PARP-1 (poly[ADP-ribose] polymerase-1) is a nuclear enzyme that processes varied stress signals and, in response, determines cell fates based on the type and strength of the stress stimulus. PAR (poly[ADP-ribose]) is generated by PARP-1 and regulates many of these stress response pathways. In this review, Kraus and colleagues discuss recent important findings that elucidate the role of PAR and PARP-1 in the regulation of stress response.
Keywords: DNA repair, inflammation, poly(ADP-ribose) (PAR), poly(ADP-ribose) polymerase-1 (PARP-1), stress, transcription
Abstract
Cellular stress responses are mediated through a series of regulatory processes that occur at the genomic, transcriptional, post-transcriptional, translational, and post-translational levels. These responses require a complex network of sensors and effectors from multiple signaling pathways, including the abundant and ubiquitous nuclear enzyme poly(ADP-ribose) (PAR) polymerase-1 (PARP-1). PARP-1 functions at the center of cellular stress responses, where it processes diverse signals and, in response, directs cells to specific fates (e.g., DNA repair vs. cell death) based on the type and strength of the stress stimulus. Many of PARP-1's functions in stress response pathways are mediated by its regulated synthesis of PAR, a negatively charged polymer, using NAD+ as a donor of ADP-ribose units. Thus, PARP-1's functions are intimately tied to nuclear NAD+ metabolism and the broader metabolic profile of the cell. Recent studies in cell and animal models have highlighted the roles of PARP-1 and PAR in the response to a wide variety of extrinsic and intrinsic stress signals, including those initiated by oxidative, nitrosative, genotoxic, oncogenic, thermal, inflammatory, and metabolic stresses. These responses underlie pathological conditions, including cancer, inflammation-related diseases, and metabolic dysregulation. The development of PARP inhibitors is being pursued as a therapeutic approach to these conditions. In this review, we highlight the newest findings about PARP-1's role in stress responses in the context of the historical data.
Poly(ADP-ribose) (PAR) polymerase-1 (PARP-1) is the most abundant and ubiquitous member of a family of 17 related mammalian proteins. Studies over the past two decades, complemented by some exciting recent reports, have begun to crystallize a clearer view of one of the most robust and consistent functions of PARP-1 across biological systems: as a stress sensor and a stress response mediator. In this review, we highlight the newest findings in this area in the context of the historical data. We did not attempt to provide a comprehensive review, but rather opted to focus on key findings that have clarified the role of PARP-1 (and related PARPs) in stress responses. The reader is directed to a variety of other excellent reviews that cover other aspects of PARP-1 biology (D'Amours et al. 1999; Shall and de Murcia 2000; Kim et al. 2005; Gagne et al. 2006; Schreiber et al. 2006; Hassa and Hottiger 2008; Kraus 2008; Ji and Tulin 2010; Krishnakumar and Kraus 2010a; Rouleau et al. 2010).
The nuts and bolts of PARP-1, PAR, and nuclear NAD+
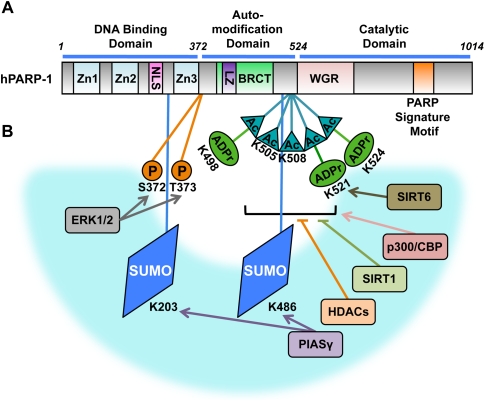
The molecular, chemical, structural, and evolutionary biology of PARP-1 as well as its enzymatic product, PAR, have been reviewed extensively elsewhere and are not repeated here (D'Amours et al. 1999; Schreiber et al. 2006; Krishnakumar and Kraus 2010a). As a starting point, we provide the following relevant background. Mammalian PARP-1 is a 116-kDa protein comprising (1) an N-terminal DNA-binding domain, which contains three zinc-binding domains (Zn1, Zn2, and Zn3) and a nuclear localization sequence (NLS); (2) a central automodification domain, which contains several glutamate, aspartate, and lysine residues as putative acceptors for auto(ADP-ribosyl)ation, a leucine zipper motif that mediates homodimerization or heterodimerization, and a BRCA1 C-terminal (BRCT) phosphopeptide-binding motif; and (3) a C-terminal catalytic domain, which contains a tryptophan-, glycine-, and arginine-rich (WGR) domain and the “PARP signature” sequence required for the catalysis of PAR synthesis (Fig. 1A; D'Amours et al. 1999; Krishnakumar and Kraus 2010a). PARP-1 belongs to a family of 17 proteins with confirmed or putative mono(ADP-ribosyl) and poly(ADP-ribosyl) transferase activity, which share the “PARP signature motif” in the homologous catalytic domain (Fig. 1A; Ame et al. 2004; Schreiber et al. 2006).
Figure 1.
Structural and functional organization of PARP-1, and site-specific post-translational modifications. (A) Human PARP-1 is a 116-kDa protein comprising (1) an N-terminal DNA-binding domain, which contains three zinc-binding domains (Zn1, Zn2, and Zn3) and a NLS; (2) a central automodification domain, which contains a leucine zipper (LZ) motif and a BRCT motif; and (3) a C-terminal catalytic domain, which contains a WGR domain and the “PARP signature” motif required for NAD+ binding and the catalysis of PAR synthesis. (B) Key post-translational modifications of PARP-1 are illustrated on the PARP-1 functional organization schematic from A. Four types of post-translational modifications are shown: phosphorylation (P), SUMOylation (SUMO), acetylation (Ac), and mono(ADP-ribosyl)ation or poly(ADP-ribosyl)ation (ADPr). The Ser (S), Lys (K), or Thr (T) residues that are the sites of covalent modification are indicated. Enzymes that add (arrows) or remove (blunt lines) the specific post-translational modifications, as described in the text, are shown in the blue arc.
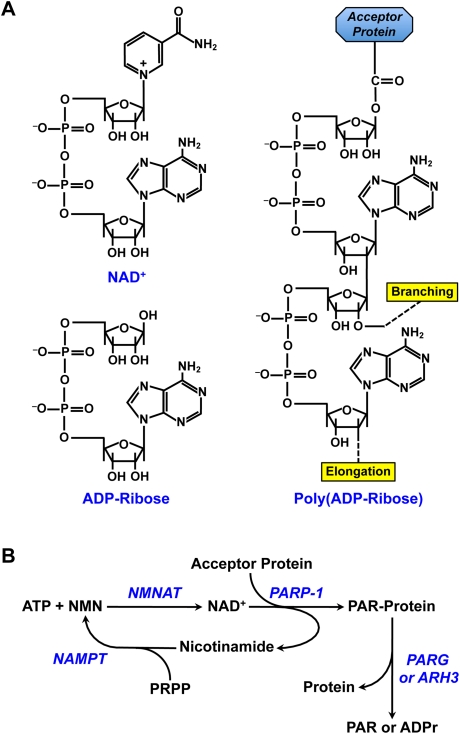
PARP-1 catalyzes the covalent attachment of PAR polymers on itself and other acceptor proteins, including histones, DNA repair proteins, transcription factors, and chromatin modulators, using NAD+ as a donor of ADP-ribose units (Fig. 2; D'Amours et al. 1999; Hassa and Hottiger 2008). Covalently attached PAR can be hydrolyzed to free PAR or mono(ADP-ribose) by PAR glycohydrolase (PARG), which possesses both endoglycosidic and exoglycosidic activities (Min and Wang 2009), and PAR hydrolase (ARH3), which also has PARG activity (Fig. 2B; Oka et al. 2006). PAR is rapidly turned over in the cell, with a half-life on the order of minutes (Gagne et al. 2006). Free or protein-bound PAR polymers work as signal transducers by binding other proteins through their conserved PAR recognition modules, including PAR-binding motifs (PBMs), PAR-binding zinc finger (PBZF) domains, and macrodomains (Kraus 2009). PAR, which is a negatively charged polymer, alters the biochemical properties of modified or interacting proteins, modulating their structure, function, and localization (Schreiber et al. 2006; Krishnakumar and Kraus 2010a).
Figure 2.
NAD+ metabolism and the synthesis of PAR. (A, counterclockwise from top left) Chemical structures of NAD+, ADP-ribose (ADRr), and PAR. The positions on the ribose moieties where the PAR polymer is elongated and branched from are indicated. In the example shown, PAR is covalently attached to an aspartic or glutamic acid residue on the acceptor proteins. PAR may also be attached to lysine residues. (B) The PARP-1-dependent NAD+ metabolic cycle. NMN is synthesized from nicotinamide (NAM) and 5-phosphoribosyl-1-pyrophosphate (PRPP) by nicotinamide phosphoribosyltransferase (NAMPT). NAD+ is synthesized from ATP and NMN by NMNAT (NMNAT-1 is the nuclear member of the NMNAT group of enzymes). PARP-1 catalyzes the covalent attachment of PAR on acceptor proteins using NAD+ as a donor of ADP-ribose units, with the concomitant release of nicotinamide. Covalently attached PAR can be hydrolyzed to free PAR or mono(ADP-ribose) (ADPr) by PARG, which possesses both endoglycosidic and exoglycosidic activities, and ARH3, which also has PARG activity.
PARP-1 plays key roles in DNA repair, chromatin modulation, and transcription (Kraus 2008; Ji and Tulin 2010; Krishnakumar and Kraus 2010a). It binds to nucleosomes containing intact DNA (Kim et al. 2004), as well as to damaged DNA structures (e.g., nicks and double-strand breaks) under conditions of genotoxic stress, leading to the activation of its enzymatic activity (D'Amours et al. 1999). PARP-1 is enriched at the promoters of actively transcribed genes, where it plays a key role in modulating the chromatin environment to regulate gene expression, especially in response to cellular signaling pathways (Krishnakumar et al. 2008; Frizzell et al. 2009). PARP-1 interacts with histone- and chromatin-modifying enzymes to control their activity at target gene promoters, ultimately affecting chromatin-dependent gene expression outcomes. PARP-1 also interacts with nicotinamide mononucleotide (NMN) adenylyltransferase-1 (NMNAT-1), a nuclear NAD+ synthase, which provides NAD+ for the nuclear functions of nuclear NAD+-consuming enzymes such as PARP-1 and the NAD+-dependent protein deacetylase SIRT1 (Berger et al. 2007; Zhang et al. 2009). As such, PARP-1 plays a key role in a number of distinct nuclear processes, integrating NAD+ availability with molecular outcomes.
PARP-1 at the crossroads of cellular stress responses
Cells are regularly exposed to a wide variety of extrinsic and intrinsic stress signals, including those initiated by oxidative, nitrosative, genotoxic, oncogenic, thermal, inflammatory, and metabolic stresses (Kourtis and Tavernarakis 2011). The propagation of these signals involves cross-talk among multiple signaling pathways that lead to defined outcomes. For example, oxidative stress can cause or amplify genotoxic stress and stimulate inflammatory responses (Kryston et al. 2011). Likewise, inflammation can promote oncogenic stress by inducing genomic instability through the induction of oxidative and nitrosative stress (Lonkar and Dedon 2011). Additionally, metabolic stress can trigger chronic inflammatory responses and oxidative stress by increasing the production of reactive oxygen species (ROS) in mitochondria and promoting endoplasmic reticulum (ER) stress (Zhang and Kaufman 2008; Wellen and Thompson 2010). Furthermore, chronic inflammation can sensitize organisms to metabolic stress signals (Medzhitov 2008). As these examples illustrate, stress-related pathological conditions typically involve cross-talk between two or more different types of stress responses.
Studies over the past two decades have placed PARP-1 and related PARP family members at the intersection of converging stress signaling pathways (Fig. 3). While the focus has been on the role of PARPs in genotoxic, oxidative, oncogenic, and inflammatory stress responses—especially their roles in DNA repair and transcriptional regulation—several recent studies have revealed novel aspects of PARPs in metabolic stress responses.
Figure 3.
PARP-1 at the crossroads of cellular stress responses. Cells are regularly exposed to a wide variety of extrinsic and intrinsic stress signals, including those initiated by oxidative, nitrosative, genotoxic, oncogenic, thermal, inflammatory, and metabolic stresses. PARP-1 senses these stresses and propagates different stress signals to execute diverse downstream molecular and cellular actions. PARP-1 and related PARP family members function at the intersection of converging stress signaling pathways. (Ac) Acetylation; (P) phosphorylation; (Su) SUMOylation; (Ub) ubiquitylation; (NAM) nicotinamide.
A new role of PARPs: metabolic stress and homeostasis
Cells detect and respond to insufficient, as well as excess, nutrients by rearranging their metabolic profiles, energy stores, and energy expenditure. The integration of the cellular responses to metabolic stress in multiple organs dictates the physiological or pathological states of the organisms, including a series of metabolic- or age-related diseases, such as obesity, diabetes, and cancer (Wellen and Thompson 2010). Several recent studies have indicated key roles for PARP-1 and PARP-2 in metabolic stress and homeostasis.
PARP-1−/− mice show strain-specific metabolic effects (Luo and Kraus 2011). For example, on an obesity-resistant (predominantly SV129) background, PARP-1 knockout leads to increased age-related weight gain and diet-induced obesity, suggesting a protective role for PARP-1 (Devalaraja-Narashimha and Padanilam 2010). In contrast, on an obesity-prone (predominantly C57BL/6J) background, PARP-1 knockout leads to lower weights and improved metabolic profiles, as well as protection against diet-induced obesity and insulin resistance, suggesting a negative role for PARP-1 (Bai et al. 2011b). Although the strain specificity of these effects are not currently understood, C57BL/6J mice are known to have a loss-of-function deletion in the gene encoding nicotinamide nucleotide transhydrogenase, an enzyme that catalyzes the production of NAD+ through the reversible reduction of NADP+ by NADH (Freeman et al. 2006). This or other genetic differences between the SV129 and the C57BL/6J strains that affect metabolic pathways may underlie the observed phenotypic differences. Some of the effects of PARP-1 on adipogenesis are due to direct effects on PPARγ-dependent adipogenic gene expression in fat cells (Erener et al. 2012), but the system-wide effects of PARP-1 suggest a broad spectrum of functions beyond that.
Interestingly, the metabolic phenotypes of the PARP-1−/− mice, whose background is predominantly C57BL/6J, resemble those in which SIRT1, an NAD+-dependent protein deacetylase and key metabolic regulator, is overexpressed or chemically activated (Herranz and Serrano 2010). These results suggest that PARP-1 depletion increases SIRT1 activity, perhaps through increased NAD+ availability, but may also do so through direct modification of the SIRT1 protein. Additional studies in mice have shown that the activity of PARP-1 oscillates in a circadian manner (Asher et al. 2010). PARP-1−/− mice display impaired food entrainment of the circadian rhythm through a mechanism that involves effects on the core circadian machinery. PARP-1 senses and integrates the feeding signal to modify and regulate CLOCK, a key circadian transcription factor, and its downstream transcriptional circuitry for rhythm generation (Asher et al. 2010). Thus, PARP-1 also elicits its metabolic effects by influencing other well-established metabolic pathways.
PARP-2−/− mice also show metabolic effects (Bai et al. 2011a), including elevated energy expenditure, oxygen consumption, lipid oxidation associated with a leaner phenotype, protection against diet-induced obesity, and a tissue-specific mitochondrial metabolic phenotype (most evident in skeletal muscle and liver). PARP-2 is the closest paralog of PARP-1 and is responsible for most of the residual PARP activity in PARP-1−/− cells. In C2C12 myocytes, RNAi-mediated knockdown of PARP-2 increases the levels of SIRT1 mRNA, protein, and activity without altering cellular NAD+ levels, suggesting a role for PARP-2 in the modulation of SIRT1 function. The increases in SIRT1 activity upon PARP-2 depletion lead to deacetylation of downstream SIRT1 targets, as well as increased mitochondrial biogenesis and oxidation. These outcomes may be due to a direct effect of PARP-2 on the SIRT1 gene; PARP-2 localizes to the SIRT1 promoter, where it functions as a negative transcriptional regulator (Bai et al. 2011a). Together, the results from these metabolic studies link PARP-1 and PARP-2 to known metabolic transcriptional regulators, such as SIRT1, and physiological outcomes in response to metabolic stress. Since metabolic phenotypes are the outcome of the action of multiple organs and PARP-1 and PARP-2 mediate distinct responses in different organs, it will be essential to develop tissue-specific conditional knockout mice to pinpoint and distinguish the functions of PARP-1 and PARP-2 in specific metabolic processes, such as adipogenesis, inflammation, lipid metabolism, glucose homeostasis, and insulin production and resistance.
Regulating the regulator: activation and deactivation of PARP-1 in response to stress signals
As might be expected for a central component of stress pathways, PARP-1 is subject to a variety of inputs that affect its stability, activity, and even localization (Fig. 3). Historically, the focus has been on effectors that alter PARP-1's NAD+-dependent catalytic activity, such as damaged DNA (i.e., single- or double-strand breaks) (D'Amours et al. 1999). More recently, studies have shown that PARP-1 can also be activated in the absence of DNA damage by (1) binding to other DNA structures (e.g., cruciforms), nucleosomes, or protein partners; (2) changing the level of metabolites or ions; and (3) post-translational modifications (Kim et al. 2005; Krishnakumar and Kraus 2010a).
Regulating PARP-1 through chromatin interactions
PARP-1-mediated modulation of chromatin structure is essential for both transcriptional regulation and DNA damage repair (Krishnakumar and Kraus 2010a). PARP-1 interacts dynamically with different chromatin domains, and its interactions with chromatin are determined by the specific nature of the nucleosomes (e.g., types of histone modifications and inclusion of histone variants). For example, in Drosophila, the histone variant H2Av (a homolog of both mammalian H2Az and H2Ax) promotes the localization of PARP-1 to H2Av-enriched nucleosomes at specific target gene promoters (Kotova et al. 2011). The phosphorylation of H2Av enhances PARP-1 activation, as well as downstream heat-shock-induced transcriptional activation and genotoxic stress-induced DNA repair (Kotova et al. 2011). In vitro and cell-based studies suggest that the replacement of H2A with H2Av changes the conformation of the core nucleosomes and facilitates the binding of PARP-1 to H3 and H4, thus directing the localization of PARP-1. Subsequent developmental signals, heat shock, or genotoxic stress induce the phosphorylation of H2Av, further exposing the H4 N-terminal tail to activate PARP-1 catalytic activity (Pinnola et al. 2007; Kotova et al. 2011). The upstream signals mediating the replacement of H2A and phosphorylation of H2Av, as well as the exact mechanism of H4-mediated activation of PARP-1, are unknown. As described in more detail below, PARP-1 also interacts with the histone variant macroH2A in a PAR-dependent manner, which facilitates the localization of PARP-1 to specific regions of damaged DNA in the genome (Kraus 2009). Understanding the cooperative effects of damaged DNA, linker DNA, core nucleosome structure, histone modifications, and histone variants will shed light on the regulation of PARP-1-mediated chromatin decondensation in both transcriptional regulation and DNA damage responses.
Regulating PARP-1 through post-translational modifications
Post-translational modifications of PARP-1 have received considerable attention in the past few years. Most of the focus has been on how PARP-1 is covalently modified. Considerably less is known about the enzymes that control the removal of the post-translational modifications of PARP-1, and more work is needed in this area. In the space below, we review the recent literature related to the modulation of PARP-1 activity and function through post-translational modifications (Fig. 1B).
ADP-ribosylation
Auto-ADP-ribosylation of PARP-1, especially extensive autopoly(ADP-ribosyl)ation in response to DNA damage, was the original post-translational modification observed for PARP-1 (D'Amours et al. 1999). Subsequent studies have shown that other PARPs, such as PARP-2, are also automodified and, by using mutagenesis (i.e., Lys to Arg alteration), have identified potential sites of automodification: Lys 498, Lys 521, and Lys 524 in PARP-1 (Fig. 1B; Altmeyer et al. 2009), and Lys 36 and Lys 37 of PARP-2 (Haenni et al. 2008). In these cases of automodification, PARP-1 mediates oligo- and poly(ADP-ribosyl)ation, while PARP-2 mediates predominantly mono(ADP-ribosyl)ation reactions (Haenni et al. 2008; Altmeyer et al. 2009). However, mutations of specific amino acids could reduce automodification without necessarily being sites for covalent attachment of ADP-ribose. As such, the field should focus on the identification of ADP-ribose adducts on target residues in vivo by mass spectrometry to conclusively address this issue. These automodification events go beyond responses to DNA damage. For example, automodification of PARP-1 plays a key role in its function as a regulated promoter-specific exchange factor required for the activation of specific neurogenic gene programs (Ju et al. 2004). Many more examples similar to this are likely to exist, placing PARP-1 firmly at the center of a variety of physiological and pathophysiological responses.
Beyond automodification, more recent studies have shown that other PARP family members can trans-modify PARP-1 and vice versa. For example, PARP-1 and PARP-2 can heterodimerize and ADP-ribosylate each other, which may play a role in mediating efficient base excision DNA repair (Schreiber et al. 2002; Menissier de Murcia et al. 2003). How the heterodimerization and cross-modification occurs, as well as their exact function in vivo, are largely unclear. PARP-3, which can interact with PARP-1, not only plays an important role in cellular response to DNA damage (Boehler et al. 2011), but can also activate the enzymatic activity of PARP-1 in the absence of DNA, although it is unclear what role the mono(ADP-ribosyl) transferase activity of PARP-3 plays in this process (Loseva et al. 2010). Removal of ADP-ribose units from PARP-1 and other proteins is mediated by PARG (Min and Wang 2009) and ARH3 (Oka et al. 2006), which promote rapid degradation and turnover of PAR polymers. The cross-talk among different PARP members suggests specific roles for individual PARP family members that go beyond simple redundancy and compensatory functions. In fact, the proliferation of PARP family members in vertebrates (e.g., two in flies and 17 in mammals) suggests an evolutionary trend toward specialized functions. The expression and localization patterns of the PARP family members ultimately determine the ways in which they interact and function.
Interestingly, the cross-modification also occurs between PARP-1 and another NAD+ consumer, SIRT6, which is a sirtuin family member that possesses both deacetylase and mono(ADP-ribosyl) transferase activities (Mao et al. 2011). Under oxidative stress, SIRT6 localizes to double-strand break sites in DNA and mono(ADP-ribosyl)ates PARP-1 at Lys 521 (Fig. 1B). This modification activates PARP-1 and subsequent PARP-1-dependent DNA repair pathways, further enhancing double-strand break repair under oxidative stress. Point mutations that eliminate both the deacetylation and mono(ADP-ribosyl)ation activities of SIRT6 abolish the function of SIRT6 in DNA damage repair (Mao et al. 2011). These interactions provide a means by which PARP-1 and SIRT6 can integrate stress signaling pathways with DNA repair. In fact, accumulating evidence points to overlapping functions of PARP-1 and SIRT6 in stress signaling, including roles in genome stability, NF-κB-mediated stress signaling, and metabolism (Tennen and Chua 2011).
Phosphorylation
A recent broad proteomic screen has identified a variety of sites on PARP-1, as well as PARG, that are phosphorylated (Gagne et al. 2009). These have been investigated in more detail in functional studies. PARP-1 is phosphorylated by ERK1/2 at Ser 372 and Thr 373 (Fig. 1B), and these modifications are required for maximal PARP-1 activation after DNA damage (Kauppinen et al. 2006). PARP-1 enzymatic activity can also be stimulated through interactions with phosphorylated ERK2, which in turn enhances ERK-mediated phosphorylation of the transcription factor Elk1 and the expression of genes involved in cell proliferation and differentiation (Cohen-Armon et al. 2007). PARP-1 is phosphorylated by JNK1 at undetermined sites, promoting sustained PARP-1 activation during H2O2-induced nonapoptotic cell death (Zhang et al. 2007). Phosphorylation of PARP-1, however, does not always promote an increase in PARP-1 activity. Earlier reports based on biochemical (Bauer et al. 1992) and cell-based (Beckert et al. 2006) assays have suggested that phosphorylation of PARP-1 mediated by protein kinase C results in decreased PARP-1 DNA-binding and catalytic activity. More studies are needed to fully explore the role of other kinases, as well as phosphatases, as upstream modulators of PARP-1 in stress responses. Activated PARP-1 is, in turn, likely to provide feedback signals by interacting with and regulating the kinases or phosphatases.
Acetylation
PARP-1 and PARP-2 are modified by acetylation, and evidence is accumulating for the specific roles for these acetylation events in stress-related responses. Acetylation of PARP-1 by the acetyltransferase p300/CBP was first identified in macrophages in the context of NF-κB-dependent transcriptional programs (Hassa et al. 2003, 2005). Upon proinflammatory stimuli, PARP-1 is acetylated by p300/CBP at Lys 498, Lys 505, Lys 508, Lys 521, and Lys 524 (Fig. 1B; Hassa et al. 2005). These acetylation events are required for interactions and functional cooperativity between PARP-1, the NF-κB subunit p50, and the Mediator complex. In this context, as well as in the context of retinoic acid receptor-mediated transcription, PARP-1 functions as an acetylation-activated coactivator independent of PARP catalytic activity (Hassa et al. 2005; Pavri et al. 2005). Another study using cardiomyocytes showed that PARP-1 is acetylated by p300/CBP and PCAF as an endpoint of stress responses (e.g., mechanical stretch or hypertrophy), resulting in the DNA damage-independent activation of PARP-1 enzymatic activity (Rajamohan et al. 2009). Like PARP-1, PARP-2 is also acetylated, which reduces its DNA-binding and enzymatic activities (Haenni et al. 2008). Interestingly, acetylation of PARP-2 occurs at the same lysine residues that are automono(ADP-ribosyl)ated (Lys 36 and Lys 37), which is likely to reduce the extent of automono(ADP-ribosyl)ation (Haenni et al. 2008).
In both macrophages and cardiomyocytes, deacetylation of PARP-1 by HDAC1, HDAC2, and HDAC3 or SIRT1 can reverse the effects of acetylation by deactivating PARP-1 (Fig. 1B; Hassa et al. 2005; Rajamohan et al. 2009). Furthermore, SIRT1 can also antagonize the action of PARP-1 through transcriptional repression of the PARP-1 gene. As such, overexpression of SIRT1 protects cells from PARP-1-dependent cell death under extensive stress stimuli (Rajamohan et al. 2009). Cellular NAD+ levels may mediate the functional interplay between PARP-1 and SIRT1 and determine the final cell fate decision. Further understanding of how the levels of NAD+ are regulated under different stress conditions and how NAD+ controls the antagonistic cross-talk between PARP-1 and SIRT1 is needed, however.
Ubiquitylation and SUMOylation
Recent studies have begun to elucidate roles for the polypeptide protein modifiers ubiquitin and SUMO (small ubiquitin-like modifier), as well as the E3 ligases that promote their covalent attachment, in the stress-related function of PARP-1. For example, the recently identified ubiquitin E3 ligase RNF146 (Iduna) protects primary cortical neurons from glutamate excitotoxicity and shock by interfering with PARP-1-dependent, PAR-induced apoptosis-inducing factor (AIF) translocation from mitochondria to the cytosol and nucleus, as well as subsequent PAR-dependent cell death (parthanatos [PAR + “thanatos,” meaning death]) (Andrabi et al. 2011; Kang et al. 2011). AIF is a mitochondrial flavoprotein with NADH oxidase activity. Extramitochondrial (ectopic) AIF promotes cell death, whereas mitochondrion-localized (eutopic) AIF does not (Cregan et al. 2004). Iduna interacts with a number of proteins that are either PARylated or bound noncovalently to PAR. The interaction of Iduna with PAR activates Iduna's E3 ubiquitin ligase activity and targets the PAR-bound or PAR-modified proteins for proteasome-dependent degradation (Andrabi et al. 2011; Kang et al. 2011). The outcome is an inhibition of excessive DNA damage responses and parthanatos that would result from sustained activation of PARP-1 (Kang et al. 2011). Moreover, Iduna may facilitate DNA repair by regulating the dynamic assembly and disassembly of PARP-1-associated DNA repair machinery at DNA lesion sites. The regulatory functions of RNF146/Iduna are not limited solely to PARP-1 targets, but may also involve tankyrase (PARP-5a/5b)-PARylated protein targets (Zhang et al. 2011). AutoPARylation of tankyrase and transPARylation of axin activate RNF146-mediated ubiquitylation and proteasomal degradation of tankyrase and axin, promoting downstream Wnt signaling (Huang et al. 2009; Callow et al. 2011). Tankyrase also PARylates and promotes the degradation of 3BP2, a component of a multiprotein signaling complex, through RNF146-mediated ubiquitylation. Interestingly, mutations in Sh3bp2, the gene encoding 3BP2, uncouple 3BP2 from tankyrase-mediated degradation, resulting in (1) the stabilization of 3BP2, (2) the subsequent hyperactivation of cellular signaling pathways, and (3) cherubism, a syndrome characterized by inflammatory destructive lesions, resulting in symmetrical deformities of the facial bones (Guettler et al. 2011; Levaot et al. 2011). The intriguing link between PARP-mediated PARylation and E3 ligase-dependent ubiquitylation is likely to be a key component in the function of PARP family members in various other physiological and pathological processes.
SUMOylation of PARP-1 mediated by the SUMO E3 ligase PIASγ facilitates PARP-1-dependent outcomes, including responses to thermal (heat shock) (Martin et al. 2009) and genotoxic (Martin et al. 2009; Stilmann et al. 2009) stresses. In HeLa cells, in response to heat shock, PARP-1 recruits and PARylates PIASγ, which in turn SUMOylates PARP-1 (Martin et al. 2009). These events are required for full heat-shock-dependent activation of the HSP70.1 gene. Mutagenesis experiments suggest that Lys 203 and Lys 486 are SUMO acceptor sites on PARP-1 (Martin et al. 2009); these sites are distinct from the automodification and acetylation sites noted above (Fig. 1B). The cascade of modification events does not end there: SUMOylated PARP-1 is bound by and ubiquitylated by the ubiquitin E3 ligase RNF4, which leads to the clearance and recycling of PARP-1 from the promoters of target genes. This cascade of PARylation, SUMOylation, and ubiquitylation promotes rapid and transient heat-shock stress responses (Martin et al. 2009). Similar players also have a key role in modulating IκB kinase (IKK)- and NF-κB-dependent apoptotic responses upon genotoxic stress (Stilmann et al. 2009). In this pathway, PARP-1 detects DNA strand breaks and promotes the assembly of a PAR-dependent “signalosome” containing IKKγ, PIASγ, and the ataxia telangiectasia-mutated (ATM) protein, a DNA damage response kinase. PAR binding by PIASγ triggers IKKγ SUMOylation, which in turn permits IKKγ and NF-κB activation, as well as NF-κB-regulated resistance to apoptosis (Stilmann et al. 2009). Given the key roles played by ubiquitylation, SUMOylation, and PARP family members in cellular stress responses, more links are likely to be revealed between PARPs and the large pool of ubiquitin and SUMO E3 ligases under diverse physiological and pathological conditions.
Multitasking as a stress modulator: role of PARP-1 in the propagation of stress signals and promoting downstream effects
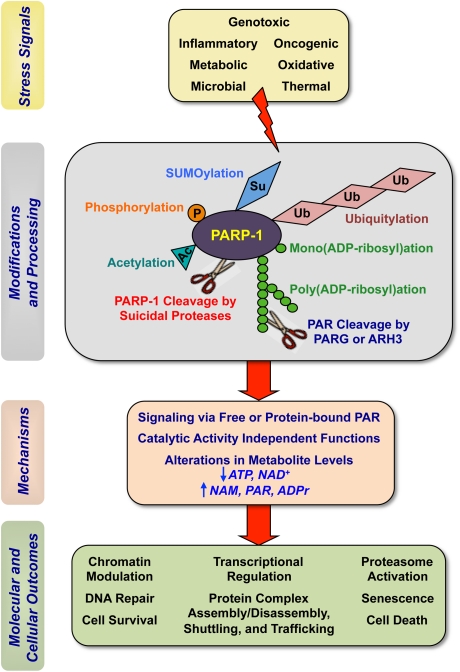
The signaling inputs to PARP-1 described above lead to a variety of downstream molecular effects that underlie biological responses (Fig. 4). These include (1) modulation of chromatin structure, transcription, and DNA repair; (2) alteration of cellular metabolic profiles through the consumption of NAD+ and the production of PAR and nicotinamide; and (3) alteration of the cellular “PARylome,” either through the covalent modification of target proteins or by ultimately changing the levels of free PAR (Fig. 3). The latter may have specific effects on (1) chromatin remodeling; (2) the recruitment of the DNA repair machinery; (3) the exchange of transcriptional regulatory complexes; (4) the redistribution of signal transduction machinery to different cellular compartments; (5) the localization, structure, and function of key stress regulatory proteins; and (6) protein stability through regulation of proteasome function (Fig. 5). In the section below, we highlight some of the most recent advances in our understanding of how PARP-1 functions to propagate stress signals and elicit downstream effects.
Figure 4.
PARP-1 modulates the molecular biology and biochemistry of stress responses at multiple levels. PARP-1 modulates cellular stress responses through a series of regulatory processes that occur at the genomic, transcriptional, post-transcriptional, translational, and post-translational levels.
Figure 5.
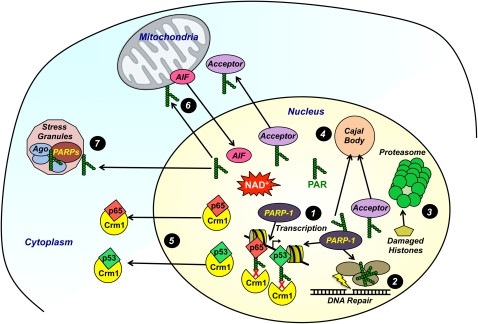
Molecular and cellular actions of PARP-1 and related PARPs. Key molecular and cellular actions of PARP-1 and related PARPs are illustrated: (1) modulating chromatin structure and regulating gene transcription, (2) facilitating the assembly and function of the DNA repair machinery, (3) activating proteasomes to remove damaged histones, (4) facilitating protein transport into Cajal bodies, (5) preventing the nuclear export of p65 and p53 through a Crm1-dependent mechanism, (6) promoting the release of AIF from mitochondria and transport to nucleus, and (7) regulating the assembly and function of stress granules (PARP family members). Details are provided in the text.
PAR-mediated signaling in protein complex assembly/disassembly, shuttling, and trafficking
PARP-1 and PAR-mediated events can drive the exchange of proteins at specific loci in the genome, the relocalization of proteins to specific nuclear structures, and the cytoplasmic–nuclear or nuclear–cytoplasmic shuttling of proteins. For example, PARP-1 has been shown to function as a promoter-specific “exchange factor” that promotes the release of inhibitory coregulators and the recruitment of stimulatory coregulators during transcriptional regulation (Ju et al. 2004, 2006; Pavri et al. 2005; Stilmann et al. 2009). More recently, automodified PARP-1 has been shown to interact noncovalently through the PAR polymers with components of Cajal bodies, spherical structures found in the nucleus of proliferating cells that have been implicated in RNA-related metabolic processes, such as snRNP biogenesis, maturation, and recycling; histone mRNA processing; and telomere maintenance (Kotova et al. 2009). This association mediates the shuttling of the PARP-1- and PAR-containing complexes from chromatin into Cajal bodies, a process that can be reversed by the actions of PARG (Fig. 5, highlight 4; Kotova et al. 2009). In contrast, PARylation of high-mobility group box 1 (HMGB1), a chromatin-associated protein, by activated PARP-1 in response to genotoxic stress disassociates HMGB1 from condensed chromatin and ultimately drives the nuclear-to-cytosolic translocation of HMGB1, where it may leak out of necrotic cells (Ditsworth et al. 2007). Extracellular HMGB1 can serve as a proinflammatory signal, which can induce macrophage activation and TNFα production.
Recent studies have implicated PARP-1 in controlling the subcellular localization of the key nuclear stress response proteins NF-κB (Zerfaoui et al. 2010) and p53 (Kanai et al. 2007) via the nuclear export receptor Crm1. Inhibition or depletion of PARP-1 prevents the nuclear accumulation of the NF-κB p65 subunit in smooth muscle cells upon TLR4 (toll-like receptor 4) stimulation. PARP-1 deficiency does not alter the expression or cytosolic localization of critical components of the nuclear import machinery (e.g., importins α3 and α4), but does increase the cytosolic levels of Crm1, which is required for the nuclear export of NF-κB. In response to proinflammatory signals, PARP-1-dependent PARylation of p65 decreases its interactions with Crm1, allowing nuclear accumulation of NF-κB and NF-κB-dependent gene regulation programs (Fig. 5, highlights 1,5; Zerfaoui et al. 2010). These PARP-1 and Crm1-mediated events may play a broader role in the assembly of the PAR-dependent signalosome containing IKKγ, PIASγ, and ATM described above (Stilmann et al. 2009), underlying PARP-1's role as a signal generator and transducer for the “cytoplasmic-to-nuclear” mode of activation of NF-κB. As with NF-κB, PARylation of p53 by PARP-1 in response to DNA damage blocks the interaction of p53 with Crm1, resulting in the nuclear accumulation of p53 (Fig. 5, highlights 1,5; Kanai et al. 2007). Thus, a model is emerging where PARylation of stress-responsive transcription factors blocks targeting by the nuclear export machinery, promoting accumulation of the transcription factors in the nucleus, where they can mediate their specific gene regulatory programs.
PARP-1 also plays a key role in a critical mitochondria-to-nuclear shuttling mechanism that triggers caspase-independent apoptotic cell death through AIF. AIF is a proapoptotic flavoprotein with NADH oxidase activity that resides in the mitochondrial intermembrane space and is a powerful trigger of apoptosis (Cregan et al. 2004). In the nucleus, AIF induces the peripheral chromatin condensation and high-molecular-weight DNA fragmentation that is a hallmark of cellular apoptotic programs (Cregan et al. 2004). PARP-1 activation in response to excessive DNA damage triggers the release of AIF from mitochondria and promotes PARP-1-dependent cell death or parthanatos (Fig. 5, highlight 6; Yu et al. 2002; David et al. 2009; Wang et al. 2011). AIF is a high-affinity PAR-binding protein. PAR binding by AIF is required for its release from the mitochondria, translocation to the nucleus, and cell death, but not for its NADH oxidase activity, its ability to bind flavin adenine dinucleotide or DNA, or its ability to induce chromatin condensation (Wang et al. 2011). Thus, AIF's bioenergetic cell survival-promoting functions are distinct from its role as a mitochondrially derived death effector in PARP-1-dependent cell death programs (Wang et al. 2011).
Signaling to chromatin through PAR
PAR production by PARP-1 can signal diverse stress stimuli to chromatin to regulate the downstream transcriptional program, modulate the action of DNA-repairing machinery, and facilitate the cooperation of transcription and DNA repair.
Transcriptional regulation
PARP-1 and PARG modulate global levels of gene expression in part through mechanisms that require the synthesis and degradation of PAR at target gene promoters through their respective enzymatic activities (Fig. 5, highlight 1; Krishnakumar et al. 2008; Frizzell et al. 2009). One target of PARP-1 in mammalian cells is the histone demethylase KDM5B (a.k.a. JARID1B). PARP-1-dependent PARylation of KDM5B helps to maintain the levels of H3K4me3, a histone mark associated with active promoters, by inhibiting KDM5B interactions with chromatin, as well as its enzymatic activity. This facilitates the recruitment of transcriptional activators and chromatin remodelers to keep an open promoter chromatin architecture conducive to RNA polymerase II binding and transcription (Krishnakumar and Kraus 2010b).
Recent studies in Drosophila embryos and cells have shown that dPARP (the Drosophila homolog of PARP-1) plays a key role in chromatin decondensation in response to heat shock (Tulin and Spradling 2003; Petesch and Lis 2008). dPARP and its enzymatic activity are essential for “puff” formation (i.e., chromatin decondensation) on salivary gland polytene chromosomes upon heat shock (Tulin and Spradling 2003). Upon heat shock, dPARP is activated at heat-shock-responsive loci, promotes nucleosome loss and chromatin decondensation in a PAR-dependent manner, and facilitates transcription by RNA polymerase II (Tulin and Spradling 2003; Petesch and Lis 2008, 2012). PARylation of the ATP-dependent nucleosome remodeler ISWI by dPARP inhibits its nucleosome binding, ATPase, and chromatin condensation activities at heat-shock loci, contributing to the chromatin decondensation observed upon heat shock (Sala et al. 2008). Heat-shock factor (HSF) plays a key role in regulating dPARP activity in the heat-shock response; upon heat shock, HSF stimulates the histone acetyltransferase dTIP60, which acetylates H2A at Lys 5, resulting in dPARP activation and release into the gene body (Petesch and Lis 2012). Furthermore, live-cell imaging and fluorescence recovery after photobleaching (FRAP) analyses in heat-shocked Drosophila salivary gland nuclei have captured the progressive recruitment of HSF, RNA polymerase II, and transcription elongation factors to the heat-shock loci to form a so-called “transcription compartment” at the Hsp70 locus. dPARP activity is essential for the maintenance of this transcription compartment (Zobeck et al. 2010). The PAR-dependent assembly of the transcription compartment provides new perspectives to PARP's function in chromatin-dependent transcriptional regulation. However, the upstream signal that mediates the redistribution and activation of dPARP, how dPARP facilitates the assembly and disassembly of the transcription compartment, and how widely applicable this model is to other stress responses and biological systems are unclear.
DNA repair
The binding of PARP-1 to damaged DNA activates its catalytic activity and stimulates the production of negatively charged PAR polymers at or adjacent to the lesion sites. The PAR polymers, covalently linked to PARP-1 itself or other chromatin-associated proteins, serve as a signal and a platform for the recruitment of proteins associated with the DNA damage response that promote chromatin remodeling, transcriptional regulation, DNA repair, cell cycle arrest, senescence, or cell death (Fig. 5, highlight 2). A series of recent studies has explored the functional connection between the PAR signal, chromatin remodeling, and DNA damage responses involving the macrodomain-containing proteins ALC1 (amplified in liver cancer 1, also known as CHD1L) (Ahel et al. 2008; Gottschalk et al. 2009) and macroH2A1.1 (a histone H2A variant) (Timinszky et al. 2009). ALC1, a SNF2 family ATPase nucleosome remodeling enzyme, binds to PAR via its macrodomain. This interaction facilitates the recruitment of ALC1 to sites of DNA damage and ALC1-dependent nucleosome remodeling, which may promote chromatin remodeling at sites of DNA repair (Ahel et al. 2008; Gottschalk et al. 2009). Likewise, macroH2A1.1 localizes to sites of PAR formation in the nuclei and promotes a transient compaction of chromatin that may play a role in regulating DNA repair responses (Timinszky et al. 2009). PBZFs have been identified in other nuclear proteins involved in DNA repair and checkpoint control, including CHFR (checkpoint protein with FHA and RING domains) and APLF (aprataxin PNK-like factor). The binding of PAR by the PBZFs in these proteins plays key roles in their localization and function (Ahel et al. 2008; Rulten et al. 2008; Mehrotra et al. 2011).
Coordination of DNA repair and transcriptional regulation
PAR-mediated signaling may also play a role in modulating and coordinating DNA damage responses and transcriptional outcomes. For example, PAR promotes the recruitment of members of the polycomb group (PcG) of chromatin-modifying, transcriptional repressor proteins (e.g., EZH1, RING1, and PCGF4) and components of the repressive NuRD nucleosome remodeling and deacetylase complex (e.g., CHD4-associated MTA1) to sites of DNA damage (Chou et al. 2010). PARP activity facilitates the removal of nascent RNA and elongating RNA polymerase at these sites to block transcription and presumably facilitate DNA repair. Extensive or persistent DNA damage causes constitutive redistribution of these transcriptional regulatory complexes from their original loci, leading to cell cycle arrest or senescence (Chou et al. 2010). The mode of recruitment by PAR and the specific PARP(s) that mediate the PcG- and NuRD-mediated responses still need to be determined.
Further connections between PARP activity, the chromatin remodeling machinery, cellular stress, and downstream transcriptional responses are illustrated in the adult heart, where stress triggers hypertrophy and a shift from the expression of adult α-myosin heavy chain (α-MHC) to fetal β-MHC. PARP-1 cooperates with the nucleosome remodeling enzyme Brg1 and a number of HDACs to drive the pathological switch in the expression pattern, which can lead to cardiac hypotrophy and failure (Hang et al. 2010). Brg1, PARP-1, and HDACs (e.g., HDAC2 and HDAC9) bind to and repress the α-MHC promoter, whereas Brg1 and PARP-1 bind to and activate the β-MHC promoter, both of which require PARP activity. Collectively, the examples cited here illustrate how PAR production by PARP-1 can drive chromatin remodeling events as an endpoint of cellular stress signals to coordinate DNA repair and transcriptional responses.
Signaling to stress granules through PAR
A recent study has extended the role of PAR to the post-transcriptional regulation of gene expression through the assembly and disassembly of stress granules (Leung et al. 2011). Stress granules are cytoplasmic structures formed when cells are stressed. They contain stalled translation preinitiation complexes and regulate the translation and stability of mRNAs by coordinating the action of the regulatory RNA-binding proteins, such as Argonaute (Ago) family members (Buchan and Parker 2009). Using a screening approach with GFP-tagged proteins and immunostaining, Leung et al. (2011) found that six PARP family members and two PARG isoforms localize to stress granules and help to modulate the kinetics of stress granule assembly and disassembly by modulating the local PAR levels. A number of stress granule proteins, including microRNA-binding Ago1, Ago2, Ago3, and Ago4, are PARylated in response to cellular stress under conditions where both microRNA-mediated translational repression and microRNA-directed mRNA cleavage are inhibited (Fig. 5, highlight 7; Leung et al. 2011). Overexpression of PARP-13 (a putative catalytically inactive PARP family member) or depletion of PARG relieves microRNA-mediated repression through a mechanism that involves PARylation of PARP-13 by another stress granule-associated PARP family member. Thus, PAR plays a role in post-transcriptional gene regulation by modulating stress granule function.
These exciting observations not only further expand the role of PARPs in stress responses, they also provide a platform for understanding how different PARP family members coordinate their activities through automodifications and transmodifications, and how PARG functions to fine-tune the PAR signal. The exact mechanisms of the PAR-dependent regulation, however, have not yet been fully addressed. One possibility is that PAR focuses the stress signals in the stress granules by modulating target RNA-binding proteins to bind mRNA or microRNA. Although PARP-1 is not directly localized in the stress granule, as the most abundant stress-detecting PARP in the cell, we cannot rule out the possibility that PARP-1 senses stress signals and transduces those signals to the cytoplasm by trans-modifying other proteins or generating free PAR. These studies raise the interesting possibility that PARPs may regulate the function of other noncoding RNAs. PARylation of the RNA-binding Ago proteins and relief of translational repression are reminiscent of the binding of hnRNPs to PAR and its effects on alternative splicing. In Drosophila, two hnRNPs, Hrb98DE/hrp38 and Squid/hrp40, bind PAR in a manner that is regulated by dPARP and dPARG in vivo. PAR binding by hrp38 and hrp40 inhibits their RNA-binding activities and alters their splicing activities (Ji and Tulin 2009). These studies highlight the expanding role for PARPs and PAR in the post-transcriptional control of gene expression.
Signaling to the proteasome through PAR
Metabolic changes during cellular stress responses and aging promote the accumulation of oxidative damage. As noted above, PARP-1 helps to re-establish cellular homeostasis in the face of oxidative damage by promoting DNA repair. However, PARP-1's role in oxidative damage responses goes beyond that, including the removal of oxidized nuclear proteins (e.g., histones). The activation of PARP-1 in response to oxidative damage (e.g., treatment with hydrogen peroxide in a cell culture model) promotes interactions with, PARylation of, and enhanced activity of the 20S proteasome (Fig. 5, highlight 3; Ullrich et al. 1999; Catalgol et al. 2010). Inhibition of PARP activity leads to an increase in the protein carbonyl content in cells (a marker of protein oxidation), as well as an increase in oxidized DNA. These results implicate PARP-1 in the proteasome-dependent removal of oxidation-damaged proteins in the nucleus. This conclusion is supported by the fact that decreasing PARP-1 activity during aging correlates with the decline of nuclear proteasome activity and accumulation of oxidized nuclear proteins (Bakondi et al. 2010). Taken together, these results support a broader role for PARP-1 as a guard against oxidative damage to both proteins and DNA.
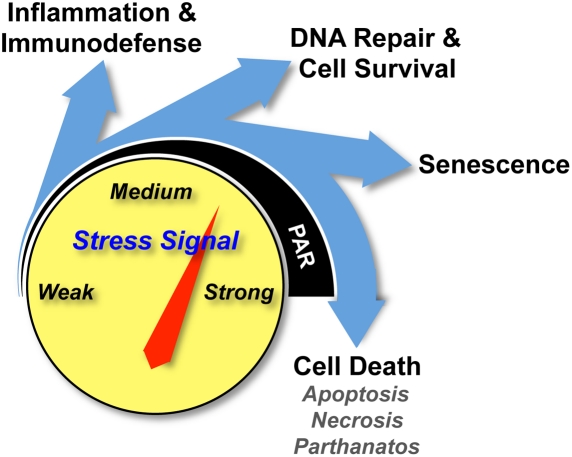
PARP-1 as a cellular rheostat: different responses for different types and levels of stress
The specific role of PARP-1 in deciding cell fate (i.e., survival or death) depends on the type, duration, and strength of the stress stimuli, as well as the metabolic and proliferative status of the cell. A large body of data has now shown that PARP-1 activation by mild or moderate stresses leads to transcription and DNA repair responses that help to maintain genome stability, promote transient innate immune and proinflammatory responses, clear and remove oxidized or damaged proteins, and re-establish homeostasis (Kim et al. 2005; Gagne et al. 2006; Hassa and Hottiger 2008). In contrast, moderate, severe, or sustained stresses cause hyperactivation of PARP-1 and graded responses that correlate with the extent of PARP-1 activation (Fig. 6). These include senescence (moderate response) or distinct cell death programs (severe responses), such as apoptosis, necrosis, and parthanatos (Koh et al. 2005; David et al. 2009). Apoptosis is programmed cell death that generally does not promote inflammation. Necrosis is cell death resulting from physical, chemical, or metabolic insults (e.g., the depletion of NAD+ and ATP pools) (Koh et al. 2005). Parthanatos (PAR-dependent cell death) was recently identified as a distinct entity, which presents a distinct cellular morphological pattern, but resembles aspects of apoptosis (e.g., DNA fragmentation and nuclear condensation) and necrosis (e.g., disrupted cell membrane and energy crisis) (David et al. 2009). A critical step of this cell death program is the PAR-induced translocation of AIF from the mitochondria to the nucleus, as mentioned above. PARP-1 has been shown to play a role in all three of these cell death programs (Fig. 6; Koh et al. 2005; David et al. 2009).
Figure 6.
PARP-1 functions as a cellular rheostat. PARP-1 promotes different cellular responses to different types and levels of stress signals. As the strength of stress stimulus increases, the levels of PARP-1 activity and PAR synthesis increase, leading to different cellular outcomes.
Activated PARP-1 helps initiate the apoptotic program by stabilizing the p53 tumor suppressor protein and facilitating its function (Wesierska-Gadek et al. 2005). Subsequently, activated caspases cleave PARP-1 into a 24-kDa fragment and a 89-kDa fragment, preventing the depletion of NAD+ (and, subsequently, ATP, a precursor of NAD+) by overactivated PARP-1 and circumventing necrotic cell death (Simbulan-Rosenthal et al. 1998). In contrast, when hyperactivated PARP-1 depletes cellular NAD+ and ATP, the cells will be directed away from an apoptotic pathway and undergo energy crisis-induced necrosis (Ha and Snyder 1999) or toxic PAR polymer-induced parthanatos (David et al. 2009). Although PARP-1 activation is required for the initiation of the above cell death programs, PARP-1 may be cleaved and inactivated by various proteases, including caspases, calpain, cathepsins, and granzymes (Chaitanya et al. 2010). The involvement of activated PARP-1, cleaved fragments of PARP-1, toxic PAR polymers, and survival or apoptotic factors such as NF-κB and p53 in the various cell death programs requires further investigation. Such information will be critical to understand the application of PARP inhibitors as a monotherapy or to enhance chemotherapy or radiotherapy in different diseases and cancer types.
Two sides of the same coin: contrasting roles of PARP-1 in genome surveillance, carcinogenesis, metabolic control, and aging
The versatile cellular functions of PARP-1 are reflected in a broad role for PARP-1 in diverse physiological and pathological processes at the organismal level. Although the original studies of genetic ablation of PARP-1 in mice showed an unremarkable phenotype (Wang et al. 1995), more specific and focused analyses, especially in response to various stresses, have revealed key roles for PARP-1 in genome surveillance, carcinogenesis, metabolic control, and aging (Shall and de Murcia 2000; Kim et al. 2005; Luo and Kraus 2011). Genetically engineered mice have proven to be some of the best tools to study the integrated functions of PARP-1 in multiple organs. A growing set of published studies with PARP-1-deficient mice have revealed increased genome instability and sensitivity to genome toxic stress, a shorter life span with accelerated aging, earlier incidence of a wide spectrum of spontaneous tumors, and higher rates of malignant tumors in the liver, uterus, and lung (Shall and de Murcia 2000; Masutani et al. 2005; Piskunova et al. 2008). These results highlight the anti-cancer and anti-aging effects of PARP-1 in these specific biological models.
In contrast, in some contexts, PARP-1 can also be considered an aging and tumor promoter, especially with respect to its cooperative action with the tumor-promoting and inflammatory effector NF-κB (Ohanna et al. 2011) and its antagonistic effects on the longevity factor and metabolic regulator SIRT1, an NAD+-dependent protein deacetylase (Rajamohan et al. 2009; Bai et al. 2011b). PARP-1 helps to propagate NFκB-mediated inflammatory responses and the secretion of protumor and prometastasis cytokines (Ohanna et al. 2011). Furthermore, hyperactivation of PARP-1 can antagonize SIRT1 functions by competing for the cellular NAD+ pool, as well as direct antagonism of SIRT1 through mutual transmodification, as described above (Rajamohan et al. 2009; Bai et al. 2011b). Thus, optimal PARP-1 expression and activity are essential to maintain a physiological, rather than pathological, level of genome surveillance, immune defense, and cellular metabolism.
Most of the previous mouse genetic studies have used PARP-1 (or PARP-2) ablation, but a recent study has shown how excess PARP-1 can lead to deleterious outcomes by examining a mouse carrying two ectopic copies of the human PARP-1 gene in addition to the two endogenous copies of the mouse PARP-1 gene (Mangerich et al. 2010). These mice exhibit impaired DNA repair, dysregulated proinflammatory cytokines, premature development of chronic inflammation and aging-related diseases, and an altered spectrum of tumor formation with a lower incidence of sarcomas and an increased incidence of carcinomas (Mangerich et al. 2010). Thus, as these studies collectively illustrate, a finely tuned level of PARP-1 expression and activity is critical for the maintenance of a healthy physiological state.
Therapeutic applications of PARP inhibitors
A priori, PARP-1 would seem to be an excellent target for small molecular therapeutics: (1) PARP-1 is not essential for life, but plays important roles in a broad array of physiological processes; (2) modulating the levels of PARP-1 protein and activity can tune physiological states in one direction or the other without necessarily blocking them completely; (3) there is a wealth of biochemical, molecular, and structural information available for PARP-1; and (4) PARP-1 has a well-defined enzymatic activity that is dependent on a metabolic cofactor. Indeed, PARP-1 and other PARP family members have proven to be effectively inhibited by a broad array of small molecules (Rouleau et al. 2010). The therapeutic application of PARP inhibitors has received a considerable amount of attention recently because of their potential utility in the treatment of cancers, but the possible therapeutic applications of PARP inhibitors extend far beyond cancer therapy to other types of stress-related diseases, such as cardiovascular diseases, stroke, metabolic disorders, diabetes, and autoimmunity, and virtually any disease or condition with acute or chronic inflammation as a root cause (Masutani et al. 2005; Mota et al. 2005; Pacher and Szabo 2007; Shevalye et al. 2010; Ford and Lee 2011; Underhill et al. 2011).
PARP inhibitors and the treatment of cancers
Most clinical trials of PARP inhibitors today are focused on cancer therapy. Genotoxic stresses are central to the etiology and progression of cancer, and as such, PARP-1 plays a critical role in determining tumor cell progression and the outcome of cancer therapy. The rationale of the therapeutic approach is to selectively induce synthetic lethality of homologous repair-deficient (e.g., BRCA1- or BRCA2-deficient) tumors by blocking PARP-1-dependent base excision repair pathways with the inhibitors (Bryant et al. 2005; Farmer et al. 2005; Fong et al. 2009). Additionally, recent studies have shown that the therapeutic effects of PARP inhibitors occur in homologous repair-sufficient tumors as well by decreasing the apoptotic threshold in cotreatments with chemotherapies and radiotherapies, regulating the expression of tumor-related genes, suppressing angiogenesis, and altering gene expression programs (Frizzell and Kraus 2009; Rouleau et al. 2010; Telli 2011; Underhill et al. 2011). Increased knowledge about the molecular actions of PARPs and PARP inhibitors in cancer has led to a greater understanding of why PARP inhibitor therapy is effective. For example, PARP-1 promotes radio resistance mediated by an NF-κB-dependent survival pathway (Hunter et al. 2012). It also functions together with NF-κB in the formation of a senescence-associated inflammatory secretome, which propagates prometastatic and protumor effects, and causes resistance to chemotherapies (Fig. 7; Ohanna et al. 2011).
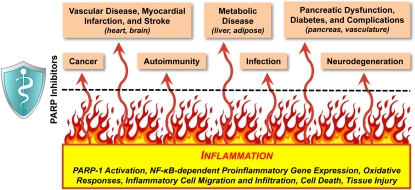
Figure 7.
Anti-inflammatory therapeutic effects of PARP inhibitors. Inflammation is a common thread underlying many diseases and conditions. The therapeutic effects of PARP inhibitors in various diseases may share a common mechanism; namely, the inhibition of NF-κB-dependent inflammatory pathways.
PARP inhibitors: beyond cancer, on to inflammation
PARP-1 serves as a global regulator of cell adaptation to a changing environment. In many disease states beyond cancers, extensive cellular stress triggers uncontrolled PARP-1 activation and subsequent inflammatory responses, cell death, and tissue damage. In this regard, PARP inhibitors have been explored as therapeutics to prevent cell death, tissue damage, and dysfunction associated with aging- or oxidative damage-related pathologies, such as cardiovascular diseases, autoimmune and inflammatory diseases, neurodegenerative diseases, stroke, and diabetes and its associated complications (Fig. 7; Masutani et al. 2005; Mota et al. 2005; Pacher and Szabo 2007; Shevalye et al. 2010; Ford and Lee 2011; Underhill et al. 2011). The recent discovery of the metabolic functions of PARP-1 and PARP-2, in concert with SIRT1, based on knockout mouse models suggests a therapeutic potential of PARP inhibitors in metabolic disorders (Bai et al. 2011a,b).
The common thread to many of the aforementioned diseases and conditions is an underlying inflammatory response (Fig. 7). Thus, the therapeutic effects of PARP inhibitors in these diseases may share a common mechanism; namely, the inhibition of inflammatory pathways (Fig. 7). Inflammation is now recognized as a hallmark of cancer (Hanahan and Weinberg 2011), and as such, the effects of PARP inhibitors on inflammatory responses may contribute to their therapeutic effects in cancers. Thus, knowledge of the regulation and function of PARP-1, as well as other PARP family members, in inflammatory pathways is critical to our ability to exploit PARP inhibitors as therapeutic agents.
Challenges and obstacles in the therapeutic application of PARP inhibitors
In spite of the promise for the therapeutic application of PARP inhibitors, a number of significant challenges and obstacles remain, which block the broad use of PARP inhibitors in cancers, as well as inflammatory and metabolic diseases. Key among these is the lack of specificity with respect to targeting both certain cell types (e.g., cancer cells versus normal cells) and specific PARP family members (e.g., even the most specific PARP inhibitors target multiple family members). A lack of specific tumor targeting can compromise genome surveillance, DNA repair, metabolic homeostasis, and immune defense functions in normal cells, potentially leading to the accumulation of additional genetic lesions and even secondary tumorigenesis. Thus, there is a need to protect against genome instability in normal tissues when using PARP inhibitors to target tumors or inflammatory diseases. The synthetic lethality approaches described above can be effective in cancers, but are dependent on a specific genetic background in the cancer cells (Bryant et al. 2005; Farmer et al. 2005; Brenner et al. 2011). Molecular profiling of tumors for specific DNA repair pathway deficiencies and the application of PARP inhibitors in combination with other inhibitors of DNA repair pathways may help. The lack of specific PARP family member targeting is complicated by the fact that we have a limited understanding of the function and cross-talk of all of the PARP family members. As such, the use of PARP inhibitors runs the risk of unintended effects and consequences mediated by the targeting of other PARP family members.
Remaining puzzles to be solved
As the studies highlighted in this review illustrate, much progress in our understanding of PARP-1 and PAR function in cellular stress responses has been made over the past two decades. Yet, many questions remain, especially with respect to the genomic localization of PARP-1, the role of PAR and other NAD+ metabolites, and the functions of other PARP family members. Some of the key remaining questions are summarized below.
PARP-1 localization and function?
PARP-1 localizes to the promoters of essentially all actively transcribed genes (and perhaps all open regions of chromatin) (Krishnakumar et al. 2008; Krishnakumar and Kraus 2010b), but affects the expression of only a subset of the genes to which it binds (Frizzell et al. 2009). Why is this so? Redundancy or compensation by other PARP family members can only partially explain this observation. Other possibilities include (1) the poising of inactive PARP-1 at promoters until an appropriate gene-specific signal is received and (2) a transcription-coupled DNA repair function of PARP-1 at promoters, which can help erase transcription-related DNA damage. Thus, investigating the upstream regulators, which dictate the specificity of PARP-1 activation, will be fundamental to address this issue. Additionally, global PARP-1-dependent transcriptome analysis and genome-wide localization studies of PARP-1 before and after stress responses will help to distinguish the genome surveillance and transcriptional functions under basal and stress-induced states. These analyses will also help to elucidate how PARP-1 mediates or coordinates responses to different types of stress stimuli.
PARylation dependence and dynamics?
As noted above, some functions of PARP-1 require its catalytic activity, while others do not. A better characterization and understanding of the catalytic-dependent and catalytic-independent functions of PARP-1 are needed. For those functions of PARP-1 that require its catalytic activity, the covalently modified (i.e., directly PARylated) targets must be distinguished from the noncovalent PAR-bound targets, and precisely how PAR affects the target proteins must be elucidated. Additional questions relate to the catabolism of PAR by PARG and ARH3. How are these enzymes brought to sites of PAR synthesis, how are their activities coordinated and regulated, and how do they regulate the half-life of PAR? Finally, questions remain about the effects of NAD+ consumption, as well as the role of NAD+ precursors (i.e., NMN and ATP) and PARP-1-dependent NAD+ metabolites (e.g., ADP-ribose and nicotinamide), in PARP-1-dependent stress signaling pathways.
PARP- and PAR-dependent processes in normal conditions?
As an increasing number of studies have shown, PAR can be accumulated in normal, steady-state, or basal conditions (e.g., during hormonal signaling, certain stages of development, circadian clock function, and mitosis) without triggering stress responses (Tulin and Spradling 2003; Bai et al. 2007; Chang et al. 2009; Asher et al. 2010; Lonn et al. 2010; Geistrikh et al. 2011; Rouleau et al. 2011; Yoo et al. 2011; Erener et al. 2012). How does the cell interpret PAR accumulation as a stress signal in one circumstance, but not another? A clear answer to this question has been elusive, but will inform us greatly about the biology of PARPs as well as the best ways to target them for therapeutic interventions. It will be informative to compare the PARP- and PAR-dependent processes in the same biological system under stress and nonstress (but perhaps stimulated) conditions to investigate whether the regulatory mechanisms and target molecules are shared or distinct. Only then can we begin to address this important question.
Answering the questions listed above and resolving these puzzles will help us to better understand the dynamic regulation and versatile functions of PARPs in stress responses, as well as guide the development and application of PARP inhibitors for different diseases.
Acknowledgments
We thank the members of the Kraus laboratory for critical comments on this review. X.L. is supported by a U.S. DOD Breast Cancer Research Program predoctoral fellowship. The PARP-related research in the Kraus laboratory is supported by grants from the NIH/NIDDK to W.L.K.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.183509.111.
References
- Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC 2008. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature 451: 81–85 [DOI] [PubMed] [Google Scholar]
- Altmeyer M, Messner S, Hassa PO, Fey M, Hottiger MO 2009. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res 37: 3723–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ame JC, Spenlehauer C, de Murcia G 2004. The PARP superfamily. Bioessays 26: 882–893 [DOI] [PubMed] [Google Scholar]
- Andrabi SA, Kang HC, Haince JF, Lee YI, Zhang J, Chi Z, West AB, Koehler RC, Poirier GG, Dawson TM, et al. 2011. Iduna protects the brain from glutamate excitotoxicity and stroke by interfering with poly(ADP-ribose) polymer-induced cell death. Nat Med 17: 692–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Reinke H, Altmeyer M, Gutierrez-Arcelus M, Hottiger MO, Schibler U 2010. Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell 142: 943–953 [DOI] [PubMed] [Google Scholar]
- Bai P, Houten SM, Huber A, Schreiber V, Watanabe M, Kiss B, de Murcia G, Auwerx J, Menissier-de Murcia J 2007. Poly(ADP-ribose) polymerase-2 controls adipocyte differentiation and adipose tissue function through the regulation of the activity of the retinoid X receptor/peroxisome proliferator-activated receptor-γ heterodimer. J Biol Chem 282: 37738–37746 [DOI] [PubMed] [Google Scholar]
- Bai P, Canto C, Brunyanszki A, Huber A, Szanto M, Cen Y, Yamamoto H, Houten SM, Kiss B, Oudart H, et al. 2011a. PARP-2 regulates SIRT1 expression and whole-body energy expenditure. Cell Metab 13: 450–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai P, Canto C, Oudart H, Brunyanszki A, Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH, et al. 2011b. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab 13: 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakondi E, Catalgol B, Bak I, Jung T, Bozaykut P, Bayramicli M, Ozer NK, Grune T 2010. Age-related loss of stress-induced nuclear proteasome activation is due to low PARP-1 activity. Free Radic Biol Med 50: 86–92 [DOI] [PubMed] [Google Scholar]
- Bauer PI, Farkas G, Buday L, Mikala G, Meszaros G, Kun E, Farago A 1992. Inhibition of DNA binding by the phosphorylation of poly ADP-ribose polymerase protein catalysed by protein kinase C. Biochem Biophys Res Commun 187: 730–736 [DOI] [PubMed] [Google Scholar]
- Beckert S, Farrahi F, Perveen Ghani Q, Aslam R, Scheuenstuhl H, Coerper S, Konigsrainer A, Hunt TK, Hussain MZ 2006. IGF-I-induced VEGF expression in HUVEC involves phosphorylation and inhibition of poly(ADP-ribose)polymerase. Biochem Biophys Res Commun 341: 67–72 [DOI] [PubMed] [Google Scholar]
- Berger F, Lau C, Ziegler M 2007. Regulation of poly(ADP-ribose) polymerase 1 activity by the phosphorylation state of the nuclear NAD biosynthetic enzyme NMN adenylyl transferase 1. Proc Natl Acad Sci 104: 3765–3770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehler C, Gauthier LR, Mortusewicz O, Biard DS, Saliou JM, Bresson A, Sanglier-Cianferani S, Smith S, Schreiber V, Boussin F, et al. 2011. Poly(ADP-ribose) polymerase 3 (PARP3), a newcomer in cellular response to DNA damage and mitotic progression. Proc Natl Acad Sci 108: 2783–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner JC, Ateeq B, Li Y, Yocum AK, Cao Q, Asangani IA, Patel S, Wang X, Liang H, Yu J, et al. 2011. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell 19: 664–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T 2005. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434: 913–917 [DOI] [PubMed] [Google Scholar]
- Buchan JR, Parker R 2009. Eukaryotic stress granules: The ins and outs of translation. Mol Cell 36: 932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callow MG, Tran H, Phu L, Lau T, Lee J, Sandoval WN, Liu PS, Bheddah S, Tao J, Lill JR, et al. 2011. Ubiquitin ligase RNF146 regulates tankyrase and Axin to promote Wnt signaling. PLoS ONE 6: e22595 doi: 10.1371/journal.pone.0022595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalgol B, Wendt B, Grimm S, Breusing N, Ozer NK, Grune T 2010. Chromatin repair after oxidative stress: Role of PARP-mediated proteasome activation. Free Radic Biol Med 48: 673–680 [DOI] [PubMed] [Google Scholar]
- Chaitanya GV, Steven AJ, Babu PP 2010. PARP-1 cleavage fragments: Signatures of cell-death proteases in neurodegeneration. Cell Commun Signal 8: 31 doi: 10.1186/1478-811X-8-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P, Coughlin M, Mitchison TJ 2009. Interaction between Poly(ADP-ribose) and NuMA contributes to mitotic spindle pole assembly. Mol Biol Cell 20: 4575–4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, Gygi SP, Colaiacovo MP, Elledge SJ 2010. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci 107: 18475–18480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Armon M, Visochek L, Rozensal D, Kalal A, Geistrikh I, Klein R, Bendetz-Nezer S, Yao Z, Seger R 2007. DNA-independent PARP-1 activation by phosphorylated ERK2 increases Elk1 activity: A link to histone acetylation. Mol Cell 25: 297–308 [DOI] [PubMed] [Google Scholar]
- Cregan SP, Dawson VL, Slack RS 2004. Role of AIF in caspase-dependent and caspase-independent cell death. Oncogene 23: 2785–2796 [DOI] [PubMed] [Google Scholar]
- D'Amours D, Desnoyers S, D'Silva I, Poirier GG 1999. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J 342: 249–268 [PMC free article] [PubMed] [Google Scholar]
- David KK, Andrabi SA, Dawson TM, Dawson VL 2009. Parthanatos, a messenger of death. Front Biosci 14: 1116–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devalaraja-Narashimha K, Padanilam BJ 2010. PARP1 deficiency exacerbates diet-induced obesity in mice. J Endocrinol 205: 243–252 [DOI] [PubMed] [Google Scholar]
- Ditsworth D, Zong WX, Thompson CB 2007. Activation of poly(ADP)-ribose polymerase (PARP-1) induces release of the pro-inflammatory mediator HMGB1 from the nucleus. J Biol Chem 282: 17845–17854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erener S, Hesse M, Kostadinova R, Hottiger MO 2012. Poly(ADP-ribose) polymerase-1 (PARP1) controls adipogenic gene expression and adipocyte function. Mol Endocrinol 26: 79–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. 2005. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434: 917–921 [DOI] [PubMed] [Google Scholar]
- Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ, et al. 2009. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 361: 123–134 [DOI] [PubMed] [Google Scholar]
- Ford AL, Lee JM 2011. Climbing STAIRs towards clinical trials with a novel PARP-1 inhibitor for the treatment of ischemic stroke. Brain Res 1410: 120–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman HC, Hugill A, Dear NT, Ashcroft FM, Cox RD 2006. Deletion of nicotinamide nucleotide transhydrogenase: A new quantitive trait locus accounting for glucose intolerance in C57BL/6J mice. Diabetes 55: 2153–2156 [DOI] [PubMed] [Google Scholar]
- Frizzell KM, Kraus WL 2009. PARP inhibitors and the treatment of breast cancer: Beyond BRCA1/2? Breast Cancer Res 11: 111 doi: 10.1186/bcr2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frizzell KM, Gamble MJ, Berrocal JG, Zhang T, Krishnakumar R, Cen Y, Sauve AA, Kraus WL 2009. Global analysis of transcriptional regulation by poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase in MCF-7 human breast cancer cells. J Biol Chem 284: 33926–33938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne JP, Hendzel MJ, Droit A, Poirier GG 2006. The expanding role of poly(ADP-ribose) metabolism: Current challenges and new perspectives. Curr Opin Cell Biol 18: 145–151 [DOI] [PubMed] [Google Scholar]
- Gagne JP, Moreel X, Gagne P, Labelle Y, Droit A, Chevalier-Pare M, Bourassa S, McDonald D, Hendzel MJ, Prigent C, et al. 2009. Proteomic investigation of phosphorylation sites in poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase. J Proteome Res 8: 1014–1029 [DOI] [PubMed] [Google Scholar]
- Geistrikh I, Visochek L, Klein R, Miller L, Mittelman L, Shainberg A, Cohen-Armon M 2011. Ca2+-induced PARP-1 activation and ANF expression are coupled events in cardiomyocytes. Biochem J 438: 337–347 [DOI] [PubMed] [Google Scholar]
- Gottschalk AJ, Timinszky G, Kong SE, Jin J, Cai Y, Swanson SK, Washburn MP, Florens L, Ladurner AG, Conaway JW, et al. 2009. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc Natl Acad Sci 106: 13770–13774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guettler S, Larose J, Petsalaki E, Gish G, Scotter A, Pawson T, Rottapel R, Sicheri F 2011. Structural basis and sequence rules for substrate recognition by tankyrase explain the basis for cherubism disease. Cell 147: 1340–1354 [DOI] [PubMed] [Google Scholar]
- Ha HC, Snyder SH 1999. Poly(ADP-ribose) polymerase is a mediator of necrotic cell death by ATP depletion. Proc Natl Acad Sci 96: 13978–13982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenni SS, Hassa PO, Altmeyer M, Fey M, Imhof R, Hottiger MO 2008. Identification of lysines 36 and 37 of PARP-2 as targets for acetylation and auto-ADP-ribosylation. Int J Biochem Cell Biol 40: 2274–2283 [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA 2011. Hallmarks of cancer: The next generation. Cell 144: 646–674 [DOI] [PubMed] [Google Scholar]
- Hang CT, Yang J, Han P, Cheng HL, Shang C, Ashley E, Zhou B, Chang CP 2010. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature 466: 62–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassa PO, Hottiger MO 2008. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci 13: 3046–3082 [DOI] [PubMed] [Google Scholar]
- Hassa PO, Buerki C, Lombardi C, Imhof R, Hottiger MO 2003. Transcriptional coactivation of nuclear factor-κB-dependent gene expression by p300 is regulated by poly(ADP)-ribose polymerase-1. J Biol Chem 278: 45145–45153 [DOI] [PubMed] [Google Scholar]
- Hassa PO, Haenni SS, Buerki C, Meier NI, Lane WS, Owen H, Gersbach M, Imhof R, Hottiger MO 2005. Acetylation of poly(ADP-ribose) polymerase-1 by p300/CREB-binding protein regulates coactivation of NF-κB-dependent transcription. J Biol Chem 280: 40450–40464 [DOI] [PubMed] [Google Scholar]
- Herranz D, Serrano M 2010. SIRT1: Recent lessons from mouse models. Nat Rev Cancer 10: 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, et al. 2009. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461: 614–620 [DOI] [PubMed] [Google Scholar]
- Hunter JE, Willmore E, Irving JA, Hostomsky Z, Veuger SJ, Durkacz BW 2012. NF-κB mediates radio-sensitization by the PARP-1 inhibitor, AG-014699. Oncogene 31: 251–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Tulin AV 2009. Poly(ADP-ribosyl)ation of heterogeneous nuclear ribonucleoproteins modulates splicing. Nucleic Acids Res 37: 3501–3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Tulin AV 2010. The roles of PARP1 in gene control and cell differentiation. Curr Opin Genet Dev 20: 512–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju BG, Solum D, Song EJ, Lee KJ, Rose DW, Glass CK, Rosenfeld MG 2004. Activating the PARP-1 sensor component of the groucho/TLE1 corepressor complex mediates a CaMKinase IIδ-dependent neurogenic gene activation pathway. Cell 119: 815–829 [DOI] [PubMed] [Google Scholar]
- Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG 2006. A topoisomerase IIβ-mediated dsDNA break required for regulated transcription. Science 312: 1798–1802 [DOI] [PubMed] [Google Scholar]
- Kanai M, Hanashiro K, Kim SH, Hanai S, Boulares AH, Miwa M, Fukasawa K 2007. Inhibition of Crm1-p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nat Cell Biol 9: 1175–1183 [DOI] [PubMed] [Google Scholar]
- Kang HC, Lee YI, Shin JH, Andrabi SA, Chi Z, Gagne JP, Lee Y, Ko HS, Lee BD, Poirier GG, et al. 2011. Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage. Proc Natl Acad Sci 108: 14103–14108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppinen TM, Chan WY, Suh SW, Wiggins AK, Huang EJ, Swanson RA 2006. Direct phosphorylation and regulation of poly(ADP-ribose) polymerase-1 by extracellular signal-regulated kinases 1/2. Proc Natl Acad Sci 103: 7136–7141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Mauro S, Gevry N, Lis JT, Kraus WL 2004. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell 119: 803–814 [DOI] [PubMed] [Google Scholar]
- Kim MY, Zhang T, Kraus WL 2005. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev 19: 1951–1967 [DOI] [PubMed] [Google Scholar]
- Koh DW, Dawson TM, Dawson VL 2005. Mediation of cell death by poly(ADP-ribose) polymerase-1. Pharmacol Res 52: 5–14 [DOI] [PubMed] [Google Scholar]
- Kotova E, Jarnik M, Tulin AV 2009. Poly (ADP-ribose) polymerase 1 is required for protein localization to Cajal body. PLoS Genet 5: e1000387 doi: 10.1371/journal.pgen.1000387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotova E, Lodhi N, Jarnik M, Pinnola AD, Ji Y, Tulin AV 2011. Drosophila histone H2A variant (H2Av) controls poly(ADP-ribose) polymerase 1 (PARP1) activation in chromatin. Proc Natl Acad Sci 108: 6205–6210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtis N, Tavernarakis N 2011. Cellular stress response pathways and ageing: Intricate molecular relationships. EMBO J 30: 2520–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus WL 2008. Transcriptional control by PARP-1: Chromatin modulation, enhancer-binding, coregulation, and insulation. Curr Opin Cell Biol 20: 294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus WL 2009. New functions for an ancient domain. Nat Struct Mol Biol 16: 904–907 [DOI] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL 2010a. The PARP side of the nucleus: Molecular actions, physiological outcomes, and clinical targets. Mol Cell 39: 8–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL 2010b. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol Cell 39: 736–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, Kraus WL 2008. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science 319: 819–821 [DOI] [PubMed] [Google Scholar]
- Kryston TB, Georgiev AB, Pissis P, Georgakilas AG 2011. Role of oxidative stress and DNA damage in human carcinogenesis. Mutat Res 711: 193–201 [DOI] [PubMed] [Google Scholar]
- Leung AK, Vyas S, Rood JE, Bhutkar A, Sharp PA, Chang P 2011. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell 42: 489–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levaot N, Voytyuk O, Dimitriou I, Sircoulomb F, Chandrakumar A, Deckert M, Krzyzanowski PM, Scotter A, Gu S, Janmohamed S, et al. 2011. Loss of tankyrase-mediated destruction of 3BP2 is the underlying pathogenic mechanism of cherubism. Cell 147: 1324–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonkar P, Dedon PC 2011. Reactive species and DNA damage in chronic inflammation: Reconciling chemical mechanisms and biological fates. Int J Cancer 128: 1999–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonn P, van der Heide LP, Dahl M, Hellman U, Heldin CH, Moustakas A 2010. PARP-1 attenuates Smad-mediated transcription. Mol Cell 40: 521–532 [DOI] [PubMed] [Google Scholar]
- Loseva O, Jemth AS, Bryant HE, Schuler H, Lehtio L, Karlberg T, Helleday T 2010. PARP-3 is a mono-ADP-ribosylase that activates PARP-1 in the absence of DNA. J Biol Chem 285: 8054–8060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Kraus WL 2011. A one and a two … expanding roles for poly(ADP-ribose) polymerases in metabolism. Cell Metab 13: 353–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangerich A, Herbach N, Hanf B, Fischbach A, Popp O, Moreno-Villanueva M, Bruns OT, Burkle A 2010. Inflammatory and age-related pathologies in mice with ectopic expression of human PARP-1. Mech Ageing Dev 131: 389–404 [DOI] [PubMed] [Google Scholar]
- Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, Seluanov A, Gorbunova V 2011. SIRT6 promotes DNA repair under stress by activating PARP1. Science 332: 1443–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin N, Schwamborn K, Schreiber V, Werner A, Guillier C, Zhang XD, Bischof O, Seeler JS, Dejean A 2009. PARP-1 transcriptional activity is regulated by sumoylation upon heat shock. EMBO J 28: 3534–3548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani M, Nakagama H, Sugimura T 2005. Poly(ADP-ribosyl)ation in relation to cancer and autoimmune disease. Cell Mol Life Sci 62: 769–783 [DOI] [PubMed] [Google Scholar]
- Medzhitov R 2008. Origin and physiological roles of inflammation. Nature 454: 428–435 [DOI] [PubMed] [Google Scholar]
- Mehrotra PV, Ahel D, Ryan DP, Weston R, Wiechens N, Kraehenbuehl R, Owen-Hughes T, Ahel I 2011. DNA repair factor APLF is a histone chaperone. Mol Cell 41: 46–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menissier de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, Dantzer F, Schreiber V, Ame JC, Dierich A, LeMeur M, et al. 2003. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J 22: 2255–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min W, Wang ZQ 2009. Poly (ADP-ribose) glycohydrolase (PARG) and its therapeutic potential. Front Biosci 14: 1619–1626 [DOI] [PubMed] [Google Scholar]
- Mota RA, Sanchez-Bueno F, Saenz L, Hernandez-Espinosa D, Jimeno J, Tornel PL, Martinez-Torrano A, Ramirez P, Parrilla P, Yelamos J 2005. Inhibition of poly(ADP-ribose) polymerase attenuates the severity of acute pancreatitis and associated lung injury. Lab Invest 85: 1250–1262 [DOI] [PubMed] [Google Scholar]
- Ohanna M, Giuliano S, Bonet C, Imbert V, Hofman V, Zangari J, Bille K, Robert C, Bressac-de Paillerets B, Hofman P, et al. 2011. Senescent cells develop a PARP-1 and nuclear factor-κB-associated secretome (PNAS). Genes Dev 25: 1245–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka S, Kato J, Moss J 2006. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J Biol Chem 281: 705–713 [DOI] [PubMed] [Google Scholar]
- Pacher P, Szabo C 2007. Role of poly(ADP-ribose) polymerase 1 (PARP-1) in cardiovascular diseases: The therapeutic potential of PARP inhibitors. Cardiovasc Drug Rev 25: 235–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R, Lewis B, Kim TK, Dilworth FJ, Erdjument-Bromage H, Tempst P, de Murcia G, Evans R, Chambon P, Reinberg D 2005. PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol Cell 18: 83–96 [DOI] [PubMed] [Google Scholar]
- Petesch SJ, Lis JT 2008. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 134: 74–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petesch SJ, Lis JT 2012. Activator-induced spread of poly(ADP-ribose) polymerase promotes nucleosome loss at Hsp70. Mol Cell 45: 64–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinnola A, Naumova N, Shah M, Tulin AV 2007. Nucleosomal core histones mediate dynamic regulation of poly(ADP-ribose) polymerase 1 protein binding to chromatin and induction of its enzymatic activity. J Biol Chem 282: 32511–32519 [DOI] [PubMed] [Google Scholar]
- Piskunova TS, Yurova MN, Ovsyannikov AI, Semenchenko AV, Zabezhinski MA, Popovich IG, Wang ZQ, Anisimov VN 2008. Deficiency in poly(ADP-ribose) polymerase-1 (PARP-1) accelerates aging and spontaneous carcinogenesis in mice. Curr Gerontol Geriatr Res 2008: 754190 doi: 10.1155/2008/754190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajamohan SB, Pillai VB, Gupta M, Sundaresan NR, Birukov KG, Samant S, Hottiger MO, Gupta MP 2009. SIRT1 promotes cell survival under stress by deacetylation-dependent deactivation of poly(ADP-ribose) polymerase 1. Mol Cell Biol 29: 4116–4129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG 2010. PARP inhibition: PARP1 and beyond. Nat Rev Cancer 10: 293–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouleau M, Saxena V, Rodrigue A, Paquet ER, Gagnon A, Hendzel MJ, Masson JY, Ekker M, Poirier GG 2011. A key role for poly(ADP-ribose) polymerase 3 in ectodermal specification and neural crest development. PLoS ONE 6: e15834 doi: 10.1371/journal.pone.0015834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulten SL, Cortes-Ledesma F, Guo L, Iles NJ, Caldecott KW 2008. APLF (C2orf13) is a novel component of poly(ADP-ribose) signaling in mammalian cells. Mol Cell Biol 28: 4620–4628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala A, La Rocca G, Burgio G, Kotova E, Di Gesu D, Collesano M, Ingrassia AM, Tulin AV, Corona DF 2008. The nucleosome-remodeling ATPase ISWI is regulated by poly-ADP-ribosylation. PLoS Biol 6: e252 doi: 10.1371/journal.pbio.0060252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber V, Ame JC, Dolle P, Schultz I, Rinaldi B, Fraulob V, Menissier-de Murcia J, de Murcia G 2002. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J Biol Chem 277: 23028–23036 [DOI] [PubMed] [Google Scholar]
- Schreiber V, Dantzer F, Ame JC, de Murcia G 2006. Poly(ADP-ribose): Novel functions for an old molecule. Nat Rev Mol Cell Biol 7: 517–528 [DOI] [PubMed] [Google Scholar]
- Shall S, de Murcia G 2000. Poly(ADP-ribose) polymerase-1: What have we learned from the deficient mouse model? Mutat Res 460: 1–15 [DOI] [PubMed] [Google Scholar]
- Shevalye H, Stavniichuk R, Xu W, Zhang J, Lupachyk S, Maksimchyk Y, Drel VR, Floyd EZ, Slusher B, Obrosova IG 2010. Poly(ADP-ribose) polymerase (PARP) inhibition counteracts multiple manifestations of kidney disease in long-term streptozotocin-diabetic rat model. Biochem Pharmacol 79: 1007–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simbulan-Rosenthal CM, Rosenthal DS, Iyer S, Boulares AH, Smulson ME 1998. Transient poly(ADP-ribosyl)ation of nuclear proteins and role of poly(ADP-ribose) polymerase in the early stages of apoptosis. J Biol Chem 273: 13703–13712 [DOI] [PubMed] [Google Scholar]
- Stilmann M, Hinz M, Arslan SC, Zimmer A, Schreiber V, Scheidereit C 2009. A nuclear poly(ADP-ribose)-dependent signalosome confers DNA damage-induced IκB kinase activation. Mol Cell 36: 365–378 [DOI] [PubMed] [Google Scholar]
- Telli ML 2011. PARP inhibitors in cancer: Moving beyond BRCA. Lancet Oncol 12: 827–828 [DOI] [PubMed] [Google Scholar]
- Tennen RI, Chua KF 2011. Chromatin regulation and genome maintenance by mammalian SIRT6. Trends Biochem Sci 36: 39–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer EH, Scheffzek K, et al. 2009. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol 16: 923–929 [DOI] [PubMed] [Google Scholar]
- Tulin A, Spradling A 2003. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science 299: 560–562 [DOI] [PubMed] [Google Scholar]
- Ullrich O, Reinheckel T, Sitte N, Hass R, Grune T, Davies KJ 1999. Poly-ADP ribose polymerase activates nuclear proteasome to degrade oxidatively damaged histones. Proc Natl Acad Sci 96: 6223–6228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill C, Toulmonde M, Bonnefoi H 2011. A review of PARP inhibitors: From bench to bedside. Ann Oncol 22: 268–279 [DOI] [PubMed] [Google Scholar]
- Wang ZQ, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, Wagner EF 1995. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev 9: 509–520 [DOI] [PubMed] [Google Scholar]
- Wang Y, Kim NS, Haince JF, Kang HC, David KK, Andrabi SA, Poirier GG, Dawson VL, Dawson TM 2011. Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos). Sci Signal 4: ra20 doi: 10.1126/scisignal.2000902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Thompson CB 2010. Cellular metabolic stress: Considering how cells respond to nutrient excess. Mol Cell 40: 323–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesierska-Gadek J, Ranftler C, Schmid G 2005. Physiological ageing: Role of p53 and PARP-1 tumor suppressors in the regulation of terminal senescence. J Physiol Pharmacol 56: 77–88 [PubMed] [Google Scholar]
- Yoo YD, Huang CT, Zhang X, Lavaute TM, Zhang SC 2011. Fibroblast growth factor regulates human neuroectoderm specification through ERK1/2-PARP-1 pathway. Stem Cells 29: 1975–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL 2002. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science 297: 259–263 [DOI] [PubMed] [Google Scholar]
- Zerfaoui M, Errami Y, Naura AS, Suzuki Y, Kim H, Ju J, Liu T, Hans CP, Kim JG, Abd Elmageed ZY, et al. 2010. Poly(ADP-ribose) polymerase-1 is a determining factor in Crm1-mediated nuclear export and retention of p65 NF-κB upon TLR4 stimulation. J Immunol 185: 1894–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Kaufman RJ 2008. From endoplasmic-reticulum stress to the inflammatory response. Nature 454: 455–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Lin Y, Kim YS, Hande MP, Liu ZG, Shen HM 2007. c-Jun N-terminal kinase mediates hydrogen peroxide-induced cell death via sustained poly(ADP-ribose) polymerase-1 activation. Cell Death Differ 14: 1001–1010 [DOI] [PubMed] [Google Scholar]
- Zhang T, Berrocal JG, Frizzell KM, Gamble MJ, DuMond ME, Krishnakumar R, Yang T, Sauve AA, Kraus WL 2009. Enzymes in the NAD+ salvage pathway regulate SIRT1 activity at target gene promoters. J Biol Chem 284: 20408–20417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu S, Mickanin C, Feng Y, Charlat O, Michaud GA, Schirle M, Shi X, Hild M, Bauer A, et al. 2011. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat Cell Biol 13: 623–629 [DOI] [PubMed] [Google Scholar]
- Zobeck KL, Buckley MS, Zipfel WR, Lis JT 2010. Recruitment timing and dynamics of transcription factors at the Hsp70 loci in living cells. Mol Cell 40: 965–975 [DOI] [PMC free article] [PubMed] [Google Scholar]