Abstract
Using live-cell confocal microscopy and particle tracking technology, the simultaneous transport of intracellular vesicles of the endo-lysosomal pathway and nonviral polyethylenimine (PEI)/DNA nanocomplexes was investigated. Due to potential problems associated with the use of acid-sensitive probes in combination with a gene vector that is hypothesized to buffer the pH of intracellular vesicles, the biological location of PEI/DNA gene vectors was revealed by probing their trafficking in cells expressing fluorescent versions of either early endosome antigen 1, a protein that localizes to early endosomes, or Niemann Pick C1, a protein that localizes to late endosomes and lysosomes. Studies directly show that PEI/DNA nanoparticles are actively transported within both early and late endosomes, and display similar overall transport rates in each. Additionally, gene vector transfer between endosomes is observed. Over time posttransfection, gene vectors accumulate in late endosomes/lysosomes; however, real-time escape of vectors from membrane-bound vesicles is not observed.
Keywords: confocal microscopy, particle tracking, endosomes, lysosomes, polyethylenimine
INTRODUCTION
Improved understanding of the intracellular trafficking of synthetic delivery vehicles may help guide the rational design of nonviral vectors used for nucleic acid delivery (Huang and others, 2011; Pack and others, 2005; Whitehead and others, 2010). The intracellular transport of a popular gene vector, polyethylenimine (PEI)/DNA nanocomplexes, involves uptake into acidic endosomes (Forrest and Pack, 2002; Kichler and others, 2001; Remy-Kristensen and others, 2001) and active transport along microtubules to the perinuclear region of cells (Suh and others, 2003). It is unclear, however, if rapid perinuclear accumulation of synthetic PEI/DNA vectors requires their retention within endosomes, or if they can actively transport through the cytoplasm following endosome escape. This question is significant since efficient endosomal escape has typically been a design criterion for improved gene delivery (Murthy and others, 1999; Wattiaux and others, 2000), but may in fact limit delivery efficiency if premature endosome escape prevents rapid gene vector transport to the perinuclear region. If gene vectors are released in the cytoplasm far from the nucleus, the cargo DNA will not likely reach the nucleus due to its hindered mobility in the cytoplasm (Dauty and Verkman, 2005; Lukacs and others, 2000). In addition, free DNA may be degraded by cytoplasmic enzymes (Lechardeur and others, 1999).
Overcoming the cytoplasmic barrier in DNA delivery is critical, as evidenced by the fact that various viruses have evolved different ways to travel to the host cell’s nucleus. For example, adenovirus serotype 5 is hypothesized to escape early endosomes and travel along microtubules to the nucleus, whereas adenovirus serotype 7 is thought to exploit the endo-lysosomal pathway and travel to perinuclear late endosomes prior to escape (Miyazawa and others, 2001). Upon escape from cellular vesicles, adenovirus-7 is then poised to have a relatively short distance to travel to reach the nucleus.
Particle tracking technology has been used to obtain quantitative transport properties of nonviral gene vectors in real-time, such as individual gene vector diffusivity, velocity, trajectory, directionality, and transport mode (e.g., random v. non-random) in live cells (Bausinger and others, 2006; de Bruin and others, 2007; Suh and others, 2003; 2004). Computational models have been developed to describe and predict the intracellular behavior of vectors (Dinh and others, 2007). Other high-resolution fluorescence techniques have also been employed to investigate the intracellular transport of nonviral vectors (Kulkarni and others, 2005). However, more studies are needed to study the motion of gene vectors in the context of the intracellular compartment in which they are located. Here, we investigated the simultaneous transport of early endosomes or late endosomes/lysosomes and nonviral PEI/DNA nanocomplexes in live cells. The combination of multiple particle tracking, multi-color confocal microscopy, and fluorescent organelle-specific proteins allowed the direct observation of polymeric gene vectors traveling within early or late endosomes, as well as transfer between vesicles. This method is widely applicable to the investigation of intracellular transport of various types of therapeutic colloids and viruses and, as such, should find wide-spread use in the development of improved gene delivery strategies.
MATERIALS AND METHODS
Fluorescent protein plasmid constructs
The plasmid construct expressing fluorescent early endosome antigen 1, EEA1-GFP, was a generous gift from Silvia Corvera (University of Massachusetts). The plasmid construct expressing fluorescent Niemann-Pick C1, NPC1-GFP, was a generous gift from Matthew Scott (Stanford University).
Cells
COS-7 cells were maintained in DMEM (Invitrogen Corp., Carlsbad, CA) + 10% FBS (Invitrogen Corp., Carlsbad, CA) + 1% penicillin/streptamycin (Invitrogen Corp., Carlsbad, CA). Unlike previous reports (Suh and others, 2003; 2004), cells used in this study were allowed to divide. Goat bone marrow derived Mesenchymal stem cells (generous gift from Jennifer Elisseeff, Johns Hopkins School of Medicine) at passage 2 to 4 were cultured in MSC Growth Medium (Cambrex, Baltimore, MD) at 37°C and 5% CO2. Cells were transfected with the various plasmids via electroporation (transfection efficiency varied between 30–50%) and used after 24–48 h. Cells were seeded between 7.5 × 104 – 1.0 × 105 cells per plate onto 35-mm glass-bottom tissue culture plates (MatTek Corp., Ashland, MA) for live cell microscopy.
Lysotracker
To fluorescently label acidic vesicles, cells were treated with 100 nM Lysotracker Red (Molecular Probes, Eugene, OR) for 30 mins prior to observation. Lysotracker is only partially protonated at neutral pH and freely enters membranes. Once protonated in acidic environments, the dye is retained within the organelles.
Gene vectors
Polyethylenimine (PEI) (25k MW, branched, Sigma, St Louis, MO) was fluorescently labeled with Oregon Green 488 or Alexa Fluor 546 (both from Molecular Probes, Eugene, OR) with reaction time extended to 2h. Unreacted dye was removed by gel filtration with Sephadex G-75 (Sigma, St Louis, MO). The concentration of PEI was measured with the 2,4,6-trinitrobenzenesulphonic acid (TNBS, Sigma, St Louis, MO) assay (Snyder and Sobocinski, 1975), which detects primary amines. Since Alexa Fluor attaches to primary amines on the PEI, the TNBS assay may underestimate the concentration of PEI in fluorescently labeled samples. Degree of labeling was estimated to be 6 or 2 dye molecules per PEI molecule for Oregon Green or Alexa Fluor labeled samples, respectively. Fluorescently labeled PEI was added drop-wise to 50 µg/ml salmon DNA (Sigma, St Louis, MO), both components in 150mM NaCl, for complexes of N/P (nitrogen to phosphate ratio) equal to 20. Reports indicate that N/P (and not necessarily DNA size) is the critical parameter affecting polyplex physicochemical and functional properties (Godbey and others, 1999a), thus at this point we do not foresee large discrepancies in the intracellular transport of polyplexes formed with salmon DNA versus plasmid DNA. The mixture was vortexed briefly and complexes were allowed to form for 30 min at room temperature. Gene vectors were then added directly to the cell culture medium.
Confocal microscopy
A confocal microscope (LSM 510 Meta, Carl Zeiss Inc., Thornwood, NY) was used to capture the transport of intracellular vesicles and gene vectors simultaneously. For dual-color confocal microscopy, samples were excited with 488 and 543 laser lines, and images were captured by multi-tracking to avoid bleed-through between the two fluorophores. Image acquisition was limited to 250 ms or less per frame to maintain accurate co-localization information and movies were obtained at 2.5 frames per second (or 400 ms intervals). To achieve the necessary acquisition speed and signal to noise ratio in the movies while obtaining the thinnest possible optical slice, the pinhole diameters were set to less than 1 airy unit. After adjustment of the pinholes of both lasers to obtain the same optical slices, the optimal optical section that fulfilled our criteria ranged from less than 0.8 to less than 0.7 µm. All samples were maintained at 37°C using an air stream stage incubator (Nevtek, Burnsville, VA) and observed under a 100X/1.4 NA oil-immersion lens.
Particle tracking and data analysis
MetaMorph software (Universal Imaging Corp., Downingtown, PA) was used to analyze the movies, as reported elsewhere (Suh and others, 2003). Briefly, the movies acquired from confocal microscopy were analyzed with the Tracking Objects application in MetaMorph to give x and y positional data of the transporting species over time. Mean-square displacement, MSD, in two dimensions, x and y, for each transporting entity is given by
| (Equation 1) |
The effective diffusivity, Deff, is calculated by
| (Equation 2) |
where τ is time scale. Additional information on particle tracking analysis, including a discussion of why 2D tracking is representative of 3D motion, can be obtained elsewhere (Saxton and Jacobson, 1997; Suh and others, 2005; Suh and others, 2003; 2004).
RESULTS & DISCUSSION
Gene vectors and Lysotracker
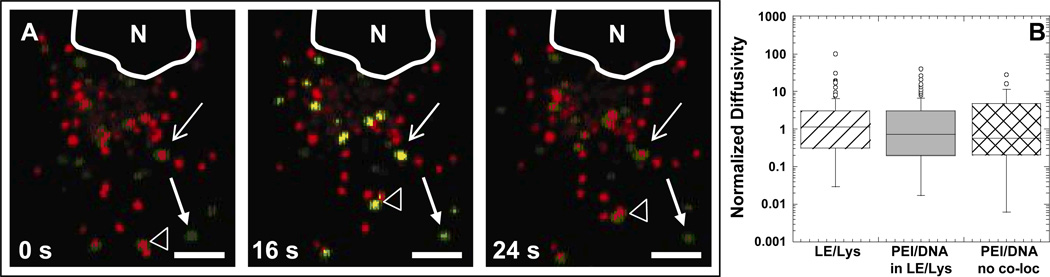
Lysotracker is alive-cell dye commonly used to fluorescently label late endosomes/lysosomes (LE/Lys) (Godbey and others, 1999b). In a pancreatic carcinoma cell line, PEI/DNA nanocomplexes were not found to co-localize with Lysotracker, but did co-localize with anti-LAMP-1 (a marker for LE/Lys) following fixation (Bieber and others, 2002). The authors suggest the buffering capacity of PEI prevents Lysotracker from staining the late endosomes and lysosomes, thereby leading to false negative results. In our studies, a population of PEI/DNA nanocomplexes co-localize with acidic vesicles stained with Lysotracker at various times post-transfection (1 h post-transfection in COS-7 cells shown in Fig. 1A).
Figure 1.
Transport of PEI/DNA complexes in Lysotracker-stained vesicles of a live COS-7 cell. (A) PEI/DNA nanocomplexes (green) are found in acidic organelles (red), where yellow indicates co-localization. Late endosomes/lysosomes (LE/Lys) are stained with the live-cell dye Lysotracker. Line arrow and solid arrow indicate gene vectors co-localizing or not co-localizing with LE/Lys, respectively, with no obvious displacements from the starting positions. Empty arrowhead indicates a gene vector inside a LE/Lys that experiences a large displacement during the same elapsed time. Images are obtained from a 40 s-movie acquired at 2.5 frames/s. Bar is 5 µm. Also see Supplementary Material Movie 1. (B) Boxplots of normalized diffusivities of gene vectors and LE/Lys at a time scale of 10 s. Values were normalized to the geometric mean diffusivity value. The number of endosomes or PEI/DNA tracked are n = 73 (LE/Lys), n = 98 (PEI/DNA in LE/Lys), and n = 43 (PEI/DNA not in LE/Lys). There is no statistically significant difference between the groups as determined by ANOVA.
Some PEI/DNA nanoparticles within Lysotracker-stained vesicles do not display large displacements (line arrow), whereas others are actively transported (unfilled arrowhead) (Supplementary Material Movie 1). Some nanocomplexes, however, do not co-localize with Lysotracker (solid arrowhead arrow). The quantitative transport of LE/Lys without PEI/DNA, LE/Lys containing PEI/DNA, and PEI/DNA not co-localizing with LE/Lys was further analyzed. We were interested to determine if gene vectors not co-localizing with LE/Lys can move as rapidly through the cytoplasm as either the vesicles alone or the gene vectors within vesicles.
The presence of PEI/DNA complexes in LE/Lys do not seem to affect the transport rate of the LE/Lys vesicles, as LE/Lys with or without PEI/DNA exhibit similar transport rates (Fig. 1B). Interestingly, PEI/DNA complexes display similar intracellular transport rates whether or not they co-localize with Lysotracker (Fig. 1B). It is possible PEI/DNA nanocomplexes that do not co-localize with Lysotracker may have escaped cellular vesicles. Since gene vectors that do not co-localize with LE/Lys display transport rates similar to vesicles alone, these vectors may be able to interact with motor proteins and/or other intracellular components in a manner similar to intracellular vesicles. If this hypothesis is correct, rapid escape of PEI/DNA complexes from endosomes may not hinder their efficient transport through the cell cytoplasm to the nucleus. However, more likely explanations are PEI/DNA complexes that do not co-localize with Lysotracker may be within early endosomes (EE), or they may be rendering Lysotracker ineffective due to the buffering capacity of PEI (Sonawane and others, 2003). Due to these possible limitations of using Lysotracker, we employed fluorescent constructs of organelle proteins to specifically label EE or LE/Lys and determined co-localization of PEI/DNA complexes with these cellular compartments in real-time in live cells.
Early endosome antigen 1 (EEA1) is a well-characterized and extensively used marker for EE (Nagamatsu and others, 2001; Sotgia and others, 2003). The Niemann Pick C1 (NPC1) protein, whose role has been linked to cholesterol trafficking in cells (Pentchev and others, 1987), has been shown to localize to late endosomal and lysosomal compartments (Garver and others, 2000; Sugii and others, 2003). NPC1-containing vesicles co-localize with Lysotracker as well as LAMP-1 (data not shown) (Sugii and others, 2003). The simultaneous real-time transport of nonviral gene vectors and EE or LE/Lys was captured to reveal the dynamic trafficking of synthetic gene vectors in live cells.
Trafficking of gene vectors over time
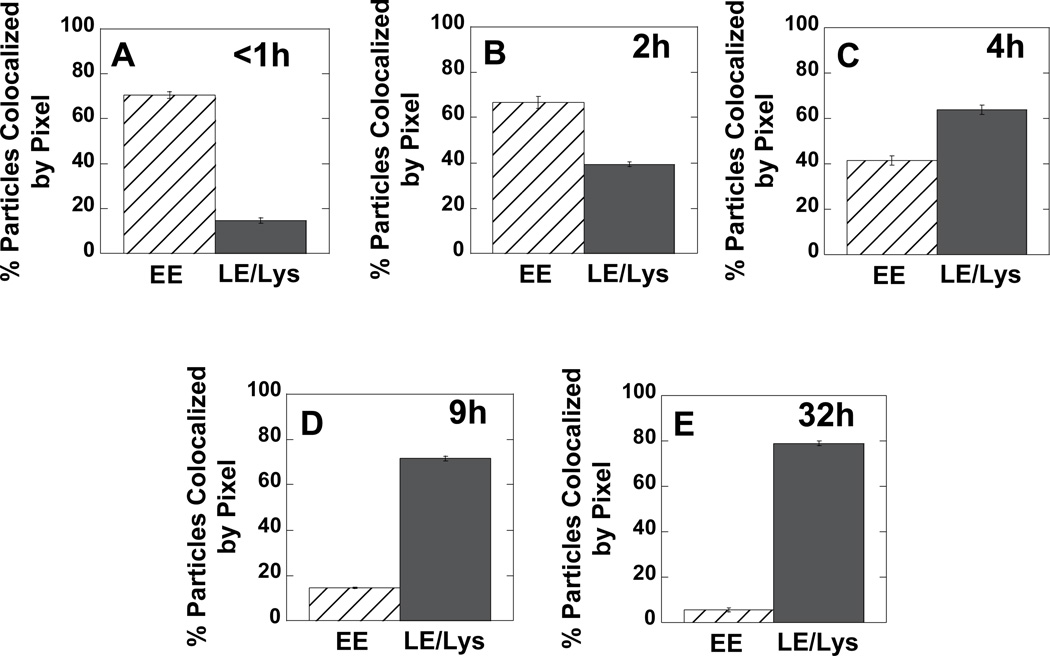
First, the extent of gene vector co-localization with EEA1-GFP (i.e., early endosomes) or NPC1-GFP (i.e., late endosomes and lysosomes) were quantified at various times (Fig. 2). At early times post-transfection, a majority of PEI/DNA nanocomplexes co-localize with the marker for EE (Fig. 2A). Over time post-transfection, increasing amounts of PEI/DNA complexes are found in LE/Lys (Fig. 2B–E). Approximately 80% of gene vectors are in LE/Lys compartments by 32 h post-transfection (Fig. 2E).
Figure 2.
Per pixel co-localization of PEI/DNA complexes with either EE or LE/Lys at various times post-transfection in MSCs. To fluorescently label endosomes, cells were pre-transfected with GFP fusion genes of EEA1 (early endosome antigen 1) for EE or NPC1 (Niemann-Pick C1) for LE/Lys. Five cells from each condition were imaged.
Our method of determining percent co-localization between gene vectors and organelles has an associated error since we measure co-localization on a per pixel basis and not on a per particle basis. EEA1 and NPC1 are both membrane-bound proteins; therefore, staining can often appear “donut-like”, since the proteins are of highest concentration along the membrane. If a gene vector is within the lumen of a LE/Lys with “donut-like” NPC1 staining, the per pixel co-localization value may be lower than that expected for a per particle basis. Visual inspection of images acquired at 32 h post-transfection indicates 100% of detectable PEI/DNA nanocomplexes are in LE/Lys and not EE.
Gene vector transport in EE and LE/Lys
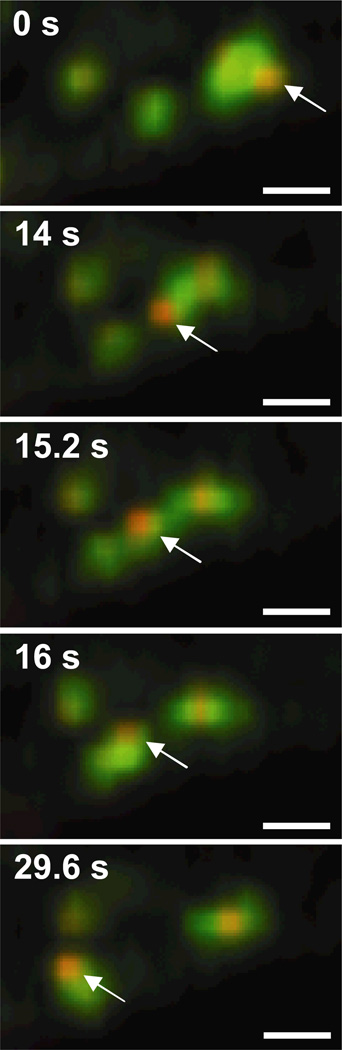
Many PEI/DNA nanocomplexes are observed to transport within EE at 1.5 h post-transfection (Fig. 3, Supplementary Material Movie 2). Most gene vectors inside EE are transported within that particular endosome for the entire duration of the movies (i.e. 40 – 60 s). Gene vectors associated with EE appear to be encased in EEA1 label, suggesting the vectors are within the vesicle lumen. Occasionally, a PEI/DNA nanocomplex is observed to transfer from one EE to another (Fig. 3). The gene vector (encased in EEA1 label) extends from the donating EE (Fig. 3, 14 s), connects to the receiving EE making a temporary bridge between the donating and receiving EEs (Fig. 3, 15.2 s), and then completes the transfer into the receiving EE (Fig. 3, 16 s).
Figure 3.
Transfer of PEI/DNA nanocomplexes between EEs of a live Mesenchymal stem cell (MSC). Stills from a 60s-movie (obtained at 2.5 frames/s) show a PEI/DNA nanocomplex (indicated with an arrow) being transferred from one EE to another. EE are green due to the expression of EEA1-GFP, and PEI/DNA nanocomplexes are fluorescently labeled red with Alexa Fluor 546. Bar is 1 µm. Also see Supplementary Material Movie 2.
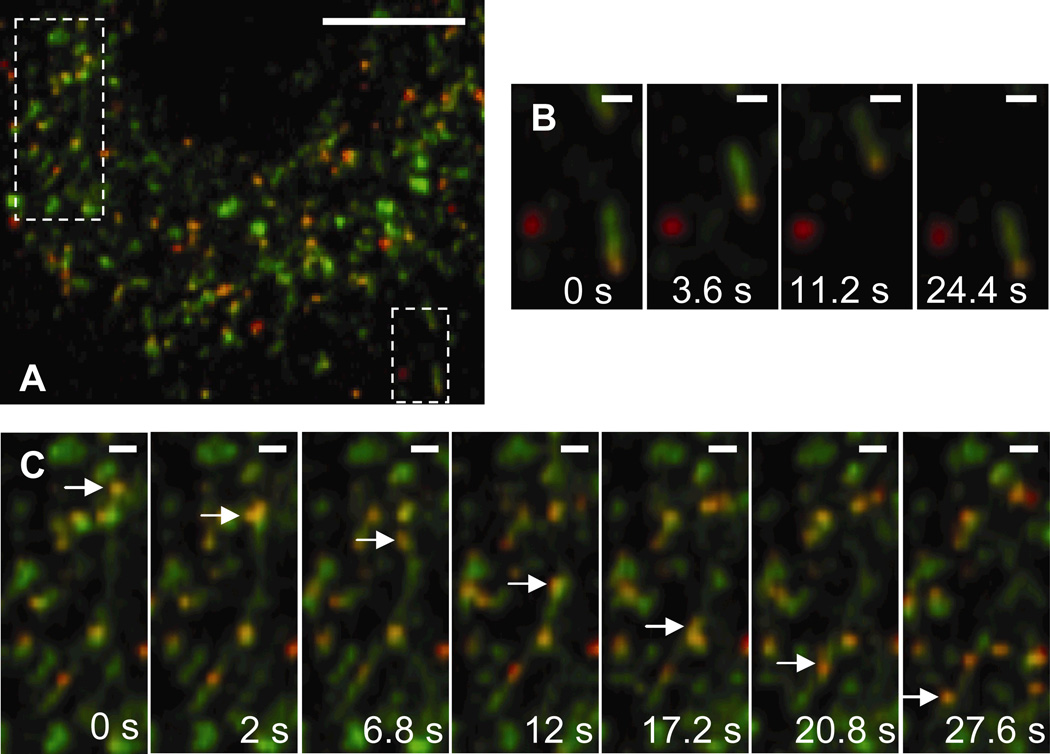
PEI/DNA complexes are also found to transport actively in LE/Lys at 1.5 h post-transfection (Fig. 4, Supplementary Material Movie 3). The NPC1 protein displays greater tubular staining compared to EEA1. Zhang et al. have also documented the presence of late endosomal tubules and have shown their transport to be dependent on microtubules (Zhang and others, 2002). PEI/DNA complexes in EE are found mainly in spherical structures, whereas complexes in LE/Lys are found in both spherical and tubular structures. The endo-lysosomal pathway is characterized by a system of tubules and vesicles, and is sometimes referred to as the tubulo-vesicular system (Munn, 2000; Nagamatsu and others, 2001). In spherical LE/Lys vesicles that are similar in size to the PEI/DNA complexes, gene vectors appear to be completely encased by the NPC1 label (suggesting the vectors are within the vesicle lumen). In Fig. 4B, a PEI/DNA complex is actively transporting in a tubular LE/Lys. On occasion, gene vectors are observed transporting actively within tubular paths lined with NPC1 in the cytoplasm (Fig. 4C).
Figure 4.
Transport of PEI/DNA nanocomplexes in late endosomes/lysosomes (LE/Lys) of a live MSC. (A) Many PEI/DNA complexes (red) co-localize with NPC-1 (green) at 2 h post-transfection. Boxes indicate regions shown in greater detail in panels B or C. Bar is 10 µm. Also see Supplementary Material Movie 3. (B) PEI/DNA complex moving within a tubular LE. Bar is 1 µm. (C) PEI/DNA (indicated by an arrow) moving actively through a tubule that is lined with NPC1. Bar is 1 µm. Movies captured at 2.5 frames/s.
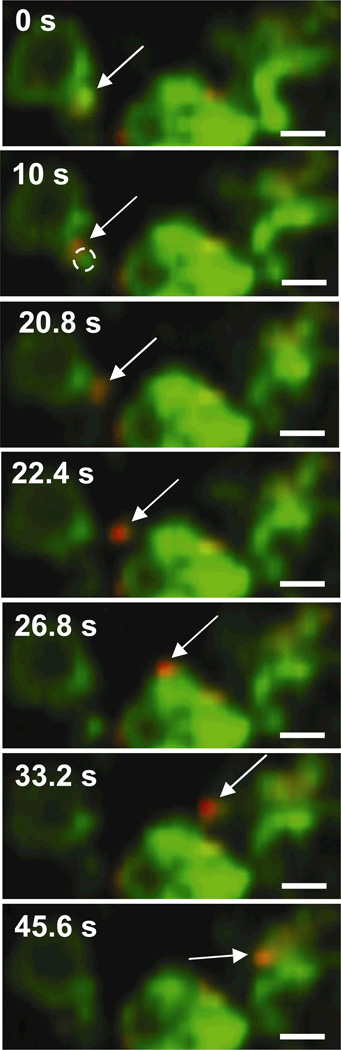
In addition to the spherical and tubular LE/Lys structures discussed above, PEI/DNA complexes are also found in association with large spherical structures with distinct lumens (Fig. 5). In these cases, the gene vector is not found in the center of the lumen, but rather associated with NPC1 staining on the side of the vesicle. Such large spherical vesicles were also observed in cells not transfected with PEI/DNA (data not shown), suggesting the presence of gene vectors is not the cause of this endosome morphology. Similar to PEI/DNA in EE, several gene vectors are observed to transfer between LE/Lys (Fig. 5, Supplementary Material Movie 4). A small section of the LE/Lys (outlined by a dotted circle in Fig. 5, 10 s) is seen displacing prior to the PEI/DNA nanocomplex leaving the donating LE/Lys. The PEI/DNA nanocomplex appears to be transferred within a small vesicle expressing NPC1-GFP and passes another LE/Lys (Fig. 5, 26.8 s) before fusing with the receiving LE/Lys (Fig. 5, 45.6 s). More in-depth investigations are necessary to enhance our understanding of the specific cellular factors regulating trafficking of PEI/DNA nanocomplexes through the endo-lysosomal pathway.
Figure 5.
Transfer of PEI/DNA between LE/Lys of a live MSC. Stills from a 60s-movie (obtained at 2.5 frames/s) showing a PEI/DNA nanocomplex (indicated by an arrow) being transferred from one LE/Lys to another. LE/Lys are green due to the expression of NPC1-GFP, and PEI/DNA nanocomplexes are fluorescently labeled red with Alexa Fluor 546. Bars are 1 µm. Also see Supplementary Material Movie 4.
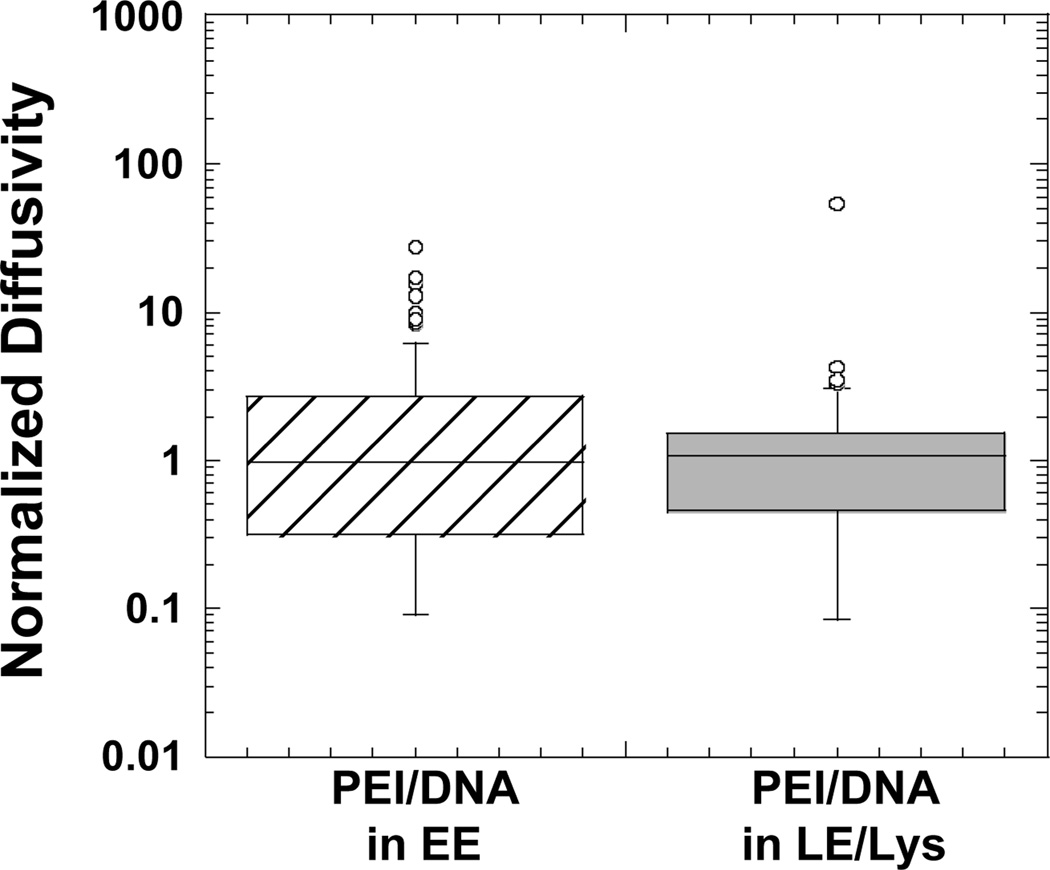
Transport rates of gene vectors in endosomes
PEI/DNA complexes within EE or LE/Lys are transported at similar rates (Fig. 6). This supports the result with Lysotracker (Fig. 1B), assuming the gene vectors that do not co-localize with the dye were complexes in EE or in vesicles not acidic enough to be stained by Lysotracker. We are not able to determine the transport rate of nanocomplexes outside of cellular vesicles since even at late times post-transfection; all PEI/DNA were found in cellular vesicles (Fig. 2). One way to determine the mobility of particles outside of vesicles would be to microinject them directly into the cytoplasm. Unfortunately, we were not able to microinject the positively-charged PEI/DNA particles due to clogging of microinjection needles. Work with commercially available polystyrene particles with diameters of 100 nm shows particles that are endocytosed are able to move up to 9-fold faster than particles microinjected directly into the cytoplasm (Suh and others, 2007). Thus, if PEI/DNA particles exit endosomes intact, their mobility is expected to be significantly reduced compared to complexes traveling within endosomes. It may, therefore, be beneficial for PEI/DNA particles to remain within the endo-lysosomal pathway until they achieve perinuclear localization.
Figure 6.
Boxplots of normalized diffusivities of PEI/DNA complexes transporting within EE or LE/Lys at a time scale of 10 s. Values were normalized to the geometric mean diffusivity value. The number of PEI/DNA tracked are n = 81 (PEI/DNA in EE) and n = 61 (PEI/DNA in LE/Lys). There is no statistically significant difference between the groups as determined by student t-test.
Lysotracker fluorescently labels LE/Lys, but not EE. Results here verify gene vectors remain within cellular vesicles of the endo-lysosomal pathway and are progressively trafficked from EE to LE/Lys. Since PEI/DNA nanocomplexes in EE or LE/Lys experience similar transport rates in live cells, gene vectors not co-localizing with Lysotracker, but still moving at similar transport rates, are likely in EE.
PEI/DNA nanocomplexes are hypothesized to escape endosomes efficiently (Klemm and others, 1998; Sonawane and others, 2003); however, we did not detect vectors outside of vesicles at 32 h post-transfection, a time point after gene transfection occurs (gene transfection is often assayed at 24 h post-transfection). Gene vectors that successfully escape vesicles may display a faint diffuse staining, instead of a bright punctate spot, due to vector unpacking in the cytoplasm. This may lead us to overlook their presence. We also did not capture PEI/DNA in the process of escaping from endosomes. This may be due to the rarity of endosomal escape by the nanoparticles or, alternatively, the process of escaping from endosomes may involve a much more subtle mechanism than previously thought. Endosome escape may not involve a sudden “burst” effect, but perhaps a more modest leakage of gene vector components during trafficking through the endo-lysosomal pathway. Given the importance of the endosomal escape step in the transfection process, it is clear an improved method is needed to detect PEI/DNA complexes that are in the process of escaping or have escaped from cellular endosomes. Utilizing fluorophores whose emission spectra change depending on cellular environment (e.g. reducing properties, presence of specific enzymes) may enable detection of complexes in the cytoplasm versus the endosome. Such an approach should further reveal insights into the intracellular transport of PEI/DNA complexes and may finally unveil the mechanism by which these nanoparticles escape vesicles.
Moreover, in our studies to date, only the polymer component was fluorescently labeled. A significant question that remains is whether the cargo DNA also experiences similar intracellular transport and processing as the polymer component. Further studies tracking the movement of fluorescently labeled cargo DNA are needed to answer this important question.
In conclusion, using confocal microscopy and multiple-particle tracking, we correlated quantitative gene vector transport rates to qualitative intracellular localization. The highly dynamic intracellular transport of PEI/DNA nanocomplexes was investigated in real-time and in live cells. Gene vectors were directly observed to transport actively in early endosomes as well as late endosomes/lysosomes. Application of this methodology to the investigation of other gene vector systems may help uncover key bottlenecks and provide critical insights that feed into the design process.
Supplementary Material
MOVIE 1: PEI/DNA nanocomplexes transporting inside Lysotracker-stained vesicles.
MOVIE 2: PEI/DNA nanocomplexes transferring between early endosomes.
MOVIE 3: PEI/DNA nanocomplexes transporting inside late endosomes/lysosomes.
MOVIE 4: PEI/DNA nanocomplexes transferring between late endosomes/lysosomes.
ACKNOWLEDGMENTS
The authors thank Christa Brown and Trina Schroer (Johns Hopkins University, Department of Biology) for their valuable guidance in the selection of the fluorescent protein constructs for live cell microscopy. This work was supported by the National Science Foundation (BES 9978160 and BES 0346716), the National Institutes of Health (T32-GM07057 and RC2GM092599), and an ARCS fellowship to J.S.
REFERENCES
- Bausinger R, von Gersdorff K, Braeckmans K, Ogris M, Wagner E, Zumbusch A, Brauchle C. The transport of nanosized gene carriers unraveled by live-cell imaging. Angewandte Chemie-International Edition. 2006;45(10):1568–1572. doi: 10.1002/anie.200503021. [DOI] [PubMed] [Google Scholar]
- Bieber T, Meissner W, Kostin S, Niemann A, Elsasser HP. Intracellular route and transcriptional competence of polyethylenimine-DNA complexes. Journal of Controlled Release. 2002;82(2–3):441–454. doi: 10.1016/s0168-3659(02)00129-3. [DOI] [PubMed] [Google Scholar]
- Dauty E, Verkman AS. Actin cytoskeleton as the principal determinant of size-dependent DNA mobility in cytoplasm: a new barrier for non-viral gene delivery. J Biol Chem. 2005;280(9):7823–7828. doi: 10.1074/jbc.M412374200. [DOI] [PubMed] [Google Scholar]
- de Bruin K, Ruthardt N, von Gersdorff K, Bausinger R, Wagner E, Ogris M, Brauchle C. Cellular dynamics of EGF receptor-targeted synthetic viruses (vol 15, pg 1297, 2007) Molecular Therapy. 2007;15(9):1735–1735. doi: 10.1038/sj.mt.6300176. [DOI] [PubMed] [Google Scholar]
- Dinh AT, Pangarkar C, Theofanous T, Mitragotri S. Understanding intracellular transport processes pertinent to synthetic gene delivery via stochastic simulations and sensitivity analyses. Biophysical Journal. 2007;92(3):831–846. doi: 10.1529/biophysj.106.095521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest ML, Pack DW. On the kinetics of polyplex endocytic trafficking: implications for gene delivery vector design. Mol Ther. 2002;6(1):57–66. doi: 10.1006/mthe.2002.0631. [DOI] [PubMed] [Google Scholar]
- Garver WS, Heidenreich RA, Erickson RP, Thomas MA, Wilson JM. Localization of the murine Niemann-Pick C1 protein to two distinct intracellular compartments. J Lipid Res. 2000;41(5):673–687. [PubMed] [Google Scholar]
- Godbey WT, Wu KK, Mikos AG. Poly(ethylenimine) and its role in gene delivery. J Control Release. 1999a;60(2–3):149–160. doi: 10.1016/s0168-3659(99)00090-5. [DOI] [PubMed] [Google Scholar]
- Godbey WT, Wu KK, Mikos AG. Tracking the intracellular path of poly(ethylenimine)/DNA complexes for gene delivery. Proc Natl Acad Sci U S A. 1999b;96(9):5177–5181. doi: 10.1073/pnas.96.9.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Dempsey C, Chona D, Suh J. Quantitative nanoparticle tracking: applications to nanomedicine. Nanomedicine (Lond) 2011;6(4):693–700. doi: 10.2217/nnm.11.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kichler A, Leborgne C, Coeytaux E, Danos O. Polyethylenimine-mediated gene delivery: a mechanistic study. J Gene Med. 2001;3(2):135–144. doi: 10.1002/jgm.173. [DOI] [PubMed] [Google Scholar]
- Klemm AR, Young D, Lloyd JB. Effects of polyethyleneimine on endocytosis and lysosome stability. Biochem Pharmacol. 1998;56(1):41–46. doi: 10.1016/s0006-2952(98)00098-7. [DOI] [PubMed] [Google Scholar]
- Kulkarni RP, Wu DD, Davis ME, Fraser SE. Quantitating intracellular transport of polyplexes by spatio-temporal image correlation spectroscopy. Proc Natl Acad Sci U S A. 2005;102(21):7523–7528. doi: 10.1073/pnas.0501950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechardeur D, Sohn KJ, Haardt M, Joshi PB, Monck M, Graham RW, Beatty B, Squire J, O'Brodovich H, Lukacs GL. Metabolic instability of plasmid DNA in the cytosol: a potential barrier to gene transfer. Gene Ther. 1999;6(4):482–497. doi: 10.1038/sj.gt.3300867. [DOI] [PubMed] [Google Scholar]
- Lukacs GL, Haggie P, Seksek O, Lechardeur D, Freedman N, Verkman AS. Size-dependent DNA mobility in cytoplasm and nucleus. J Biol Chem. 2000;275(3):1625–1629. doi: 10.1074/jbc.275.3.1625. [DOI] [PubMed] [Google Scholar]
- Miyazawa N, Crystal RG, Leopold PL. Adenovirus serotype 7 retention in a late endosomal compartment prior to cytosol escape is modulated by fiber protein. J Virol. 2001;75(3):1387–1400. doi: 10.1128/JVI.75.3.1387-1400.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn AL. The yeast endocytic membrane transport system. Microsc Res Tech. 2000;51(6):547–562. doi: 10.1002/1097-0029(20001215)51:6<547::AID-JEMT5>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Murthy N, Robichaud JR, Tirrell DA, Stayton PS, Hoffman AS. The design and synthesis of polymers for eukaryotic membrane disruption. J Control Release. 1999;61(1–2):137–143. doi: 10.1016/s0168-3659(99)00114-5. [DOI] [PubMed] [Google Scholar]
- Nagamatsu S, Nakamichi Y, Watanabe T, Matsushima S, Yamaguchi S, Ni J, Itagaki E, Ishida H. Localization of cellubrevin-related peptide, endobrevin, in the early endosome in pancreatic beta cells and its physiological function in exo-endocytosis of secretory granules. J Cell Sci. 2001;114(Pt 1):219–227. doi: 10.1242/jcs.114.1.219. [DOI] [PubMed] [Google Scholar]
- Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4(7):581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- Pentchev PG, Comly ME, Kruth HS, Tokoro T, Butler J, Sokol J, Filling-Katz M, Quirk JM, Marshall DC, Patel S, et al. Group C Niemann-Pick disease: faulty regulation of low-density lipoprotein uptake and cholesterol storage in cultured fibroblasts. Faseb J. 1987;1(1):40–45. doi: 10.1096/fasebj.1.1.3609608. [DOI] [PubMed] [Google Scholar]
- Remy-Kristensen A, Clamme JP, Vuilleumier C, Kuhry JG, Mely Y. Role of endocytosis in the transfection of L929 fibroblasts by polyethylenimine/DNA complexes. Biochim Biophys Acta. 2001;1514(1):21–32. doi: 10.1016/s0005-2736(01)00359-5. [DOI] [PubMed] [Google Scholar]
- Saxton MJ, Jacobson K. Single-particle tracking: applications to membrane dynamics. Annu Rev Biophys Biomol Struct. 1997;26:373–399. doi: 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- Snyder SL, Sobocinski PZ. An improved 2,4,6-trinitrobenzenesulfonic acid method for the determination of amines. Anal Biochem. 1975;64(1):284–288. doi: 10.1016/0003-2697(75)90431-5. [DOI] [PubMed] [Google Scholar]
- Sonawane ND, Szoka FC, Jr, Verkman AS. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. J Biol Chem. 2003;278(45):44826–44831. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- Sotgia F, Bonuccelli G, Bedford M, Brancaccio A, Mayer U, Wilson MT, Campos-Gonzalez R, Brooks JW, Sudol M, Lisanti MP. Localization of phospho-beta-dystroglycan (pY892) to an intracellular vesicular compartment in cultured cells and skeletal muscle fibers in vivo. Biochemistry. 2003;42(23):7110–7123. doi: 10.1021/bi0271289. [DOI] [PubMed] [Google Scholar]
- Sugii S, Reid PC, Ohgami N, Du H, Chang TY. Distinct endosomal compartments in early trafficking of low density lipoprotein-derived cholesterol. J Biol Chem. 2003;278(29):27180–27189. doi: 10.1074/jbc.M300542200. [DOI] [PubMed] [Google Scholar]
- Suh J, Choy KL, Lai S, Suk JS, Tang BC, Prabhu S, Hanes J. PEGylation of Nanoparticles Improves Their Cytoplasmic Transport. Int J Nanomed. 2007;2(4):1–7. [PMC free article] [PubMed] [Google Scholar]
- Suh J, Dawson M, Hanes J. Real-time multiple-particle tracking: applications to drug and gene delivery. Adv Drug Deliv Rev. 2005;57(1):63–78. doi: 10.1016/j.addr.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Suh J, Wirtz D, Hanes J. Efficient active transport of gene nanocarriers to the cell nucleus. Proc Natl Acad Sci U S A. 2003;100(7):3878–3882. doi: 10.1073/pnas.0636277100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh J, Wirtz D, Hanes J. Real-time intracellular transport of gene nanocarriers studied by multiple particle tracking. Biotechnol Prog. 2004;20(2):598–602. doi: 10.1021/bp034251y. [DOI] [PubMed] [Google Scholar]
- Wattiaux R, Laurent N, Wattiaux-De Coninck S, Jadot M. Endosomes, lysosomes: their implication in gene transfer. Adv Drug Deliv Rev. 2000;41(2):201–208. doi: 10.1016/s0169-409x(99)00066-6. [DOI] [PubMed] [Google Scholar]
- Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery (vol 8, pg 129, 2009) Nature Reviews Drug Discovery. 2010;9(5):412–412. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Liu P, Dwyer NK, Christenson LK, Fujimoto T, Martinez F, Comly M, Hanover JA, Blanchette-Mackie EJ, Strauss JF., 3rd MLN64 mediates mobilization of lysosomal cholesterol to steroidogenic mitochondria. J Biol Chem. 2002;277(36):33300–33310. doi: 10.1074/jbc.M200003200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MOVIE 1: PEI/DNA nanocomplexes transporting inside Lysotracker-stained vesicles.
MOVIE 2: PEI/DNA nanocomplexes transferring between early endosomes.
MOVIE 3: PEI/DNA nanocomplexes transporting inside late endosomes/lysosomes.
MOVIE 4: PEI/DNA nanocomplexes transferring between late endosomes/lysosomes.