Embryonic stem cell (ESC) self-renewal and differentiation is a highly regulated process. In this study by Vardy and colleagues, the authors identify Amd1, an enzyme in the polyamine synthesis pathway, as being translationally down-regulated during differentiation of ESCs to neural precursor cells (NPCs), and this down-regulation is required for efficient NPC conversion. In addition, knockdown and overexpression of Amd1 demonstrates that Amd1 is necessary for ESC self-renewal. These results delineate a novel ESC factor that controls ESC self-renewal and differentiation.
Keywords: translational regulation, stem cells, neural precursor cells, miRNA, Amd1, polyamines
Abstract
The gene expression networks governing embryonic stem cell (ESC) pluripotency are complex and finely regulated during differentiation toward specific lineages. We describe a new role for Amd1 (adenosyl methionine decarboxylase), a key enzyme in the polyamine synthesis pathway, in regulating both ESC self-renewal and differentiation to the neural lineage. Amd1 is highly expressed in ESCs and is translationally down-regulated by the neural precursor cell (NPC)-enriched microRNA miR-762 during NPC differentiation. Overexpression of Amd1 or addition of the polyamine spermine blocks ESC-to-NPC conversion, suggesting Amd1 must be down-regulated to decrease the levels of inhibitory spermine during differentiation. In addition, we demonstrate that high levels of Amd1 are required for maintenance of the ESC state. We show that forced overexpression of Amd1 in ESCs results in maintenance of high Myc levels and a delay in differentiation on removal of LIF. We propose that Amd1 is a major regulator of ESC self-renewal and that its essential role lies in its regulation of Myc levels within the cell.
Embryonic stem cells (ESCs) possess the unique ability to self-renew and differentiate into multiple cell lineages, giving them unique therapeutic potential. Differentiation involves an orchestrated cascade of events, the precise timing and location of which is coordinated at multiple levels. Signaling pathways feed into gene expression regulatory networks at many levels, enabling rapid and tightly modulated changes to a cell's repertoire of proteins. Transcription factors have been shown to play a major role in reorganizing the transcriptome of ESCs in response to differentiation signals (Chambers and Tomlinson 2009), but it is the post-transcriptional controls that really determine the final rate of protein production. Genome-wide studies comparing the transcriptome and the proteome of ESCs have illustrated the significant noncorrelation between mRNA and protein levels for many mRNAs. It is likely that regulation of protein stability and translational control are responsible for this phenomenon (Williamson et al. 2008; Lu et al. 2009).
The importance of translational control is well documented in lower eukaryotes, and its role in mammalian development has recently begun to be addressed (Smirnova et al. 2005; Wienholds and Plasterk 2005; Sampath et al. 2008). Translational control allows the production of proteins to be finely controlled both temporally and spatially at critical developmental time points. The major coordinators of such regulation are RNA-binding proteins and microRNAs (miRNAs), both of which can regulate a plethora of mRNA targets in response to signaling pathways (Miranda et al. 2006; Proud 2007; Sonenberg and Hinnebusch 2009; Thomas et al. 2010). miRNAs function by promoting both transcript destabilization and translational repression of target mRNAs (Baek et al. 2008; Selbach et al. 2008; Hendrickson et al. 2009; Huntzinger and Izaurralde 2011). In ESCs, miRNAs play a pivotal role in differentiation through targeting mRNAs for post-transcriptional control (Sinkkonen et al. 2008; Wang et al. 2008; Xu et al. 2009; Zovoilis et al. 2009; Tiscornia and Izpisua Belmonte 2010). ESCs carrying mutations in Dicer or Drosha, components of the miRNA pathway, fail to differentiate into most lineages (Kanellopoulou et al. 2005; Murchison et al. 2005; Wang et al. 2007), highlighting the importance of post-transcriptional control in ESC differentiation.
Mouse ESCs can be efficiently differentiated into neural precursor cells (NPCs), and this has been used in the identification of regulators of neural differentiation (Cai and Grabel 2007; Johnson et al. 2008; Tay et al. 2008). miRNAs play a role in many aspects of neuronal development and function (Saba and Schratt 2010). Only a small number of post-transcriptionally controlled mRNAs have been studied in differentiating ESCs, but it is clear that there are many more whose regulation and function are not yet known (Sampath et al. 2008; Tay et al. 2008). The same is true for the miRNA and RNA-binding protein regulators. Despite the many miRNAs and RNA-binding proteins known to be differentially expressed in ESCs, the targets of only a few have been described (Houbaviy et al. 2003; Suh et al. 2004).
By taking advantage of the sedimentation differences in sucrose gradients of mRNAs that are differentially loaded with ribosomes, we used microarray analysis to identify mRNAs that are translationally regulated on differentiation of ESCs to NPCs. Here, we describe the mechanism and functional significance of translational control of one such mRNA: Amd1 (adenosyl methionine decarboxylase). Amd1 is a critical enzyme required for the synthesis of the polyamines spermine and spermidine. Polyamines are required for a wide range of cellular processes, including differentiation and cell proliferation, and their levels are tightly regulated (Persson 2009; Igarashi and Kashiwagi 2010). We demonstrate that Amd1 is targeted for translational repression by miR-762, whose expression increases during NPC differentiation. We show that manipulation of Amd1 levels can influence both the ESC state and NPC differentiation and that this phenotype is mimicked by regulation of spermine levels. We suggest that precise regulation of polyamine levels by Amd1 is critical for both ESC self-renewal and directed differentiation. Finally, we demonstrate that in the absence of the self-renewal factor LIF, high levels of Amd1 promote elevated Myc levels, which allows ESC self-renewal independent of STAT3 activation.
Results
Identification of translationally regulated mRNAs on differentiation of ESCs to NPCs
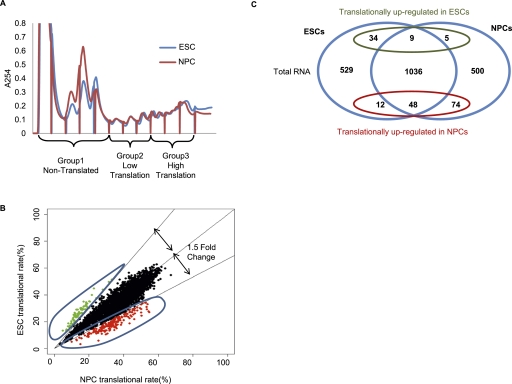
We sought to determine the translational controls operating during the conversion of ESCs to NPCs. We used an established monolayer differentiation protocol with an ESC line carrying Sox1, an NPC marker, tagged with GFP (Aubert et al. 2003; Ying et al. 2003; Pollard et al. 2006). After 6 d of differentiation, we obtained >80% Sox1-positive NPCs (Supplemental Fig. S1A–E). To identify the translational changes occurring during ESC differentiation, we used microarrays to interrogate the ribosomal load of individual mRNAs isolated from sucrose gradients from ESCs and NPCs (Fig. 1A). mRNAs were isolated into three groups—nonttranslated (NT), low translation (LT), and high translation (HT)—dependent on their sedimentation through the sucrose gradient. These groups of mRNAs, along with the total cytoplasmic RNA, were analyzed on Illumina microarrays. The microarray data were validated using quantitative RT–PCR (qRT–PCR) for a selection of 40 primers and showed a high rate of correlation, suggesting the microarrays give an accurate representation of mRNA distribution within the gradient (Supplemental Fig. S2).
Figure 1.
Global analysis of translational control in ESCs and NPCs. (A) Representative polysome profiles of ESCs and NPCs after 6 d of differentiation. Nontranslated (NT), low-translation (LT), and high-translation (HT) groups are indicated. (B) Scatter plot of the translation rates between NPCs and ESCs. Red dots represent mRNAs translationally repressed in ESCs with a fold change of >1.5. Green dots represent mRNAs translationally repressed in NPCs with a fold change of >1.5. (C) Overlay of the transcriptional and translational changes occurring on differentiation of ESCs to NPCs.
Analysis of the data showed that the majority of gene expression changes occur at the total mRNA level (Fig. 1C). We identified >182 mRNAs that are translationally controlled on NPC differentiation, with 134 mRNAs being up-regulated and 48 mRNAs down-regulated (Fig. 1B,C). When overlaid with the transcriptome data set, we saw that many of the translationally controlled mRNAs are also under transcriptional regulation. Over 55% of mRNAs that are translationally up-regulated in NPCs are also up-regulated at the total mRNA level. Over 70% of mRNAs that are translationally down-regulated in NPCs also show a decrease in total mRNA (Fig. 1C). This suggests there could be coordination between transcriptional and translational controls in these cells.
Validation of translationally regulated candidates
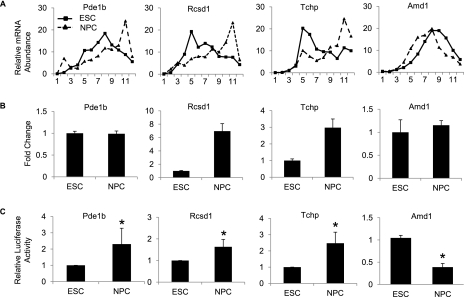
We selected a range of translationally regulated mRNAs for further validation. Polysome gradients were separated into 12 fractions, and the percentage of each mRNA within these fractions was determined by qRT–PCR. mRNA shifts to the heavier polysome fractions in NPCs were confirmed for Pdelb, Rcsd1, and Tchp as shown. Amd1 showed a shift to the lighter polysome fractions in NPCs, confirming the microarray results (Fig. 2A). We also measured changes in total mRNA levels for each of the genes. Rcsd1 and Tchp both showed corresponding increases in total mRNA in NPCs, suggesting coordination between transcription and translation. Pde1b and Amd1 showed no significant change in mRNA levels (Fig. 2B), suggesting these mRNAs may be under exclusive translational control.
Figure 2.
Validation of translationally regulated mRNAs. (A) qRT–PCR analysis showing the percentage of mRNA in each polysomal fraction from ESCs (squares) and NPCs (triangles). All samples are normalized to internal spike-in controls. Fraction 1 represents the top of the gradient and fraction 12 represents the bottom of the gradient. (B) qRT–PCR of candidate mRNAs from total RNA from ESCs and NPCs. (C) Relative luciferase levels in ESCs and NPCs of vectors containing the 3′ UTRs of candidate genes downstream from the luciferase ORF. Values are normalized to control luciferase, and ESC activities are set to 1. Values are means ± SD. (*) P < 0.05.
The regulatory sequences dictating translational activation or repression are generally located within the untranslated regions (UTRs) of an mRNA. To demonstrate the translation changes resulting from the ribosomal shifts, we used the luciferase reporter plasmid pGL3 carrying the luciferase mRNA under the control of the 3′ UTR from the translationally regulated mRNAs. These were transfected into ESCs and NPCs, and the levels of luciferase were measured as an indication of translation efficiency (Fig. 2C). Pde1b, Rcsd1, and Tchp show decreased luciferase activity in ESCs compared with NPCs, while Amd1 shows a decrease in luciferase activity in NPCs compared with ESCs. Luciferase mRNA levels were measured in all cases and were found to be not significantly different between ESCs and NPCs, suggesting the decrease in luciferase activity is a result of translational repression (Supplemental Fig. S3). Taken together, these validations demonstrate that a significant number of mRNAs are translationally regulated on differentiation of ESCs to NPCs and that polysome profiling provides an effective means of identifying them.
Amd1 is translationally down-regulated during NPC differentiation
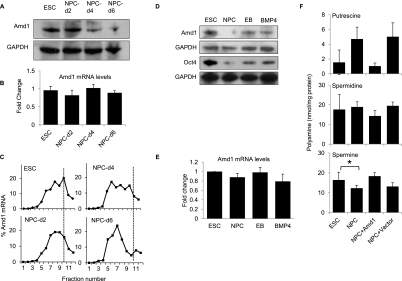
Amd1 is a highly conserved enzyme required for the production of the polyamines spermine and spermidine. Polyamines are important for growth and proliferation, but their role in ESCs is currently unknown. To determine the kinetics of translational control of Amd1, we performed Western blots of ESCs and NPCs 2, 4, and 6 d after induction of differentiation. Amd1 protein levels are high in ESCs and are reduced by day 4 and further reduced by day 6 of NPC differentiation (Fig. 3A). Total Amd1 transcript levels did not change significantly during the 6 d of differentiation to NPCs (Fig. 3B), suggesting the change in protein level is independent of mRNA abundance. High-resolution sucrose density gradient fractionation showed that Amd1 mRNA is associated with the heavier polysome fractions (fractions 8–10) in ESCs. After 4 d of differentiation, Amd1 mRNA shifts partly to the lighter fractions and by day 6 is found almost exclusively in the lighter fractions (fractions 5–7) (Fig. 3C). These data strongly suggest that Amd1 starts to be translationally repressed from day 4 and is most dramatically repressed by day 6 of NPC differentiation.
Figure 3.
Amd1 is translationally down-regulated in NPCs. (A) Western blot showing Amd1 protein levels in ESCs and NPCs after 2, 4, and 6 d of differentiation. GAPDH is shown as a loading control. (B) qRT–PCR showing Amd1 mRNA levels in ESCs and cells differentiated for 2, 4, or 6 d in N2B27 medium. (C) Amd1 mRNA distribution in polysome gradients from ESCs and cells differentiated for 2, 4, or 6 d into NPCs. Fraction 1 represents the top of the gradient and fraction 12 represents the bottom of the gradient. (D) Western blot showing Amd1 and Oct4 protein levels in ESCs, NPCs, EBs, and BMP4-treated cells. GAPDH was used as a loading control. (E) qRT–PCR showing Amd1 mRNA levels in ESCs, NPCs, EBs, and BMP4-treated cells. (F) Quantification of the polyamine levels in ESC and NPCs. Polyamine levels are also shown for NPCs transfected with the pTracer Amd1 vector or empty vector. Values are means ± SD. (*) P < 0.05.
To address the specificity of Amd1 down-regulation, ESCs were differentiated into embryoid bodies (EBs) or in the presence of N2B27 medium with BMP4. BMP4 functions to inhibit the differentiation of NPCs (Ying et al. 2003). Western blot analysis showed that Amd1 protein levels were most dramatically reduced in NPCs compared with EBs or BMP4-treated cells (Fig. 3D). Amd1 mRNA levels remain the same (Fig. 3E). This suggests that the translational repression seen on NPC differentiation is specific for this lineage.
The Amd1 enzyme functions to decarboxylate S-adenosylmethionine (AdoMet) to produce AdoMet-DC. This provides the aminopropyl donor for the synthesis of spermidine from putrescine and of spermine from spermidine. We predicted that in NPCs with lower levels of Amd1 protein, the ratio of putrescine to spermidine and spermine would change. We measured the levels of all three polyamines in ESCs and NPCs. As expected, the ratio of the three polyamines changes dramatically in NPCs. The levels of putrescine are increased threefold. Interestingly, the levels of spermine are decreased in NPCs (Fig. 3F). These data suggest that the functional consequence of decreased Amd1 levels may be a change in the ratio of the three polyamines and a decrease in the levels of spermine.
Down-regulation of Amd1 protein is required for NPC differentiation
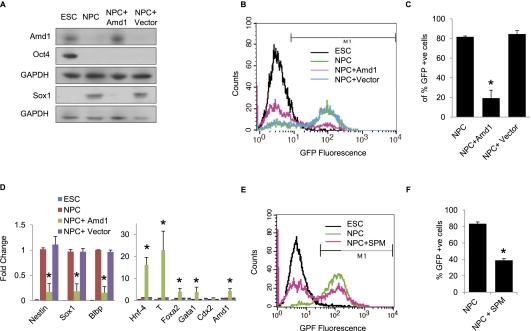
To establish the significance of Amd1 down-regulation for NPC differentiation, we overexpressed the Amd1 ORF using the EF-1a promoter in the pTracer vector. Amd1 protein levels start to decrease between 2 and 4 d of NPC differentiation (Fig. 3A), so we transfected NPCs that had been differentiating for 3 d with the pTracer-Amd1 or empty vector. Overexpression of Amd1 was demonstrated using Western blot, and after 6 d of differentiation, Amd1 levels were similar to those found in ESCs, while cells transfected with the empty vector showed decreased protein levels, similar to the untransfected cells (Fig. 4A). Interestingly, continued expression of Amd1 resulted in a failure in the accumulation of the NPC marker Sox1. This was further demonstrated using flow cytometry. In the presence of the empty vector alone, >80% of Sox1-GFP-positive cells were recovered after 6 d of differentiation. In the presence of high Amd1 protein levels, however, <30% of cells were Sox1-positive (Fig. 4B,C). These data were further confirmed with immunofluorescence, showing that overexpression of Amd1 resulted in a failure of Nestin and Sox1 accumulation after 6 d of differentiation (Supplemental Fig. S4). qRT–PCR confirmed that on overexpression of Amd1 in differentiated NPCs, NPC markers Nestin and Sox1 failed to accumulate. Differentiation markers Hnf4, T, Foxa2, and Gata1 were elevated on Amd1 overexpression, suggesting the cells are differentiating into alternative lineages (Fig. 4D).
Figure 4.
Amd1 translational down-regulation is required for NPC differentiation. (A) Differentiating ESCs were transfected with the pTracer-Amd1 vector or empty vector after 3 d in N2B27 medium and allowed to differentiate for a further 3 d. Western blots showing protein levels in ESCs and NPCs of Amd1, Oct4, and Sox1 after 6 d of differentiation. GAPDH is shown as a loading control. (B) Flow cytometry analysis showing accumulation of Sox1-GFP cells after differentiation of ESCs expressing pTracer-Amd1 or empty vector after 6 d in N2B27 medium. (C) Quantification of Sox1-GFP cells from B is shown. (D) qRT–PCR showing mRNA levels from cells differentiated to NPCs after transfection with the Amd1-overexpressing vector. (E) Flow cytometry data of NPCs differentiated with and without the addition of 100 μM spermine (SPM). (F) Quantification of Sox1-GFP cells from E is shown. Values are means ± SD. (*) P < 0.05.
We showed that NPCs with low Amd1 protein levels have reduced levels of the polyamine spermine and an altered polyamine ratio (Fig. 3F). We show that overexpression of Amd1 in differentiating NPCs restored the polyamine ratio and that spermine levels are elevated to those seen in ESCs (Fig. 3F). To determine whether NPC differentiation requires lower levels of spermine, we added spermine to NPCs after 3 d of differentiation, the same time that Amd1 starts to be down-regulated, and assessed the effect on accumulation of Sox1-positive cells after 6 d. Strikingly, we saw a reduction in the number of Sox1-positive cells, suggesting that spermine is inhibitory to NPC differentiation (Fig. 4E,F). Taken together, these data suggest that down-regulation of Amd1 during NPC differentiation may be required to decrease the levels of inhibitory spermine in the cells.
Amd1 is translationally regulated by miR-762
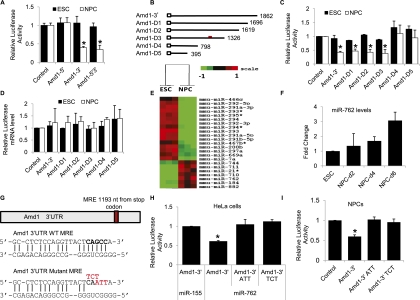
To identify the cis-acting elements responsible for the translational repression of Amd1, we constructed a firefly luciferase construct carrying the Amd1 5′ UTR, 3′ UTR, or both the 5′ and 3′ UTRs and assessed their ability to regulate translation in a dual luciferase reporter assay. Luciferase constructs were transfected into ESCs and NPCs after 4 d of differentiation (Fig. 5A). The 5′ UTR had no effect on luciferase activity in either cell type, while the 3′ UTR promoted strong luciferase repression in NPCs. This suggests that the cis-acting elements responsible for the translational repression seen on NPC differentiation reside within the 3′ UTR. To map these cis-acting elements, we made a series of 3′ UTR deletions, Amd1-D1–5 (Fig. 5B), and tested them for their repressive effect on luciferase levels (Fig. 5C). Amd1-D1, Amd1-D2, and Amd1-D3 retained their ability to repress luciferase activity in NPCs, while this was lost in D4 and D5, suggesting the cis-element lies within the 528 nucleotides (nt) retained in D3 but lost on D4. There was no significant effect on the levels of luciferase mRNA between the two cell types, suggesting that these 528 nt are promoting translational repression and not transcript destabilization (Fig. 5D).
Figure 5.
Amd1 translational down-regulation is mediated by miR-762. (A) A bar chart showing the effect on luciferase activity of the Amd1 5′ UTR, 3′ UTR, or 5′ and 3′ UTRs in ESCs and NPCs. Luciferase values are normalized to cotransfected Renilla luciferase and empty vector. (B) Diagram showing the deletion constructs made to map the Amd1 3′ UTR cis-acting elements. The numbers refer to the length of the 3′ UTR remaining in each construct. The red mark indicates the location of the miR-762 target site. (C) Relative luciferase activities for the constructs shown in B after transfection into ESCs and NPCs. Values are normalized to firefly luciferase and empty vector. (D) qRT–PCR analysis of the relative luciferase mRNA levels after transfection into ESCs and NPCs. The Renilla luciferase is normalized to the firefly luciferase mRNA levels. (E) Heat map showing differentially expressed miRNAs from ESCs and NPCs. (F) qRT–PCR showing up-regulation of miR-762 in NPCs 2, 4, and 6 d after differentiation. (G) Predicted miR-762 miRNA response element (MRE) sequences within the Amd1 3′ UTR and its mutant counterpart. (H) Relative luciferase activity of PsiCHECK-2 constructs carrying the Amd1 wild-type 3′ UTR or Amd1 mutant 3′ UTR cotransfected into HeLa cells with miR-762. Values are normalized to control luciferase and empty vectors. (I) Relative luciferase activity of PsiCHECK-2 constructs carrying the Amd1 wild-type 3′ UTR or Amd1 mutant 3′ UTR cotransfected into ESCs and NPCs. Values are normalized to control luciferase and empty vector. Values are means ± SD. (*) P < 0.05.
We performed miRNA microarrays on ESCs and NPCs to identify miRNAs that are differentially expressed between the two cells lines. The microarray showed that a number of miRNAs were up-regulated in NPCs compared with ESCs (Fig. 5E). One of these, miR-762, a little-studied but conserved miRNA, has a target site within the 528 nt responsible for Amd1 translational repression (Miranda et al. 2006). miR-762 was up-regulated threefold in NPCs compared with ESCs, making it a likely candidate for translational repression of Amd1 (Fig. 5F). miR-762 up-regulation was specific to NPCs, as no increase in levels was seen in EBs or BMP4-treated NPCs (Supplemental Fig. S5A). We coexpressed the Amd1 3′ UTR luciferase construct with the miR-Vec vector carrying the miR-762 miRNA or nontargeting miR-155 in HeLa cells and saw a decrease in luciferase activity of >40% in the presence of miR-762 (Fig. 5H). To confirm the 528-nt region was being targeted by miR-762, we tested the ability of miR-762 to repress luciferase activities of the five Amd1 3′ UTR deletion constructs (Fig. 5B). Constructs carrying the full-length Amd1 3′ UTR or D1, D2, or D3 showed strong repression of luciferase activity, while constructs D4 and D5, which do not contain the miR-762 target site, were not repressed in the presence of the miRNA (Supplemental Fig. S5B). There was no significant decrease in normalized luciferase mRNA levels between the constructs in the presence or absence of the miRNA (Supplemental Fig. S5C). To confirm that the miR-762 target site was solely responsible for the translational repression, we made mutants in the miRNA target site (Fig. 5G). Constructs carrying two different triplet target site mutations showed a complete restoration of luciferase activity when cotransfected with miR-762 into HeLa cells, confirming that the Amd1 3′ UTR is a direct target of miR-762 (Fig. 5H). To further confirm that miR-762 targets Amd1 in NPCs, we transfected the mutated Amd1 3′ UTR luciferase constructs into NPCs and again saw a complete restoration of luciferase activity (Fig. 5I). To confirm the specificity of Amd1 repression in NPCs, we transfected BMP4-treated NPCs with the Amd1 reporter vector. We saw no luciferase repression in these cells, confirming that the translational repression is specific for NPCs (Supplemental Fig. S5D). These data demonstrate that miR-762 targets the Amd1 3′ UTR for translational repression in NPCs and that miR-762 is sufficient for the translational repression of Amd1 on NPC differentiation.
Amd1 is required for ESC self-renewal
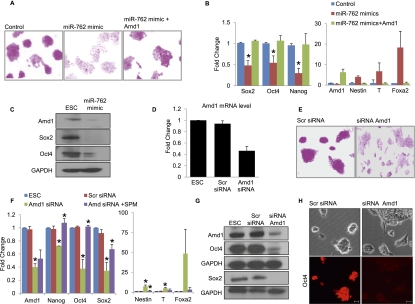
miR-762 is expressed at low levels in mouse ESCs compared with NPCs, so we determined whether its overexpression could promote differentiation of ESCs. ESCs were transfected with control or miR-762 mimics and maintained in ESC culture conditions. Cells were stained with alkaline phosphatase (AP) 4 d after transfection, and cells transfected with the miR-762 mimic showed a clear decrease in AP-stained colonies, suggesting that the cells are differentiating and losing their self-renewal capacity (Fig. 6A). qRT–PCR showed a decrease in the pluripotency markers Oct4, Sox2, and Nanog and an increase in the differentiation markers Nestin, T, and FoxA2 (Fig. 6B). Decreased pluripotency markers were confirmed by Western blot (Fig. 6C). The miR-762 mimic also promoted the down-regulation of Amd1 protein. We postulated that miR-762 targeting of Amd1 could be responsible for the differentiation phenotype observed. To test this, we expressed the Amd1 ORF, which cannot be targeted by miR-762, in ESCs with the miR-762 mimic and assessed the effect on pluripotency. We saw an almost complete rescue of the differentiation phenotype in the presence of the Amd1 ORF. AP staining is retained in the presence of the Amd1 ORF (Fig. 6A), and qRT–PCR shows that pluripotency markers Oct4, Sox2, and Nanog are not affected and that differentiation markers Nestin, T, and FoxA2 remain low (Fig. 6B). These data suggest that the differentiation phenotype seen on expression of miR-762 in ESCs is due to the reduction of Amd1 protein.
Figure 6.
Amd1 is required for ESC maintenance. (A) ESCs transfected with negative control mimics or with miR-762 mimics with or without the pTracer-Amd1 vector. Cells were stained with AP 4 d after transfection. (B) qRT–PCR analysis of mRNAs 4 d after transfection as in A. (C) Western blot analysis showing Amd1 and ESC markers 4 d after transfection with miR-762 mimics. GAPDH was used as loading control. (D) qRT–PCR showing Amd1 knockdown with siRNAs. Scrambled siRNAs are shown as a control. (E) AP staining of ESCs transfected with siRNAs to Amd1. (F) qRT–PCR showing mRNA expression levels following transfection of ESCs with siRNAs targeting Amd1 with and without the addition of spermine (SPM). P-values show significant differences between ESCs and siAmd1 samples or between siAmd1 plus and minus spermine. (G) Western blot showing Amd1, Oct4, and Sox2 levels following transfection of ESCs with siRNAs targeting Amd1. GAPDH is shown as a loading control. (H) Immunofluorescence staining for ESC marker Oct4 in ESCs transfected with pools of siRNA targeting Amd1 or scrambled siRNA. Bars, 20 μm. Values are means ± SD. (*) P < 0.05.
To confirm a role for Amd1 in the maintenance of ESC pluripotency, we used a pool of siRNAs to transiently knock down Amd1 mRNA in ESCs (Fig. 6D). Following knockdown of Amd1, ESC colonies showed a reduction in AP staining, indicative of a differentiated state (Fig. 6E). To confirm the loss of pluripotency, we compared the levels of the pluripotency markers Oct4, Sox2, and Nanog after Amd1 knockdown (Fig. 6F). We saw significant reductions in mRNA levels for all three transcripts and a corresponding increase in the differentiation markers Nestin, T, and FoxA2 (Fig. 6F). This was further confirmed by Western blot for Oct4 and Sox2 (Fig. 6G). Immunofluorescence analysis shows that Oct4 and Sox2 are reduced in the majority of ESCs after Amd1 knockdown (Fig. 6H; Supplemental Fig. S6), suggesting that all ESCs are responsive to decreased Amd1 levels and not just a subset. To determine whether Amd1 is required in ESCs to maintain high levels of spermine, we added it to cells that had been treated with siRNAs to Amd1. Strikingly, we saw that the addition of spermine can rescue the differentiation phenotype seen on knockdown of Amd1, as shown by PCR of the pluripotency markers (Fig. 6F). We also demonstrated that addition of spermine can suppress the differentiation phenotype seen on addition of the miR-762 mimic to ESCs. The effect is specific to spermine, however, as spermidine had no effect (Supplemental Fig. S6A). These data clearly demonstrate that high Amd1 protein levels are required to maintain ESC self-renewal, implicating Amd1 as an essential self-renewal factor. Spermine can rescue the Amd1 knockdown phenotype, suggesting that Amd1 is required to promote the levels of spermine for ESC self-renewal.
Forced expression of Amd1 can delay differentiation of ESCs
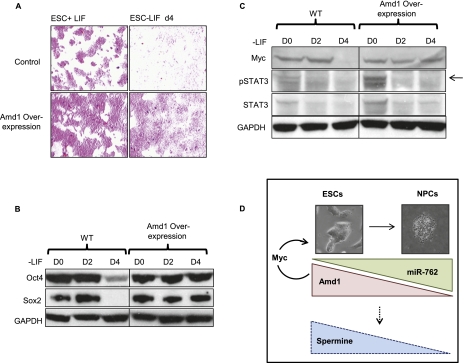
Having established that Amd1 is required for maintenance of the ESC state, we wanted to determine whether its overexpression could delay differentiation. We overexpressed Amd1 in ESCs and differentiated the cells in the absence of LIF for 4 d. Control cells lose AP staining rapidly, but strikingly, AP staining remains high on Amd1 overexpression (Fig. 7A). This is further demonstrated by the maintenance of pluripotency markers Oct4 and Sox2 on removal of LIF when Amd1 is overexpressed (Fig. 7B). This demonstrates that increased Amd1 levels can delay ESC differentiation. Polyamines have been shown to regulate gene expression of a number of different proteins, one of which is Myc (Wang et al. 1993; Patel and Wang 1997; Liu et al. 2009), and maintenance of high levels of Myc promotes ESC self-renewal in the absence of LIF (Cartwright et al. 2005). We show that on removal of LIF, Myc protein levels are reduced dramatically by day 4 of differentiation, whereas when Amd1 is overexpressed, Myc levels are maintained at high levels (Fig. 7C). LIF signaling promotes the phosphorylation of STAT3, which in turn activates Myc transcription (Niwa et al. 1998; Matsuda et al. 1999; Cartwright et al. 2005).We demonstrate that Amd1 promotes elevated Myc levels in the absence of STAT3 activation, as shown by a decrease in pSTAT3 on removal of LIF in both control and Amd1-overexpressing cells (Fig. 7C).
Figure 7.
Overexpression of Amd1 promotes the ESC state. (A) AP staining of ESCs and cells differentiated for 4 d in the absence of LIF. Control and Amd1-overexpressing cells are shown. (B) Western blot showing pluripotency markers Oct4 and Sox2 in cells treated as in A. (C) Western blot showing Myc, STAT3, and phosphorylated STAT3 (arrow) in cells treated as in A. (D) Model depicting the role of Amd1 in ESC self-renewal and NPC differentiation. High levels of Amd1 are required to maintain the ESC state by promoting high levels of Myc. On differentiation to NPCs, miR-762 is up-regulated, resulting in translational down-regulation of Amd1. Decreased Amd1 protein levels result in lower levels of the inhibitory polyamine spermine, allowing NPCs to differentiate.
We propose that regulation of Amd1 protein levels by miR-762 is essential to control polyamine ratios in ESCs and in differentiating NPCs (Fig. 7D). In undifferentiated ESCs, high levels of Amd1 are required to maintain the ESC state through promotion of Myc levels. During NPC differentiation, up-regulation of miR-762 results in targeting of Amd1 for translational repression. This leads to decreased levels of spermine, allowing for efficient NPC differentiation. Taken together, our data demonstrate that Amd1 is an essential enzyme for ESC self-renewal and that its translational regulation is critical for early neural specification.
Discussion
In this study, we undertook a genome-wide screen to identify new regulators of ESC differentiation to NPCs. Using polysome profiling coupled to microarray analysis, we identified a range of mRNAs that are under translational control. We focused our studies on one candidate, Amd1, that has been implicated in early mammalian development (Nishimura et al. 2002). The role of Amd1 and polyamines in ESC differentiation had not been addressed, prompting us to investigate the significance of Amd1 regulation in ESC maintenance. We demonstrate that Amd1 is translationally repressed by miR-762 on NPC differentiation. Amd1 has been shown to be translationally controlled by an uORF (Law et al. 2001; Ivanov et al. 2010) in its 5′ UTR; however, our luciferase data suggest that the regulation seen on NPC differentiation is mediated entirely through the 3′ UTR. miR-762 is a recently identified, conserved miRNA, and Amd1 is the first validated target to be described. Mutation of the miRNA target site completely restores luciferase activity in NPCs, suggesting that targeting by miR-762 is sufficient for Amd1 down-regulation. Our data clearly show that miR-762 promotes the translational repression of Amd1. We demonstrate this by showing both a shift in ribosomal load for Amd1 in NPCs and a miR-762-induced reduction in luciferase activity that is independent of RNA levels. miR-762 is up-regulated in NPCs, but it is currently not clear what drives this. Further analysis is required to understand the transcriptional controls governing miR-762 expression in ESCs and NPCs.
The levels of polyamines in the cell are regulated on many levels, including synthesis, uptake, and export. The two rate-limiting enzymes in their synthesis, Odc1 and Amd1, are under tight control, and Amd1 has been shown to be regulated at the levels of transcription, RNA stability, translation, enzyme activity, and protein degradation (Pegg et al. 1998). Our demonstration that Amd1 is regulated by the 3′ UTR targeting miRNA miR-762 is the first demonstration of miRNA-mediated regulation of the polyamine pathway. It is currently not clear how widespread this miRNA-mediated translational control is for polyamine homeostasis and whether it is specific to early differentiation of neural progenitors. The levels of polyamines are regulated at such a tight level in all cells that in order to override that control in a given cellular context, an additional dominant level of regulation may be required. This may be the reason that this mechanism of control exists during NPC differentiation in an already tightly regulated network. Increased polyamine levels have been shown to be coincident with cancer progression, and Amd1 has been targeted for the development of cancer treatments. A more detailed understanding of the role of miRNA-mediated regulation of polyamine levels in cancer cells could lead to novel therapeutics aimed at targeting polyamine levels through down-regulation of its enzymatic regulators.
Analysis of the levels of the three polyamines in ESCs and NPCs suggests that down-regulation of Amd1 results in a change in the ratio of polyamines; notably, spermine levels are decreased. The selective effect on spermine and not spermidine levels likely reflects additional regulation of the enzymes controlling spermine and spermidine synthesis, degradation, and export. Interestingly, Amd1 down-regulation only becomes apparent after 3 d of NPC differentiation, suggesting that its down-regulation is not required for the early stages of ES differentiation. We demonstrate that overexpression of Amd1 after 3 d of differentiation results in a dramatic decrease in NPCs and an increase in differentiation markers for other lineages. This is accompanied by restoration of the polyamine ratios seen in ESCs. In addition, we mimic this inhibitory effect though the addition of spermine after 3 d of NPC differentiation. These data suggest that high levels of spermine are inhibitory to NPC differentiation and that Amd1 down-regulation may be required to promote a change in the polyamine ratio and a decrease in intracellular spermine levels.
Amd1 is expressed at high levels in ESCs, and we demonstrated that it is essential for maintenance of the ESC state. Knockdown of Amd1 by siRNA resulted in a loss of AP staining and a decrease in the mRNA and protein levels of the pluripotency markers (Fig. 6). Markers for all three lineages were increased on Amd1 knockdown, suggesting that the differentiation may not be lineage-specific. Addition of spermine can rescue the differentiation phenotype seen on Amd1 knockdown, suggesting the essential role of Amd1 lies in maintaining high levels of spermine. Previous reports have demonstrated that embryos devoid of Amd1 die during early gastrulation and that ESCs can only be recovered from Amd1−/− blastocystes in the presence of exogenous spermidine (Nishimura et al. 2002). This is in agreement with our finding that Amd1 is essential for ESC self-renewal.
To further demonstrate the critical role of Amd1 in self-renewal, we show that forced overexpression of Amd1 can delay differentiation, as shown by the prolonged expression of pluripotency markers in conditions that promote differentiation (Fig. 7). Notably, Myc levels are maintained in the absence of LIF when Amd1 is overexpressed. It has been demonstrated that nondegradable Myc can maintain ESCs in an ESC state with high Oct4 levels in the absence of LIF (Cartwright et al. 2005). We propose that the mechanism by which Amd1 maintains ESC self-renewal in the absence of LIF is through promoting high Myc levels. LIF signaling is required in part to activate STAT3, which in turn promotes Myc transcription (Niwa et al. 1998; Matsuda et al. 1999; Cartwright et al. 2005). Phosphorylated STAT3 is diminished equally in control and Amd1-overexpressing cells in the absence of LIF, showing that Amd1 promotes Myc levels independent of STAT3. Reports in the literature have demonstrated that polyamines can enhance Myc translation by promoting phosphorylation of the RNA-binding protein HuR (Liu et al. 2009). It is possible that a similar level of activation is operating on overexpression of Amd1 in differentiating ESCs. On removal of LIF, Myc is also regulated at the protein level following phosphorylation on Thr 58, which triggers protein degradation mediated by GSK3β (Sato et al. 2004). While it is possible that Amd1 is functioning to promoting Myc protein stabilization, it is less likely, as polyamines are believed to largely exist in RNA and DNA complexes regulating gene expression at the transcription or RNA level (Igarashi and Kashiwagi 2010).
Our data demonstrate that on removal of LIF, increased Amd1 can sustain the high levels of Myc needed for self-renewal. It is therefore likely that the differentiation phenotype we see on down-regulation of Amd1 is due to decreased Myc levels. We propose that in order for LIF to promote self-renewal, it requires appropriate levels of the polyamines. The levels of Amd1, and thus the polyamines, are likely tightly regulated during ESC self-renewal and differentiation. Small changes in the ratio of the polyamines could function to signal the cell to differentiate or self-renew. Our data suggest that maintaining an appropriate ratio of polyamines and ESC signaling networks maintains ESC self-renewal, but disruption of any of these will result in differentiation of the cells.
Polyamines function in a wide range of cellular processes, and each of the polyamines can have different functions in different cell types. Manipulation of polyamine levels have been linked to differentiation of F9 teratocarcinoma cells and the enhancement of cardiac differentiation (Frostesjo et al. 1997; Sasaki et al. 2008). There have been reports of both neuroprotective and neurotoxic effects of all three polyamines on neurons (de Vera et al. 2008; Igarashi and Kashiwagi 2010), and disruption of polyamine homeostasis has been linked to a number of neurological disorders (Casero and Pegg 2009). The role of polyamines in ESC self-renewal and differentiation is clearly complex, and it is likely that they function in different ways depending on the developmental window and the cellular context.
Our identification of Amd1 as a crucial regulator of ESC self-renewal highlights the importance of polyamine regulation in ESC biology. While Amd1 regulation and polyamine levels have been linked to cancer and proliferation, their significance had never been appreciated in ESCs. Here we demonstrate that Amd1 has an essential role in maintaining ESC self-renewal through promoting high Myc levels. Polyamines have the capacity to regulate a wide range of targets, and this work opens the door to exploring a new level of regulation in ESC self-renewal and differentiation. Understanding the role of polyamines in ESCs will greatly broaden our knowledge of the regulatory pathways operating to maintain or direct the differentiation of ESCs.
Materials and methods
Cell culture
ESCs were cultured on 0.1% gelatin in DMEM (GIBCO) supplemented with 15% fetal bovine serum (FBS) (GIBCO), 0.2 mM β-mercaptoethanol, 2 mM L-glutamine (GIBCO), 1× MEM nonessential amino acids (GIBCO), and LIF. EBs were formed by culturing ESCs in the absence of LIF on low-attachment plates as described (Sampath et al. 2008). NPC differentiation was performed in N2B27 medium as described (Ying et al. 2003). BMP4 (10 ng/mL; R&D, catalog no. 5020) was used to block NPC differentiation (Ying et al. 2003). Spermine (Sigma, S3256) and spermidine (Sigma, S0266) were used at a concentration of 100 μM for all experiments. For neuronal differentiation, NPCs were replated onto laminin-coated plates for 3 d in N2B27 medium. For astrocyte differentiation, NPCs were replated on gelatin-coated plates for 8 d in N2B27 plus 1% FBS (Ying et al. 2003).
Polysome fractionation
For polysome fractionations, 20 million cells were incubated with 100 μg/mL cycloheximide (Sigma, catalog no. C4859) for 10 min. The cell pellets were resuspended in 2× RSB buffer (20 mM Tris-HCl at pH 7.4, 20 mM NaCl, 30 mM MgCl2, 200 μg/mL cycloheximide, 0.2 mg/mL heparin [Sigma, catalog no. H4787], 1000 U/mL RNasin), lysed with 2× lysis buffer (20 mM Tris-HCl at pH 7.4, 20 mM NaCl, 300 mM MgCl2, 1% Triton X-100, 2% Tween-20, 1% deoxycholate), and incubated on ice for 8 min, followed by centrifugation at 12,000g for 3 min to remove the nuclei. Supernatants were further centrifuged at 12,000g for 8 min at 4°C. Equal OD units were loaded onto 10%–50% linear sucrose gradients (prepared in 10 mM Tris-HCl at pH 7.4, 75 mM KCl, 1.5 mM MgCl2) and centrifuged at 36,000 rpm for 120 min at 8°C in a SW41 rotor (Beckman Coulter). Twelve fractions were collected from the top of the gradient using a piston gradient fractionator (BioComp Instruments). The absorbance at 254 nm was measured with a UV-M II monitor (Bio-Rad). Following fractionation, 110 μL of 10% SDS and 12 μL of proteinase K (10 mg/mL; Invitrogen) were added to each fraction and incubated for 30 min at 42°C. Fractions 1–5, 6–8, and 9–11 were combined as groups NT (nontranslated), LT (low translation), and HT (high translation), respectively.
Unfractionated cytoplasmic RNA and different groups or fractions of polysomal RNA were purified with phenol chloroform isoamyl extraction, followed by purification on an RNeasy column with on-column DNase digestion. Affymetrix GeneChip spike-in poly(A) RNAs (Ambion, catalog no. 900433) were added to each group of RNA. For the microarray, 300 ng of RNA from each group was amplified using the TargetAmp Nano-g Biotin-aRNA labeling kit (Epicentre Biotechnologies, catalog no. TAN07924), and the resultant cRNA was purified using the Picopure RNA isolation kit (Invitrogen, catalog no. 0202). Illumina Mouse Ref-8 v2 BeadChip arrays were loaded according to manufacturer's instructions. For the miRNA microarray, total RNA from ESCs and NPCs was purified using Trizol. Triplicate samples of RNA were profiled using the miRCURY LNA microarray (Exiqon).
Microarray analysis
For total cytoplasmic RNA, the background-subtracted intensities were first normalized using the cross-correlation method (Chua et al. 2006) and then log2-transformed. Identification of differentially expressed genes coding for cytoplasmic proteins was performed using the following criteria, with a P-value of ≤0.05 and a fold change of ≥1.5. For miRNAs, a cutoff mean of a twofold change and P-value of <0.001 were flagged as differentially expressed. The data for ribosomal profiling were first scaled using the external spike-in controls (thrB, pheA, and lysA) to account for the variations between different groups isolated based on sucrose gradients. As the volume of each isolation group varied, the scaled intensities were adjusted accordingly with their respective group volume. Genes coding for cytoplasmic proteins were assumed to be translationally differentially regulated when the translational rate between NPCs and ESCs was a ≥1.5-fold change and a P-value of <0.05. The translation rate is defined as the maximum of the ratios of scaled intensities between any two isolation groups.
qPCR analysis
cDNA was synthesized using a reverse transcription kit (SuperScript III, Invitrogen) according to the manufacturer's instruction. For fractionated RNA, the same volume of RNA was used, and for total RNA analysis, the same quantity of RNA was used. SYBR Green was used with gene-specific primers, or TaqMan probes were used for qRT–PCR on an ABI PRISM 7900 sequence detection system. For polysome fractions, CT values were normalized to spike in control RNAs dap and thr. Quantification of miRNA levels was done using the TaqMan miRNA reverse transcription kit. The expression level of miRNAs was measured using gene-specific predesigned TaqMan primers. U6 was used as normalization control.
Western blot and polyamine quantification
Thirty micrograms of protein extract was separated on a NuPAGE 4%–12% Bis-Tris gel and transferred to a PVDF membrane. Amd1 (Santa Cruz Biotechnology, catalog no. sc-98569), Oct4 (Santa Cruz Biotechnology, catalog no. sc-5279), Sox1 (Abcam, catalog no. ab-22572), GAPDH (Abcam, catalog no. ab-9484), STAT3 (Santa Cruz Biotechnology, catalog no. sc-8019), pSTAT3 (Santa Cruz Biotechnology, catalog no. sc-8059), and Sox2 (R&D Systems, catalog no. MAb2018) antibodies were used at 1:1000 dilution for overnight primary antibody incubation; c-Myc (Santa Cruz Biotechnology, catalog no. sc-764) was used at 1:50. Polyamine levels were measured using high-performance liquid chromatography (HPLC) as previously described (Igarashi et al. 1986).
Immunofluorescence
Cells were fixed with 4% PFA for 20 min and then blocked (2% BSA, 0.5% Triton X-100 in PBS) for 15 min at room temperature. The primary antibody Sox2 (Mab2018) or Oct4 (sc-5279) was used at a 1:200 dilution and incubated for 1 h at room temperature. Alexa488- or Alexa568-coupled secondary antibody (1 mg/mL; Invitrogen) was added to the cells (1:1000 in 0.2% BSA, 0.05% Triton X-100 in PBS) for 30 min. Cells were washed three times with 0.2% BSA and 0.05% Triton X-100 in PBS between each step. For AP staining, cells were stained using the Alkaline Phosphatase Detection kit (Millipore, SCR004) according to the manufacturer's instructions. For flow cytometry analysis, ESCs and NPCs were trypsinized and washed with PBS. The cells were analyzed for the intensity of GFP expression. FACS analysis was carried out using a BD FACSCalibur machine.
Luciferase assays
The 3′ UTR of candidate mRNAs were cloned at the 3′ end of the luciferase gene in PGL3 vector. pTK was used as a cotransfection control. The 3′ UTR of Amd1 was cloned into PsiCHECK-2 vector (Promega) at the 3′ end of the Renilla luciferase coding sequence. Firefly luciferase was used for normalization. ESCs or NPCs after 4 d of differentiation were transfected using FuGENE HD (Roche) according to manufacturer's instructions. Two days after transfection, cells were lysed, and Renilla and firefly luciferase activities were determined using the Dual-Luciferase Reporter Assay system (Promega). Amd1 3′ UTR deletion constructs were cloned into PsiCHECK-2. HeLa cells were transfected as for ESCs, and samples were collected 2 d later for luciferase assay. For miRNA analysis, miR-762 (genomic sequence NCBIM37:7:134851801:134852276) was cloned into the miR-Vec miRNA expression vector; miR-Vec carrying hsa-miR-155 was used as a control (kind gifts from Dr. Mathijs Voorhoeve) (Voorhoeve et al. 2006). To generate the Amd1 3′ UTR miR-762 target site mutant, PCR-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions.
Overexpression and siRNA treatment of Amd1
The Amd1 ORF was cloned into pTracer-EF vector (Invitrogen) under the control of the EF1α promoter. Cells were transfected after 3 d of NPC differentiation with FuGENE HD according to the manufacturer's instructions. Cells were analyzed at day 6 of NPC differentiation. For overexpression of Amd1 in differentiating ESCs, stable cell lines were made expressing Amd1 using the pCAG-GFP-IRESPuro vector (a gift from Dr. Stuart Avery) (Liew et al. 2007). Cells were induced to differentiate through withdrawal of LIF. For the Amd1 knockdown experiments, ESCs were transfected with 100 nM siRNAs using DharmaFECT transfection reagent 1 d and 3 d after plating according to the manufacturer's instructions. At day 6, cells were harvested. Amd1 siRNAs were purchased from Ambion (Amd1 siRNA AM 16708A, ID nos. 65502, 65596, and 160655). Scrambled siRNA was used as a control (Thermo Scientific, D-001810-10-05). miR762 mimics (Thermo Scientific, catalog no. 310780-01-0005) were transfected in a manner similar to the siRNAs.
Acknowledgments
We thank Drs. Colin Stewart, Davor Solter, Prabha Sampath, and Lim Chin Yan for critical reading of the manuscript; Dr. Austin Smith for providing the Sox1-GFP cell line; and Dr. Mathijs Voorhoeve for the miR-Vec constructs and critical discussion on the work. This work was supported by the Agency for Science, Technology, and Research (A*STAR) and the Singapore Stem Cell Consortium.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.182998.111.
References
- Aubert J, Stavridis MP, Tweedie S, O'Reilly M, Vierlinger K, Li M, Ghazal P, Pratt T, Mason JO, Roy D, et al. 2003. Screening for mammalian neural genes via fluorescence-activated cell sorter purification of neural precursors from Sox1-gfp knock-in mice. Proc Natl Acad Sci 100: 11836–11841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP 2008. The impact of microRNAs on protein output. Nature 455: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C, Grabel L 2007. Directing the differentiation of embryonic stem cells to neural stem cells. Dev Dyn 236: 3255–3266 [DOI] [PubMed] [Google Scholar]
- Cartwright P, McLean C, Sheppard A, Rivett D, Jones K, Dalton S 2005. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development 132: 885–896 [DOI] [PubMed] [Google Scholar]
- Casero RA, Pegg AE 2009. Polyamine catabolism and disease. Biochem J 421: 323–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Tomlinson SR 2009. The transcriptional foundation of pluripotency. Development 136: 2311–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua SW, Vijayakumar P, Nissom PM, Yam CY, Wong VV, Yang H 2006. A novel normalization method for effective removal of systematic variation in microarray data. Nucleic Acids Res 34: e38 doi: 10.1093/nar/qk1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vera N, Martinez E, Sanfeliu C 2008. Spermine induces cell death in cultured human embryonic cerebral cortical neurons through N-methyl-D-aspartate receptor activation. J Neurosci Res 86: 861–872 [DOI] [PubMed] [Google Scholar]
- Frostesjo L, Holm I, Grahn B, Page AW, Bestor TH, Heby O 1997. Interference with DNA methyltransferase activity and genome methylation during F9 teratocarcinoma stem cell differentiation induced by polyamine depletion. J Biol Chem 272: 4359–4366 [DOI] [PubMed] [Google Scholar]
- Hendrickson DG, Hogan DJ, McCullough HL, Myers JW, Herschlag D, Ferrell JE, Brown PO 2009. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol 7: e1000238 doi: 10.1371/journal.pbio.1000238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaviy HB, Murray MF, Sharp PA 2003. Embryonic stem cell-specific microRNAs. Dev Cell 5: 351–358 [DOI] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E 2011. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat Rev Genet 12: 99–110 [DOI] [PubMed] [Google Scholar]
- Igarashi K, Kashiwagi K 2010. Modulation of cellular function by polyamines. Int J Biochem Cell Biol 42: 39–51 [DOI] [PubMed] [Google Scholar]
- Igarashi K, Kashiwagi K, Hamasaki H, Miura A, Kakegawa T, Hirose S, Matsuzaki S 1986. Formation of a compensatory polyamine by Escherichia coli polyamine-requiring mutants during growth in the absence of polyamines. J Bacteriol 166: 128–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Atkins JF, Michael AJ 2010. A profusion of upstream open reading frame mechanisms in polyamine-responsive translational regulation. Nucleic Acids Res 38: 353–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Teh CH, Kunarso G, Wong KY, Srinivasan G, Cooper ML, Volta M, Chan SS, Lipovich L, Pollard SM, et al. 2008. REST regulates distinct transcriptional networks in embryonic and neural stem cells. PLoS Biol 6: e256 doi: 10.1371/journal.pbio.0060256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K 2005. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 19: 489–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law GL, Raney A, Heusner C, Morris DR 2001. Polyamine regulation of ribosome pausing at the upstream open reading frame of S-adenosylmethionine decarboxylase. J Biol Chem 276: 38036–38043 [DOI] [PubMed] [Google Scholar]
- Liew CG, Draper JS, Walsh J, Moore H, Andrews PW 2007. Transient and stable transgene expression in human embryonic stem cells. Stem Cells 25: 1521–1528 [DOI] [PubMed] [Google Scholar]
- Liu L, Rao JN, Zou T, Xiao L, Wang PY, Turner DJ, Gorospe M, Wang JY 2009. Polyamines regulate c-Myc translation through Chk2-dependent HuR phosphorylation. Mol Biol Cell 20: 4885–4898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Markowetz F, Unwin RD, Leek JT, Airoldi EM, MacArthur BD, Lachmann A, Rozov R, Ma'ayan A, Boyer LA, et al. 2009. Systems-level dynamic analyses of fate change in murine embryonic stem cells. Nature 462: 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Nakamura T, Nakao K, Arai T, Katsuki M, Heike T, Yokota T 1999. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J 18: 4261–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I 2006. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell 126: 1203–1217 [DOI] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ 2005. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci 102: 12135–12140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Nakatsu F, Kashiwagi K, Ohno H, Saito T, Igarashi K 2002. Essential role of S-adenosylmethionine decarboxylase in mouse embryonic development. Genes Cells 7: 41–47 [DOI] [PubMed] [Google Scholar]
- Niwa H, Burdon T, Chambers I, Smith A 1998. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes & Dev 12: 2048–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AR, Wang JY 1997. Polyamines modulate transcription but not posttranscription of c-myc and c-jun in IEC-6 cells. Am J Physiol 273: C1020–C1029 [DOI] [PubMed] [Google Scholar]
- Pegg AE, Xiong H, Feith DJ, Shantz LM 1998. S-adenosylmethionine decarboxylase: Structure, function and regulation by polyamines. Biochem Soc Trans 26: 580–586 [DOI] [PubMed] [Google Scholar]
- Persson L 2009. Polyamine homoeostasis. Essays Biochem 46: 11–24 [DOI] [PubMed] [Google Scholar]
- Pollard SM, Benchoua A, Lowell S 2006. Neural stem cells, neurons, and glia. Methods Enzymol 418: 151–169 [DOI] [PubMed] [Google Scholar]
- Proud CG 2007. Signalling to translation: How signal transduction pathways control the protein synthetic machinery. Biochem J 403: 217–234 [DOI] [PubMed] [Google Scholar]
- Saba R, Schratt GM 2010. MicroRNAs in neuronal development, function and dysfunction. Brain Res 1338: 3–13 [DOI] [PubMed] [Google Scholar]
- Sampath P, Pritchard DK, Pabon L, Reinecke H, Schwartz SM, Morris DR, Murry CE 2008. A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell 2: 448–460 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Matsuoka H, Saito M 2008. Generation of a multi-layer muscle fiber sheet from mouse ES cells by the spermine action at specific timing and concentration. Differentiation 76: 1023–1030 [DOI] [PubMed] [Google Scholar]
- Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH 2004. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med 10: 55–63 [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63 [DOI] [PubMed] [Google Scholar]
- Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG, Zavolan M, Svoboda P, Filipowicz W 2008. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol 15: 259–267 [DOI] [PubMed] [Google Scholar]
- Smirnova L, Grafe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG 2005. Regulation of miRNA expression during neural cell specification. Eur J Neurosci 21: 1469–1477 [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG 2009. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, Cha KY, Chung HM, Yoon HS, Moon SY, et al. 2004. Human embryonic stem cells express a unique set of microRNAs. Dev Biol 270: 488–498 [DOI] [PubMed] [Google Scholar]
- Tay YM, Tam WL, Ang YS, Gaughwin PM, Yang H, Wang W, Liu R, George J, Ng HH, Perera RJ, et al. 2008. MicroRNA-134 modulates the differentiation of mouse embryonic stem cells, where it causes post-transcriptional attenuation of Nanog and LRH1. Stem Cells 26: 17–29 [DOI] [PubMed] [Google Scholar]
- Thomas C, Lieberman J, Lal A 2010. Desperately seeking microRNA targets. Nat Struct Mol Biol 17: 1169–1174 [DOI] [PubMed] [Google Scholar]
- Tiscornia G, Izpisua Belmonte JC 2010. MicroRNAs in embryonic stem cell function and fate. Genes Dev 24: 2732–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, et al. 2006. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 124: 1169–1181 [DOI] [PubMed] [Google Scholar]
- Wang JY, McCormack SA, Viar MJ, Wang H, Tzen CY, Scott RE, Johnson LR 1993. Decreased expression of protooncogenes c-fos, c-myc, and c-jun following polyamine depletion in IEC-6 cells. Am J Physiol 265: G331–G338 [DOI] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R 2007. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet 39: 380–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R 2008. Embryonic stem cell-specific microRNAs regulate the G1–S transition and promote rapid proliferation. Nat Genet 40: 1478–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E, Plasterk RH 2005. MicroRNA function in animal development. FEBS Lett 579: 5911–5922 [DOI] [PubMed] [Google Scholar]
- Williamson AJ, Smith DL, Blinco D, Unwin RD, Pearson S, Wilson C, Miller C, Lancashire L, Lacaud G, Kouskoff V, et al. 2008. Quantitative proteomics analysis demonstrates post-transcriptional regulation of embryonic stem cell differentiation to hematopoiesis. Mol Cell Proteomics 7: 459–472 [DOI] [PubMed] [Google Scholar]
- Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS 2009. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 137: 647–658 [DOI] [PubMed] [Google Scholar]
- Ying QL, Stavridis M, Griffiths D, Li M, Smith A 2003. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol 21: 183–186 [DOI] [PubMed] [Google Scholar]
- Zovoilis A, Smorag L, Pantazi A, Engel W 2009. Members of the miR-290 cluster modulate in vitro differentiation of mouse embryonic stem cells. Differentiation 78: 69–78 [DOI] [PubMed] [Google Scholar]