
We constructed mutants of TadZ-EGFP that lacked either the first putative amphipathic helix (tadZ-egfpΔA1) or the second (tadZ-egfpΔA2). Although Western blot analysis showed that both proteins are stably produced (data not shown), the ΔA1 and ΔA2 proteins showed altered localization patterns (Fig. 5A and 5B). Unlike wild-type AaTadZ-EGFP, where the majority of fluorescent cells had polar foci, the ΔA1protein localized to the cytoplasm (46.7%), to the poles (18.5%), and to both the poles and cytoplasm (13.5%) (Fig. 5A) The ΔA2 protein showed predominantly cytoplasmic localization (81.6%), with uniform fluorescence throughout the cell (Fig. 5B). In both experiments, approximately 20% of the cells lacked a measurable GFP signal. Our results indicate that both putative amphipathic helices are involved in polar localization of AaTadZ, although the A2 region may be more important.
Figure 5. Foci and adherence conferred by AaTadZ-EGFP mutants deleted for the putative amphipathic helices.
Shown are fluorescence images of the A. actinomycetemcomitans tadZ mutant strain treated with TMA-DPH and expressing (A) AaTadZΔA1-EGFP (Aa3331) and (B) AaTadZΔA2-EGFP (Aa3332). Cells were collected for fluorescence microscopy, as described in Materials and Method. Arrows note typical cells with non-polar fluorescence. Size bars are ~1.5 μm. (C) Shown are the results from quantitative adherence assays. The amount of crystal violet (CV) released (A590) is proportional to biofilm formation. In the strain names, the relevant strain genotype is indicated before the slash; plasmid-borne genes are indicated after the slash. Error bars represent standard deviation.
To test if regions A1 and A2 are important for targeting AaTadZ to the inner membrane, we did subcellular fractionation of A. actinomycetemcomitans as done previously (Clock et al., 2008). We first established the localization pattern of AaTadZ by fractionation of wild-type A. actinomycetemcomitans strain CU1000N. The fractionation involved ultracentrifugation and selective solubilization of inner membrane proteins with Sarkosyl (see SI Material and Methods). Immunoblotting of subcellular fractions with polyclonal antiserum specific for AaTadZ showed that a majority of AaTadZ localizes to the inner membrane; a small amount of AaTadZ was also present in the cytoplasmic fraction (Fig. 6A). For analysis of the requirement of the A1 and A2 regions in the localization of AaTadZ, we used the tadZ mutant strain Aa0886 with pBP346 to express the ΔA1 protein and with pBP352 to express the ΔA2 protein. Subcellular fractions prepared from these strains and immunoblotting revealed that deletion of region A1 had no obvious effect on the localization pattern. The AaTadZΔA1 protein localized normally to the inner membrane. In contrast, the putative A2 helix had a clear impact on AaTadZΔA2 in the inner membrane. Deletion of the A2 region resulted in an enrichment of AaTadZΔA2 in both the soluble and inner membrane fractions, with more AaTadZΔA2 protein in the soluble fraction than in the inner membrane fraction (Fig 6B).
To examine the requirement of the A1 and A2 regions for biofilm formation, we tested if tadZ mutants with the ΔA1 or ΔA2 mutations complement the tadZ mutant strain for restoration of adherence-related phenotypes: biofilm formation, autoaggregation, and rough-colony morphology (Kachlany et al., 2000) (Planet et al., 2003). Despite Western blot analysis that showed that both mutant proteins were expressed and accumulated to levels equivalent to that of the wild-type protein (data not shown), the mutants were unable to restore any of the adherence-related phenotypes (adherence shown in Fig. 5C; other data not shown). Increasing expression by the addition of 100 μM IPTG to the growth medium allowed the ΔA2 mutant to complement partially, but the ΔA1 mutant was unable to complement under any condition tested (data not shown).
Localization of AaTadA in A. actinomycetemcomitans cells
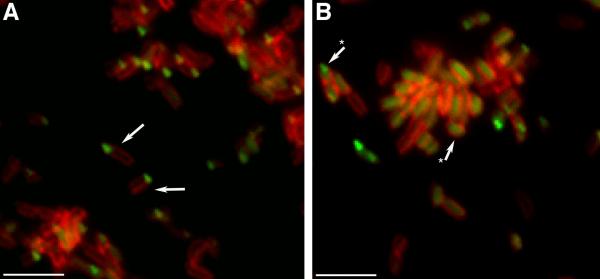
AaTadA is an ATPase required for Flp pili biogenesis (Bhattacharjee et al., 2001). TadZ and TadA are the only Tad proteins of A. actinomycetemcomitans that are predicted to be soluble. We wanted to know if AaTadA also localizes to a polar region of the cell. The coding region for EGFP was fused to the 3'-end of tadA. The tadA-egfp fusion was expressed from tac promoter. To visualize AaTadA-EGFP, the strains were grown in the presence of 5 μM IPTG. AaTadA-EGFP partially complements biofilm formation, autoaggregation, and rough-colony formation in a tadA mutant (Fig. S4). Localization was by fluorescence microscopy, as described previously. Due to the aggregation problem from the complementation of the tadA mutant, we expressed AaTadA-EGFP in a flp-1 mutant, which does not produce pili and is defective in autoaggregation. Western blot analysis verified that a flp-1 defect does not affect the abundance of TadA (Fig. S5). AaTadA-EGFP showed single foci at poles in flp-1 mutant cells (Fig. 7A). Measurements showed that among cells with measurable GFP signal, 81.6% of the cells had a polar focus; 14.8% showed diffuse localization in the cytoplasm; and 3.6% of the cells had localization in both the poles and cytoplasm. However, when AaTadA-EGFP was expressed in the tadZ mutant, we observed a defect in polar localization (Fig. 7B), as 54.7% displayed diffuse cytoplasmic localization (vs 14.8% in tadZ+ cells), and 32.8% showed a single polar focus. In addition, 12.4% displayed localization to both the poles and the cytoplasm. In summary, our analysis of AaTadA-EGFP localization patterns indicated that most cells with a focus displayed cytoplasmic localization in the absence of TadZ. Western blot analysis of TadA production in the tadZ mutant appears to be slightly reduced (Fig. S5). This observation may indicate that TadZ stabilizes the TadA protein. These results indicate that TadZ is important for the polar localization of TadA.
Figure 7. AaTadA-EGFP localization in A. actinomycetemcomitans.
Shown are fluorescence images of the A. actinomycetemcomitans (A) flp-1 mutant strain expressing AaTadA-EGFP (Aa3337) and (B) tadZ mutant strain expressing AaTadA-EGFP (Aa3333). To localize proteins in A. actinomycetemcomitans, all strains were stained with TMA-DPH. Size bars are ~1.5 μm. Arrows in panel A show polar foci, whereas arrows with (*) in panel B note typical cells with mislocalized AaTadA-EGFP.
The AaTadZ Walker-like A box is required for biofilm formation
To examine if the alanine residue at AaTadZ155 [(A6) Walker-like A box position 6] (Fig.1C) is important for function, we used site-directed PCR mutagenesis to replace it with a lysine to restore the canonical Walker-like A box of the ParA and MinD proteins. This Walker-like A box mutant, designated AatadZ A155K, was cloned under the control of the tac promoter in an IncQ broad-host vector. Western blot analysis showed that under our growth conditions the AaTadZ A155K protein was produced at wild-type AaTadZ levels (data not shown).
To determine if AaTadZ A155K is functional, we tested its ability to restore adherence, rough-colony morphology, and autoaggregation to a tadZ− mutant strain of A. actinomycetemcomitans. A mutant strain expressing AaTadZ A155K was defective for all these adherence-related properties. The A155K allele was unable to promote formation of a biofilm, as determined by the CV assay (Fig. 8). Massive overexpression did not help (data not shown). Substitutions of A155 with chemically distinct amino acids (valine, asparagine, glycine, and serine) resulted in mutants of AaTadZ that complemented a tadZ− mutant strain (data not shown). We found that the alanine residue at position 6 of the AaTadZ Walker-like A box is not conserved among other TadZ proteins. Our results indicate that, except for lysine, the identity of the amino acid at position 155 is not important to AaTadZ function.
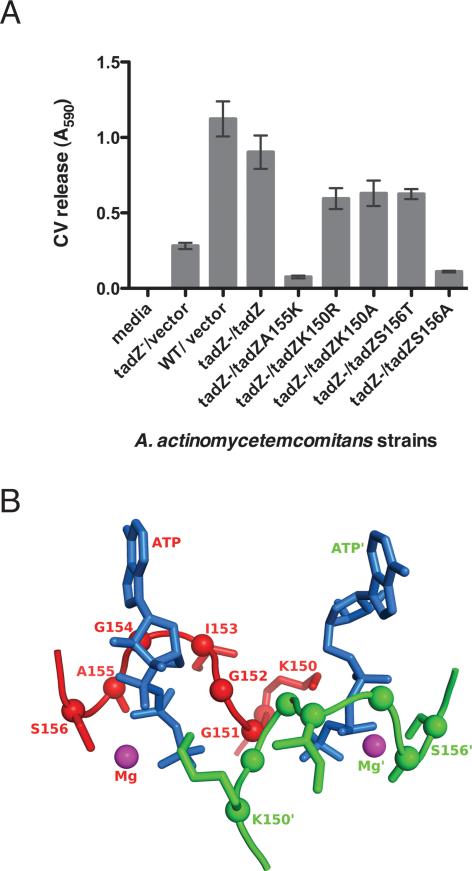
Figure 8. Biofilm formation by AaTadZ Walker-like A box mutants and the predicted structure of the ATP-binding pocket of AaTadZ.
(A) Results from quantitative adherence assays. A. actinomycetemcomitans wild-type CU1000N strain with vector (Aa3048) and tadZ mutant strains with vector (Aa3132), tadZ (Aa3133), tadZ A155K (Aa3097), tadZ K150R (Aa3146), tadZ K150A (Aa3224), tadZ S156T (Aa3120), and tadZ S156A (Aa3219) were grown for ~16 hrs. Adherence was measured in the wells of 96-well microtiter plates. In the strain names, the relevant host genotype is indicated before the slash; plasmids are indicated after the slash. The amount of crystal violet released is proportional to biofilm formation. (B) A homology model of the AaTadZ Walker-like A box regions in a dimer based on the structure of ErTadZ from the accompanying paper (Xu et al., 2012). The pocket of one monomer is shown in red with all residues fully labeled; that of the second monomer is shown in green with some residues labeled. ATP is shown in marine blue and Mg+2 in magenta. The asterisks denote components that are part of or interact with the second monomer.
The conservation of the other residues in the Walker-like A boxes of proteins of TadZ homologs indicated that the rest of the Walker-like A box is important for the function. To examine the role of the Walker-like A box in AaTadZ, we replaced K150 (K1, Walker-like A box position 1) with alanine and arginine; and we replaced S156 (S7, Walker-like A box position 7) with alanine and threonine. To test if the mutant proteins are functional, we examined their ability to complement the tadZ mutant of A. actinomycetemcomitans. The K150R, K150A, and S156T mutants of AaTadZ corrected the defects for rough-colony morphology and autoaggregation (data not shown). However, these mutants showed a significant reduction in biofilm formation (Fig. 8A), indicating that biofilm formation is more sensitive than the other phenotypes and that AaTadZ proteins having the K150R, K150A, or S156T changes are only partially functional. Changing residue S156 to alanine (AatadZ S156A) resulted in the complete loss of adherence and rough-colony morphology (Fig. 8A). Overexpression did not help. The results indicate that the Walker-like A box in AaTadZ is important for function.
The AaTadZ Walker-like A box is not involved in localization
We examined the involvement of the Walker-like A box in the localization of AaTadZ. EGFP was added to the C-termini of the Walker-like A box mutants (AaTadZ K150R, K150A, A155K, S156A, and S156T). We localized the proteins in the tadZ mutant strain of A. actinomycetemcomitans by fluorescence microscopy, as described above. All proteins appeared as distinct condensed foci at the cell poles. We did not observe any localization defects for any of the mutants (data not shown and Fig. S6).
AaTadZ form dimers in vivo
Because of the similarity between the predicted products of the parA/minD genes and the tadZ/cpaE clade, we were interested in determining whether or not AaTadZ can dimerize. We used a bacterial reporter system for protein dimerization in vivo based on the bacteriophage λ cI repressor. We fused the N-terminal DNA-binding domain of λ cI (λcIDB), which cannot repress because it cannot dimerize, to AaTadZ. Since dimers are needed to repress the λ pR promoter, dimerization was determined by repression of a λpR-lacZ reporter construct in an E. coli strain. An immunoblot revealed that the λcIDB-AaTadZ fusion protein is expressed and stable (data not shown). Expression of λcIDB-AatadZ results in partial restoration of rough-colony morphology and adherence in the tadZ mutant (data not shown). We found that the λcIDB-AaTadZ protein was also able to repress λpR-lacZ, as measured by β-galactosidase assays (Table 4). These results indicate that AaTadZ can interact with itself.
Table 4.
TadZ dimerization and repression of λpR-lacZ
| Plasmid | Repressor | % Repression of λPR-lacZa ± %SD |
|---|---|---|
| pZ150 | Empty vector | 0.0 ± 0.0 |
| pJH391 | λcIDB | 9.7 ± 4.0 |
| pJH157 | λcI | 66.7 ± 1.4 |
| pBP294 | λcIDB-AaTadZ | 60.1 ± 0.7 |
| pBP323 | λcIDB-AaTadZ K150R | 49.0 ± 1.9 |
| pBP303 | λcIDB-AaTadZ K150A | 60.3 ± 1.5 |
| pBP319 | λcIDB-AaTadZ A155K | 49.7 ± 2.1 |
| pBP305 | λcIDB-AaTadZ S156T | 53.7 ± 0.8 |
| pBP318 | λcIDB-AaTadZ S156A | 57.6 ± 4.2 |
Repression (%) of λpR-lacZ = 1- (β-Galactosidase units with repressor/(β-Galactosidase units without repressor) × 100%. SD, standard deviation.
Mutant AaTadZ proteins altered in residues of the Walker-like A box can dimerize
To investigate a possible role for the Walker-like A box in AaTadZ dimerization, we fused the Walker-like A box mutant proteins AaTadZ A155K, K150A, K150R, S156T, and S156A to λcIDB. As before, we confirmed that these Walker-like A box fusion proteins were expressed and stable by immunoblots with α–AaTadZ and α–λcI antibodies. All fusions repressed λpR-lacZ (Table 4). Therefore, individual changes of the highly conserved residues at positions 1 and 7 of the canonical Walker-like A box are not essential for AaTadZ dimerization.
Discussion
We have shown that the AaTadZ-EGFP fusion protein forms a single focus at a pole in A. actinomycetemcomitans. The AaTadZ-EGFP focus requires no other tad locus protein for localization. In contrast, we found that the AaTadA ATPase fused to EGFP also forms a focus at a pole; but AaTadZ is needed for proper AaTadA-EGFP localization. We hypothesize that AaTadZ localizes the Tad secretion apparatus in the cell and that Flp pili are secreted at a pole.
We have also demonstrated that the tadZ gene family belongs to the parA/minD superfamily. Our analysis showed that the TadZ proteins are predicted to have a Walker-like A box similar to the one in the ParA and MinD family of proteins (Motallebi-Veshareh et al., 1990, Leonard et al., 2005a, Hayashi et al., 2001). The TadZ protein from A. actinomycetemcomitans (AaTadZ) is predicted to have two amphipathic helices at its carboxy-terminus; we have shown that both domains are important for spatial localization of TadZ and biofilm formation. Several other predicted TadZ proteins from other organisms also have putative amphipathic helices at their carboxy-termini (supplementary data).
The atypical Walker-like A box found in TadZ proteins is different from the one in ParA and MinD proteins at position 6 in the A box. In the TadZ family of proteins, another residue substitutes for lysine. Restoration of the ParA/MinD Walker-like A box by putting lysine at position 6 inactivates AaTadZ. Substitution of other amino acids at position 6 did not fully inactivate TadZ. Therefore, the absence of lysine at position 6 of the Walker-like A box of AaTadZ is important for the assembly and secretion of Flp pili and its phenotypes in A. actinomycetemcomitans (biofilm formation, autoaggregation, and rough-colony morphology).
Recently crystal structures were determined for the TadZ homologs from E. rectale (Xu et al., 2012) and Chlorobium tepidum (PDB 3ea0, uniprot ID Q8KF94, unpublished results). These analyses confirmed that the overall structure of the ATPase domain of TadZ is very similar in MinD and in other known members of the family, despite sequence divergence. More unexpectedly, ATP and Mg+2 were found in both structures, indicating that TadZ is still capable of binding nucleotide. While it appears that ErTadZ can hydrolyze ATP at low levels (Xu et al., 2012), the functional implications of this activity remain unclear. All tadZ sequences predict proteins that lack lysine at Walker box A position 6. This lysine is predicted to play a role in ATP hydrolysis. Our A155K mutation in AaTadZ restored the canonical Walker-like A box but inactivated the protein. Since the MinD ATPase domain is needed for MinD to oscillate in the cell (Hu & Lutkenhaus, 2001), it is possible that the inability of the TadZ Walker-like A box to promote the hydrolysis of ATP is important for TadZ proteins to remain stationary. AaTadZ localization may be necessary for assembly of the apparatus for the secretion of Flp pili by A. actinomycetemcomitans.
The fact that the remainder of the residues that constitute the Walker-like A box are highly conserved among TadZ proteins indicates that these residues are important for function. Our results are consistent with this idea. We constructed AaTadZ mutants in which substitutions were made at positions 1 and 7 of the TadZ Walker-like A box. These changes disrupted biofilm formation indicating that the atypical Walker-like A box is important for function.
We have shown by the cI in vivo assay that, as in the cases of ParA and MinD, AaTadZ can dimerize. Studies have shown that the Walker-like A box of the E. coli MinD is required for dimerization (Zhou & Lutkenhaus, 2004). Structure data of the E. rectale TadZ protein (ErTadZ) indicate that the ATP pocket is at the dimerization interface, and ATP itself is involved in the dimer interaction (Xu et al., 2012). Surprisingly, three AatadZ Walker-like A box mutations (K150A, A155K, and S156A) failed to disrupt the AaTadZ dimer in our in vivo studies. It may be possible that a change in a single residue may only serve to weaken the dimer interface without disrupting it, as some of above mutants are still partially functional in biofilm formation. A weakness of the in vivo assay is that relative strengths of the mutant dimers cannot be determined, nor do we know what strength is needed for repression of the λpr-lacZ reporter. Alternatively, our results could indicate that the Walker-like A box of AaTadZ and ATP binding are not essential for dimerization. Perhaps, the Walker-like A box of TadZ proteins is important for the binding of another protein.
The mechanism by which TadZ localizes to the poles is unknown. We have isolated N-terminal deletion mutants of AaTadZ that no longer localize to poles (B. Perez-Cheeks and D. Figurski, unpublished results) but rather to the periphery of the cytoplasm. One possibility that takes into account these results is that a host factor captures and concentrates TadZ at a pole through interactions at the N-terminal ARD domain (Xu et al., 2012). The C. crescentus PodJ and PleC proteins interact with the product of the tadZ homolog cpaE. PodJ localizes CpaE to a specific pole of the swarmer cell, and PleC modulates the distribution of CpaE in the cell (Viollier et al., 2002b, Viollier et al., 2002a). Although no PodJ- or PleC-like proteins are encoded by A. actinomycetemcomitans, other proteins may be involved.
Structural prediction indicated that two putative amphipathic helices exist in the C-terminal region of AaTadZ. An amphipathic helix is thought to be a membrane-targeting domain in MinD (Szeto et al., 2002, Szeto et al., 2003). We tested both predicted amphipathic helices (A1 and A2) for membrane-binding activity by deleting each domain from AaTadZ. We believe that the ΔA1 and ΔA2 proteins fold properly because they are abundant (data not shown). Both putative amphipathic helices are required for membrane association and for biofilm formation by A. actinomycetemcomitans. However, they do not seem to be sufficient for targeting AaTadZ-EGFP to the membrane because N-terminal deletion mutants also failed to localize properly. Both microscopy and subcellular fractionation experiments revealed that A1 appears to be less important than A2 for membrane localization. Although A2 is important for AaTadZ inner membrane localization, a significant portion of the protein still accumulates to the inner membrane fraction in the absence of A2.
Together these results suggest a model in which another domain (possibly ARD) targets AaTadZ to a pole of the cell, where A2 (and possibly A1) acts to stabilize an association with the inner membrane. E. rectale TadZ has a single helix at its carboxy-terminus (Xu et al., 2012). It is possible that this region is sufficient to stabilize the molecule at the inner membrane. Alternatively ErTadZ may have other interactions that stabilize it at the inner membrane.
Our results indicate that AaTadZ is targeted to an “older” pole, as opposed to the septum-proximal pole. The mechanism is unknown, but we have shown that the Walker-like A box is not involved. The pole may be distinct from other cellular sites. Blocking cell division in A. actinomycetemcomitans with piperacillin, which interferes with FtsI in E. coli (Botta & Park, 1981, Pogliano et al., 1997), occasionally resulted in the formation of AaTadZ-EGFP foci at both poles and at future sites of division. Apparently, AaTadZ is able to localize to septum-proximal poles. Perhaps, a newly forming end can mature (e.g., by changes in lipids or in protein composition) (Rudner & Losick, 2010) to lead to the recruitment of AaTadZ. While we have no data to suggest it, we are aware of the possibility that AaTadZ localization could be cell-cycle regulated. There is a precedent. Previous studies in C. crescentus have shown that the polar localization of the CpaE (CcTadZ), the secretin CpaC, and the production of polar pili are linked to the cell division cycle (Skerker & Shapiro, 2000, Viollier et al., 2002b).
Because both AaTadZ and AaTadA form polar foci, we believe that the Tad apparatus in A. actinomycetemcomitans is located at a pole. Polar localization of other proteins involved organelle biogenesis has also been observed previously (Christie & Cascales, 2005, Hahn et al., 2005, Janakiraman & Goldberg, 2004, Mattick, 2002, Russel, 1998). Many Type IV pili, including pili of Pseudomonas aeruginosa, Neisseria gonorrhoeae, Myxococcus xanthus, and C. crescentus are assembled and secreted at poles (Viollier et al., 2002a) (Mattick, 2002). Polar localization of the Tad secretion system in A. actinomycetemcomitans predicts that Flp pili are secreted at a pole.
Experimental Procedures
Bacterial strains and growth conditions
Bacterial strains are described in Tables 1 and 2. A. actinomycetemcomitans strains were cultured in AAGM with 10% CO2, as previously described (Thomson et al., 1999). The following concentrations of antibiotics were used: chloramphenicol (Cm), 2 μg ml−1; kanamycin (Km), 40 μg ml−1; nalidixic acid (Nal), 20 μg ml−1.
Table 1.
A. actinomycetemcomitans strains
| Strains | Relevant characteristicsa | Reference or source |
|---|---|---|
| Aa0886 | CU1000N tadZ∷IS903φkan; Kmr | (Planet et al., 2003) |
| Aa1332 | CU1000N tadB∷IS903φkan; Kmr | (Kachlany et al., 2000) |
| Aa1347 | CU1000N tadE∷IS903φkan; Kmr | (Kachlany et al., 2000) |
| Aa1354 | CU1000N rcpA∷IS903φkan; Kmr | (Planet et al., 2003) |
| Aa1359 | CU1000N tadC∷IS903φkan; Kmr | (Kachlany et al., 2000) |
| Aa1360 | CU1000N tadA∷IS903φkan; Kmr | (Kachlany et al., 2000) |
| Aa1361 | CU1000N rcpC∷IS903φkan; Kmr | (Planet et al., 2003) |
| Aa1511 | CU1000N tadF∷IS903φkan; Kmr | (Kachlany et al., 2000) |
| Aa1561 | CU1000N tadG∷IS903φkan; Kmr | (Kachlany et al., 2000) |
| Aa1577 | CU1000N tadD∷IS903φkan; Kmr | (Kachlany et al., 2000) |
| Aa3048 | CU1000N(pJAK16) | This study |
| Aa3097 | Aa0886(pBP154) | This study |
| Aa3100 | CU1000N(pJAK 17) | This study |
| Aa3103 | Aa0886(pJAK 16) | This study |
| Aa3120 | Aa0886(pBP187) | This study |
| Aa3132 | Aa0886(pJAK 17) | This study |
| Aa3133 | Aa0886(pPP29) | This study |
| Aa3136 | Aa0886(pGS-GFP-04) | This study |
| Aa3146 | Aa0886(pBP197) | This study |
| Aa3194 | Aa1360(pGS-GFP-04) | This study |
| Aa3219 | Aa0886(pBP219) | This study |
| Aa3224 | Aa0886(pBP200) | This study |
| Aa3224 | Aa0886(pBP200) | This study |
| Aa3242 | Aa0886(pBP225) | This study |
| Aa3244 | Aa0886(pBP226) | This study |
| Aa3247 | Aa0886(pBP231) | This study |
| Aa3313 | JK1009 (flp-1 mutant) (pBP225) | This study |
| Aa3314 | Aa3237 (flp-2 mutant) (pBP225) | This study |
| Aa3315 | Aa3074 (tadV mutant) (pBP225) | This study |
| Aa3316 | Aa1361 (rcpC mutant) (pBP225) | This study |
| Aa3317 | Aa1354 (rcpA mutant) (pBP225) | This study |
| Aa3318 | Aa1360 (tadA mutant) (pBP225) | This study |
| Aa3319 | Aa1332 (tadB mutant) (pBP225) | This study |
| Aa3320 | Aa1359 (tadC mutant) (pBP225) | This study |
| Aa3321 | Aa1577 (tadD mutant) (pBP225) | This study |
| Aa3322 | Aa1347 (tadE mutant) (pBP225) | This study |
| Aa3323 | Aa1511 (tadF mutant) (pBP225) | This study |
| Aa3324 | Aa1561 (tadG mutant) (pBP225) | This study |
| Aa3325 | Aa0886(pBP310) | This study |
| Aa3327 | Aa1360(pJAK16) | This study |
| Aa3328 | Aa1360(pEK2) | This study |
| Aa3329 | Aa1360(pBP321) | This study |
| Aa3331 | Aa0886(pBP325) | This study |
| Aa3332 | Aa0886(pBP327) | This study |
| Aa3333 | Aa0886(pBP321) | This study |
| Aa3335 | CU1000N(pBP321) | This study |
| Aa3337 | JK1009(pBP321) | This study |
| Aa3341 | Aa0886(pBP346) | This study |
| Aa3346 | Aa0886(pBP352) | This study |
| CU1000N | Spontaneous Nalr variant of CU1000 (serotype f) | (Fine et al., 1999a, Fine et al., 1999b) |
| DF2200N | Spontaneous Nalr variant of DF2200 (serotype a) | (Kaplan et al., 2002) |
| JK1009 | CU1000N flp-1∷IS903φkan; Kmr | (Kachlany et al., 2001b) |
Nalr, nalidixic acid resistance; Kmr, kanamycin resistance; Cmr, chloramphenicol resistance.
Table 2.
E. coli strains
| E. coli strains | Relevant characteristics | Reference or source |
|---|---|---|
| AB1457 | JH372(pJH157) | (Bhattacharyya & Figurski, 2001) |
| AB1499 | JH372(pJH391) | (Bhattacharyya & Figurski, 2001) |
| BL21(DE3) | F− ompT gal dcm lon hsdSB (rB− mB−) (DE3); an E. coli B strain with DE3, a λ prophage carrying the T7 RNA polymerase gene controlled by laclq. | Novagen |
| BP267 | BL21(DE3)(pBP169) | This study |
| BP269 | BL21(DE3)(pBP171) | This study |
| BP317 | JMB9.1(pGS-GFP-04) | This study |
| BP542 | Top10(pBP287) | This study |
| BP557 | JH372(pBP294) | This study |
| BP563 | JMB9.1(pBP225) | This study |
| BP594 | JMB9.1(pBP325) | This study |
| BP595 | JMB9.1(pBP327) | This study |
| BP615 | JH372(pBP303) | This study |
| BP616 | JH372(pBP305) | This study |
| BP617 | JH372(pBP318) | This study |
| BP618 | JH372(pBP319) | This study |
| BP619 | JH372(pBP323) | This study |
| JH372 | AG1688(λ202) = MC1061(λ202)(F'128 lacl lacZ∷Tn5) = λpRoR1+oR2−-lacZ reporter strain | (Hu et al., 1990) |
| JH380 | JH372(pZ150) | (Hu et al., 1990) |
| JMB9.1 | F− leu thi gal1,2 lac xyl ara hsdRK (rK− mK+) ΔtrpE5 Su+ | From C. Yanofsky |
| SC1036 | BL21(DE3)(pET30a) | S. Clock, unpublished |
| TOP10 | F− λ− mcrA Δ(mrr-hsdRMS-mcrBC) ΔlacX74 deoR nupG recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 (φ80lacZΔM15) | Invitrogen |
E. coli strain TOP10 (Invitrogen) was used for DNA cloning. Transformants were cultured in Luria-Bertani (LB) medium and grown at 37 °C. TOP10 strains carrying the plasmid RK2 derivative pRK21761 were used as conjugative donors for the mobilization of broad-host-range IncQ plasmids into A. actinomycetemcomitans, as described previously (Thomson et al., 1999). When an antibiotic was needed, either for cloning or for conjugation, the following concentrations were used: Cm, 50 μg ml−1; Km, 50 μg ml−1; ampicillin (Ap), 50 μg ml−1; penicillin (Pn), 150 μg ml−1.
DNA procedures
Plasmids (Table 3) were constructed by standard genetic procedures. Plasmid DNA was prepared with Miniprep Spin Kits (Qiagen). DNA manipulations with restriction endonucleases and T4 DNA ligase were done according to the manufacturer's (New England Biolabs) recommendations. PCR amplification was done with either high fidelity Triplemaster Taq polymerase or Taq polymerase (Eppendorf) for 30 cycles. A SpinX tube (Corning) or the QiaexII kit (Qiagen) was used for agarose gel extraction. All plasmids were confirmed by nucleotide sequencing at the Columbia University DNA sequencing facility. The oligonucleotides are described in SI and Table S1.
Table 3.
Plasmids
| Plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| pBP141 | pCR2.1 containing tadZ A155K Af/II fragment | This study |
| pBP149 | pCR2.1 containing tadZ A155K | This study |
| pBP154 | pJAK17 containing tadZ A155K | This study |
| pBP169 | pET30a containing tadZ | This study |
| pBP171 | pET30a containing tadA | This study |
| pBP174 | pCR2.1 containing tadZ S156T Af/II fragment | This study |
| pBP180 | pCR2.1 containing tadZ S156T | This study |
| pBP182 | pCR2.1 containing tadZ K150A Af/II fragment | This study |
| pBP184 | pCR2.1 containing tadZ K150R Af/II fragment | This study |
| pBP187 | pJAK17 containing tadZ S156T | This study |
| pBP193 | pCR2.1 containing tadZ K150R | This study |
| pBP195 | pCR2.1 containing tadZ K150A | This study |
| pBP196 | pCR2.1 containing tadZ S156A Af/II fragment | This study |
| pBP197 | pJAK17 containing tadZ K150R | This study |
| pBP200 | pJAK17 containing tadZ K150A | This study |
| pBP209 | pCR2.1 containing tadZ S156A | This study |
| pBP214 | pCR4 containing tadZ | This study |
| pBP215 | pCR4 containing tadZ A155K (BamHI-NcoI fragment) | This study |
| pBP216 | pCR4 containing tadZ S156A (BamHI-NcoI fragment) | This study |
| pBP219 | pJAK17 containing tadZ S156A | This study |
| pBP222 | pCR4 containing tadZ S156T (BamHI-NcoI fragment) | This study |
| pBP225 | pJAK17 containing tadZ-egfp | This study |
| pBP226 | pJAK17 containing tadZ S156A-egfp | This study |
| pBP227 | pEGFP containing tadZ | This study |
| pBP228 | pEGFP containing tadZ S156A | This study |
| pBP230 | pEGFP containing tadZ A155K | This study |
| pBP231 | pJAK17 containing tadZ A155K-egfp | This study |
| pBP236 | pEGFP containing tadZ S156T | This study |
| pBP237 | pCR4 containing tadZ K150A (BamHI-NcoI fragment) | This study |
| pBP238 | pCR4 containing tadZ K150R (BamHI-NcoI fragment) | This study |
| pBP244 | pJAK17 containing tadZ S156T-egfp | This study |
| pBP246 | pEGFP containing tadZ K150A | This study |
| pBP248 | pEGFP containing tadZ K150R | This study |
| pBP252 | pJAK17 containing tadZ K150A-egfp | This study |
| pBP254 | pJAK17 containing tadZ K150R-egfp | This study |
| pBP287 | pCR2.1 containing tadZ (BamHI-Sa/I fragment) | This study |
| pBP294 | pJH391 containing tadZ | This study |
| pBP296 | pCR2.1 containing tadZ K150R (BamHI-Sa/I fragment) | This study |
| pBP297 | pCR2.1 containing tadZ K150A (BamHI-Sa/I fragment) | This study |
| pBP298 | pCR2.1 containing tadZ S156T (BamHI-Sa/I fragment) | This study |
| pBP299 | pCR2.1 containing tadZ S156A (BamHI-Sa/I fragment) | This study |
| pBP300 | pCR2.1 containing tadZ A155K (BamHI-Sa/I fragment) | This study |
| pBP301 | pCR2.1 containing λcIDB-tadZ | This study |
| pBP303 | pJH391 containing tadZ K150A | This study |
| pBP305 | pJH391 containing tadZ S156T | This study |
| pBP307 | pCR2.1 containing tadA | This study |
| pBP308 | pEGFP containing tadZΔA1 | This study |
| pBP310 | pJAK16 containing λcIDB-tadZ | This study |
| pBP312 | pEGFP containing tadA | This study |
| pBP318 | pJH391 containing tadZ S156A | This study |
| pBP319 | pJH391 containing tadZ A155K | This study |
| pBP320 | pEGFP containing tadZΔA2 | This study |
| pBP321 | pJAK17 containing tadA-Δegfp | This study |
| pBP323 | pJH391 containing tadZ K150R | This study |
| pBP325 | pJAK17 containing tadZΔA1-egfp | This study |
| pBP327 | pJAK17 containing tadZΔA2-egfp | This study |
| pBP339 | pCR2.1 containing tadZΔA1 | This study |
| pBP340 | pCR2.1 containing tadZΔA2 | This study |
| pBP346 | pJAK17 containing tadZΔA1 | This study |
| pBP352 | pJAK17 containing tadZΔA2 | This study |
| pCR2.1 | E. coli cloning vector; Kmr Apr | Invitrogen |
| pCR4 | E. coli cloning vector; Kmr Apr | Invitrogen |
| pEGFP | E. coli EGFP cloning vector; Apr | Clontech |
| pEK2 | pJAK16 containing tadA | (Bhattacharjee et al.,2001) |
| pET30a | E. coli protein expression vector; Kmr | Novagen |
| pGS-GFP | IncQ broad-host-range expression vector pMMB207 encoding GFP; Cmr | (Hilbietal., 2001) |
| pJAK16 | IncQ broad-host-range expression vector; tacp+, Cmr | J. Kornacki, unpublished |
| pJAK17 | IncQ broad-host-range expression vector; tacp+, Cmr | J. Kornacki, unpublished |
| pPP26 | pCR2.1 containing tadZ | This study |
| pPP29 | pJAK16 containing tadZ | (Planet et al., 2003) |
| pZ150 | Vector control for pJH391 and derivatives; Apr | (Hu et al., 1990) |
Apr, ampicillin resistance; Kmr, Kanamycin resistance; Cmr, chloramphenicol resistance.
Localization of AaTadZ and AaTadA
Construction of all EGFP fusion proteins is described in SI. Colonies of A. actinomycetemcomitans were scraped into 500 μl fresh AAGM broth with 2 μg ml-1 Cm and incubated at 37°C for one hour. When used, isopropyl β-D-thiogalactoside (IPTG) was added to the cultures at a final concentration of 10 μM (for tadZ-egfp) and 5 μM (for tadA-egfp); and the cultures were incubated for an additional 2–3 hours.
To stain the bacterial cell membranes, 100 μl of the cultures were either stained directly with 2 μM TMA-DPH [1-(4-trimethylammoniumphenyl)-6-phenyl-1,3,5-hexatriene p-toluenesulfonate] (Invitrogen) or first harvested by centrifugation at 4,000 × g in a microcentrifuge (Eppendorf) and then stained with TMA-DPH. When necessary, the cell pellets were resuspended in PBS. The suspensions were dropped onto coverslips coated with 0.1% poly-L-lysine (Sigma). The cells were visualized with a Nikon 90i fluorescence microscope mounted with a Hamamatsu Orca-ER camera. Nikon Elements software and ImageJ software (National Institutes of Health) was used for quantification of localization data and on average a minimum of 300 cells were analyzed.
Elongation of A. actinomycetemcomitans cells
A. actinomycetemcomitans strains with tadZ-egfp were grown as described above. Colonies were scraped into 500 μl fresh AAGM broth with 2 μg ml−1 Cm and 2 μg ml−1 piperacillin (Sigma). The strains were then prepared for microscopy.
AaTadZ expression and detection
Strains of A. actinomycetemcomitans were grown overnight in 5 ml AAGM before being harvested by centrifugation. Cell pellets were resuspended and boiled for 5 min in Laemmli buffer. Total protein was measured at OD280. Equivalent amounts of total protein were loaded onto SDS-PAGE gels for Western (immunoblot) analysis. Samples were analyzed by reacting with α-AaTadZ polyclonal rabbit antibody (1:5,000). A goat polyclonal IgG α-rabbit antibody (Sigma) conjugated to HRP was used as the secondary antibody at 1:50,000. Femto Western chemiluminescence reagent (Pierce) was used as substrate for HRP. A description of AaTadZ antisera production is found in SI.
Genetic complementation with tadZ derivatives
Construction of AaTadZ amphipathic deletion mutants and Walker-like A box AaTadZ mutants are described in SI. To assay biofilm formation, we used a previously described 96-well polystyrene crystal violet (CV) assay (O'Toole & Kolter, 1998) with some modifications (Kaplan et al., 2003, Perez et al., 2006). The amount of released CV was measured at OD590 in a plate reader (Molecular Devices SpectraMAX 340pc) to assay adherence. Adherence was assayed in triplicate. Details are described in SI.
Dimerization assay
Construction of AatadZ-λcI fusions are described in SI. The assay for β-galactosidase activity was done, as described previously (Miller, 1972). JH372 strains expressing the λcI-AaTadZ fusions were grown overnight and then diluted 50-fold. The cultures were grown for 1 hour at 37°C in the presence or absence of IPTG, and an absorbance OD600 reading was determined after the incubation. Cells were chilled on ice and assayed for β-galactosidase activity, as described (Miller, 1972). Briefly, 300 μl of the suspension was added to 700 μl of Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, pH 7.0); and the cells were lysed with 50 μl of chloroform and 1% SDS. Two hundred μl of 0.4 mg ml−1 o-nitrophenyl β-D-galactopyranoside (ONPG) was added to the cell suspension, and the samples were incubated at 35°C. The reaction was stopped by the addition of 500 μl of 1M Na2CO3. The cell debris was removed by centrifugation, and the optical density of supernatant was measured at 420 nM in a 96-well plate reader (Molecular Devices SpectraMAX 340pc). Samples were tested in triplicate wells, and error bars indicate standard error. Miller units were calculated as follows: (1000 × OD420 reading) / (reaction time × volume of sample × OD600 reading).
Homology of tadZ genes and phylogenetic analysis
Despite weak overall similarity scores, a systematic BLAST-based approach strongly suggested that tadZ-like genes are homologs of minD and parA (see SI). To determine further the relationship of tadZ genes to parA and minD relatives, we collected as many possible parA/minD homologs as possible for phylogenetic analysis. We started with 25 “seed” sequences as queries for our search (see SI table). Seed sequences were selected based on an extensive literature review with an emphasis on collecting as diverse a collection of putative homologs as possible. We used the 25 seeds to search protein sequence databases at NCBI, keeping genes that had an e-value score that was at least 1×10−20 or better to any one of the seed sequences. After eliminating redundant hits, this pool of putative homologs contained 4,074 genes.
To obtain a computationally tractable phylogenetic dataset, we sampled further from this pool. To obtain a broad sampling of diversity from this superfamily, we first performed an all-against-all BLAST and used a single linkage clustering algorithm (SLC) with an e-value threshold of 1×10−20 to divide the pool of sequences into more similar groups. This yielded 142 SLC groups. We then selected 2 sequences at random from each of the 142 groups for further analysis. The 25 seed sequences were added back to the resulting 284 sequences (total=309), and all were aligned using default parameters in the program MUSCLE (Edgar, 2004). To control for bias introduced during our random sampling procedure, we repeated the sampling procedure 10 times, yielding ten separate datasets. All 10 resulting alignments were subjected to a rigorous parsimony-based phylogenetic analysis using the Ratchet technique as implemented by PAUPRat (Sikes & Lewi, 2001) in conjunction with PAUP* (Swofford, 1998).
Supplementary Material
Acknowlegdments
We thank Jonathan Dworkin, Daniel Fine, Aaron Mitchell, and Howard Shuman for helpful discussions. We are grateful to the Dworkin laboratory for use of equipment and reagents, to Jianyuan Hua for technical help, and to the Ian A. Wilson laboratory for sharing unpublished results. D.H.F. gives special thanks to Saul Silverstein and Vincent Racaniello for their support. This work was funded by a grant from the National Institutes of Health to D.H.F (DE014713) and a National Institutes of Health Kirschstein–NRSA Individual Fellowship to B.A.P.
References
- Abrahams JP, Leslie AG, Lutter R, Walker JE. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- Atmakuri K, Cascales E, Burton OT, Banta LM, Christie PJ. Agrobacterium ParA/MinD-like VirC1 spatially coordinates early conjugative DNA transfer reactions. EMBO J. 2007;26:2540–2551. doi: 10.1038/sj.emboj.7601696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee MK, Kachlany SC, Fine DH, Figurski DH. Nonspecific adherence and fibril biogenesis by Actinobacillus actinomycetemcomitans: TadA protein is an ATPase. J Bacteriol. 2001;183:5927–5936. doi: 10.1128/JB.183.20.5927-5936.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya A, Figurski DH. A small protein-protein interaction domain common to KlcB and global regulators KorA and TrbA of promiscuous IncP plasmids. J Mol Biol. 2001;310:51–67. doi: 10.1006/jmbi.2001.4729. [DOI] [PubMed] [Google Scholar]
- Botta GA, Park JT. Evidence for involvement of penicillin-binding protein 3 in murein synthesis during septation but not during cell elongation. J Bacteriol. 1981;145:333–340. doi: 10.1128/jb.145.1.333-340.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie PJ, Cascales E. Structural and dynamic properties of bacterial type IV secretion systems (review) Mol Membr Biol. 2005;22:51–61. doi: 10.1080/09687860500063316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clock SA, Planet PJ, Perez BA, Figurski DH. Outer membrane components of the Tad (tight adherence) secreton of Aggregatibacter actinomycetemcomitans. J Bacteriol. 2008;190:980–990. doi: 10.1128/JB.01347-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell SC, Lowe J. Crystal structure of the bacterial cell division regulator MinD. FEBS Lett. 2001;492:160–165. doi: 10.1016/s0014-5793(01)02216-5. [DOI] [PubMed] [Google Scholar]
- Das M, Badley AD, Cockerill FR, Steckelberg JM, Wilson WR. Infective endocarditis caused by HACEK microorganisms. Annu Rev Med. 1997;48:25–33. doi: 10.1146/annurev.med.48.1.25. [DOI] [PubMed] [Google Scholar]
- Davis MA, Radnedge L, Martin KA, Hayes F, Youngren B, Austin SJ. The P1 ParA protein and its ATPase activity play a direct role in the segregation of plasmid copies to daughter cells. Mol Microbiol. 1996;21:1029–1036. doi: 10.1046/j.1365-2958.1996.721423.x. [DOI] [PubMed] [Google Scholar]
- de Boer PA, Crossley RE, Hand AR, Rothfield LI. The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. EMBO. J. 1991;10:4371–4380. doi: 10.1002/j.1460-2075.1991.tb05015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersbach G, Gerdes K. The double par locus of virulence factor pB171: DNA segregation is correlated with oscillation of ParA. Proc Natl Acad Sci U S A. 2001;98:15078–15083. doi: 10.1073/pnas.261569598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersbach G, Gerdes K. Bacterial mitosis: partitioning protein ParA oscillates in spiral-shaped structures and positions plasmids at mid-cell. Mol Microbiol. 2004;52:385–398. doi: 10.1111/j.1365-2958.2004.04002.x. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine DH, Furgang D, Kaplan J, Charlesworth J, Figurski DH. Tenacious adhesion of Actinobacillus actinomycetemcomitans strain CU1000 to salivary-coated hydroxyapatite. Arch Oral Biol. 1999a;44:1063–1076. doi: 10.1016/s0003-9969(99)00089-8. [DOI] [PubMed] [Google Scholar]
- Fine DH, Furgang D, Schreiner HC, Goncharoff P, Charlesworth J, Ghazwan G, Fitzgerald-Bocarsly P, Figurski DH. Phenotypic variation in Actinobacillus actinomycetemcomitans during laboratory growth: implications for virulence. Microbiology. 1999b;145(Pt 6):1335–1347. doi: 10.1099/13500872-145-6-1335. [DOI] [PubMed] [Google Scholar]
- Fine DH, Kaplan JB, Kachlany SC, Schreiner HC. How we got attached to Actinobacillus actinomycetemcomitans: a model for infectious diseases. Periodontol 2000. 2006;42:114–157. doi: 10.1111/j.1600-0757.2006.00189.x. [DOI] [PubMed] [Google Scholar]
- Fung E, Bouet JY, Funnell BE. Probing the ATP-binding site of P1 ParA: partition and repression have different requirements for ATP binding and hydrolysis. EMBO. J. 2001;20:4901–4911. doi: 10.1093/emboj/20.17.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Howard M, Szardenings F. Pushing and pulling in prokaryotic DNA segregation. Cell. 2010;141:927–942. doi: 10.1016/j.cell.2010.05.033. [DOI] [PubMed] [Google Scholar]
- Hahn J, Maier B, Haijema BJ, Sheetz M, Dubnau D. Transformation proteins and DNA uptake localize to the cell poles in Bacillus subtilis. Cell. 2005;122:59–71. doi: 10.1016/j.cell.2005.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi I, Oyama T, Morikawa K. Structural and functional studies of MinD ATPase: implications for the molecular recognition of the bacterial cell division apparatus. EMBO. J. 2001;20:1819–1828. doi: 10.1093/emboj/20.8.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson B, Nair SP, Ward JM, Wilson M. Molecular pathogenicity of the oral opportunistic pathogen Actinobacillus actinomycetemcomitans. Annu Rev Microbiol. 2003;57:29–55. doi: 10.1146/annurev.micro.57.030502.090908. [DOI] [PubMed] [Google Scholar]
- Henderson B, Wilson M, Sharp L, Ward JM. Actinobacillus actinomycetemcomitans. J Med Microbiol. 2002;51:1013–1020. doi: 10.1099/0022-1317-51-12-1013. [DOI] [PubMed] [Google Scholar]
- Hilbi H, Segal G, Shuman HA. Icm/dot-dependent upregulation of phagocytosis by Legionella pneumophila. Mol Microbiol. 2001;42:603–617. doi: 10.1046/j.1365-2958.2001.02645.x. [DOI] [PubMed] [Google Scholar]
- Hu JC, O'Shea EK, Kim PS, Sauer RT. Sequence requirements for coiled-coils: analysis with lambda repressor-GCN4 leucine zipper fusions. Science. 1990;250:1400–1403. doi: 10.1126/science.2147779. [DOI] [PubMed] [Google Scholar]
- Hu Z, Gogol EP, Lutkenhaus J. Dynamic assembly of MinD on phospholipid vesicles regulated by ATP and MinE. Proc Natl Acad Sci U S A. 2002;99:6761–6766. doi: 10.1073/pnas.102059099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Lutkenhaus J. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol Microbiol. 1999;34:82–90. doi: 10.1046/j.1365-2958.1999.01575.x. [DOI] [PubMed] [Google Scholar]
- Hu Z, Lutkenhaus J. Topological regulation of cell division in E. coli. spatiotemporal oscillation of MinD requires stimulation of its ATPase by MinE and phospholipid. Mol Cell. 2001;7:1337–1343. doi: 10.1016/s1097-2765(01)00273-8. [DOI] [PubMed] [Google Scholar]
- Hu Z, Lutkenhaus J. A conserved sequence at the C-terminus of MinD is required for binding to the membrane and targeting MinC to the septum. Mol Microbiol. 2003;47:345–355. doi: 10.1046/j.1365-2958.2003.03321.x. [DOI] [PubMed] [Google Scholar]
- Inouye T, Ohta H, Kokeguchi S, Fukui K, Kato K. Colonial variation and fimbriation of Actinobacillus actinomycetemcomitans. FEMS Microbiol Lett. 1990;57:13–17. doi: 10.1016/0378-1097(90)90405-f. [DOI] [PubMed] [Google Scholar]
- Janakiraman A, Goldberg MB. Recent advances on the development of bacterial poles. Trends Microbiol. 2004;12:518–525. doi: 10.1016/j.tim.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Kachlany SC, Planet PJ, Bhattacharjee MK, Kollia E, DeSalle R, Fine DH, Figurski DH. Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in bacteria and archaea. J Bacteriol. 2000;182:6169–6176. doi: 10.1128/jb.182.21.6169-6176.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachlany SC, Planet PJ, DeSalle R, Fine DH, Figurski DH. Genes for tight adherence of Actinobacillus actinomycetemcomitans: from plaque to plague to pond scum. Trends Microbiol. 2001a;9:429–437. doi: 10.1016/s0966-842x(01)02161-8. [DOI] [PubMed] [Google Scholar]
- Kachlany SC, Planet PJ, Desalle R, Fine DH, Figurski DH, Kaplan JB. flp-1, the first representative of a new pilin gene subfamily, is required for non-specific adherence of Actinobacillus actinomycetemcomitans. Mol Microbiol. 2001b;40:542–554. doi: 10.1046/j.1365-2958.2001.02422.x. [DOI] [PubMed] [Google Scholar]
- Kaplan JB, Ragunath C, Ramasubbu N, Fine DH. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous beta-hexosaminidase activity. J Bacteriol. 2003;185:4693–4698. doi: 10.1128/JB.185.16.4693-4698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan JB, Schreiner HC, Furgang D, Fine DH. Population structure and genetic diversity of Actinobacillus actinomycetemcomitans strains isolated from localized juvenile periodontitis patients. J Clin Microbiol. 2002;40:1181–1187. doi: 10.1128/JCM.40.4.1181-1187.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkaria CE, Chen CM, Rosen BP. Mutagenesis of a nucleotide-binding site of an anion-translocating ATPase. J Biol Chem. 1990;265:7832–7836. [PubMed] [Google Scholar]
- Koonin EV. A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J Mol Biol. 1993;229:1165–1174. doi: 10.1006/jmbi.1993.1115. [DOI] [PubMed] [Google Scholar]
- Kram KE, Hovel-Miner GA, Tomich M, Figurski DH. Transcriptional regulation of the tad locus in Aggregatibacter actinomycetemcomitans: a termination cascade. J Bacteriol. 2008 doi: 10.1128/JB.00128-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumoto A, Kamisaka K, Yakushi T, Terashima H, Shinohara A, Homma M. Regulation of polar flagellar number by the flhF and flhG genes in Vibrio alginolyticus. J Biochem. 2006;139:113–121. doi: 10.1093/jb/mvj010. [DOI] [PubMed] [Google Scholar]
- Le Quere B, Ghigo JM. BcsQ is an essential component of the Escherichia coli cellulose biosynthesis apparatus that localizes at the bacterial cell pole. Mol Microbiol. 2009;72:724–740. doi: 10.1111/j.1365-2958.2009.06678.x. [DOI] [PubMed] [Google Scholar]
- Lemonnier M, Bouet JY, Libante V, Lane D. Disruption of the F plasmid partition complex in vivo by partition protein SopA. Mol Microbiol. 2000;38:493–505. doi: 10.1046/j.1365-2958.2000.02101.x. [DOI] [PubMed] [Google Scholar]
- Leonard TA, Butler PJ, Lowe J. Bacterial chromosome segregation: structure and DNA binding of the Soj dimer--a conserved biological switch. EMBO. J. 2005a;24:270–282. doi: 10.1038/sj.emboj.7600530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard TA, Moller-Jensen J, Lowe J. Towards understanding the molecular basis of bacterial DNA segregation. Philos Trans R Soc Lond B Biol Sci. 2005b;360:523–535. doi: 10.1098/rstb.2004.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Dabrazhynetskaya A, Youngren B, Austin S. The role of Par proteins in the active segregation of the P1 plasmid. Mol Microbiol. 2004;53:93–102. doi: 10.1111/j.1365-2958.2004.04111.x. [DOI] [PubMed] [Google Scholar]
- Libante V, Thion L, Lane D. Role of the ATP-binding site of SopA protein in partition of the F plasmid. J Mol Biol. 2001;314:387–399. doi: 10.1006/jmbi.2001.5158. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J. Dynamic proteins in bacteria. Curr Opin Microbiol. 2002;5:548–552. doi: 10.1016/s1369-5274(02)00376-4. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J, Sundaramoorthy M. MinD and role of the deviant Walker A motif, dimerization and membrane binding in oscillation. Mol Microbiol. 2003;48:295–303. doi: 10.1046/j.1365-2958.2003.03427.x. [DOI] [PubMed] [Google Scholar]
- Marston AL, Errington J. Selection of the midcell division site in Bacillus subtilis through MinD-dependent polar localization and activation of MinC. Mol Microbiol. 1999;33:84–96. doi: 10.1046/j.1365-2958.1999.01450.x. [DOI] [PubMed] [Google Scholar]
- Mattick JS. Type IV pili and twitching motility. Annu Rev Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- Miller J. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press; Cold Springs Harbor, NY: 1972. [Google Scholar]
- Motallebi-Veshareh M, Rouch DA, Thomas CM. A family of ATPases involved in active partitioning of diverse bacterial plasmids. Mol Microbiol. 1990;4:1455–1463. doi: 10.1111/j.1365-2958.1990.tb02056.x. [DOI] [PubMed] [Google Scholar]
- O'Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- Perez BA, Planet PJ, Kachlany SC, Tomich M, Fine DH, Figurski DH. Genetic Analysis of the Requirement for flp-2, tadV, and rcpB in Actinobacillus actinomycetemcomitans Biofilm Formation. J Bacteriol. 2006;188:6361–6375. doi: 10.1128/JB.00496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planet PJ, Kachlany SC, Fine DH, DeSalle R, Figurski DH. The Widespread Colonization Island of Actinobacillus actinomycetemcomitans. Nat Genet. 2003;34:193–198. doi: 10.1038/ng1154. [DOI] [PubMed] [Google Scholar]
- Pogliano J, Pogliano K, Weiss DS, Losick R, Beckwith J. Inactivation of FtsI inhibits constriction of the FtsZ cytokinetic ring and delays the assembly of FtsZ rings at potential division sites. Proc Natl Acad Sci U S A. 1997;94:559–564. doi: 10.1073/pnas.94.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preus HR, Namork E, Olsen I. Fimbriation of Actinobacillus actinomycetemcomitans. Oral Microbiol Immunol. 1988;3:93–94. doi: 10.1111/j.1399-302x.1988.tb00090.x. [DOI] [PubMed] [Google Scholar]
- Quisel JD, Lin DC, Grossman AD. Control of development by altered localization of a transcription factor in B. subtilis. Mol Cell. 1999;4:665–672. doi: 10.1016/s1097-2765(00)80377-9. [DOI] [PubMed] [Google Scholar]
- Raskin DM, de Boer PA. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J Bacteriol. 1999;181:6419–6424. doi: 10.1128/jb.181.20.6419-6424.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosan B, Slots J, Lamont RJ, Listgarten MA, Nelson GM. Actinobacillus actinomycetemcomitans fimbriae. Oral Microbiol Immunol. 1988;3:58–63. doi: 10.1111/j.1399-302x.1988.tb00082.x. [DOI] [PubMed] [Google Scholar]
- Rothfield L, Taghbalout A, Shih YL. Spatial control of bacterial division-site placement. Nat Rev Microbiol. 2005;3:959–968. doi: 10.1038/nrmicro1290. [DOI] [PubMed] [Google Scholar]
- Rudner DZ, Losick R. Protein subcellular localization in bacteria. Cold Spring Harb Perspect Biol. 2010;2:a000307. doi: 10.1101/cshperspect.a000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel M. Macromolecular assembly and secretion across the bacterial cell envelope: type II protein secretion systems. J Mol Biol. 1998;279:485–499. doi: 10.1006/jmbi.1998.1791. [DOI] [PubMed] [Google Scholar]
- Schindelin H, Kisker C, Schlessman JL, Howard JB, Rees DC. Structure of ADP × AIF4(−)-stabilized nitrogenase complex and its implications for signal transduction. Nature. 1997;387:370–376. doi: 10.1038/387370a0. [DOI] [PubMed] [Google Scholar]
- Schreiner HC, Sinatra K, Kaplan JB, Furgang D, Kachlany SC, Planet PJ, Perez BA, Figurski DH, Fine DH. Tight-adherence genes of Actinobacillus actinomycetemcomitans are required for virulence in a rat model. Proc Natl Acad Sci U S A. 2003;100:7295–7300. doi: 10.1073/pnas.1237223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikes DS, Lewi PO. PAUPRat: A tool to implement Parsimony Ratchet searches using PAUP* Storrs, CT; 2001. [Google Scholar]
- Skerker JM, Shapiro L. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO. J. 2000;19:3223–3234. doi: 10.1093/emboj/19.13.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slots J. Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in periodontal disease: introduction. Periodontol 2000. 1999;20:7–13. doi: 10.1111/j.1600-0757.1999.tb00155.x. [DOI] [PubMed] [Google Scholar]
- Stepanovic S, Tosic T, Savic B, Jovanovic M, K'Ouas G, Carlier JP. Brain abscess due to Actinobacillus actinomycetemcomitans. Apmis. 2005;113:225–228. doi: 10.1111/j.1600-0463.2005.apm1130312.x. [DOI] [PubMed] [Google Scholar]
- Story RM, Steitz TA. Structure of the recA protein-ADP complex. Nature. 1992;355:374–376. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: Phylogenetic analysis using parsimony (*and other methods) Sinauer; Sunderland, Massachussetts: 1998. [Google Scholar]
- Szeto TH, Rowland SL, Habrukowich CL, King GF. The MinD membrane targeting sequence is a transplantable lipid-binding helix. J Biol Chem. 2003;278:40050–40056. doi: 10.1074/jbc.M306876200. [DOI] [PubMed] [Google Scholar]
- Szeto TH, Rowland SL, Rothfield LI, King GF. Membrane localization of MinD is mediated by a C-terminal motif that is conserved across eubacteria, archaea and chloroplasts. Proc Natl Acad Sci U S A. 2002;99:15693–15698. doi: 10.1073/pnas.232590599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler M. Synchronization of chromosome dynamics and cell division in bacteria. Cold Spring Harb Perspect Biol. 2010;2:a000331. doi: 10.1101/cshperspect.a000331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson VJ, Bhattacharjee MK, Fine DH, Derbyshire KM, Figurski DH. Direct selection of IS903 transposon insertions by use of a broad-host-range vector: isolation of catalase-deficient mutants of Actinobacillus actinomycetemcomitans. J Bacteriol. 1999;181:7298–7307. doi: 10.1128/jb.181.23.7298-7307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomich M, Planet PJ, Figurski DH. The tad locus: postcards from the widespread colonization island. Nat Rev Microbiol. 2007;5:363–375. doi: 10.1038/nrmicro1636. [DOI] [PubMed] [Google Scholar]
- Viollier PH, Sternheim N, Shapiro L. A dynamically localized histidine kinase controls the asymmetric distribution of polar pili proteins. EMBO. J. 2002a;21:4420–4428. doi: 10.1093/emboj/cdf454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollier PH, Sternheim N, Shapiro L. Identification of a localization factor for the polar positioning of bacterial structural and regulatory proteins. Proc Natl Acad Sci U S A. 2002b;99:13831–13836. doi: 10.1073/pnas.182411999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO. J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Christen B, Chiu H, Jaroszewski L, Klock HE, Knuth MW, Miller MD, Elsliger M, Deacon AM, Godzik A, Lesley SA, Figurski DH, Shapiro L, Wilson IA. Structure of pilus assembly protein TadZ from Eubacterium rectale: Implications for polar localization of Tad pili. Mol. Microbiol. 2012;0:0. doi: 10.1111/j.1365-2958.2011.07954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambon JJ. Actinobacillus actinomycetemcomitans in human periodontal disease. J Clin Periodontol. 1985;12:1–20. doi: 10.1111/j.1600-051x.1985.tb01348.x. [DOI] [PubMed] [Google Scholar]
- Zhou H, Lutkenhaus J. Membrane binding by MinD involves insertion of hydrophobic residues within the C-terminal amphipathic helix into the bilayer. J Bacteriol. 2003;185:4326–4335. doi: 10.1128/JB.185.15.4326-4335.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Lutkenhaus J. The switch I and II regions of MinD are required for binding and activating MinC. J Bacteriol. 2004;186:1546–1555. doi: 10.1128/JB.186.5.1546-1555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Radaev S, Rosen BP, Gatti DL. Structure of the ArsA ATPase: the catalytic subunit of a heavy metal resistance pump. EMBO. J. 2000;19:4838–4845. doi: 10.1093/emboj/19.17.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.