The tRNA splicing endonuclease (SEN) complex has different cellular localizations in yeast and vertebrates. In this study, Hopper and colleagues use yeast that have been re-engineered to mimic vertebrate tRNA splicing to show that yeast SEN has a novel role in the cytoplasm, unrelated to its known role in tRNA splicing.
Keywords: tRNA biogenesis, rRNA processing, mitochondria, nucleus
Abstract
Pre-tRNA splicing is an essential process in all eukaryotes. In yeast and vertebrates, the enzyme catalyzing intron removal from pre-tRNA is a heterotetrameric complex (splicing endonuclease [SEN] complex). Although the SEN complex is conserved, the subcellular location where pre-tRNA splicing occurs is not. In yeast, the SEN complex is located at the cytoplasmic surface of mitochondria, whereas in vertebrates, pre-tRNA splicing is nuclear. We engineered yeast to mimic the vertebrate cell biology and demonstrate that all three steps of pre-tRNA splicing, as well as tRNA nuclear export and aminoacylation, occur efficiently when the SEN complex is nuclear. However, nuclear pre-tRNA splicing fails to complement growth defects of cells with defective mitochondrial-located splicing, suggesting that the yeast SEN complex surprisingly serves a novel and essential function in the cytoplasm that is unrelated to tRNA splicing. The novel function requires all four SEN complex subunits and the catalytic core. A subset of pre-rRNAs accumulates when the SEN complex is restricted to the nucleus, indicating that the SEN complex moonlights in rRNA processing. Thus, findings suggest that selection for the subcellular distribution of the SEN complex may reside not in its canonical, but rather in a novel, activity.
tRNAs deliver amino acids to ribosomes during translation. Transcribed as precursors (pre-tRNAs), they undergo numerous post-transcriptional processing steps, including 5′ and 3′ maturation and nucleoside modifications (for reviews, see Hopper and Shaheen 2008; Hopper et al. 2010; Phizicky and Hopper 2010). For tRNAs encoded by intron-containing genes, removal of the introns (pre-tRNA splicing) is an essential, multistep process (Trotta et al. 1997; Hopper and Shaheen 2008; Hopper et al. 2010; Phizicky and Hopper 2010). Of the 274 genes that encode tRNAs in the yeast Saccharomyces cerevisiae, 61 contain an intron (Phizicky and Hopper 2010).
In yeast and vertebrates, the heterotetrameric tRNA splicing endonuclease (SEN) complex catalyzes pre-tRNA cleavage at the intron–exon junctions (Peebles et al. 1983; Trotta et al. 1997; Paushkin et al. 2004). The catalytic subunits Sen2 and Sen34 are conserved from Archaea (Kleman-Leyer et al. 1997; Lykke-Andersen and Garrett 1997; Tocchini-Valentini et al. 2005) to vertebrates, whereas the other two subunits, Sen15 and Sen54, are absent from Archaea and are not highly conserved between yeast and vertebrates (Paushkin et al. 2004). Intron removal generates two tRNA half-molecule exons: The 5′ half possesses a terminal 2′,3′-cyclic phosphate, and the 3′ half possesses a 5′-hydroxyl (Peebles et al. 1983). In vertebrates and Archaea, the tRNA half exons are ligated by a tRNA ligase that joins 2′,3′-cyclic phosphate and 5′-hydroxyl (Salgia et al. 2003; Popow et al. 2011). In contrast, the plant and yeast tRNA ligase Trl1 possesses phosphodiesterase, polynucleotide kinase, and ligase domains that convert the 2′,3′-cyclic phosphate to a 2′-phosphate and activate and phosphorylate the 5′ half prior to ligation (Phizicky et al. 1986; Wang et al. 2006). The resulting 2′-phosphate at the ligation junction is transferred to NAD+ by a 2′-phosphotransferase, Tpt1, in the third step of splicing (Culver et al. 1997).
Although the SEN complex is conserved among eukaryotes, its spatial distribution is not. First demonstrated in Xenopus oocytes (Melton et al. 1980; Lund and Dahlberg 1998) and supported by studies in human cells (Paushkin et al. 2004), pre-tRNA splicing was shown to occur solely in the nucleus, leading to the expectation that it would be nuclear in all eukaryotes. However, the S. cerevisiae SEN complex subunits are not nuclear, but rather are located on the cytoplasmic surface of mitochondria (Yoshihisa et al. 2003). Moreover, the S. cerevisiae tRNA ligase is distributed throughout the cytoplasm, consistent with its additional unessential function in the cytoplasmic unfolded protein response (UPR) (Nikawa et al. 1996; Sidrauski et al. 1996; Mori et al. 2010). The location of the endonuclease in the cytoplasm has been verified (Huh et al. 2003; Shaheen and Hopper 2005), and genetic and biochemical studies showed that yeast pre-tRNA splicing occurs in the cytoplasm (Yoshihisa et al. 2007). Data also indicate that pre-tRNA splicing in Schizosaccharomyces pombe, a distantly related fungus, may also occur in the cytoplasm (Intine et al. 2002; Matsuyama et al. 2006).Therefore, pre-tRNA splicing is nuclear in vertebrates but cytoplasmic in yeast.
Here we investigated three possible nonexclusive explanations for the cytoplasmic location of pre-tRNA splicing in yeast: (1) Yeast may have evolved cytoplasmic tRNA splicing to evade harm caused by cleavage of putative off-target nuclear substrates; (2) the involvement of yeast tRNA ligase in UPR may have provided a selection for cytoplasmic pre-tRNA splicing; and (3) the yeast tRNA splicing machinery or pre-tRNA processing intermediates may serve additional unknown cytoplasmic functions. There is precedence for an alternative function for the tRNA SEN, as it was reported that Sen34 depletion results in delayed pre-rRNA processing (Volta et al. 2005), and the vertebrate SEN complex has been proposed to affect pre-mRNA 3′ processing (Paushkin et al. 2004). To address the possibilities, we genetically engineered yeast that conditionally lack endogenous mitochondrially located tRNA SEN subunits (the mito SEN complex) and instead express nuclear tRNA SEN subunits (the nuc SEN complex). We show that the nuc SEN complex is functional and is not detrimental to yeast, arguing against possibility 1. Even though pre-tRNA splicing by the nuc SEN complex occurs efficiently, yeast are not viable without the functional mito SEN complex. Our data provide evidence that all four subunits of the mito SEN complex and the catalytic core of Sen2 are required for a novel and essential function unrelated to pre-tRNA splicing, thereby supporting possibility 3. One such possible role for the mito SEN complex is in pre-rRNA processing.
Results
Redistribution of yeast tRNA splicing machinery from the cytoplasm to the nucleus
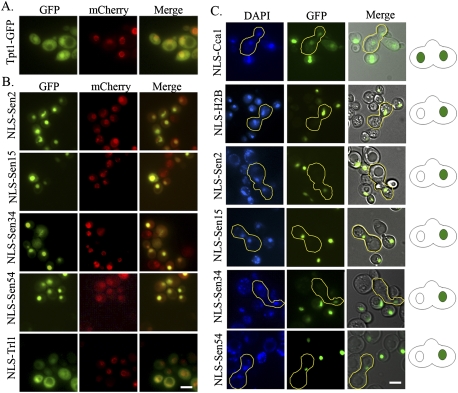
As previously reported (Huh et al. 2003; Yoshihisa et al. 2003; Shaheen and Hopper 2005), each of the four tRNA SEN subunits is located on the mitochondrial surface (Supplemental Fig. S1), and the tRNA ligase (Trl1) is cytoplasmic (Mori et al. 2010). The location of the 2′-phosphotransferase (Tpt1), previously not determined, is in both the nucleus and the cytoplasm (Fig. 1A).
Figure 1.
Subcellular location of tRNA splicing machinery in yeast. (A) Location of Tpt1-GFP and Nup49-mCherry, and merged images. (B) Nuclear redistribution of tRNA SEN subunits and tRNA ligase. Location of plasmid-encoded NLS-tagged and 2GFP-tagged tRNA SEN subunits (NLS-Sen) or tRNA ligase (NLS-Trl1) in cells harboring Nup49-mCherry. (C) Heterokaryon analyses of nuc SEN complex subunits. Yeast expressing GFP-tagged NLS-Sen, NLS-H2B, or NLS-Cca1 were mated with the kar1-1 mutant to generate heterokaryons (outlined). Cartoons of heterokaryons on the right depict either shuttling (two green nuclei) or nonshuttling (one green nucleus) results. DAPI staining shows visualization of nuclear DNA. More than 20 heterokaryons were counted, and the same results were obtained as depicted in the figure. Bar, 4 μm.
To assess whether yeast pre-tRNA splicing can occur in the nucleus instead of on the mitochondrial surface, we directed the mito SEN complex and the cytoplasmic tRNA ligase (cyt ligase) to the nucleoplasm, mimicking the distribution in vertebrates. Each SEN protein and tRNA ligase was tagged with a nuclear localization sequence (NLS) at the N terminus and with two tandem GFPs at the C terminus. Since Tpt1 localizes to both the nucleus and cytoplasm, its distribution was not altered. Recombinant proteins were expressed under a galactose (Gal)-inducible promoter, which is both inducible and repressible. The presence of glucose in the growth medium represses expression and thereby prevents potential harm by ectopic expression. The localizations of the recombinant proteins were assessed following a 2-h induction by addition of Gal. All four NLS-tagged tRNA SEN subunits (the nuc SEN complex)—NLS-Sen2-2GFP, NLS-Sen15-2GFP, NLS-Sen34-2GFP, and NLS-Sen54-2GFP—are nuclear, as evidenced by the GFP-localized signal within the nuclear margins of the nucleoporin Nup49-mCherry signal (Fig. 1B; Lai et al. 2009). The NLS-tagged tRNA ligase NLS-Trl1-2GFP is localized to both the nucleus and the cytoplasm (Fig. 1B).
Although no steady-state cytoplasmic pools of the nuc SEN complex were visualized, it is possible that such pools dynamically exist. To address this concern, we analyzed heterokaryons formed after mating strains expressing nuclear-redirected SEN complex proteins to karyogamy-deficient kar1-1 mutants that are unable to fuse nuclei when mated, resulting in a shared cytoplasm and two parental nuclei in a single heterokaryon. As expected, control experiments using cells with the shuttling NLS-Cca1-GFP (Feng and Hopper 2002) protein mated to kar1-1cells resulted in both nuclei of the heterokaryon exhibiting GFP signals (Fig. 1C). The control nonshuttling protein NLS-H2B-GFP (Peng and Hopper 2000) was located in only one nucleus of the heterokaryon (Fig. 1C). When strains expressing each of the four nuc SEN complex subunits were mated with kar1-1, the resulting heterokaryons exhibited only one nucleus with GFP signals (Fig. 1C). The data show that none of the nuc SEN complex subunits shuttle between the nucleus and cytoplasm; the data also indicate that it is unlikely that there are low cytoplasmic levels of the tRNA SEN subunits.
Pre-tRNA splicing in the nucleus
Since the nuc SEN complex subunits are modified at both the N and C termini, it is possible that their catalytic activities were compromised. To address this possibility, we assessed the abilities of the recombinant nuc SEN complex and nuclear and cytoplasmic tRNA ligase (nuc-cyt ligase) to complement los1Δ, rna1-1, sen2-42, and rlg1-4 mutations. Los1 is the only known nuclear export protein for intron-containing pre-tRNAs (Hopper and Shaheen 2008; Murthi et al. 2010). Deletion of LOS1 (los1Δ) results in nuclear retention of tRNAs (Sarkar and Hopper 1998) so that intron-containing tRNAs have reduced access to the mitochondrial splicing machinery and are not spliced (Hopper et al. 1980). To test whether the nuc SEN complex and the nuc-cyt ligase are catalytically active, we transformed the complete set of plasmids expressing the Gal-inducible nuc SEN complex and nuc-cyt ligase into los1Δ cells. Accumulation of intron-containing pre-tRNAIleUAU in los1Δ cells is suppressed by the nuc SEN complex and the nuc-cyt ligase (Supplemental Fig. S2A). Similar observations were obtained using a strain that has a temperature-sensitive (ts) mutation in the gene encoding Ran GAP, rna1-1 (Supplemental Fig. S2B; Corbett et al. 1995). The data show that the recombinant nuc SEN complex is catalytically active.
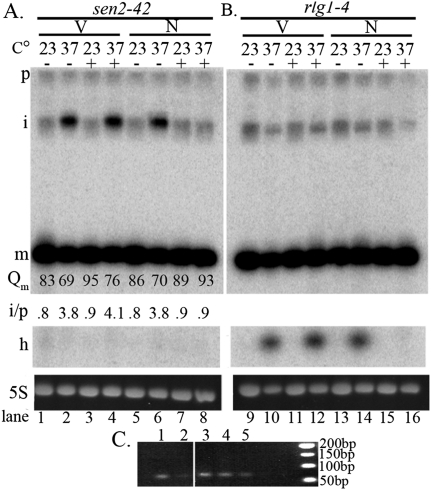
We also used a yeast strain expressing the rapid and reversible ts SEN2 allele sen2-42 (Yoshihisa et al. 2007) to assess tRNA splicing. At the nonpermissive (np) temperature, sen2-42 causes accumulation of intron-containing pre-tRNA in the cytoplasm (Yoshihisa et al. 2007). We transformed sen2-42 cells with the set of plasmids encoding the nuc SEN complex and the nuc-cyt tRNA ligase and assessed whether the pre-tRNA splicing defect of the sen2-42 mutant can be complemented. sen2-42 cells transformed with vectors (V) accumulate intron-containing tRNALeuCAA after a 2-h shift to the np temperature (37°C) in the presence (+) or absence (−) of Gal (Fig. 2A, lanes 2,4). A small compensatory decrease of mature tRNA levels is detectable at 37°C (Fig. 2A, m). As anticipated, the ratio of the signal intensity of the end-matured intron-containing pre-tRNA (Fig. 2A, i) to the initial transcript (Fig. 2A, p) is higher at 37°C than at the permissive temperature (23°C). In contrast, sen2-42 cells expressing the nuc SEN complex and nuc-cyt ligase (N) do not accumulate intron-containing tRNALeuCAA after a 2-h shift to 37°C in the presence of Gal (Fig. 2A, lane 8), but do so in its absence (Fig. 2A, lane 6). When pre-tRNA accumulation is corrected by the nuc SEN complex and nuc-cyt ligase, a compensatory increase in the mature tRNA results (Fig. 2A). Similarly, the other nine families of tRNAs encoded by intron-containing tRNA genes are also processed by nuclear-localized tRNA splicing machinery (Supplemental Fig. S3). The data show that all pre-tRNAs serve as substrates for nuclear-localized tRNA SEN and also confirm our finding that the modified splicing proteins are catalytically active (Supplemental Fig. S2).
Figure 2.
Ability of nuclear-localized tRNA splicing machinery to catalyze pre-tRNALeuCAA splicing. (A) Correction of splicing defect of sen2-42 by the nuc SEN complex and nuc-cyt ligase. Northern analysis of sen2-42 cells containing either vectors (V) or the set of plasmids encoding the nuc SEN complex and nuc-cyt ligase (N) in the presence (+) or absence (−) of Gal induction at the permissive temperature (23°C) or after 2 h at the np temperature (37°C). (B) Correction of tRNA ligation defects of rlg1-4 by the nuc SEN complex and nuc-cyt ligase. Northern analysis of rlg1-4 cells containing the same set of plasmids as A under the same conditions. (P) Initial pre-tRNA transcript; (i) end-matured intron-containing pre-tRNA; (m) mature tRNA; (h) half tRNAs; (5S) loading control; (Qm) quantifications of m signal intensity. Ratios of the signal intensities of i/p are shown. (C) 2′-Phosphotransferase activity. RT–PCR was used to assess whether spliced tRNA possess a 2′-phosphate at the splice junction of tRNAIle. (Lane 1) tpt1Δ cells with Gal-inducible TPT1 in the presence of Gal. (Lane 2) The same cells as lane 1 without Gal induction. (Lanes 3,4) sen2-42 cells with the set of plasmids encoding nuclear-localized splicing machinery in the presence of Gal at 37°C. (Lane 5) Wild-type cells.
The apparent lack of half tRNAs accumulating in sen2-42 cells expressing the engineered nuc SEN complex and nuc-cyt ligase (Fig. 2A, h) suggests that NLS-Trl1-2GFP-mediated tRNA ligation is not defective. To confirm this, we transformed the same set of plasmids into a strain with a ts mutation of the tRNA ligase gene rlg1-4 (Phizicky et al. 1992). As expected, at the np temperature (37°C), rlg1-4 cells with vectors, with (+) or without (−) Gal induction, accumulate species that migrate at the anticipated position for tRNA halves (Fig. 2B, V, lanes 10,12, h). The half molecules do not accumulate at the permissive temperature (23°C) (Fig. 2B, lanes 9,11,13,15). rlg1-4 cells transformed with plasmids encoding the Gal-inducible nuc SEN complex and nuc-cyt ligase without Gal induction (−) accumulate the tRNA half molecules at 37°C (Fig. 2B, N, lane 14), whereas tRNA half molecules do not accumulate in cells after 2 h of Gal induction (+) (Fig. 2B, N, lane 16). The data demonstrate that upon removal of introns by the nuc SEN complex, NLS-Trl1-2GFP efficiently ligates tRNA halves.
We devised a RT–PCR strategy to assess 2′-phosphate removal, the third step of pre-tRNA splicing. The strategy was based on the report that 2′-phosphates at RNA ligation junctions inhibit reverse transcription, prohibiting PCR amplification (Schutz et al. 2010). In a strain lacking 2′-phosphotransferase (tpt1Δ) complemented with Gal-inducible TPT1, cDNA production from tRNAIleUAU is hampered when Tpt1 production is inhibited by glucose (Fig. 2C, lane 2), but cDNA production and subsequent PCR amplification occur when the medium contains Gal (Fig. 2C, lane 1). Thus, Tpt1 activity can be assessed by this method. After a shift to the np temperature for 2 h, tRNAIleUAU extracted from sen2-42 cells expressing the nuc SEN complex and nuc-cyt ligase generated full-length cDNA at levels (Fig. 2C, lanes 3,4) similar to wild-type cells (Fig. 2C, lane 5). The data show that upon removal of introns by the nuclear-localized endonuclease subunits, the endogenous 2′-phosphotransferase Tpt1 catalyzes 2′-phosphate removal from ligated tRNAIleUAU. Thus, intron-containing pre-tRNAs are fully processed in sen2-42 cells with the nuclear tRNA splicing machinery.
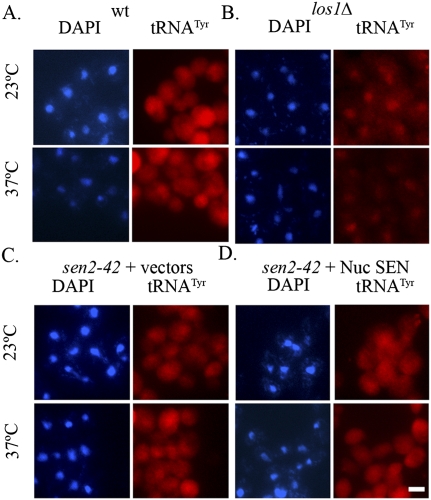
To determine whether tRNAs cleaved in the nucleus are exported to the cytoplasm, we monitored the location of tRNAs by fluorescence in situ hybridization (FISH) analyses. FISH of intron-containing tRNATyr was carried out employing sen2-42 cells transformed with either vectors or the set of plasmids encoding the Gal-inducible nuc SEN complex and nuc-cyt ligase using a probe complementary to mature tRNATyr. Wild-type cells, with an equal distribution of tRNATyr throughout the nucleus and cytoplasm (Fig. 3A; Sarkar and Hopper 1998), and los1Δ cells, with nuclear tRNATyr pools (Fig. 3B; Sarkar and Hopper 1998), served as controls. Nuclear accumulation of tRNATyr was not detected in sen2-42 cells at 23°C or 37°C whether the cells expressed vectors alone or the nuc SEN complex and nuc-cyt ligase (Fig. 3C,D). The data show that tRNA spliced in the nucleus are normally exported from the nucleus.
Figure 3.
FISH analyses of tRNATyrGUA in yeast expressing nuclear tRNA splicing machinery. (A) Wild type (wt). (B) los1Δ. (C) sen2-42 cells expressing vectors. (D) sen2-42 mutants with the plasmids encoding the nuc SEN complex and nuc-cyt ligase. Cells were assessed after growth at 23°C or after a 2-h shift to 37°C, and the location of tRNATyr was determined. DAPI was used to locate nuclear DNA. Bar, 5 μm.
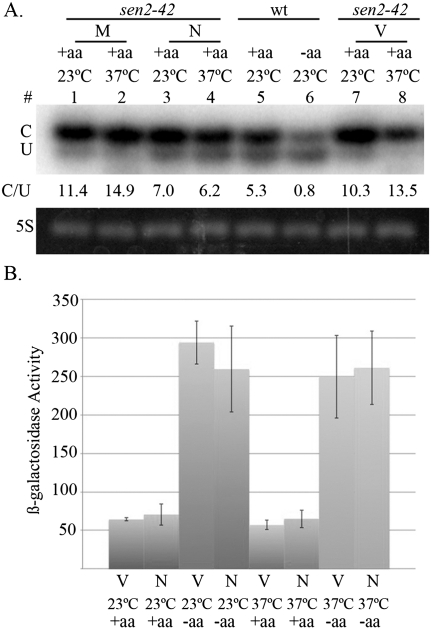
We performed Northern analyses to assess whether aminoacylation of tRNA is affected in sen2-42 cells expressing the nuc SEN complex and nuc-cyt ligase. We first assessed tRNALeuUAG, one of the families tRNAs encoded by intron-containing tRNA genes (Fig. 4A). For a control, we extracted tRNAs from wild-type cells grown in fed conditions (+aa) and starved conditions (−aa). As expected, the majority of tRNALeuUAG from fed cells is aminoacylated (Fig. 4A, lane 5, C). Also as expected, the majority of tRNALeuUAG from wild-type cells grown in starved conditions (−aa) was not aminoacylated (Fig. 4A, lane 6, U), demonstrating that aminoacylation defects can be detected by this analysis. sen2-42 cells—whether expressing vectors (V), the mito SEN complex (M), or the nuc SEN complex and nuc-cyt ligase (N) at the permissive (23°C) or np (37°C) temperature—have the same levels of aminoacylated tRNAs as wild-type fed cells (Fig. 4A). We also assessed tRNATyr (data not shown) and obtained the same results. Thus, by this Northern assay, there is no defect in tRNA aminoacylation when pre-tRNAs are spliced in the nucleus. To address all of the tRNA families by a different assay, we used a lacZ reporter fused in-frame to the 5′ upstream regulatory region of GCN4 (Hinnebusch 1985), which responds to the global tRNA aminoacylation status in cells. Binding of nonaminoacylated tRNAs to a domain in Gcn2 activates its kinase activity and ultimately results in derepression of Gcn4 translation (for review, see Hinnebusch 1997).We transformed the plasmid expressing the GCN4 reporter into sen2-42 cells expressing either vectors (V) or the nuc SEN complex and nuc-cyt ligase (N) and monitored the β-galactosidase activity at the permissive temperature (23°C) or after a 2-h incubation at the np temperature (37°C). Control experiments assessed β-galactosidase activity in cells incubated in medium lacking amino acids (−aa). As expected, when cells were transferred to medium lacking amino acids, β-galactosidase activity was significantly higher than when cells were maintained in fed conditions at both 23°C and 37°C (Fig. 4B, −aa/+aa). The results verify that the assay detects nonaminoacylated tRNAs. Under fed conditions (+aa), sen2-42 cells expressing either vectors (V) or the nuc SEN complex and nuc-cyt ligase (N) exhibit significantly lowered β-galactosidase activity, indicating that there is no detectable defect in tRNA aminoacylation when pre-tRNAs are spliced in the nucleus (Fig. 4B, +aa, V, N). The cumulative data show that when distribution of the tRNA SEN is changed from exclusively mitochondrial to nuclear, all three steps of tRNA splicing, tRNA nuclear export, and tRNA aminoacylation occur.
Figure 4.
Efficient tRNA aminoacylation in cells possessing nuclear tRNA splicing. (A) Assessment of tRNALeuUAG aminoacylation by Northern analysis of tRNA from W303 (wt) Leu− cells or sen2-42 cells containing either vectors (V), the set of plasmids encoding the nuc SEN complex and nuc-cyt ligase (N), or the set of plasmids encoding the mito SEN complex and cytoplasmic ligase (M) in the presence of Gal induction at the permissive temperature (23°C) or after 2 h at the np temperature (37°C). (C) Aminoacylated tRNALeuUAG; (U) nonaminoacylated tRNALeuUAG. (B) Assessment of tRNA aminoacylation with Gcn4 translation using a β-galactosidase reporter assay. sen2-42 cells containing either vectors (V) or the set of plasmids encoding the nuc SEN complex and nuc-cyt ligase (N) in the presence (+aa) or absence (−aa) of amino acids.
Novel cytoplasmic function for the tRNA SEN
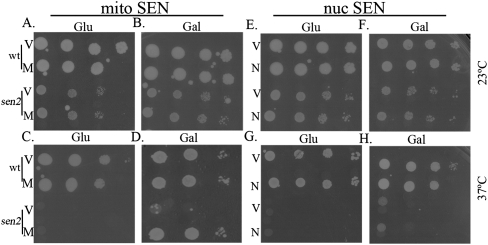
Since tRNA splicing can occur in the nucleus of yeast, we hypothesized that nuclear tRNA splicing would substitute for the mitochondrial/cytoplasmic pre-tRNA splicing machinery. We assessed whether ts growth of sen2-42 cells at 37°C was alleviated by the nuc SEN complex and the nuc-cyt ligase (Fig. 5). Cells possessing the set of plasmids encoding the Gal-inducible mitochondrially located tRNA SEN (the mito SEN complex) are able to grow at 37°C in selective medium containing Gal (Fig. 5D, M) but not in selective medium lacking Gal (Fig. 5C, M) and thus complement the sen2-42 growth defect. sen2-42 cells transformed with vectors are unable to grow at 37°C, but grow similarly to wild type at 23°C in medium with or without Gal (Fig. 5, V). Likewise, sen2-42 mutants transformed with plasmids encoding the nuc SEN complex and nuc-cyt ligase grow comparably to wild-type cells when propagated at 23°C (Fig. 5E,F). sen2-42 mutant cells with the nuc SEN complex and nuc-cyt ligase in the absence of Gal fail to grow at 37°C (Fig. 5G, N). Surprisingly, sen2-42 mutants expressing the nuc SEN complex and nuc-cyt tRNA ligase, which results in efficient pre-tRNA splicing, are unable to grow at 37°C (Fig. 5H, N). The inability to grow in medium containing Gal is not due to high-level continuous expression of the nuc SEN complex and nuc-cyt ligase, since wild-type cells transformed with the same plasmids grow well at both 23°C and 37°C (Fig. 5F,H). Furthermore, we also conducted a growth assay of the ts tRNA ligase mutant rlg1-4 expressing nuclear Trl1 to ensure that the lack of viability is not due to defective NLS-Trl1 and showed that NLS-Trl1 expressed in rlg1-4 rescues the ts growth phenotype of rlg1-4 cells (Supplemental Fig. S4). Since pre-tRNA splicing is efficient in sen2-42 cells expressing the nuc SEN complex and nuc-cyt ligase (Fig. 2), and availability of the mito SEN complex can rescue the ts growth defect associated with the mutation (Fig. 5D), the inability of sen2-42 cells to grow at the np temperature when expressing nuclear splicing machinery (Fig. 5H) indicates that the mito SEN complex serves an essential cytoplasmic function.
Figure 5.
The mitochondrial tRNA SEN complex and cytoplasmic tRNA ligase can rescue the ts growth phenotype of sen2-42, whereas the nuclear tRNA splicing machinery does not. Wild-type (wt) or sen2-42 (sen2) cells with the set of vectors (V) or the set of Gal-inducible plasmids encoding the nuc SEN complex and nuc-cyt ligase (N) or the mito SEN complex and cytoplasmic ligase (M). The spot assay was accomplished by placing an equal number of serially diluted cells on selective medium containing Gal (Gal) or lacking Gal (Glu) and incubated at the permissive temperature (23°C) (A,B,E,F) or the np temperature (37°C) (C,D,G,H).
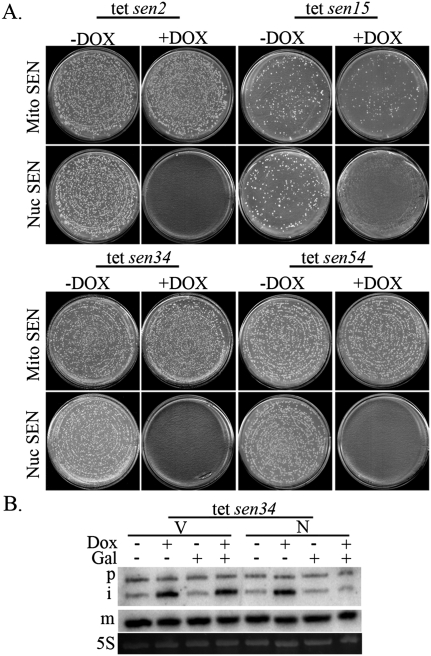
The above experiments address only the requirement for Sen2 in the cytoplasm. To determine whether the other three SEN subunits are required in the cytoplasm when tRNA endonucleolytic cleavage occurs in the nucleus, we assessed whether depletion of each individual subunit can be complemented by the nuc SEN complex and nuc-cyt ligase. We generated yeast strains with endogenous tRNA SEN subunits expressed from tetracycline (tet)-regulatable promoters (Mnaimneh et al. 2004). The presence of doxycycline (+DOX) in the growth medium represses expression of individual tRNA SEN subunits, which prevents cell growth, whereas the absence of doxycycline (−DOX) allows cell growth (Fig. 6A). We transformed each tet-regulatable strain—tet sen2, tet sen15, tet sen34, and tet sen54—with the set of plasmids encoding either the mito SEN complex and cyt ligase or the nuc SEN complex and nuc-cyt ligase. The strains were grown in +Gal −DOX medium and were replica-plated to +Gal +DOX or +Gal −DOX medium. As expected, all four tet-regulatable yeast strains expressing the mito SEN complex and cyt ligase were able to grow in +DOX or −DOX medium (Fig. 6A, Mito SEN). Also as anticipated, each of the strains expressing plasmids encoding the nuc SEN complex and nuc-cyt tRNA ligase grows in −DOX medium (Fig. 6A, −DOX, Nuc SEN). In contrast, none of the tet-regulatable yeast strains expressing the nuc SEN complex and nuc-cyt ligase grows on medium containing DOX (Fig. 6A, +DOX, Nuc SEN).
Figure 6.
All four SEN subunits are required in the cytoplasm for viability. (A) Assessment of whether depletion of the endogenous mitochondrial tRNA SEN can be complemented with nuclear tRNA splicing. Yeast expressing tet-regulated tRNA SEN subunits (tet sen) were transformed with sets of plasmids encoding either the Nuc SEN complex and nuc-cyt ligase or the mito SEN complex and cyt ligase. Growth after 5 d was assessed on Gal-containing medium in which cells were depleted (+DOX) or not depleted (−DOX) of the endogenous tet-regulated tRNA SEN subunit. Where colonies are absent, no colonies appeared after >7-d incubation. (B) Assessment of whether the nuclear tRNA splicing defect of tet sen34 can be complemented by nuclear tRNA splicing. The yeast tet sen34 strain containing either vectors (V) or the set of plasmids encoding the nuc SEN complex and nuc-cyt ligase (N) were assessed for nuclear tRNA splicing by northern analysis in the presence (+) or absence (−) of either Gal or DOX. Subunits were depleted by addition of DOX for 9 h. (P) Initial pre-tRNA transcript; (i) end-matured intron-containing pre-tRNA; (m) mature tRNA; (5S) loading control.
To ensure that plasmids expressing nuc SEN complex and nuc-cyt ligase were functional in tet sen strains, we performed Northern analysis on the tet sen34 strain (Fig. 6B). When the strain is expressing the nuc SEN complex and nuc-cyt ligase (N), intron-containing pre-tRNAIleUAU accumulates after 9 h of growth in +DOX(+) medium in the absence of Gal, whereas in the presence of Gal, the pre-tRNAIleUAU accumulation is suppressed to levels that are comparable with − DOX medium. Controls expressing vectors (V), do not exhibit lowered intron-containing pre-tRNA levels even in the presence of Gal. The data indicate that the nuc SEN complex and nuc-cyt ligase are catalytically active in tet sen strains and further demonstrate that the requirement of the endogenous mitochondrial tRNA SEN subunits are unrelated to tRNA splicing. Thus, each of the four endogenous mitochondrial SEN subunits is required for viability even if pre-tRNA splicing occurs efficiently in the nucleus.
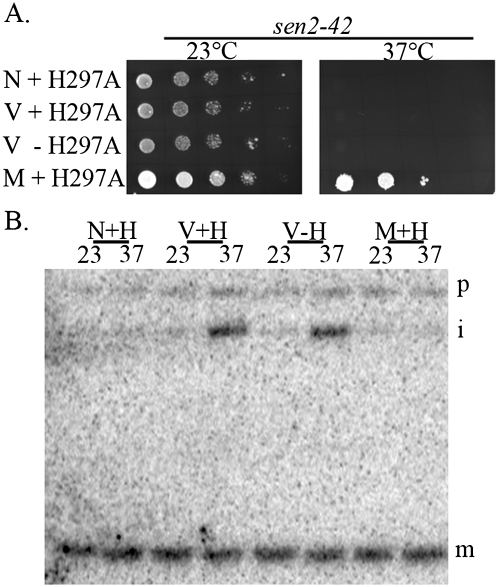
To determine whether the catalytic core of Sen2 for tRNA splicing is required in the cytoplasm when efficient pre-tRNA splicing occurs in the nucleus, we expressed a catalytically inactive sen2 allele, sen2 H297A (Trotta et al. 2006), in sen2-42 mutants harboring the set of plasmids encoding either the mito SEN complex and cyt ligase or the nuc SEN complex and nuc-cyt ligase. We assessed whether the ts phenotype of the sen2-42 transformants was alleviated with the catalytically inactive sen2 H297A, which we confirmed localizes to the endogenous mitochondrial location (data not shown). As anticipated, sen2-42 transformants coexpressing empty vectors as well as the catalytically inactive sen2 H297A are unable to grow at the np temperature (Fig. 7A, V+H297A). Also as expected, sen2-42 expressing plasmids encoding the entire set of mito SEN complex and cyt ligase at the endogenous location was able to complement the ts phenotype of sen2-42 when coexpressing the catalytically inactive sen2 H297A (Fig. 7, M+H297A). However, expression of the entire set of plasmids encoding the nuc SEN complex and nuc-cyt ligase in sen2-42 coexpressing sen2 H297A was unable to complement the ts phenotype (Fig. 7, N+H297A). The data indicate that in addition to the requirement of each of the subunits in the cytoplasm (Fig. 6), Sen2 protein with a normal catalytic core, required for tRNA splicing, must also be present in the cytoplasm even when splicing of tRNAs occurs efficiently in the nucleus. To assess the possibility that nuclear splicing is inhibited by the presence of the catalytically inactive sen2H297A, thereby leading to growth defects, we performed Northern analysis and demonstrated that the presence of sen2 H297A does not inhibit nuclear splicing carried out by the nuclear endonuclease subunits and ligase after a shift to the np temperature (Fig. 7B, N+H). Taken together, the results show that the catalytic core of the SEN is required for an essential cytoplasmic function unrelated to tRNA splicing.
Figure 7.
The catalytic core of Sen2 for tRNA splicing is required for the novel function of the tRNA SEN complex at cytoplasm and does not affect nuclear tRNA splicing by the nuc SEN complex. sen2-42 cells were transformed with the set of plasmids encoding either the mito SEN complex (M), vectors (V), or the Nuc SEN complex (N) in addition to the presence (+) or absence (−) of catalytically inactive sen2 (H297A). (A) Transformants were assessed for growth in a spot assay in selective medium containing Gal at the permissive temperature (23°C) or np temperature (37°C). (B) Transformants were assessed for tRNA splicing defects. (p) Precursor tRNA; (i) end-processed intron-containing precursor tRNA; (m) mature tRNA; (H) catalytically inactive sen2 H297A.
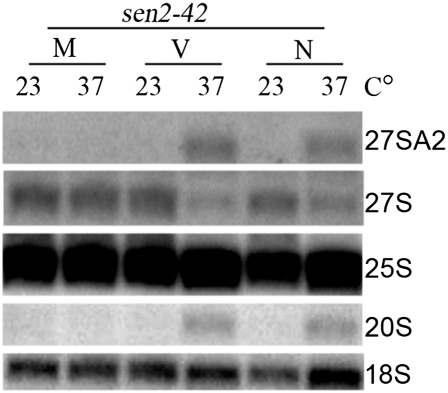
A previous study reported that pre-rRNA processing is delayed upon Sen34 depletion for 8–16 h (Volta et al. 2005). To confirm the results and extend them to another SEN subunit, we investigated pre-rRNA processing by Northern analysis in sen2-42 cells transformed with the set of plasmids encoding the nuc SEN complex and nuc-cyt ligase (Fig. 8, N). For controls, we assessed rRNA processing in sen2-42 mutants expressing the mito SEN complex and cyt ligase (Fig. 8, M) and mutants expressing the set of vectors (Fig. 8, V). sen2-42 cells with the set of vectors displayed normal pre-rRNA processing at 23°C. After a 2-h shift to 37°C, the cells have increased steady-state levels of 20S and 27SA2 pre-rRNAs; there is a compensatory decrease in the level of 27S, the next intermediate in the 25S rRNA processing pathway (Fig. 8). The defects are specific to sen2-42, rather than mere slow growth of sen2-42 cells, since another mutant, cdc6-1, inhibited for growth at high temperature, does not accumulate pre-rRNA precursors at the np temperature (Supplemental Fig. S5, cdc6-1). The pre-rRNA accumulation in sen2-42 cells at 37°C is complemented by expression of the mito SEN complex and cyt ligase (Fig. 8, M), but not expression of the nuc SEN complex and nuc-cyt ligase (Fig. 8, N). We also performed experiments with the catalytically inactive sen2 H297A at the endogenous mitochondrial location and showed that it does not complement pre-rRNA accumulation (data not shown). The data indicate that pre-rRNA processing requires functional mitochondrial tRNA SEN and show that the nuclear tRNA splicing machinery cannot complement the rRNA processing defect of sen2-42 cells.
Figure 8.
Northern analyses of pre-rRNA processing in sen2-42 mutants expressing nuclear tRNA splicing machinery. sen2-42 cells were transformed with either the set of plasmids encoding the mito SEN complex and cyt ligase (M), the vectors (V), or the set of plasmids encoding the nuc SEN complex and nuc-cyt ligase (N). RNAs from cells induced with Gal for 2 h and incubated at 23°C or for 2 h at 37°C were resolved on an agarose–formaldehyde gel and transferred to a membrane, and the membrane was probed for 27SA2, 27S, and 20S pre-rRNAs and 25S and 18S rRNAs.
Discussion
Historically, pre-tRNA splicing was thought to occur in the nucleus in all eukaryotes. However, the surprising discovery that yeast pre-tRNA splicing occurs on the cytoplasmic surface of mitochondria (Yoshihisa et al. 2003, 2007) raised the question of why the cellular location of pre-tRNA splicing is not conserved. To address this question, we re-engineered yeast to express the nuclear tRNA SEN complex, essentially mimicking the vertebrate cellular organization of pre-tRNA splicing. Redistribution of the yeast tRNA SEN complex to the nucleus results in efficient pre-tRNA splicing, as each splicing step—endonucleolytic cleavage, ligation of tRNA halves, and 2′ phosphate removal from the splice junction—occurs efficiently in the re-engineered yeast cells; moreover, pre-tRNAs cleaved in the nucleus are efficiently exported to the cytoplasm and efficiently aminoacylated. In our experiments, yeast cells have both nuclear and cytoplasmic pools of tRNA ligase and 2′-phosphotransferase, and thus the results do not address the cellular locations of ligation and 2′-phosphate removal. However, it seems likely that these steps also occur in the nucleus, because Los1 binds to the tertiary tRNA backbone (Cook et al. 2009) and it is unlikely that Los1 would export tRNA halves prior to ligation unless the halves remain together after intron cleavage. Since there are functional nuclear pools of the ligase and 2′-phosphotransferase in the re-engineered cells, the most likely model is that pre-tRNA introns are cleaved, ligated, and dephosphorylated prior to nuclear export.
Expression of tRNA SEN subunits in the nucleus of wild-type cells has no detectable deleterious affect on yeast growth, arguing against the possibility that yeast evolved mitochondrial splicing to avoid off-target harmful cleavages by nuclear tRNA SEN. Given the efficient splicing, aminoacylation, and nuclear export of tRNAs in cells with the nuc SEN complex, we were surprised that the ts growth phenotype of the sen-42 mutant is not complemented in the re-engineered cells. Furthermore, all four endogenous mito SEN complex subunits, including the catalytic activity of Sen2, are required even when efficient pre-tRNA splicing occurs in the nucleus.
It appears that all four SEN subunits of the heterotetramer are required for an essential function in the cytoplasm that is unrelated to tRNA processing, making it a bifunctional enzyme. Interestingly, tRNA ligase also has an extra function in the unessential UPR process (Nikawa et al. 1996; Sidrauski et al. 1996). Likewise, since 2′-phoshotransferase exists in cells that do not encode tRNA introns or cells that ligate tRNA without generating a 2′-phosphate at the splice junction (Spinelli et al. 1998; Harding et al. 2008), 2′-phosphotransferase likely serves an additional function. Therefore, each of the three pre-tRNA splicing enzymes serves at least one function beyond tRNA splicing.
Why are cells not viable when complete tRNA splicing occurs in the nucleus? One possibility we considered is that ectopic splicing in the nucleus results in tRNAs lacking essential modifications. However, this is unlikely for the following reasons: (1) There are no known essential intron-dependent tRNA modifications (Jiang et al. 1997; Phizicky and Hopper 2010), and there are no reports of negative synthetic interactions among the known intron-dependent modification enzymes, Pus1 and Trm4 (SGD). (2) Two of the four essential tRNA modification enzymes, Tad2, and Tad3 (Gerber and Keller 1999), are located in both the nucleus and cytoplasm (Huh et al. 2003) and thus likely accesses tRNAs whether spliced in the nucleus or the cytoplasm. (3) The other two essential tRNA modification enzymes, the Trm6/Trm61 complex, and Thg1, are only necessary for modifying the intronless tRNAMeti and tRNAHis tRNAs, respectively (Anderson et al. 1998; Gu et al. 2003).
Another possibility for growth defects is that introns, once cleaved from pre-tRNA, may play an essential role in the cytoplasm, possibly in rRNA processing. If so, upon nuclear pre-tRNA splicing, perhaps the resulting introns are prevented from accessing their cytoplasmic substrates. However, a recent study demonstrated that the absence of introns from one of the families of tRNAs encoded by intron-containing genes, tRNATrp, does not exhibit an adverse phenotype in yeast (Mori et al. 2011), thus partially arguing against this possibility.
In an attempt to identify functions for the tRNA SEN independent of tRNA splicing, we learned that mitochondrial-located Sen2 is required for efficient pre-rRNA processing, consistent with a previous report for Sen34 (Volta et al. 2005). Despite accumulation of the pre-rRNAs, we did not detect reduced 25S and 18S mature RNA levels, perhaps due to the short incubation at the np temperature. The data demonstrate that the mito SEN complex is likely involved in pre-rRNA processing and cannot be replaced by the nuc SEN complex. As processing of 20S to mature 18S rRNA occurs in the cytoplasm (for review, see Fatica and Tollervey 2002), the mito SEN complex may be directly involved in this endonucleolytic step, perhaps in parallel to Nob1 (Lamanna and Karbstein 2009; Pertschy et al. 2009). However, direct endonucleolytic activity cannot account for the other intermediates, as they are processed in the nucleolus (Fatica and Tollervey 2002). Although 20S pre-rRNA accumulation in the cytoplasm might signal for slowed pre-rRNA processing in the nucleus, this is unlikely given that there are mutants with 20S-to-18S processing defects that do not affect 25S processing (Vanrobays et al. 2001). Another possibility is that the mito SEN complex is involved in the biogenesis of gene products required for pre-rRNA processing, thus indirectly affecting rRNA processing. For example, the mito SEN complex may function to process the RNase MRP RNA NME1 (Schmitt and Clayton 1992) or other snoRNAs involved in pre-rRNA processing, even though the nuclear–cytoplasmic shuttling of snoRNAs is controversial (Hopper 2006; Watkins et al. 2007). Our preliminary experiments provide no evidence for defective NME1 biogenesis (data not shown), and in the future, we will explore this hypothesis further by assessing the large number of yeast snoRNAs.
In this study, we provide evidence that tRNA splicing can occur in the nucleus of yeast if the tRNA SEN complex is located there. However, we also show that the mito SEN complex possesses a function independent of pre-tRNA intron cleavage that requires all four subunits of the mitochondrial tRNA SEN complex, including the requirement of the Sen2 catalytic core. We hypothesize that in yeast, the selection for the tRNA SEN at the mitochondrial surface is not a result of its canonical activity in tRNA splicing, but rather a result of this noncanonical, yet essential, activity. One possible function that may or may not be essential for the tRNA SEN is in pre-rRNA processing, either directly or indirectly. Our studies of the yeast tRNA splicing machinery may be applicable to vertebrate pre-tRNA splicing, since in mammalian cells, an alternative tRNA SEN complex with a Sen2 splice variant has been postulated to function in the catalysis of unknown RNA substrates (Paushkin et al. 2004).
Materials and methods
Probes and primers
All probes and primers used are listed in the Supplemental Material.
Yeast strains
All yeast strains were constructed in the W303 background (MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15). sen2-42 was modified from Yoshihisa et al. (2007) by replacing sen2Δ::LEU2 with sen2Δ::LEU2::NATR(W303 with sen2Δ::LEU2::NATR/pTYSC017[CEN6-ARSH4 URA3 sen2-42]); los1Δis W303 with los1Δ::NATR.
Tet-regulatable SEN subunit strains
The tet-regulatable promoter region, carrying a kanR-tet07-TATA (Tet-operon), for each SEN gene was cloned from the Tet-Promoter Strain Collection (OpenBio Systems) by using primers (Supplemental Material) ND0058 and ND0059 (SEN2), ND0054 and ND0055 (SEN15), ND0052 and ND0053 (SEN34), and ND0056 and ND0057 (SEN54). The resulting fragments containing 5′ and 3′ flanking sequences of the SEN gene 5′ region (carrying the Tet-operon cassette) were integrated, replacing the endogenous 5′ region as described in Mnaimneh et al. (2004). The tet activator Tet-A, required for tet-inducible SEN expression, was expressed from a plasmid (OpenBio Systems) prior to creating tet-regulatable tRNA SEN strains. Each strain—tet sen2 (W303 with PtetO7−SEN2), tet sen15 (W303 with PtetO7–SEN15), tet sen34 (W303 with PtetO7–SE34), and tet sen54 (W303 with PtetO7–SEN54)—was transformed with the set of plasmids described below.
Construction of plasmids encoding mito SEN complex subunits
The coding sequence for each gene—SEN2, SEN15, SEN34, SEN54, TRL1, and TPT1—was cloned by PCR from the start to the termination codon using primers (Supplemental Material) that included restriction sites BamHI or EcoRI. PCR products were ligated into pGEM-T (Promega). DNA sequences were confirmed, and the coding sequences were digested with BamHI and EcoRI and ligated into pGP54a plasmid (XhoI, PGAL, BamHI, EcoRI, GFP, terminator, and EagI) (Murthi and Hopper 2005), generating six plasmids encoding each Gal-inducible and GFP-tagged mito SEN subunit and cytoplasmic ligase. XhoI–EagI fragments containing Gal-inducible GFP-tagged proteins were ligated into vectors to allow expression of SEN genes from multiple plasmids in a single yeast strain. PGAL-Sen2-GFP and PGAL-Sen15-GFP were cloned into pRS412 and pRS422 (to generate pND00219 and pND00220, respectively), and two other fragments, PGAL-Sen34-GFP and PGAL-Sen54-GFP, were cloned into pRS415 and pRS425 (to generate pND00221 and pND00222, respectively). XhoI–EagI fragments containing PGAL-Trl1-GFP and PGAL-Tpt1-GFP were cloned into pRS416 to generate pND00223. To reduce the numbers of plasmids required to express all five genes encoding mitochondrial/cytoplasmic tRNA splicing machinery, plasmids were opened with HindIII and ligated together pairwise. pND00219 and pND00220 were digested with HindIII, and the resulting two fragments were ligated to form the plasmid pND00225, expressing both PGAL-Sen2-GFP and PGAL-Sen15-GFP. pND00221 and pND00222 were digested with HindIII, and the resulting two fragments were ligated to form plasmid pND00226, expressing both PGAL-Sen34-GFP and PGAL-Sen54-GFP. Cells transformed with the set of plasmids (pND00225, pND00226, and pND00223) result in yeast expressing plasmids encoding all of the recombinant tRNA SEN subunits and tRNA ligase (the mito SEN complex and cytoplasmic ligase).
Construction of plasmids encoding nuc SEN complex subunits
The coding sequence for each gene—SEN2, SEN15, SEN34, SEN54, TRL1, and TPT1—was cloned as described above and ligated into pGEM-T (Promega). Sequences of the genes were confirmed, and coding sequences were digested with BamHI and EcoRI and ligated into pIGOutA plasmid (XhoI, PGAL, N-terminal NLS, BamHI, EcoRI, 2-GFPs, terminator, and EagI) (Butterfield-Gerson et al. 2006), generating six plasmids encoding each Gal-inducible, NLS-tagged, and GFP-tagged nuc SEN subunit and nuc-cyt ligase. XhoI–EagI fragments containing Gal-inducible GFP-tagged proteins were ligated into vectors to allow expression of SEN genes from multiple plasmids in a single yeast strain. PGAL-NLS-Sen2-2GFP and PGAL-NLS-Sen15-2GFP were cloned into pRS412 and pRS422 (to generate pND00227 and pND00228, respectively), and two other fragments, PGAL-NLS-Sen34-2GFP and PGAL-NLS-Sen54-2GFP, were cloned into pRS415 and pRS425 (to generate pND00229 and pND00230, respectively). XhoI–EagI fragments containing PGAL-Trl1-GFP were cloned into pRS416 (to generate pND00233). To reduce the numbers of plasmids required to express all five genes encoding nuclear tRNA splicing machinery, plasmids were opened with HindIII and ligated together pairwise. pND00227 and pND00228 were digested with HindIII, and the resulting two fragments were ligated to form the plasmid pND00231, expressing both PGAL-NLS-Sen2-2GFP and PGAL-NLS-Sen15-2GFP. pND00229 and pND00230 were digested with HindIII, and the resulting two fragments were ligated to form plasmid pND00232, expressing both PGAL-NLS-Sen34-2GFP and PGAL-NLS-Sen54-2GFP. Cells transformed with the set of plasmids (pND00231, pND00232, and pND00233) result in yeast expressing plasmids encoding all of the recombinant nuclear tRNA SEN subunits and nuclear–cytoplasmic tRNA ligase (the nuc SEN complex and nuc-cyt ligase). For β-galactosidase activity assays, XhoI–EagI fragments containing PGAL-Trl1-GFP were cloned into pRS413 (Trp marker) instead of into pRS416 (to generate pND00234) such that cells could be transformed with plasmid p130 (Hinnebush 1985), the Gcn4 reporter plasmid containing the URA3 gene for selection.
Construction of the catalytically inactive sen2 H297A plasmid
A plasmid encoding wild-type Sen2 (Yoshihisa et al. 2007) was used as a template for site-directed mutagenesis of His297 to Ala using the QuickChange Mutagenesis kit (Stratagene). Briefly, primers ND0070 and ND0071 (Supplemental Material), harboring mutations for His297Ala, were used in conjunction with high-fidelity Pfu polymerase to amplify the entire plasmid, digested with DpnI to purify plasmids containing the His297Ala mutation, and transformed into Escherichia coli to collect plasmids now able to express sen2 H297A.
Northern analysis
tRNA Northerns
Small RNAs were extracted from yeast cultures grown at the permissive temperature (23°C) to 0.6 OD600nm as in Whitney et al. (2007). ts cultures of sen2-42 cells were either Gal-induced for 2 h or not Gal-induced prior to transfer to the np temperature (37°C) for 2 h before RNA extraction. Five micrograms of RNA was electrophoretically separated and transferred onto Hybond N+ membranes as described (Whitney et al. 2007). Aminoacylated tRNAs were extracted from log phase yeast cells under acidic conditions (0.3 M NaOAc at pH 4.5, 10 mM EDTA) via glass bead lysis (Sarkar et al. 1999). RNAs were separated by electrophoresis on a 10% polyacrylamide (pH 4.5), 8 M urea gel. tRNAs were detected employing appropriate complementary γ-32P-terminally labeled probes (Supplemental Material) as described (Whitney et al. 2007) and were exposed to phosphor screens (Amersham Pharmacia). Phosphor screens were scanned and quantified with a Typhoon System using ImageQuant software.
Tet sen34 Northerns
Small RNAs were extracted from yeast cultures grown at 23°C to 0.6 OD600nm that were transferred to selective medium containing or lacking 8 μg/mL DOX for 9 h and induced or not induced with 2% Gal for 2 h. Subsequent electrophoresis and transfer were done as described above.
rRNA Northerns
rRNAs were extracted using the Qiagen RNeasy Mini kit (Qiagen) using the supplier's protocol. RNA (2.5 μg) was loaded and resolved on 1.2% agarose–formaldehyde gels as described (Kressler et al. 1997). RNAs were transferred and probed (Supplemental Material) as above, except hybridization was at 42°C.
Microscopic analyses
Microscopy
All fluorescence imaging employed a Nikon Eclipse E1000 microscope. Images were captured by using a SenSys charge-coupled device camera (Photometrics) using Nikon Elements software (Nikon). Images were arranged with Photoshop CS3 (Adobe Systems).
Live-cell imaging
Strains harboring plasmids encoding the Gal-inducible GFP-tagged SEN subunit Trl1 or Tpt1 were grown overnight in medium lacking Gal and then induced in the presence of 2% Gal for 2 h. Strains were also transformed with plasmids encoding either mCherry-tagged Tom20 to visualize mitochondria or mCherry-tagged Nup49 to visualize the nuclear membrane (Lai et al. 2009).
FISH studies
A published (Sarkar and Hopper 1998) probe (SRIM15) and procedures were used to monitor the cellular location of tRNATyr.
β-Galactosidase activity assay
Cells expressing GFP-tagged nuc SEN complex subunits or empty vectors were transformed with plasmid p180 (Hinnebusch 1985) encoding β-galactosidase (lacZ) regulated by the GCN4 promoter and translation regulatory sequences to assess starvation signaling in cells. Quantitative determination of β-galactosidase activity of cells in the presence or absence of amino acids was performed as described in the Yeast β-Galactosidase Assay kit using the microcentrifuge protocol (Thermo Scientific). β-Galactosidase activity was assessed as absorbance at A420/volume X concentration of cells.
Tpt1 activity assay
To assay Tpt1 activity, we employed a procedure based on Schutz et al. (2010), who demonstrated that 2′-phosphates inhibit reverse transcriptase activity. Small RNAs were extracted, and RT–PCR was used for first strand cDNA synthesis of tRNAIleUAU using primer IVY2. Subsequent PCR reactions (10 cycles) were carried out using primer IVY1 to amplify the small RNAs. Reactions were run on 2% agarose gel and assessed for amplification of tRNAIle cDNA. RT–PCR reactions employing small RNAs from yeast tpt1Δ strains complemented with Gal-inducible TPT1 plasmid (Spinelli et al. 1997) after either Gal induction or Glu repression for 4 h served as a positive control.
Growth assays
Aliquots (5 μL) from serial dilutions of the indicated yeast cultures were spotted onto selective solid medium. Plates were incubated for 2–5 d at the indicated temperatures.
Tet-off studies
tet sen2, tet sen15, tet sen34, and tet sen54 expressing the set of plasmids encoding either the nuc SEN complex or the mito SEN complex were grown overnight at 23°C in selective liquid medium lacking doxycycline (−DOX) and Gal. Cultures were diluted, grown overnight on selective −DOX liquid medium containing 2% Gal, plated to selective +Gal −DOX plates, and incubated for 3 d. Plates were replica-plated to selective +Gal plates containing 8 μg/mL DOX or selective +Gal −DOX plates.
Heterokaryon analyses
Heterokaryon analysis was performed as described in Shaheen and Hopper (2005), except after allowing mating, heterokaryons were fixed in 1/10 volume formaldehyde and stained with DAPI before microscopic analysis. The nuclear karyogamy-deficient kar1-1 mutant (Shaheen and Hopper 2005) was mated with yeast cells expressing GFP-tagged nuc SEN complex subunits. H2B-GFP (Feng and Hopper 2002) was employed as a nonshuttling control, and Cca1-GFP (Peng and Hopper 2000) was employed as a shuttling control. kar1-1 cells were mated with Gal-induced (2 h with 2% Gal) W303 strains with KAR1 harboring plasmids encoding either of the Gal-inducible nuclear-localized tRNA SEN subunits tagged with GFP (pND00227, pND00228, pND00229, or pND00230) or with W303 strains with KAR1 harboring plasmids encoding NLS-H2B-GFP or NLS-CCA1-GFP (Feng and Hopper 2002).
Acknowledgments
We thank Dr. I. Meier and A.K.H. laboratory members, especially Drs. E. Kramer and R. Hurto, for valuable scientific interactions and comments on the manuscript. We also thank Drs. E.M. Phizicky, V. Gopalan, J. Larson, and J. Alfonzo for comments on the manuscript, and Drs. E.M. Phizicky and T. Yoshihisa for reagents. This work was supported by National Institutes of Health Grant GM27930 (to A.K.H.) and The Ohio State University Pelotonia Fellowship (to N.D.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.183004.111.
References
- Anderson J, Phan L, Cuesta R, Carlson BA, Pak M, Asano K, Bjork GR, Tamame M, Hinnebusch AG 1998. The essential Gcd10p–Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev 12: 3650–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield-Gerson KL, Scheifele LZ, Ryan EP, Hopper AK, Parent LJ 2006. Importin-β family members mediate alpharetrovirus gag nuclear entry via interactions with matrix and nucleocapsid. J Virol 80: 1798–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook AG, Fukuhara N, Jinek M, Conti E 2009. Structures of the tRNA export factor in the nuclear and cytosolic states. Nature 461: 60–65 [DOI] [PubMed] [Google Scholar]
- Corbett AH, Koepp DM, Schlenstedt G, Lee MS, Hopper AK, Silver PA 1995. Rna1p, a Ran/TC4 GTPase activating protein, is required for nuclear import. J Cell Biol 130: 1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver GM, McCraith SM, Consaul SA, Stanford DR, Phizicky EM 1997. A 2′-phosphotransferase implicated in tRNA splicing is essential in Saccharomyces cerevisiae. J Biol Chem 272: 13203–13210 [DOI] [PubMed] [Google Scholar]
- Fatica A, Tollervey D 2002. Making ribosomes. Curr Opin Cell Biol 14: 313–318 [DOI] [PubMed] [Google Scholar]
- Feng W, Hopper AK 2002. A Los1p-independent pathway for nuclear export of intronless tRNAs in Saccharomyces cerevisiae. Proc Natl Acad Sci 99: 5412–5417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Keller W 1999. An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science 286: 1146–1149 [DOI] [PubMed] [Google Scholar]
- Gu W, Jackman JE, Lohan AJ, Gray MW, Phizicky EM 2003. tRNAHis maturation: An essential yeast protein catalyzes addition of a guanine nucleotide to the 5′ end of tRNAHis. Genes Dev 17: 2889–2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Lackey JG, Hsu HC, Zhang Y, Deng J, Xu RM, Damha MJ, Ron D 2008. An intact unfolded protein response in Trpt1 knockout mice reveals phylogenic divergence in pathways for RNA ligation. RNA 14: 225–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG 1985. A hierarchy of trans-acting factors modulates translation of an activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol 5: 2349–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG 1997. Translational regulation of yeast GCN4. A window on factors that control initiator-trna binding to the ribosome. J Biol Chem 272: 21661–21664 [DOI] [PubMed] [Google Scholar]
- Hopper AK 2006. Cellular dynamics of small RNAs. Crit Rev Biochem Mol Biol 41: 3–19 [DOI] [PubMed] [Google Scholar]
- Hopper AK, Shaheen HH 2008. A decade of surprises for tRNA nuclear-cytoplasmic dynamics. Trends Cell Biol 18: 98–104 [DOI] [PubMed] [Google Scholar]
- Hopper AK, Schultz LD, Shapiro RA 1980. Processing of intervening sequences: A new yeast mutant which fails to excise intervening sequences from precursor tRNAs. Cell 19: 741–751 [DOI] [PubMed] [Google Scholar]
- Hopper AK, Pai DA, Engelke DR 2010. Cellular dynamics of tRNAs and their genes. FEBS Lett 584: 310–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Intine RV, Dundr M, Misteli T, Maraia RJ 2002. Aberrant nuclear trafficking of La protein leads to disordered processing of associated precursor tRNAs. Mol Cell 9: 1113–1123 [DOI] [PubMed] [Google Scholar]
- Jiang HQ, Motorin Y, Jin YX, Grosjean H 1997. Pleiotropic effects of intron removal on base modification pattern of yeast tRNAPhe: An in vitro study. Nucleic Acids Res 25: 2694–2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleman-Leyer K, Armbruster DW, Daniels CJ 1997. Properties of H. volcanii tRNA intron endonuclease reveal a relationship between the archaeal and eucaryal tRNA intron processing systems. Cell 89: 839–847 [DOI] [PubMed] [Google Scholar]
- Kressler D, de la Cruz J, Rojo M, Linder P 1997. Fal1p is an essential DEAD-box protein involved in 40S-ribosomal-subunit biogenesis in Saccharomyces cerevisiae. Mol Cell Biol 17: 7283–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai TP, Stauffer KA, Murthi A, Shaheen HH, Peng G, Martin NC, Hopper AK 2009. Mechanism and a peptide motif for targeting peripheral proteins to the yeast inner nuclear membrane. Traffic 10: 1243–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamanna AC, Karbstein K 2009. Nob1 binds the single-stranded cleavage site D at the 3′-end of 18S rRNA with its PIN domain. Proc Natl Acad Sci 106: 14259–14264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Dahlberg JE 1998. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science 282: 2082–2085 [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J, Garrett RA 1997. RNA-protein interactions of an archaeal homotetrameric splicing endoribonuclease with an exceptional evolutionary history. EMBO J 16: 6290–6300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, Kobayashi Y, Hashimoto A, Hamamoto M, Hiraoka Y, et al. 2006. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol 24: 841–847 [DOI] [PubMed] [Google Scholar]
- Melton DA, De Robertis EM, Cortese R 1980. Order and intracellular location of the events involved in the maturation of a spliced tRNA. Nature 284: 143–148 [DOI] [PubMed] [Google Scholar]
- Mnaimneh S, Davierwala AP, Haynes J, Moffat J, Peng WT, Zhang W, Yang X, Pootoolal J, Chua G, Lopez A, et al. 2004. Exploration of essential gene functions via titratable promoter alleles. Cell 118: 31–44 [DOI] [PubMed] [Google Scholar]
- Mori T, Ogasawara C, Inada T, Englert M, Beier H, Takezawa M, Endo T, Yoshihisa T 2010. Dual functions of yeast tRNA ligase in the unfolded protein response: Unconventional cytoplasmic splicing of HAC1 pre-mRNA is not sufficient to release translational attenuation. Mol Biol Cell 21: 3722–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, Kajita T, Endo T, Yoshihisa T 2011. The intron of tRNA-TrpCCA is dispensable for growth and translation of Saccharomyces cerevisiae. RNA 17: 1760–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthi A, Hopper AK 2005. Genome-wide screen for inner nuclear membrane protein targeting in Saccharomyces cerevisiae: Roles for N-acetylation and an integral membrane protein. Genetics 170: 1553–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthi A, Shaheen HH, Huang HY, Preston MA, Lai TP, Phizicky EM, Hopper AK 2010. Regulation of tRNA bidirectional nuclear-cytoplasmic trafficking in Saccharomyces cerevisiae. Mol Biol Cell 21: 639–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikawa J, Akiyoshi M, Hirata S, Fukuda T 1996. Saccharomyces cerevisiae IRE2/HAC1 is involved in IRE1-mediated KAR2 expression. Nucleic Acids Res 24: 4222–4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paushkin SV, Patel M, Furia BS, Peltz SW, Trotta CR 2004. Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3′ end formation. Cell 117: 311–321 [DOI] [PubMed] [Google Scholar]
- Peebles CL, Gegenheimer P, Abelson J 1983. Precise excision of intervening sequences from precursor tRNAs by a membrane-associated yeast endonuclease. Cell 32: 525–536 [DOI] [PubMed] [Google Scholar]
- Peng G, Hopper JE 2000. Evidence for Gal3p's cytoplasmic location and Gal80p's dual cytoplasmic-nuclear location implicates new mechanisms for controlling Gal4p activity in Saccharomyces cerevisiae. Mol Cell Biol 20: 5140–5148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertschy B, Schneider C, Gnadig M, Schafer T, Tollervey D, Hurt E 2009. RNA helicase Prp43 and its co-factor Pfa1 promote 20 to 18 S rRNA processing catalyzed by the endonuclease Nob1. J Biol Chem 284: 35079–35091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky EM, Hopper AK 2010. tRNA biology charges to the front. Genes Dev 24: 1832–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky EM, Schwartz RC, Abelson J 1986. Saccharomyces cerevisiae tRNA ligase. Purification of the protein and isolation of the structural gene. J Biol Chem 261: 2978–2986 [PubMed] [Google Scholar]
- Phizicky EM, Consaul SA, Nehrke KW, Abelson J 1992. Yeast tRNA ligase mutants are nonviable and accumulate tRNA splicing intermediates. J Biol Chem 267: 4577–4582 [PubMed] [Google Scholar]
- Popow J, Englert M, Weitzer S, Schleiffer A, Mierzwa B, Mechtler K, Trowitzsch S, Will CL, Luhrmann R, Soll D, et al. 2011. HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science 331: 760–764 [DOI] [PubMed] [Google Scholar]
- Salgia SR, Singh SK, Gurha P, Gupta R 2003. Two reactions of Haloferax volcanii RNA splicing enzymes: Joining of exons and circularization of introns. RNA 9: 319–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Hopper AK 1998. tRNA nuclear export in Saccharomyces cerevisiae: In situ hybridization analysis. Mol Biol Cell 9: 3041–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Azad AK, Hopper AK 1999. Nuclear tRNA aminoacylation and its role in nuclear export of endogenous tRNAs in Saccharomyces cerevisiae. Proc Natl Acad Sci 96: 14366–14371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt ME, Clayton DA 1992. Yeast site-specific ribonucleoprotein endoribonuclease MRP contains an RNA component homologous to mammalian RNase MRP RNA and essential for cell viability. Genes Dev 6: 1975–1985 [DOI] [PubMed] [Google Scholar]
- Schutz K, Hesselberth JR, Fields S 2010. Capture and sequence analysis of RNAs with terminal 2′,3′-cyclic phosphates. RNA 16: 621–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen HH, Hopper AK 2005. Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc Natl Acad Sci 102: 11290–11295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Cox JS, Walter P 1996. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell 87: 405–413 [DOI] [PubMed] [Google Scholar]
- Spinelli SL, Consaul SA, Phizicky EM 1997. A conditional lethal yeast phosphotransferase (tpt1) mutant accumulates tRNAs with a 2′-phosphate and an undermodified base at the splice junction. RNA 3: 1388–1400 [PMC free article] [PubMed] [Google Scholar]
- Spinelli SL, Malik HS, Consaul SA, Phizicky EM 1998. A functional homolog of a yeast tRNA splicing enzyme is conserved in higher eukaryotes and in Escherichia coli. Proc Natl Acad Sci 95: 14136–14141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocchini-Valentini GD, Fruscoloni P, Tocchini-Valentini GP 2005. Structure, function, and evolution of the tRNA endonucleases of Archaea: An example of subfunctionalization. Proc Natl Acad Sci 102: 8933–8938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta CR, Miao F, Arn EA, Stevens SW, Ho CK, Rauhut R, Abelson JN 1997. The yeast tRNA splicing endonuclease: A tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell 89: 849–858 [DOI] [PubMed] [Google Scholar]
- Trotta CR, Paushkin SV, Patel M, Li H, Peltz SW 2006. Cleavage of pre-tRNAs by the splicing endonuclease requires a composite active site. Nature 441: 375–377 [DOI] [PubMed] [Google Scholar]
- Vanrobays E, Gleizes PE, Bousquet-Antonelli C, Noaillac-Depeyre J, Caizergues-Ferrer M, Gelugne JP 2001. Processing of 20S pre-rRNA to 18S ribosomal RNA in yeast requires Rrp10p, an essential non-ribosomal cytoplasmic protein. EMBO J 20: 4204–4213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volta V, Ceci M, Emery B, Bachi A, Petfalski E, Tollervey D, Linder P, Marchisio PC, Piatti S, Biffo S 2005. Sen34p depletion blocks tRNA splicing in vivo and delays rRNA processing. Biochem Biophys Res Commun 337: 89–94 [DOI] [PubMed] [Google Scholar]
- Wang LK, Schwer B, Englert M, Beier H, Shuman S 2006. Structure-function analysis of the kinase-CPD domain of yeast tRNA ligase (Trl1) and requirements for complementation of tRNA splicing by a plant Trl1 homolog. Nucleic Acids Res 34: 517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Lemm I, Luhrmann R 2007. Involvement of nuclear import and export factors in U8 box C/D snoRNP biogenesis. Mol Cell Biol 27: 7018–7027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney ML, Hurto RL, Shaheen HH, Hopper AK 2007. Rapid and reversible nuclear accumulation of cytoplasmic tRNA in response to nutrient availability. Mol Biol Cell 18: 2678–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihisa T, Yunoki-Esaki K, Ohshima C, Tanaka N, Endo T 2003. Possibility of cytoplasmic pre-tRNA splicing: The yeast tRNA splicing endonuclease mainly localizes on the mitochondria. Mol Biol Cell 14: 3266–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihisa T, Ohshima C, Yunoki-Esaki K, Endo T 2007. Cytoplasmic splicing of tRNA in Saccharomyces cerevisiae. Genes Cells 12: 285–297 [DOI] [PubMed] [Google Scholar]