Abstract
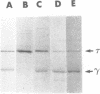
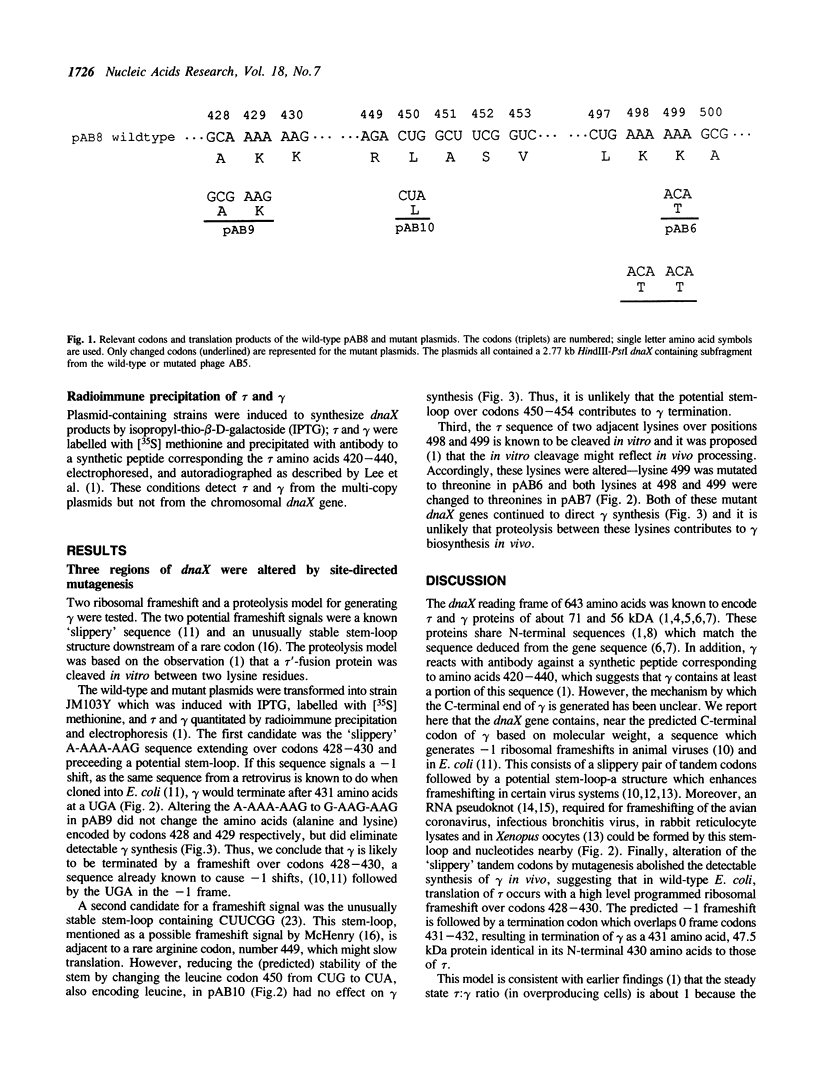
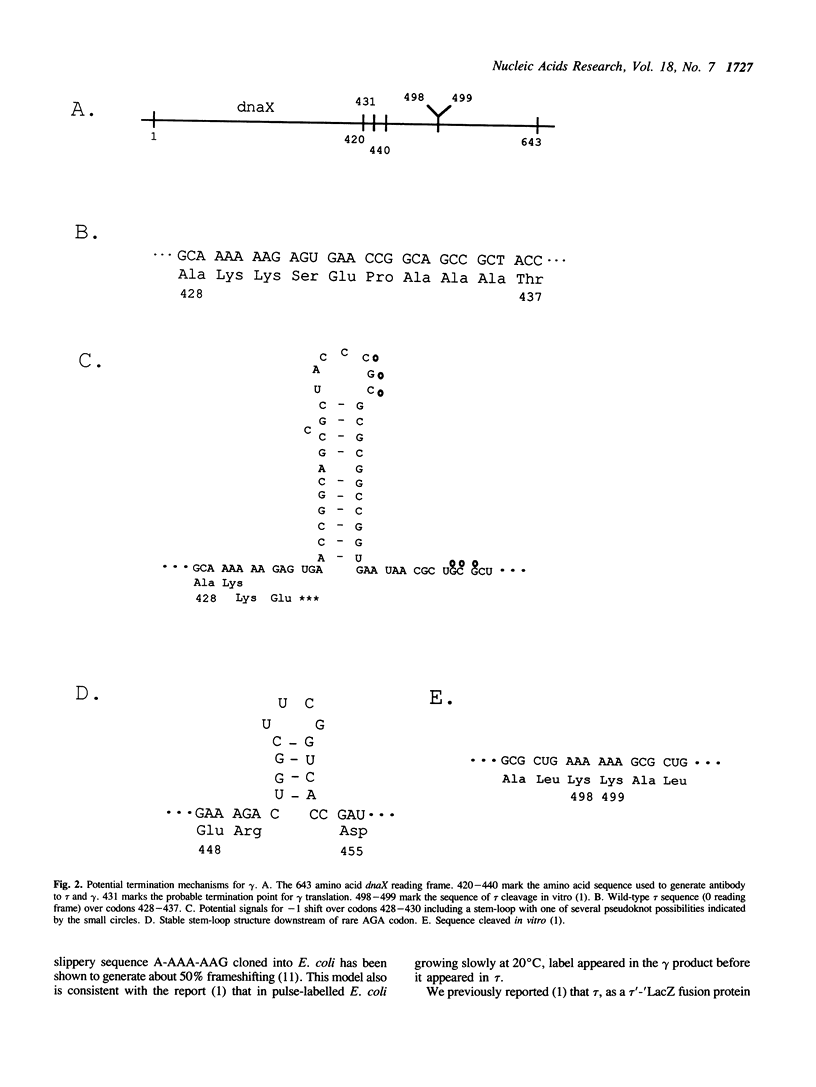
The Escherichia coli dnaX gene encodes both the tau and gamma subunits of DNA polymerase III holoenzyme in one reading frame. The 71.1 kDa tau and the shorter gamma share N-terminal sequences. Mutagenesis of a potential ribosomal frameshift signal located at codons 428-430 without changing the amino acid sequence of the tau product, eliminated detectable synthesis of the gamma subunit, suggesting that the reading frame is shifted at that sequence and gamma is terminated by a nonsense codon located in the -1 frame 3 nucleotides downstream of the signal. This seems to be the first known case of a frameshift which is used, along with the termination codon in the -1 frame, to terminate a peptide within a reading frame. [Mutagenesis of a dibasic peptide (lys-lys) at codons 498-499, the site at which a tau'-'LacZ fusion protein was cleaved in vitro (1) had no effect on gamma formation in vivo, suggesting that cleavage observed in vitro is not the mechanism of gamma formation in vivo.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkins J. F., Gesteland R. F., Reid B. R., Anderson C. W. Normal tRNAs promote ribosomal frameshifting. Cell. 1979 Dec;18(4):1119–1131. doi: 10.1016/0092-8674(79)90225-3. [DOI] [PubMed] [Google Scholar]
- Brierley I., Digard P., Inglis S. C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989 May 19;57(4):537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare J., Farabaugh P. Nucleotide sequence of a yeast Ty element: evidence for an unusual mechanism of gene expression. Proc Natl Acad Sci U S A. 1985 May;82(9):2829–2833. doi: 10.1073/pnas.82.9.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigen W. J., Caskey C. T. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature. 1986 Jul 17;322(6076):273–275. doi: 10.1038/322273a0. [DOI] [PubMed] [Google Scholar]
- Craigen W. J., Cook R. G., Tate W. P., Caskey C. T. Bacterial peptide chain release factors: conserved primary structure and possible frameshift regulation of release factor 2. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3616–3620. doi: 10.1073/pnas.82.11.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Flower A. M., McHenry C. S. The adjacent dnaZ and dnaX genes of Escherichia coli are contained within one continuous open reading frame. Nucleic Acids Res. 1986 Oct 24;14(20):8091–8101. doi: 10.1093/nar/14.20.8091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey H. V., Taylor M. W. Nucleotide sequence and deduced amino acid sequence of Escherichia coli adenine phosphoribosyltransferase and comparison with other analogous enzymes. Gene. 1986;43(3):287–293. doi: 10.1016/0378-1119(86)90218-0. [DOI] [PubMed] [Google Scholar]
- Icho T., Wickner R. B. The double-stranded RNA genome of yeast virus L-A encodes its own putative RNA polymerase by fusing two open reading frames. J Biol Chem. 1989 Apr 25;264(12):6716–6723. [PubMed] [Google Scholar]
- Jacks T., Madhani H. D., Masiarz F. R., Varmus H. E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988 Nov 4;55(3):447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson K. O., McHenry C. S. Adenosine 5'-O-(3-thiotriphosphate) can support the formation of an initiation complex between the DNA polymerase III holoenzyme and primed DNA. J Biol Chem. 1984 Apr 10;259(7):4589–4595. [PubMed] [Google Scholar]
- Kastelein R. A., Remaut E., Fiers W., van Duin J. Lysis gene expression of RNA phage MS2 depends on a frameshift during translation of the overlapping coat protein gene. Nature. 1982 Jan 7;295(5844):35–41. doi: 10.1038/295035a0. [DOI] [PubMed] [Google Scholar]
- Kodaira M., Biswas S. B., Kornberg A. The dnaX gene encodes the DNA polymerase III holoenzyme tau subunit, precursor of the gamma subunit, the dnaZ gene product. Mol Gen Genet. 1983;192(1-2):80–86. doi: 10.1007/BF00327650. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lee S. H., Kanda P., Kennedy R. C., Walker J. R. Relation of the Escherichia coli dnaX gene to its two products--the tau and gamma subunits of DNA polymerase III holoenzyme. Nucleic Acids Res. 1987 Oct 12;15(19):7663–7675. doi: 10.1093/nar/15.19.7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki H., Maki S., Kornberg A. DNA Polymerase III holoenzyme of Escherichia coli. IV. The holoenzyme is an asymmetric dimer with twin active sites. J Biol Chem. 1988 May 15;263(14):6570–6578. [PubMed] [Google Scholar]
- Maki S., Kornberg A. DNA polymerase III holoenzyme of Escherichia coli. I. Purification and distinctive functions of subunits tau and gamma, the dnaZX gene products. J Biol Chem. 1988 May 15;263(14):6547–6554. [PubMed] [Google Scholar]
- Maki S., Kornberg A. DNA polymerase III holoenzyme of Escherichia coli. II. A novel complex including the gamma subunit essential for processive synthesis. J Biol Chem. 1988 May 15;263(14):6555–6560. [PubMed] [Google Scholar]
- Maki S., Kornberg A. DNA polymerase III holoenzyme of Escherichia coli. III. Distinctive processive polymerases reconstituted from purified subunits. J Biol Chem. 1988 May 15;263(14):6561–6569. [PubMed] [Google Scholar]
- Maki S., Kornberg A. DNA polymerase III holoenzyme of Escherichia coli. III. Distinctive processive polymerases reconstituted from purified subunits. J Biol Chem. 1988 May 15;263(14):6561–6569. [PubMed] [Google Scholar]
- McHenry C. S. DNA polymerase III holoenzyme of Escherichia coli. Annu Rev Biochem. 1988;57:519–550. doi: 10.1146/annurev.bi.57.070188.002511. [DOI] [PubMed] [Google Scholar]
- McHenry C., Kornberg A. DNA polymerase III holoenzyme of Escherichia coli. Purification and resolution into subunits. J Biol Chem. 1977 Sep 25;252(18):6478–6484. [PubMed] [Google Scholar]
- Mellor J., Fulton S. M., Dobson M. J., Wilson W., Kingsman S. M., Kingsman A. J. A retrovirus-like strategy for expression of a fusion protein encoded by yeast transposon Ty1. Nature. 1985 Jan 17;313(5999):243–246. doi: 10.1038/313243a0. [DOI] [PubMed] [Google Scholar]
- Moore R., Dixon M., Smith R., Peters G., Dickson C. Complete nucleotide sequence of a milk-transmitted mouse mammary tumor virus: two frameshift suppression events are required for translation of gag and pol. J Virol. 1987 Feb;61(2):480–490. doi: 10.1128/jvi.61.2.480-490.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullin D. A., Woldringh C. L., Henson J. M., Walker J. R. Cloning of the Escherichia coli dnaZX region and identification of its products. Mol Gen Genet. 1983;192(1-2):73–79. doi: 10.1007/BF00327649. [DOI] [PubMed] [Google Scholar]
- O'Donnell M. E. Accessory proteins bind a primed template and mediate rapid cycling of DNA polymerase III holoenzyme from Escherichia coli. J Biol Chem. 1987 Dec 5;262(34):16558–16565. [PubMed] [Google Scholar]
- O'Donnell M., Studwell P. S. Total reconstitution of DNA polymerase III holoenzyme reveals dual accessory protein clamps. J Biol Chem. 1990 Jan 15;265(2):1179–1187. [PubMed] [Google Scholar]
- Pleij C. W., Rietveld K., Bosch L. A new principle of RNA folding based on pseudoknotting. Nucleic Acids Res. 1985 Mar 11;13(5):1717–1731. doi: 10.1093/nar/13.5.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice N. R., Stephens R. M., Burny A., Gilden R. V. The gag and pol genes of bovine leukemia virus: nucleotide sequence and analysis. Virology. 1985 Apr 30;142(2):357–377. doi: 10.1016/0042-6822(85)90344-7. [DOI] [PubMed] [Google Scholar]
- Saigo K., Kugimiya W., Matsuo Y., Inouye S., Yoshioka K., Yuki S. Identification of the coding sequence for a reverse transcriptase-like enzyme in a transposable genetic element in Drosophila melanogaster. Nature. 1984 Dec 13;312(5995):659–661. doi: 10.1038/312659a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine Y., Ohtsubo E. Frameshifting is required for production of the transposase encoded by insertion sequence 1. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4609–4613. doi: 10.1073/pnas.86.12.4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha N. K., Morris C. F., Alberts B. M. Efficient in vitro replication of double-stranded DNA templates by a purified T4 bacteriophage replication system. J Biol Chem. 1980 May 10;255(9):4290–4293. [PubMed] [Google Scholar]
- Studnicka G. M., Rahn G. M., Cummings I. W., Salser W. A. Computer method for predicting the secondary structure of single-stranded RNA. Nucleic Acids Res. 1978 Sep;5(9):3365–3387. doi: 10.1093/nar/5.9.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchihashi Z., Kornberg A. ATP interactions of the tau and gamma subunits of DNA polymerase III holoenzyme of Escherichia coli. J Biol Chem. 1989 Oct 25;264(30):17790–17795. [PubMed] [Google Scholar]
- Tuerk C., Gauss P., Thermes C., Groebe D. R., Gayle M., Guild N., Stormo G., d'Aubenton-Carafa Y., Uhlenbeck O. C., Tinoco I., Jr CUUCGG hairpins: extraordinarily stable RNA secondary structures associated with various biochemical processes. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1364–1368. doi: 10.1073/pnas.85.5.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Atkins J. F., Gesteland R. F. Slippery runs, shifty stops, backward steps, and forward hops: -2, -1, +1, +2, +5, and +6 ribosomal frameshifting. Cold Spring Harb Symp Quant Biol. 1987;52:687–693. doi: 10.1101/sqb.1987.052.01.078. [DOI] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Shuh M., Atkins J. F., Gesteland R. F. E. coli ribosomes re-phase on retroviral frameshift signals at rates ranging from 2 to 50 percent. New Biol. 1989 Nov;1(2):159–169. [PubMed] [Google Scholar]
- Wickner S. Mechanism of DNA elongation catalyzed by Escherichia coli DNA polymerase III, dnaZ protein, and DNA elongation factors I and III. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3511–3515. doi: 10.1073/pnas.73.10.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin K. C., Blinkowa A., Walker J. R. Nucleotide sequence of the Escherichia coli replication gene dnaZX. Nucleic Acids Res. 1986 Aug 26;14(16):6541–6549. doi: 10.1093/nar/14.16.6541. [DOI] [PMC free article] [PubMed] [Google Scholar]