Abstract
The discovery of functional MRI (fMRI), with the first papers appearing in 1992, gave rise to new categories of data that drove the development of new signal-processing strategies. Workers in the field were confronted with image time courses, which could be reshuffled to form pixel time courses. The waveform in an active pixel time-course was determined not only by the task sequence but also by the hemodynamic response function. Reference waveforms could be cross-correlated with pixel time courses to form an array of cross-correlation coefficients. From this array of numbers, colorized images could be created and overlaid on anatomical images. An early paper from the authors’ laboratory is extensively reviewed here (Bandettini et al. 1993. Magn. Reson. Med. 30:161–173). That work was carried out using the vocabulary of vector algebra. Cross-correlation methodology was central to the discovery of functional connectivity MRI (fcMRI) by Biswal et al. (1995. Magn. Reson. Med. 34:537–541). In this method, a whole volume time course of images is collected while the brain is nominally at rest and connectivity is studied by cross-correlation of pixel time courses.
Keywords: cross-correlation coefficient, fMRI/fcMRI, autocorrelation, vector algebra, signal processing
INTRODUCTION
In January 1993, Bandettini, Jesmanowicz, Wong, and Hyde submitted a paper for publication in Magnetic Resonance in Medicine titled, “Processing Strategies for Time-Course Data Sets in Functional MRI of the Human Brain (Bandettini et al., 1993).” This paper, which has been cited more than 1,000 times, reaches this conclusion: “The most effective method for image processing involves thresholding by shape as characterized by the correlation coefficient of the data with respect to a reference function followed by formation of a cross-correlation image.” In the present article, we attempt to reconstruct the early history of the cross-correlation method in fMRI—noting that we were at the same time developing image processing tools based on cross-correlation that played a central role in the discovery of functional connectivity MRI (fcMRI) in 1995 by Biswal et al. (1995).
The story begins with an abstract that we presented at that wonderful San Francisco meeting of ISMRM in 1991 where fMRI suddenly appeared (Jesmanowicz et al., 1991). In this abstract, Andrzej Jesmanowicz addressed the problem of computation of T1, T2, and diffusion coefficient images using the primitive computers of the day. Trial vectors were produced from trial exponential functions. Each exponential curve was represented as a vector. Each point of each curve was represented as one component in N dimensional space. About 200 normalized trial vectors were predefined, representing different time constants in a predetermined range. For each experimental vector, which need not be normalized and which is very sparse, the scalar, or dot, product was formed with each of the 200 predefined vectors. The best match was that particular trial vector that yielded the maximum value of the scalar product—or, graphically, the maximum value of the projection of the experimental vector onto a predefined vector. The text of the abstract goes on to state that one can show this procedure is mathematically equivalent to minimization of the least square difference between a normalized experimental exponential curve and the trial exponential. The point of the abstract was that the new method was computationally efficient. Andrzej later came to realize that the method was also equivalent to maximization of the cross-correlation of an experimental vector with a reference vector, but the path took a few turns since we were dealing not with a cleanly posed mathematical problem but with novel and poorly understood data from the human brain.
AN EARLY EXPERIMENT
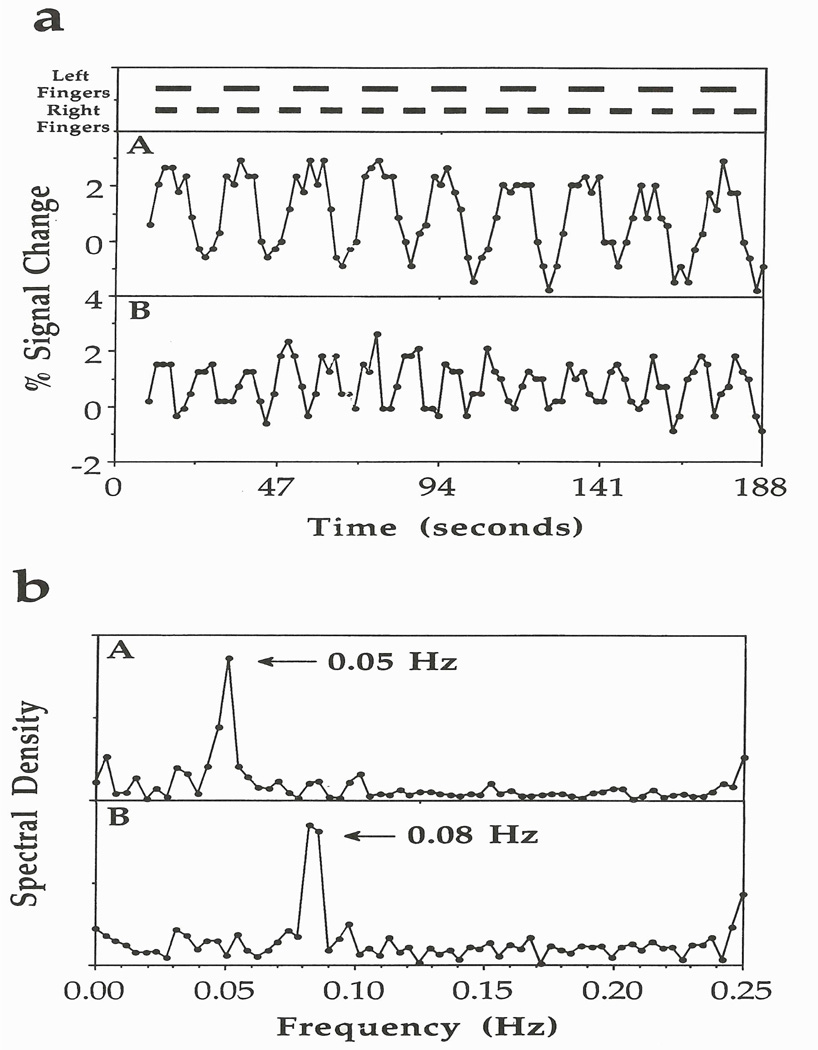
One of the elegant experiments that Peter A. Bandettini and Eric C. Wong did was asynchronous bilateral finger-tapping. The experiment is described in Bandettini et al. (1993) and is summarized here. The paradigm involves on/off frequencies of 0.05 Hz for finger movement of the left hand and 0.08 Hz for finger movement of the right hand! Figure 1 shows the timing at the top and representative pixel time courses from the finger representations of the right and left cortices (Fig. 1a). It also shows, in Fig. 1b, the Fourier transforms (FT) of the waveforms in Fig. 1a. As expected, strong peaks are seen at 0.05 and 0.08 Hz in the FT displays. Images were formed from the intensities of these peaks, which are shown in Fig. 2.
Figure 1.
Plots from the left and right pixels of the motor cortex in the asynchronous bilateral finger-tapping experiment. At the top is the activation paradigm that involves on/off frequencies of 0.05 Hz for finger movement of the left hand and of 0.08 Hz for finger movement of the right hand. (a) Plots in boxes A and B correspond closely with left and right finger movements, respectively. (b) Plots of the spectral density versus frequency from the pixels in boxes A and B, which reveal peaks at the activation frequencies that correspond also to the left-and right-hand activation frequencies, respectively. Figure 1 reprinted from Bandettini et al. 1993. Magn. Reson. Med. 30:161–173, where it originally appeared as Figure 13.
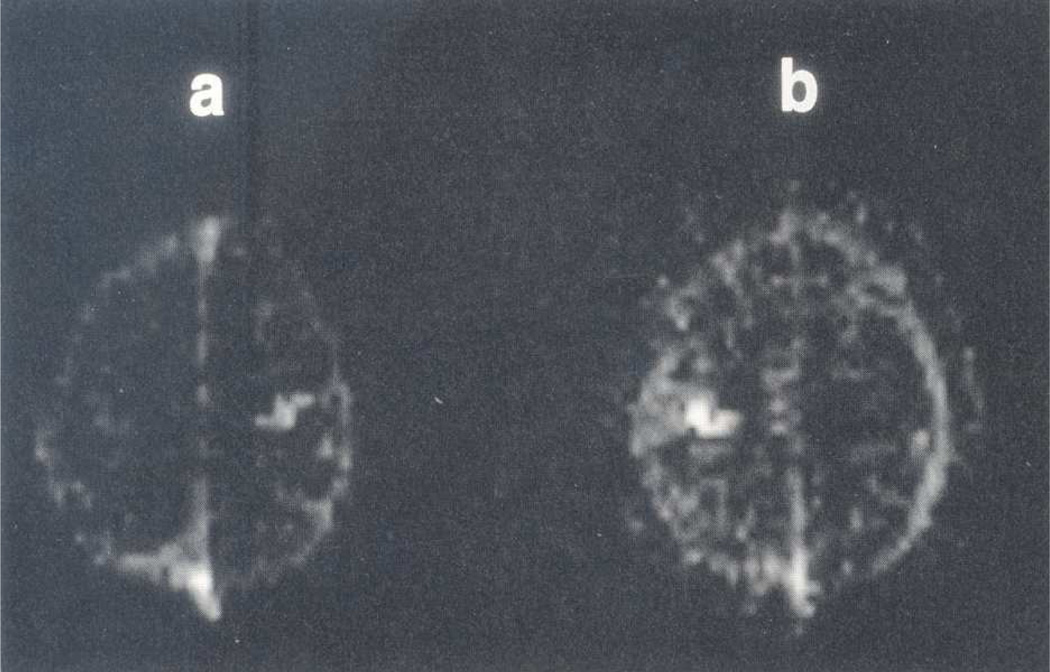
Figure 2.
Functional images obtained from the Fourier transform data set of Fig. 1 in which finger movement of the left and right hands was at on/off frequencies of 0.05 and 0.08 Hz, respectively. (a) Spectral density image at 0.08 Hz reveals high signal intensity in the left motor cortex. (b) Spectral density image at 0.05 Hz reveals high signal intensity in the right motor cortex. Figure 2 reprinted from Bandettini et al. 1993. Magn. Reson. Med. 30:161–173, where it originally appeared as Figure 14.
It is known that plotting the intensity of one frequency of the FT of a time series is equivalent to phase sensitive detection of the time series at the specified frequency, and phase sensitive detection had been of interest to Jim Hyde since his earliest years of research in EPR spectroscopy. That would be 1954. Jim was truly delighted with the experiment.
In fact, the phase sensitive detector was invented by R.H. Dicke in 1946 (Dicke, 1946), the year that NMR was discovered. The paper, however, has very little detail. Dr. E.M. Purcell, who shared the Nobel Prize with F. Bloch for the discovery of NMR, was acknowledged in Dicke’s paper. Magnetic field modulation followed by RF detection and phase sensitive detection were used in the early NMR experiments at Harvard. The full circuit was provided in N. Bloembergen’s dissertation two years later (Bloembergen, 1948, 1961). One can also find indications that phase sensitive detection was used not only by the Bloch group at Stanford (Anderson, 1960) but also two years earlier in 1944 by Zavoisky in the discovery of EPR in Kazan (Kochelaev and Yablokov, 1995). Modulation followed by phase sensitive detection is deeply embedded in the history of magnetic resonance. Square-wave modulation in a block-design fMRI experiment was not unlike field modulation in an NMR or EPR experiment. The fMRI block design experiment is actually “amplitude modulation,” and the field modulation experiment is actually “frequency modulation.” Phase sensitive detection is blind to the difference. It just picks out one frequency in the time series.
Phase sensitive detection originally provided an output for graphic display of the correlation of an experimental waveform with a sinusoidal waveform. In fMRI, we have an image time course composed of thousands of pixel time courses. Data are digitized and available for analysis in a seemingly endless variety of ways. We can cross-correlate if we wish, in the way that Peter and Eric did in the experiment described above, but more was possible, which was the thrust of Bandettini et al. (1993).
THE CROSS-CORRELATION METHOD
During 1993, we finally recognized that we were developing the “cross-correlation method” for analysis of fMRI. We were drowning in data, and automated image processing tools were desperately needed. Each pixel time course was represented as a vector. The cross-correlation coefficient (CC) was defined (Eq. (1)) and recast in vector notation (Eq. (2)).
| (1) |
| (2) |
Bandettini et al. (1993) provides intermediate steps linking Eqs. (1) and (2).
In a first step in signal processing, the constant average value of the reference vector was removed by the process of vector orthogonalization, and a projection was made on the normalized reference vector. In this way, a time course was obtained for each pixel that was a measure of the amount of neuronal activity in a given voxel. The correlation coefficient itself was used as a threshold to make a decision about displaying the functional value. In addition, a computer program was developed that could remove unwanted ramps (i.e., linear drifts) from the time course—yes, again by a process of vector orthogonalization. It was called FIM for functional imaging. Eric Wong followed up on a suggestion of Jim Hyde and developed the functional display (FD) program: an array of squares that mapped into an array of pixels. In each square, the pixel time course was displayed. Bob Cox combined FIM and FD and called it “FD2”. Bob notes in his article in this issue that he almost called AFNI “FD3” until he came to his senses.
A controversy arose over what reference vector should be used. Hypothesis-driven research would mandate that the reference vector be a square wave since the task was always on and off, in equal periods, and we were testing the hypothesis that the brain was responding in accordance with the task. It was soon recognized, however, that the response of the brain was filtered through the somewhat sluggish hemodynamic response function. Quite beautiful images could be made using a reference vector that was created from this function. But to make an image using a reference vector formed from the data itself seemed illogical, and statistically unsound.
Consider, as an example, synchronous bilateral finger-tapping data. If we cross-correlate with a boxcar, images from troughs and peaks of the fMRI response are simply subtracted. The dot product of a box-car waveform and a pixel time course is essentially the same as averaging all images during the interleaved activation periods and subtracting from an average of the corresponding interleaved resting-state periods. Because all images in the time course are used, the contrast-to-noise increases, even though the actual pixel responses do not represent box-car waveforms.
As an alternative to use of a boxcar, one can use a reference waveform based on the experimental hemodynamic response function of one strongly responding pixel. The image quality is improved compared with use of a simple boxcar because the reference vector more closely approximates the actual response vector and the dot products are higher. A potential difficulty with this approach, however, is that various artifacts related to task-correlated motion or vessel pulsatility may be enhanced.
Still another approach is to create a time-averaged hemodynamic response function from one or more strongly responding pixel time courses, and then replicate it as many times as necessary to match the pixel time-course vectors. The image quality is found to be slightly improved.
It is also possible to use a non-periodic boxcar since the cross-correlation method does not require a periodic reference waveform to create the reference vector.
In preparation of this article, we reviewed the history of cross-correlation. The correlation coefficient itself was introduced by Sir Francis Galton, an English doctor, explorer, and statistician in 1869 (see Upton and Cook, 2008). He was a cousin of Charles Darwin. The primary use was studying random-like processes that exhibit similarity in their behavior or occurrence. A good example would be the temperature of the air, which is no doubt correlated with seasons. The question was, by how much? This correlation was known as long as human history but no number had been given until the 19th century. The formal definition of the correlation coefficient was given in the form:
| (3) |
It seems somewhat obscure. It would take a half page to explain Cov(X,Y) and Var(X) terms. One can find the details in Oxford’s A Dictionary of Statistics (Upton and Cook, 2008).
Correlation statistics applies well to processes that are independent of time. Finding the correlation between the forearm and a child’s age can be done in any order of time and any order of age. By contrast, time-ordered correlation cannot be studied randomly. Time-ordered correlation was introduced in the surprisingly late year of 1926 by the Scottish statistician George Udny Yule (see Upton and Cook, 2008). He was concerned with a periodic time series that was obscured by noise. Yule introduced the concept of autocorrelation, which found immediate application, even before computers became popular. If a priori information exists about the periodicity, the autocorrelation method is appropriate.
DISCUSSION
The principles of bandwidth management, data collection, and digital filtering were discussed in an early publication by Klein and Barton (1963). To paraphrase this work: if noise is white and two spectra are compared, the first acquired in a single scan in time T with an integrating time constant τ and the second acquired by summing n spectra, each acquired in time T/n with integrating time constant τ/n, the SNRs will be the same. However, if the noise has a 1/f character, the latter method will exhibit lower noise. In the present context, it is important that digital filters be applied prior to calculation of the cross-correlation coefficient. It should be recognized that the data are inherently complex valued, and complex valued digital filters are therefore appropriate.
The cross-correlation method is widely applied in fcMRI, but the problems are daunting. If there are N pixel time courses in an image time course data set, there are N2 cross-current coefficient images that can be formed. Strategies to reduce the size of functional connectivity data sets include the following: restriction of the pixels-of-interest to gray matter; restriction to networks-of-interest; restriction to areas defined by an fMRI task, which we call fMRI-driven fcMRI; and restriction to histologically defined regions as reported in an atlas. The latter approach is particularly helpful: regions are defined, average resting-state time courses are formed, and the regional pairwise correlation-coefficient (RPCC) matrix is formed (Pawela et al., 2008).
ACKNOWLEDGMENT
The long-running grant from the National Institutes of Health to James S. Hyde, fMRI Technology and Analysis (EB000215), provided support for the work that is reviewed here. We are grateful.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Anderson WA. NMR and EPR Spectroscopy: Papers Presented at Varian’s Third Annual Workshop on Nuclear Magnetic Resonance and Electron Paramagnetic Resonance, held at Palo Alto, California. Oxford: Pergamon Press; 1960. Measurement of relaxation times; pp. 168–175. [Google Scholar]
- Bandettini PA, Jesmanowicz A, Wong EC, Hyde JS. Processing strategies for time-course data sets in functional MRI of the human brain. Magn. Reson. Med. 1993;30:161–173. doi: 10.1002/mrm.1910300204. [DOI] [PubMed] [Google Scholar]
- Biswal BB, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn. Reson. Med. 1995;34:537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Bloembergen N. Nuclear Magnetic Relaxation [dissertation] Nijhoff, The Hague; 1948. [Google Scholar]
- Bloembergen N. Nuclear Magnetic Relaxation. In: Pines D, editor. Frontiers in Physics, A Lecture Note and Reprint Series. New York, NY: Benjamin, Inc; 1961. [Google Scholar]
- Dicke RH. The measurement of thermal radiation at microwave frequencies. Rev. Sci. Instrum. 1946;17:268–276. doi: 10.1063/1.1770483. [DOI] [PubMed] [Google Scholar]
- Jesmanowicz A, Wong EC, Wu JC, Hyde JS. Fast algorithm for computation of T1, T2, and diffusion coefficient images [abstract] Proc. Soc. Magn. Reson. Med. 1991;1:740. [Google Scholar]
- Klein MP, Barton GW., Jr Enhancement of signal-to-noise ratio by continuous averaging: Application to magnetic resonance. Rev. Sci. Instrum. 1963;34:754–759. [Google Scholar]
- Kochelaev BI, Yablokov YV. The Beginning of Paramagnetic Resonance. Singapore: World Scientific; 1995. [Google Scholar]
- Pawela CP, Biswal BB, Cho YR, Kao DS, Li R, Jones SR, Schulte ML, Matloub HS, Hudetz AG, Hyde JS. Resting-state functional connectivity of the rat brain. Magn. Reson. Med. 2008;59:1021–1029. doi: 10.1002/mrm.21524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton GJG, Cook I. A dictionary of statistics (Oxford Paperback Reference) Oxford University Press; 2008. [Google Scholar]