The histone variant macroH2A is generally associated with heterochromatin domains, but how it is incorporated into chromatin is unknown. Bernstein and colleagues now identify ATRX as a novel binding partner for macroH2A that negatively regulates macroH2A association with chromatin in a manner independent of its H3.3-related role. The study also sheds light on α-globin gene regulation and how ATRX mutation can cause α-thalassemia.
Keywords: macroH2A, histone variant, ATRX, α-globin, chromatin remodeling, histone chaperone
Abstract
The histone variant macroH2A generally associates with transcriptionally inert chromatin; however, the factors that regulate its chromatin incorporation remain elusive. Here, we identify the SWI/SNF helicase ATRX (α-thalassemia/MR, X-linked) as a novel macroH2A-interacting protein. Unlike its role in assisting H3.3 chromatin deposition, ATRX acts as a negative regulator of macroH2A's chromatin association. In human erythroleukemic cells deficient for ATRX, macroH2A accumulates at the HBA gene cluster on the subtelomere of chromosome 16, coinciding with the loss of α-globin expression. Collectively, our results implicate deregulation of macroH2A's distribution as a contributing factor to the α-thalassemia phenotype of ATRX syndrome.
The replacement of canonical histones with histone variants contributes to the dynamic nature of chromatin. Due to amino acid differences and, in turn, unique post-translational modifications, histone variants can alter nucleosome structure, stability, and binding of effector proteins. Histone variants have unique genomic localization patterns, and thus specialized roles such as regulating gene expression or chromosome segregation during cell division (Banaszynski et al. 2010). Therefore, the differential genomic incorporation of histone variants directly impacts critical cellular functions.
The histone variant macroH2A (mH2A) is a vertebrate-specific member of the H2A family and is unusual due to the presence of a C-terminal macro domain (Pehrson and Fried 1992). Two different genes encode mH2A1 and mH2A2 (H2AFY and H2AFY2, respectively), and two splice forms of mH2A1 exist: mH2A1.1 and mH2A1.2 (Costanzi and Pehrson 2001). mH2A is abundant in heterochromatin, including senescence-associated heterochromatic foci (SAHF) and the inactivated X chromosome (Xi) (Costanzi and Pehrson 1998; Zhang et al. 2005). In vitro studies suggest that the macro domain sterically hinders access of transcription factors to DNA, while mH2A's L1 loop produces inflexible nucleosomes (Angelov et al. 2003; Chakravarthy et al. 2005).
Our group has recently demonstrated a role for mH2A isoforms in suppressing melanoma progression, and others have linked mH2A expression or its splice patterns to breast and lung cancer (Sporn et al. 2009; Kapoor et al. 2010; Novikov et al. 2011). However, the factors that regulate the association of mH2A with chromatin remain obscure. Therefore, identifying regulators of the incorporation of histone variants at distinct genomic loci is key to understanding how chromatin domains are established and maintained and how these may go awry in disease.
A second group of factors contributing to chromatin dynamics are ATP-dependent chromatin remodeling complexes that rearrange or mobilize nucleosomes. Deregulation of members of the SWI/SNF family is implicated in various cancers and mental retardation (MR) syndromes, including ATRX (α-thalassemia/MR, X-linked), (Wilson and Roberts 2011). Mutations in ATRX, predominantly found in the H3K9me3-binding ADD (ATRX–DNMT3–DNMT3L) and/or helicase domains, are associated with ATRX syndrome (Higgs et al. 2005; Iwase et al. 2011). This syndrome is characterized by MR and α-thalassemia—a loss of α-globin gene production (Higgs et al. 2005). However, the mechanisms by which HBA (hemoglobin α) gene repression occurs are unknown (Higgs et al. 2005).
In addition to its role in regulating gene expression, ATRX acts in concert with Daxx to deposit the H3 variant H3.3 specifically at telomeres (Drane et al. 2010; Goldberg et al. 2010; Lewis et al. 2010), and ATRX deficiency results in loss of telomere integrity (Goldberg et al. 2010; Wong et al. 2010; Heaphy et al. 2011). However, it remains unclear how loss of functional ATRX protein affects the global chromatin landscape of ATRX patients, which may have tissue-specific effects (Berube 2011).
Here, we sought to discover factors involved in regulating mH2A's chromatin association. By isolating mH2A in its chromatin-free state, we identified ATRX as a novel mH2A partner. Unlike H3.3, mH2A does not interact with Daxx in chromatin-free extracts, suggesting that these two variants interact with unique ATRX complexes. As such, we observed a mutual exclusion between mH2A1.2 and H3.3 in the nucleosome. We further demonstrate that ATRX negatively regulates mH2A1 chromatin incorporation. Loss of ATRX results in increased mH2A1 levels at telomeres, as well as at the α-globin locus in erythroleukemic cells, concomitant with reduced transcription of the HBA genes. These data implicate dysregulation of mH2A's chromatin incorporation as a novel facet of the α-thalassemia phenotype of ATRX syndrome.
Results and Discussion
mH2A interacts with ATRX in a chromatin-free cellular fraction
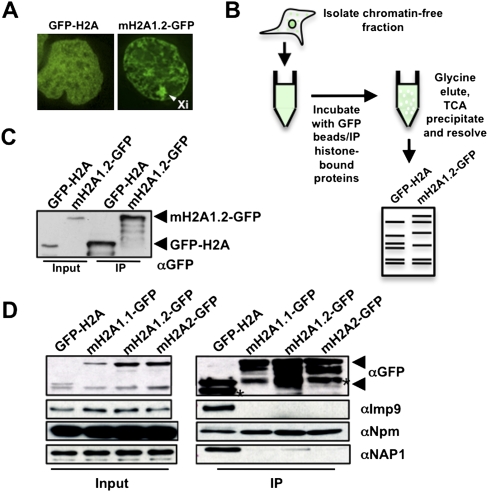
To identify factors involved in the regulation of mH2A chromatin association, we reasoned they would associate in the soluble nuclear and/or cytoplasmic fractions (“chromatin-free”). Due to their different cellular localization patterns (Fig. 1A; Costanzi and Pehrson 1998) and differential mobility in chromatin, as assayed by fluorescence recovery after photobleaching (FRAP) analysis (Supplemental Fig. 1), we hypothesized that mH2A and H2A have unique regulatory factors. To this end, we employed a large-scale biochemical strategy to purify chromatin-free GFP-H2A or mH2A1.2-GFP, similar to that used for the identification of the CENPA chaperone HJURP (Fig. 1B; Foltz et al. 2009). Confirmation of immunoprecipitated histones was performed by immunoblotting (Fig. 1C). Following extensive washing and TCA precipitation of the entire immunoprecipitated material, proteins were resolved on a gradient gel, both lanes were excised (10 slices per lane), and mass spectrometry (MS) analysis on all gel slices was performed (Fig. 1B; Supplemental Fig. 2A).
Figure 1.
Identification of mH2A1.2 chromatin-free interacting factors. (A) Fluorescence microscopy of HEK293 cells stably expressing GFP-H2A and mH2A1.2-GFP. Arrowhead indicates Xi. (B) Procedure used to isolate chromatin-free H2A- and mH2A1.2-interacting factors. (C) αGFP immunoblot confirms expression and immunoprecipitation of histones in stable cell lines. (D) Immunoblots of Imp9, Npm, and NAP1 association with GFP-H2A or mH2A-GFP isoforms. Arrows on all αGFP blots indicate GFP-H2A (bottom) and mH2A-GFP (top), and asterisks indicate degradation products.
From the proteins retrieved (Supplemental Table 1), we focused on factors that regulate chromatin association of histones, including nuclear import factors (Supplemental Fig. 2B). For example, kap114p mediates the nuclear import of H2A/H2B in Saccharomyces cerevisiae (Mosammaparast et al. 2005), and accordingly, we identified the mammalian homolog Importin 9 (Imp9) as an H2A import factor. We validated Imp9 by immunoblots and independent MS experiments where specific bands were excised (Fig. 1D; Supplemental Fig. 2C,D). These data suggest that the mechanism of histone import is evolutionarily conserved and, importantly, validate our technical approach.
As we were interested in factors that directly regulate chromatin association of mH2A1, we further focused on histone chaperones (De Koning et al. 2007; Park and Luger 2008). We identified peptides from NAP1, SET/TAF-I, nucleolin (Ncl), and nucleophosmin (Npm) in our MS analysis (Supplemental Figs. 2A, 3A). Some of these factors, such as Ncl and Npm, were present in both the GFP-H2A and mH2A1.2-GFP immunoprecipitations, and we reasoned they were general histone-interacting proteins. Indeed, this is the case (Dunleavy et al. 2009; Gaume et al. 2011), as confirmed by our immunoblots (Fig. 1D). We further confirmed specificity of NAP1 for H2A by immunoblot (Fig. 1D), as reported (Park and Luger 2008). Due to the lack of specificity for mH2A, these factors were unlikely candidates for regulating its chromatin association.
Of the potential histone chaperones identified by MS, the SWI/SNF chromatin remodeling protein ATRX interacted uniquely with mH2A1.2 (Supplemental Fig. 3A). Nineteen peptides spanning the entire ATRX protein were identified (Supplemental Fig. 3B). We confirmed this interaction using the immunoprecipitation protocol as performed for MS analysis (Fig. 2A) and via an alternative chromatin-free approach with similar results (Supplemental Fig. 3C; Mendez and Stillman 2000). Of note, we did not detect H3 in the immunoprecipitation from either protocol, suggesting that the tagged histones in our extracts were indeed chromatin-free (nonnucleosomal) and that the mH2A–ATRX interaction is independent of H3 binding (Fig. 2A). H2B peptides were detected via MS, suggesting the presence of (m)H2A–H2B dimers.
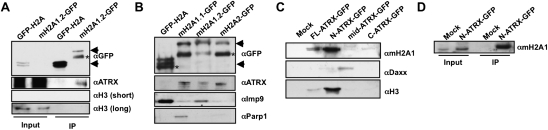
Figure 2.
ATRX interacts with mH2A isoforms in chromatin-free extracts. (A,B) Immunoblots of GFP and ATRX from chromatin-free extracts. The absence of H3 in immunoprecipitations confirms chromatin-free interactions; see the long exposure. (B) Co-IP of GFP-H2A and all mH2A isoforms in chromatin-free extracts for ATRX. Immunoblots for Imp9 and Parp1 demonstrate chromatin-free interactions. (C) Whole-cell immunoprecipitations of GFP-tagged ATRX constructs (full-length, N-terminal [1–841], middle region [800–1670], and C-terminal [1670–2492]), followed by mH2A1, H3, and Daxx immunoblots. (D) Chromatin-free co-IP of N-ATRX-GFP with mH2A1.
We next examined the interaction between ATRX and mH2A isoforms in chromatin-free extracts. ATRX interacts with all mH2A isoforms (Fig. 2B), suggesting the interaction occurs through the highly conserved H2A domain. As expected, Imp9 preferentially associated with H2A, and Parp1 uniquely interacted with mH2A1.1, as previously described (Timinszky et al. 2009). We then performed reverse coimmunoprecipitation (co-IP) experiments in HEK293 cells transfected with GFP-tagged ATRX fragments (Supplemental Fig. 4). Using whole-cell extracts, we narrowed down the region required for mH2A1 binding to amino acids 1–841 of ATRX (Fig. 2C). As positive controls, this fragment also bound H3, while a construct spanning the middle region of ATRX (800–1670) bound Daxx (Tang et al. 2004). We next validated this interaction in chromatin-free immunoprecipitations (Fig. 2D). Interestingly, the N terminus of ATRX contains an ADD domain, an HP1-binding “PxVxL” motif, an α-helical region, and an acid-rich motif (B Xiang and H Li, unpubl.). It will be key to decipher whether mH2A–ATRX binding is direct and, if so, which domains mediate this interaction.
mH2A and H3.3 are in distinct ATRX complexes
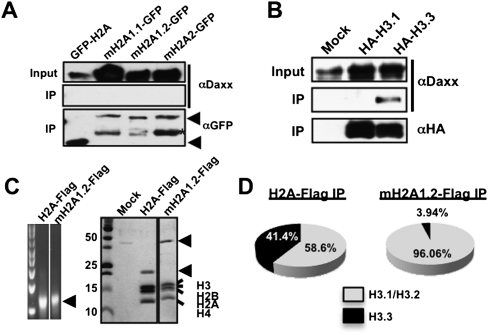
While ATRX deposits H3.3 at telomeres via its interaction with Daxx (Drane et al. 2010; Goldberg et al. 2010; Lewis et al. 2010), we were unable to detect an interaction between mH2A isoforms and Daxx via immunoblot (Fig. 3A), and Daxx peptides were not detected in our MS analysis (Supplemental Table 1). However, Daxx indeed interacts specifically with H3.3 in chromatin-free extracts (Fig. 3B). These results suggest that H3.3 and mH2A are in distinct ATRX complexes, distinguished by the presence of Daxx.
Figure 3.
mH2A and H3.3 are in distinct ATRX complexes. Co-IPs of chromatin-free association of H2A and mH2A isoforms (A) or H3 variants (B) with Daxx. Immunoblots detected the presence of Daxx specifically with H3.3. (C) Ethidium bromide-stained (left) and Coomassie-stained (right) mononucleosomes from H2A-Flag and mH2A1.2-Flag immunoprecipitations. (D) H3 composition of H2A-Flag and mH2A1.2-Flag mononucleosomes as analyzed by MS. The pie chart depicts the abundance of H3.1/H3.2 (gray) and H3.3 (black) in immunoprecipitated nucleosomes.
Based on the above, we hypothesized that mH2A1 and H3.3 exist in mutually exclusive nucleosomes. To test this directly, we immunoprecipitated mononucleosomes from H2A- and mH2A1.2-Flag-tagged HeLa cells (Fig. 3C) and performed MS analysis to determine their H3 variant composition. While H2A-containing nucleosomes contain >40% H3.3, those of mH2A1.2 contain ∼4% (Fig. 3D). This suggests that distinct factors or complexes regulate chromatin association of mH2A1 and H3.3, and we hypothesized that while ATRX–Daxx deposits H3.3 into chromatin, ATRX inhibits mH2A chromatin incorporation.
Loss of ATRX results in altered levels of mH2A1 in chromatin
To test this hypothesis, we probed the effects of ATRX depletion on mH2A chromatin association. We engineered HEK293 cells to stably express shRNAs targeting ATRX or luciferase (control). We selected two shRNA lines that induced significant knockdown (sh90 and sh92) (Fig. 4A; Supplemental Fig. 5A) for further experiments and ensured that mH2A levels were unaffected (Fig. 4A). This knockdown may mimic ATRX syndrome, as patients with ATRX mutations have reduced protein levels or decreased enzymatic activity of the helicase domain (Berube 2011; Mitson et al. 2011).
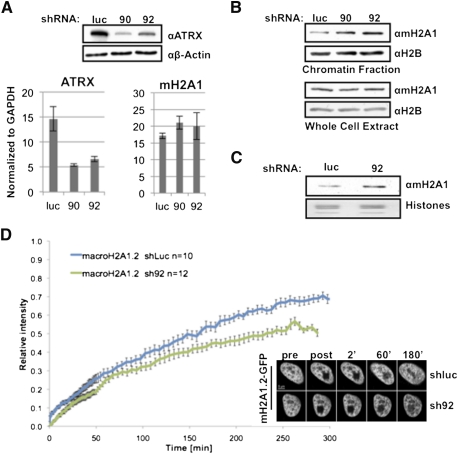
Figure 4.
ATRX knockdown results in increased levels and stability of mH2A1 in chromatin. (A) shRNA-mediated knockdown of ATRX (sh90 and sh92) in HEK293 cells results in the loss of ATRX protein and mRNA, compared with shluc, without affecting mH2A1 mRNA. β-Actin was used for loading. (B) Loss of ATRX results in accumulation of mH2A1 in chromatin (top panel), and the whole-cell content remains unaffected (bottom panel). H2B was used for loading. (C) mH2A1 immunoblot of chromatin-extracted histones analyzed by qMS. (D, left) Quantitation of FRAP experiments indicates slower recovery of mH2A1.2-GFP in HeLa1.2.11 cells depleted of ATRX (sh92; green line) compared with control (shluc; blue line). (Right) Representative images of FRAP time series pre- and post-bleach are shown.
We next inquired whether loss of ATRX altered mH2A chromatin association. Loss of ATRX resulted in a global increase of mH2A1 in chromatin, while total cellular levels remained constant (Fig. 4B), implicating ATRX as a negative regulator of mH2A's chromatin incorporation. Quantitative MS (qMS) analysis on histones extracted from chromatin (Plazas-Mayorca et al. 2009) of shluc and sh92 HEK293 cells revealed that loss of ATRX results in ∼30% more mH2A1 in chromatin (Fig. 4C; Supplemental Fig. 6). To examine the dynamics of mH2A1.2-GFP in the absence of ATRX in vivo, we performed FRAP using stable shRNA lines generated in HeLa1.2.11 cells (Supplemental Fig. 5B,C). FRAP studies revealed a decrease in fluorescence recovery of mH2A1.2 in sh92 cells, suggesting a more stable association of mH2A1.2 with chromatin upon loss of ATRX (Fig. 4D).
ATRX regulates mH2A1 incorporation at telomeres and the α-globin cluster
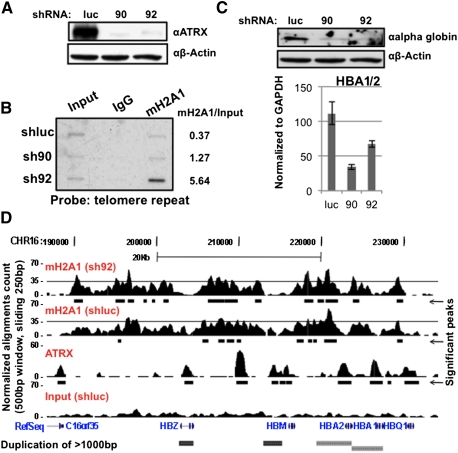
As mH2A1 chromatin association increased in ATRX-depleted cells, we inquired which genomic regions are enriched in mH2A1. Because ATRX localizes to telomeres (Goldberg et al. 2010; Wong et al. 2010), we hypothesized that global increase of mH2A1 might, in part, be a result of telomeric accumulation. Chromatin immunoprecipitation (ChIP) followed by Southern blot demonstrated increased association of mH2A1 with telomeres in ATRX knockdown lines of HEK293 and erythroleukemic K562 cells, which express α-globin (Fig 5B; Supplemental Fig. 7; see below). The weaker of the two ATRX knockdowns (sh92) revealed >10-fold mH2A1 enrichment in K562 cells (Fig. 5B), suggesting partial loss of function may have significant consequences, possibly akin to the syndrome (Berube 2011; Mitson et al. 2011). While currently unclear, ATRX-mediated regulation of mH2A levels at telomeres may help to ensure telomeric integrity.
Figure 5.
ATRX loss results in increased association of mH2A1 at telomeres and the α-globin cluster, concomitant with loss of α-globin expression. (A) shRNA-mediated knockdown of ATRX (sh90 and sh92) in K562 cells. β-Actin was used for loading. (B) ChIP reveals increased association of mH2A1 with telomeres in the absence of ATRX. (Right) Densitometry quantitation. One of two biological replicates is shown. (C) Loss of ATRX results in decreased α-globin protein and HBA mRNA. (D) Capture of the University of California at Santa Cruz Genome Browser showing an ∼50-kb region around the α-globin locus. ChIP-seq-enriched peaks are shown for mH2A1 (sh92 and shluc), ATRX (Law et al. 2010), and input (shluc). Significant peaks (MACS) are shown below each panel as black bars. (Bottom) RefSeq annotated genes. The threshold line was set at 35 to facilitate visualization.
Next, we queried whether loss of ATRX affects expression of the subtelomeric α-globin gene cluster on human chromosome 16 in erythroid cells. Upon ATRX knockdown in K562 cells (Fig. 5A; Supplemental Fig. 8A), HBA mRNA and protein levels were dramatically reduced (Fig. 5C). Other genes in this region, including the ATRX target NME4 (Law et al. 2010), were also transcriptionally decreased, while CDK8 on chromosome 13 was unaffected (Supplemental Fig. 8B; Kapoor et al. 2010).
As mH2A is generally transcriptionally repressive, we hypothesized that deregulation of mH2A nucleosome occupancy represses the HBA genes, which are silenced in ATRX patients by undefined mechanisms (Higgs et al. 2005; Berube 2011). To examine mH2A1 distribution across the α-globin cluster, we performed native ChIP-seq in shluc and sh92 K562 cells (Supplemental Fig. 9A). We obtained 56,540,184 reads for shluc, 67,219,237 for sh92, and 148,165,330 for input DNA using Illumina Hi-Seq (Supplemental Fig. 9B). Analyses were performed on normalized alignments (to the total number of alignments) to account for the different number of reads between samples. We found mH2A1 to be generally (1) excluded from transcriptional start sites (TSSs) and (2) in broad domains both upstream of and downstream from the TSS, particularly at genes transcribed at low levels. This supports its role as a repressive variant and is consistent with ChIP–chip studies (Supplemental Fig. 10A; Buschbeck et al. 2009; Gamble et al. 2010).
While K562 cells express α-globin, levels are lower than primary erythroblasts (D Higgs, pers. comm.). In accordance, we observed a distinct mH2A1 domain at the α-globin cluster in K562 cells, however, with more significant peaks of enrichment in sh92 cells (Fig. 5D; Supplemental Fig. 11A). These data strongly suggest that mH2A1 is enriched at this gene cluster in the absence of ATRX. Globally, the total number of base pairs covered by mH2A1 significant peaks is 20% higher in sh92 cells than shluc cells. This is likely due to mH2A1 redistribution, as only ∼45% of the peaks are shared between shluc and sh92 (Supplemental Fig. 9B,C). Interestingly, we observed a global anti-correlation of mH2A1 domains and ATRX peaks, which are generally concentrated around TSSs (Fig. 5D; Supplemental Fig 10B,C; Law et al. 2010), suggesting that ATRX prevents mH2A1 chromatin incorporation. Finally, by quantitative PCR (qPCR) analysis of native ChIP DNA and cross-linked ChIP DNA, we observed marked increase at regions in the α-globin cluster previously reported to be mH2A1-enriched (Supplemental Fig. 11B,C; Gamble et al. 2010). Taken together, these data suggest that mH2A1 is specifically deposited at the α-globin gene cluster in an ATRX-mediated fashion.
Here, we took an unbiased approach to identify factors that specifically associate with mH2A in its chromatin-free state. We identify ATRX as a negative regulator of mH2A1 chromatin incorporation, particularly at telomeres and the α-globin locus. Such regulation of histone incorporation via inhibitory factors remains relatively unexplored. A recent study identified INO80, also an ATP-dependent chromatin remodeling enzyme, as a negative regulator of H2A.Z nucleosomal incorporation (Papamichos-Chronakis et al. 2011). In the absence of INO80, genomic distribution of H2A.Z is perturbed, resulting in a reduced response to transcriptional changes. Whether ATRX directly interacts with mH2A to evict this histone variant from chromatin or inhibit its deposition remains unclear. If the interaction is direct, dissecting the surfaces that mediate binding will be important. In addition, it remains formally possible that alternatively spliced or modified forms of ATRX differentially interact with mH2A and H3.3 (Berube et al. 2000; Garrick et al. 2004). We look forward to future studies addressing the positive and negative regulation of histone variants within the chromatin template, the factors involved, and the underlying mechanisms.
Finally, our data point toward a novel mechanism by which the histone variant mH2A1 is involved in the α-thalassemia phenotype of ATRX patients. While the ATRX–Daxx complex has been shown to deposit H3.3, the genomic localization and function of H3.3 have yet to be explored in the context of ATRX syndrome. Here, we implicate ATRX in nucleosomal association of mH2A1, which may be important for establishing and/or maintaining chromatin states.
Materials and methods
Cell culture, plasmids, and shRNA
HEK293 and HeLa cells were grown in DMEM, and K562 cells were suspension-cultured in RPMI (10% FBS, 1% penicillin/streptomycin). GFP-tagged H2A or mH2A isoforms were expressed in HEK293 cells (pEGFP-C1 or N1, respectively), HA-tagged H3.1 and H3.3 were expressed in HeLa cells (Wiedemann et al. 2010), and Flag-tagged H2A or mH2A1.2 were infected into HeLa cells (pQXCIP). Selection was carried out in either 800 μg/mL neomycin or 1 μg/mL puromycin. For shRNA studies, HEK293, HeLa1.2.11, and K562 cells were infected with lentiviral plasmids encoding ATRX shRNAs (Open Biosystems, RHS4533-NM_000489) or luciferase shRNA by standard procedures and grown in 1 μg/mL puromycin. GFP-tagged plasmids of ATRX (N, mid, C, and full-length; gift of D. Picketts) were transiently transfected into HEK293 cells.
Chromatin-free immunoprecipitation
Chromatin-free fractions were isolated essentially as described (Foltz et al. 2009). Material from large-scale chromatin-free immunoprecipitations was glycine-eluted, TCA-precipitated, and resolved by 4%–12% gel (NuPAGE, Invitrogen). Immunoprecipitations were carried out with αGFP beads (Vector Laboratories or Chromotek) for 3 h (at 250–375 mM salt). For immunoblots, beads were washed and boiled in Laemmli loading buffer, and proteins were resolved by PAGE.
Chromatin fractionation, histone acid extraction, and immunoblots
Chromatin fractionation was performed as described (Mendez and Stillman 2000). Histone acid extraction was performed as described (Kapoor et al. 2010), with the exception that isolated chromatin was treated with H2SO4. Immunoblots were performed with the following antibodies: αGFP (Roche, 11 814 460 001), αATRX (Santa Cruz Biotechnology, sc15408), αIMP9 (Abcam, ab52605), αNap1L1 (Abcam, ab33076), αNpm (Chemicon, MAB4500), αH3 (Millipore, 05-928), αDaxx (Cell Signaling, 25C12), αParp1 (Active Motif, 39561), αβActin (Sigma, A5316), αH2B (Millipore, 07-371), αHemoglobin α (Santa Cruz Biotechnology, sc-21005), and αmH2A1 (Abcam, ab37264).
LC-MS/MS and qMS
Protein identification via LC-MS/MS was carried out essentially as described (Kaneko et al. 2010). qMS of mH2A1 was performed as described (Kapoor et al. 2010).
Mononucleosome immunoprecipitation and MS quantification of H3 variants
Mononucleosome immunoprecipitation and qMS of the histone peptides were carried out essentially as described (Viens et al. 2006; Kapoor et al. 2010). In order to quantify the ratio of H3.3 to H3.1 and H3.2, residue 31 was used (serine in place of an alanine, respectively). All methylated and acetylated forms of the peptides were considered.
cDNA isolation and qPCR
qPCR and mRNA analysis were carried out as described (Kapoor et al. 2010). cDNA expression was normalized to GAPDH levels. Primer sequences are provided in the Supplemental Material.
FRAP
Live-cell imaging and long-term FRAP experiments were carried out essentially as reported (Wiedemann et al. 2010). For visualization of the results, single-cell measurements were averaged and plotted together with the respective standard error for every time point.
Native ChIP-seq
Native mH2A1 ChIP (Abcam, ab37264) and Input DNA were prepared from K562 cells, and subsequent sequencing was performed using Illumina Hi-Seq. See the Supplemental Material for full details.
Telomere Southern blot
DNA was isolated following mH2A1 ChIP, and telomere Southern blot was carried out as described (Goldberg et al. 2010), with the exception that the probe used was T2AG3 from pSty-11.
Acknowledgments
We thank Doug Higgs, Marco De Gobbi, and the Bernstein laboratory for discussions and assistance; Aurelian Radu, Peter Warburton, Dung-Fang Lee, Eros Lazzerini Denchi, Titia de Lange, Yvette Yien, Jim Bieker, David Picketts, Dan Foltz, and Stuart Aaronson's laboratory for advice and reagents; and the Mount Sinai Institute for Genomics, Amin Mazloom, Heinrich Leonhardt, and the BioImaging Network Munich for support. This work is supported by NCI T32-CA078207 to L.F.D.; NIH MSTP T32-GM007280 to D.Y.Z.; Major State Basic Research Development Program, China (2011CB965300), to H.L.; National Natural Science Foundation of China (60905013, 60934004, and 91019016) to X.W.; DFG SFB TR5 to S.B.H. and SCHE1596/2-1 to L.S.; CIPSM to S.B.H.; an NIH Innovator award (DP2OD007447) and NSF Faculty Early CAREER award to B.A.G.; and The Ellison Medical Foundation New Scholar Award and NCI/NIH R01CA154683 to E.B.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.179416.111.
References
- Angelov D, Molla A, Perche PY, Hans F, Cote J, Khochbin S, Bouvet P, Dimitrov S 2003. The histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol Cell 11: 1033–1041 [DOI] [PubMed] [Google Scholar]
- Banaszynski LA, Allis CD, Lewis PW 2010. Histone variants in metazoan development. Dev Cell 19: 662–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berube NG 2011. ATRX in chromatin assembly and genome architecture during development and disease. Biochem Cell Biol 89: 435–444 [DOI] [PubMed] [Google Scholar]
- Berube NG, Smeenk CA, Picketts DJ 2000. Cell cycle-dependent phosphorylation of the ATRX protein correlates with changes in nuclear matrix and chromatin association. Hum Mol Genet 9: 539–547 [DOI] [PubMed] [Google Scholar]
- Buschbeck M, Uribesalgo I, Wibowo I, Rue P, Martin D, Gutierrez A, Morey L, Guigo R, Lopez-Schier H, Di Croce L 2009. The histone variant macroH2A is an epigenetic regulator of key developmental genes. Nat Struct Mol Biol 16: 1074–1079 [DOI] [PubMed] [Google Scholar]
- Chakravarthy S, Gundimella SK, Caron C, Perche PY, Pehrson JR, Khochbin S, Luger K 2005. Structural characterization of the histone variant macroH2A. Mol Cell Biol 25: 7616–7624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzi C, Pehrson JR 1998. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature 393: 599–601 [DOI] [PubMed] [Google Scholar]
- Costanzi C, Pehrson JR 2001. MACROH2A2, a new member of the MACROH2A core histone family. J Biol Chem 276: 21776–21784 [DOI] [PubMed] [Google Scholar]
- De Koning L, Corpet A, Haber JE, Almouzni G 2007. Histone chaperones: An escort network regulating histone traffic. Nat Struct Mol Biol 14: 997–1007 [DOI] [PubMed] [Google Scholar]
- Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A 2010. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev 24: 1253–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G 2009. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137: 485–497 [DOI] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR 3rd, Bassett EA, Wood S, Black BE, Cleveland DW 2009. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 137: 472–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble MJ, Frizzell KM, Yang C, Krishnakumar R, Kraus WL 2010. The histone variant macroH2A1 marks repressed autosomal chromatin, but protects a subset of its target genes from silencing. Genes Dev 24: 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrick D, Samara V, McDowell TL, Smith AJ, Dobbie L, Higgs DR, Gibbons RJ 2004. A conserved truncated isoform of the ATR-X syndrome protein lacking the SWI/SNF-homology domain. Gene 326: 23–34 [DOI] [PubMed] [Google Scholar]
- Gaume X, Monier K, Argoul F, Mongelard F, Bouvet P 2011. In vivo study of the histone chaperone activity of nucleolin by FRAP. Biochem Res Int 2011: 187624 doi: 10.1155/2011/187624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. 2010. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140: 678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, Bettegowda C, Rodriguez FJ, Eberhart CG, Hebbar S, et al. 2011. Altered telomeres in tumors with ATRX and DAXX mutations. Science 333: 425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs DR, Garrick D, Anguita E, De Gobbi M, Hughes J, Muers M, Vernimmen D, Lower K, Law M, Argentaro A, et al. 2005. Understanding α-globin gene regulation: Aiming to improve the management of thalassemia. Ann N Y Acad Sci 1054: 92–102 [DOI] [PubMed] [Google Scholar]
- Iwase S, Xiang B, Ghosh S, Ren T, Lewis PW, Cochrane JC, Allis CD, Picketts DJ, Patel DJ, Li H, et al. 2011. ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome. Nat Struct Mol Biol 18: 769–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA, Reinberg D 2010. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev 24: 2615–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A, Goldberg MS, Cumberland LK, Ratnakumar K, Segura MF, Emanuel PO, Menendez S, Vardabasso C, Leroy G, Vidal CI, et al. 2010. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature 468: 1105–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law MJ, Lower KM, Voon HP, Hughes JR, Garrick D, Viprakasit V, Mitson M, De Gobbi M, Marra M, Morris A, et al. 2010. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell 143: 367–378 [DOI] [PubMed] [Google Scholar]
- Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD 2010. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci 107: 14075–14080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez J, Stillman B 2000. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: Assembly of prereplication complexes in late mitosis. Mol Cell Biol 20: 8602–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitson M, Kelley LA, Sternberg MJ, Higgs DR, Gibbons RJ 2011. Functional significance of mutations in the Snf2 domain of ATRX. Hum Mol Genet 20: 2603–2610 [DOI] [PubMed] [Google Scholar]
- Mosammaparast N, Del Rosario BC, Pemberton LF 2005. Modulation of histone deposition by the karyopherin kap114. Mol Cell Biol 25: 1764–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikov L, Park JW, Chen H, Klerman H, Jalloh AS, Gamble MJ 2011. Qki-mediated alternative splicing of the histone variant Macroh2a1 regulates cancer cell proliferation. Mol Cell Biol 31: 4244–4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL 2011. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell 144: 200–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YJ, Luger K 2008. Histone chaperones in nucleosome eviction and histone exchange. Curr Opin Struct Biol 18: 282–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehrson JR, Fried VA 1992. MacroH2A, a core histone containing a large nonhistone region. Science 257: 1398–1400 [DOI] [PubMed] [Google Scholar]
- Plazas-Mayorca MD, Zee BM, Young NL, Fingerman IM, LeRoy G, Briggs SD, Garcia BA 2009. One-pot shotgun quantitative mass spectrometry characterization of histones. J Proteome Res 8: 5367–5374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporn JC, Kustatscher G, Hothorn T, Collado M, Serrano M, Muley T, Schnabel P, Ladurner AG 2009. Histone macroH2A isoforms predict the risk of lung cancer recurrence. Oncogene 28: 3423–3428 [DOI] [PubMed] [Google Scholar]
- Tang J, Wu S, Liu H, Stratt R, Barak OG, Shiekhattar R, Picketts DJ, Yang X 2004. A novel transcription regulatory complex containing death domain-associated protein and the ATR-X syndrome protein. J Biol Chem 279: 20369–20377 [DOI] [PubMed] [Google Scholar]
- Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer EH, Scheffzek K, et al. 2009. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol 16: 923–929 [DOI] [PubMed] [Google Scholar]
- Viens A, Mechold U, Brouillard F, Gilbert C, Leclerc P, Ogryzko V 2006. Analysis of human histone H2AZ deposition in vivo argues against its direct role in epigenetic templating mechanisms. Mol Cell Biol 26: 5325–5335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann SM, Mildner SN, Bonisch C, Israel L, Maiser A, Matheisl S, Straub T, Merkl R, Leonhardt H, Kremmer E, et al. 2010. Identification and characterization of two novel primate-specific histone H3 variants, H3.X and H3.Y. J Cell Biol 190: 777–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BG, Roberts CW 2011. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer 11: 481–492 [DOI] [PubMed] [Google Scholar]
- Wong LH, McGhie JD, Sim M, Anderson MA, Ahn S, Hannan RD, George AJ, Morgan KA, Mann JR, Choo KH 2010. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res 20: 351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Poustovoitov MV, Ye X, Santos HA, Chen W, Daganzo SM, Erzberger JP, Serebriiskii IG, Canutescu AA, Dunbrack RL, et al. 2005. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev Cell 8: 19–30 [DOI] [PubMed] [Google Scholar]