Contact-dependent growth inhibition (CDI) in bacteria plays an important role in competition for environmental niches. CDI is mediated by cell surface CdiA protein, which contains a toxic effector domain at its C terminus (CdiA-CT). In this study by Hayes and colleagues, the authors demonstrate that the UPEC strain of E. coli inhibits growth of target cells by using a cytotoxic tRNase that becomes activated when it binds to the CysK protein by mimicking CysE, a subunit of the cysteine synthase complex. In addition, the authors show that E. coli ΔcysK target cells are resistant the UPEC536 CDI system. Thus, this study provides insights into novel mechanisms regulating prokaryotic growth inhibition.
Keywords: CDI, contact-dependent growth inhibition, CysK, tRNase activity, UPEC536
Abstract
Bacterial contact-dependent growth inhibition (CDI) is mediated by the CdiB/CdiA family of two-partner secretion proteins. CdiA effector proteins are exported onto the surface of CDI+ inhibitor cells, where they interact with susceptible bacteria and deliver effectors/toxins derived from their C-terminal regions (CdiA-CT). CDI+ cells also produce an immunity protein that binds the CdiA-CT and blocks its activity to prevent autoinhibition. Here, we show that the CdiA-CT from uropathogenic Escherichia coli strain 536 (UPEC536) is a latent tRNase that requires activation by the biosynthetic enzyme CysK (O-acetylserine sulfhydrylase A). UPEC536 CdiA-CT exhibits no nuclease activity in vitro, but cleaves within transfer RNA (tRNA) anti-codon loops when purified CysK is added. CysK and CdiA-CT form a stable complex, and their binding interaction appears to mimic that of the CysK/CysE cysteine synthase complex. CdiA-CT activation is also required for growth inhibition. Synthesis of CdiA-CT in E. coli cysK+ cells arrests cell growth, whereas the growth of ΔcysK mutants is unaffected by the toxin. Moreover, E. coli ΔcysK cells are completely resistant to inhibitor cells expressing UPEC536 CdiA, indicating that CysK is required to activate the tRNase during CDI. Thus, CysK acts as a permissive factor for CDI, providing a potential mechanism to modulate growth inhibition in target cells.
Bacteria possess a variety of systems that facilitate cooperation and competition with other organisms in the environment. One such mechanism is contact-dependent growth inhibition (CDI), which allows some bacteria to inhibit the growth of neighboring cells upon direct cell-to-cell contact (Aoki et al. 2005, 2010; Hayes et al. 2010). CDI systems are widely distributed amongst the α-, β-, and γ-proteobacteria and have been shown to play a significant role in intraspecies growth competition (Aoki et al. 2010). CDI is mediated by the CdiB/CdiA family of two-partner secretion proteins. CdiB is a predicted outer-membrane β-barrel protein that is required for the export and assembly of the CdiA toxic effector protein onto the cell surface. CdiA proteins are very large (250–600 kDa) and contain numerous hemagglutinin peptide repeats throughout their N-terminal regions. This extensive repeat region is predicted to form a β-helical structure projecting several hundred angstroms from the surface of CDI+ inhibitor cells (Kajava et al. 2001). Presumably, the long extended CdiA filaments facilitate interaction with neighboring bacteria to induce growth inhibition. All CDI loci also contain a small cdiI immunity gene located immediately downstream from cdiA (Aoki et al. 2005, 2010). The CdiI protein provides immunity to the CdiA-derived toxin, thereby protecting CDI+ cells from autoinhibition.
We recently localized the CDI growth inhibition activity to the C-terminal region of CdiA (CdiA-CT) (Aoki et al. 2010). The CdiA-CT is a highly variable region of ∼230–360 residues at the extreme C terminus of CdiA proteins. This variable C-terminal region is sharply demarcated by the conserved VENN peptide motif, which is part of the DUF638 (Pfam, PF04829) domain of unknown function (Aoki et al. 2010). The polymorphic nature of the CdiA-CT region implies that a variety of distinct toxins are deployed by different CDI systems. The CdiA-CTEC93 from Escherichia coli isolate EC93 may form a pore in the inner membrane of target cells, leading to dissipation of the proton motive force and a concomitant decrease in respiration and ATP production (Aoki et al. 2009). Other CdiA-CTs possess distinct nuclease activities (Aoki et al. 2010). The CdiA-CTIDd3937 from Dickeya dadantii 3937 has DNase activity, and the CdiA-CTUPEC536 from uropathogenic E. coli strain 536 (UPEC536) cleaves transfer RNA (tRNA) (Aoki et al. 2010). Like the CdiA-CT region, the CdiI immunity proteins from different CDI systems also share little sequence identity with one another and, accordingly, only confer immunity to their cognate CdiA-CT toxins (Aoki et al. 2010). In vitro experiments have demonstrated that the CdiIIDd3937 and CdiIUPEC536 immunity proteins bind specifically to their cognate CdiA-CTs and block nuclease activity (Aoki et al. 2010), suggesting that these enzymatic activities are responsible for growth inhibition during CDI.
Here we identify an additional step in the CDI pathway in which a CdiA-CT toxin is activated by a protein cofactor in the cytoplasm of target cells. Although CdiA-CTUPEC536 (hereafter referred to as CdiA-CT) exhibits tRNase activity when expressed inside E. coli cells, the purified protein does not cleave tRNA in vitro. Using a biochemical approach, we identified a biosynthetic enzyme, CysK (O-acetylserine sulfhydrylase A), which is required to activate the CdiA-CT nuclease. CysK is one of two O-acetylserine sulfhydrylase isoenzymes (along with CysM) that catalyze the second and final step of cysteine synthesis from serine. Purified CysK is sufficient to activate the CdiA-CT tRNase in vitro, and the resulting nuclease activity is blocked specifically by CdiIUPEC536 immunity protein (hereafter referred to as CdiI). CysK and CdiA-CT form a stable complex, which is directly associated with tRNase activity. Remarkably, the CysK/CdiA-CT-binding interaction appears to mimic that of the cysteine synthase complex formed between CysK and CysE (serine O-acetyltransferase) in some bacteria and higher plants (Kredich et al. 1969; Droux et al. 1998; Zhu et al. 1998; Mino et al. 2000b; Kumaran et al. 2009). The cysteine synthase complex interaction is mediated by the C-terminal GDGI peptide of CysE, which binds in the active site cleft of CysK (Mino et al. 1999, 2000a; Zhao et al. 2006; Kumaran and Jez 2007). UPEC536 CdiA contains a similar C-terminal GYGI peptide motif, and these residues are required for binding to CysK. In accord with the biochemical data, we found that CysK is also required for tRNase activity when CdiA-CT is expressed inside E. coli cells. Moreover, E. coli ΔcysK mutants are completely resistant to cell-mediated CDIUPEC536, demonstrating that CysK is a critical player in this pathway. Based on these findings, we propose that CysK acts as a specific permissive factor for CDI mediated by UPEC536. These results reveal unanticipated complexity in the CDI pathway and raise the possibility that CdiA-CTs may play roles in communication between CDI+ cells.
Results
CdiA-CT-associated tRNase activity requires a cofactor
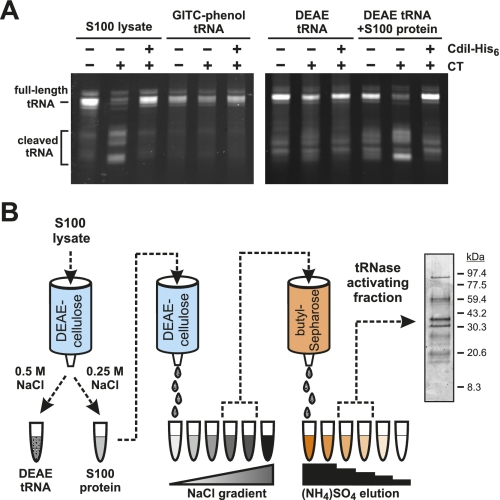
We recently reported that purified CdiA-CT from UPEC536 (corresponding to residues Val 3016–Ile 3242 of CdiA) cleaves tRNA in the S100 fraction of cell lysates (Aoki et al. 2010). The S100 fraction is the supernatant from a 100,000g centrifugation and contains a number of soluble proteins in addition to substrate tRNA. To characterize this tRNase activity under defined conditions, we extracted E. coli S100 fractions with guanidinium isothiocyanate-phenol (GITC-phenol) to isolate tRNAs for in vitro assays. Surprisingly, CdiA-CT had no activity on purified tRNA (Fig. 1A). In contrast, CdiA-CT induced tRNA cleavage when added to S100 lysates, and this tRNase activity was neutralized with equimolar CdiI-His6 immunity protein (Fig. 1A). We reasoned that tRNA could be denatured during GITC-phenol extraction, rendering them resistant to cleavage. Therefore, we purified E. coli tRNA under nondenaturing conditions using DEAE-cellulose chromatography and found this substrate was also refractory to cleavage (Fig. 1A,B). Together, these results suggest that a factor (or factors) present in the S100 fraction is required to support tRNase activity. To test this hypothesis, we removed nucleic acids from the S100 fraction using DEAE-cellulose in buffer containing 250 mM NaCl (Fig. 1B). Most of the proteins present in the S100 fraction were not retained on the DEAE-cellulose column under these conditions and were recovered in the flowthrough fraction (Fig. 1B; data not shown). This S100 protein fraction supported tRNase activity on purified tRNA isolated by both DEAE-cellulose and GITC-phenol methods (Fig. 1A; data not shown), suggesting that a protein in this fraction activates CdiA-CT.
Figure 1.
CdiA-CT tRNase activity requires a cofactor. (A) In vitro tRNase assays. S100 lysates or isolated tRNA (purified by either GITC-phenol or DEAE-cellulose chromatography) were incubated with CdiA-CT and CdiI-His6 (where indicated) as described in the Materials and Methods. The samples labeled DEAE tRNA+S100 protein contained a reconstituted S100 lysate in which the separated tRNA and protein fractions were recombined. Reactions were run on denaturing polyacrylamide gels, and tRNAs were visualized by ethidium bromide staining. The migration positions of full-length and cleaved tRNAs are indicated. (B) Purification scheme for enrichment of the CdiA-CT-activating factor. Proteins from E. coli S100 lysates were fractionated by anion exchange (DEAE-cellulose) and hydrophobic interaction (butyl-Sepharose) chromatography as illustrated in the flow chart. The final peak activating fraction was analyzed by SDS-PAGE, and proteins were visualized by staining with Coomassie blue.
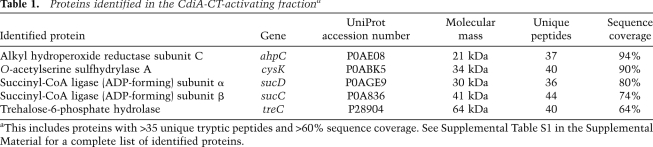
To identify the factor required for CdiA-CT activation, we fractionated S100 lysates using DEAE-cellulose chromatography and a linear salt gradient (Fig. 1B). The activator eluted from the column at ∼100–125 mM NaCl, and these fractions were pooled for further purification by butyl-Sepharose chromatography. SDS-PAGE analysis of the resulting active fraction revealed several proteins (Fig. 1B), which were identified by mass spectrometry following digestion with trypsin. This analysis identified several dozen E. coli proteins in the CdiA-CT-activating fraction, although most were present at very low levels (Supplemental Table S1). To identify the activating factor, we screened S100 lysates prepared from E. coli strains deleted for genes that encode candidate proteins. We focused on the most abundant candidate proteins, testing those with >35 unique tryptic peptides and >60% sequence coverage in the mass analysis (Table 1). The S100 lysates from ΔsucC, ΔsucD, ΔahpC, and ΔtreC mutants all supported tRNase activity to a level similar to that of the lysate prepared from wild-type cells (Fig. 2A). In contrast, there was dramatically less tRNase activity in the S100 lysate from E. coli ΔcysK cells (Fig. 2A). These results suggest that CysK plays a role in activating the CdiA-CT tRNase.
Table 1.
Proteins identified in the CdiA-CT-activating fractiona
Figure 2.
CysK is necessary and sufficient for tRNase activity. (A) CysK is required for tRNase activity. E. coli S100 lysates from the indicated genetic backgrounds were incubated with purified CdiA-CT (and CdiI-His6) and then analyzed by gel electrophoresis and ethidium bromide staining to assay tRNase activity. The migration positions of full-length and cleaved tRNAs are indicated. (B) CysK and CdiA-CT are sufficient for tRNase activity in vitro. GITC-phenol-extracted tRNA was treated with purified CysK-His6, CysK(K42A)-His6, and CysM-His6 in combination with CdiA-CT and CdiI-His6 where indicated. Buffer was added to the mock treatment control in place of CysK/CysM. Reactions were analyzed by denaturing gel electrophoresis as described for A.
CysK activates the CdiA-CT tRNase in vitro
To determine whether CysK is sufficient to stimulate tRNase activity, we overproduced and purified His6-tagged CysK protein for in vitro assays. Equimolar mixtures of CysK-His6 and CdiA-CT were able to cleave purified tRNA, and this activity was blocked by the CdiI immunity protein (Fig. 2B). CysK is a PLP-dependent enzyme that catalyzes the second and final step of cysteine synthesis from serine. To address whether O-acetylserine sulfhydrylase activity is required for tRNase activation, we tested a mutated version of CysK in which Lys 42 had been changed to an alanine residue (K42A). Lys 42 forms a Schiff base with PLP and is required for enzymatic activity (Rabeh and Cook 2004). Although catalytically inactive, purified CysK(K42A)-His6 protein still supported tRNase activity in vitro (Fig. 2B). E. coli contains an additional O-acetylserine sulfhydrylase isoenzyme encoded by the cysM gene (Tai et al. 1993). We tested purified E. coli CysM-His6 and found that it did not support tRNase activity (Fig. 2B). Together, these results show that nuclease activation requires CysK, but is not dependent on its O-acetylserine sulfhydrylase activity.
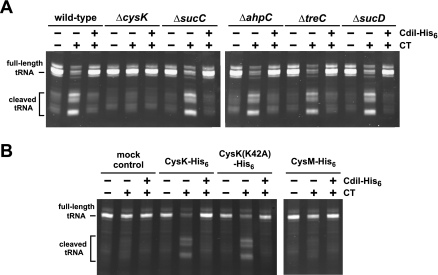
Because both CysK and CdiA-CT are required for tRNA cleavage in vitro, it is formally possible that CysK is in fact the tRNase and that CdiA-CT is the activating factor. We used BLAST analysis to identify CdiA-CT homologs, reasoning that multiple sequence alignment with related proteins could identify conserved residues required for tRNase activity. This analysis revealed that the CdiA-CT is bipartite, with the N-terminal and C-terminal regions sharing sequence identity with two distinct sets of proteins (Fig. 3A; Supplemental Figs. S1, S2). The N-terminal region of CdiA-CT (residues Val 1–Tyr 82) is homologous to the corresponding region in CdiA proteins from Yersinia pestis, Photorhabdus luminescens, and Enterobacter cloacae, but the extreme C-terminal sequences of these proteins are divergent (Fig. 3A; Supplemental Fig. S1). Because the CDI systems from these other bacteria encode distinct CdiI immunity proteins that are unrelated to the UPEC536 CdiI, we concluded that the shared N-terminal region of CdiA-CT is not required for tRNase activity. To test this prediction, we purified a truncated version of CdiA-CT lacking the N-terminal 82 residues and tested its nuclease activity in vitro. Remarkably, the truncated CdiA-CT cleaved tRNA in a CysK-independent fashion, and its activity was completely blocked by equimolar CdiI immunity protein (Fig. 3B). Multiple sequence alignment of the C-terminal tRNase domain with related sequences from γ-proteobacteria and Gram-positive bacteria revealed a handful of completely conserved residues that could be involved in catalysis (Fig. 3A; Supplemental Fig. S2). We changed the conserved His 178 residue in CdiA-CT (His 3193 in full-length CdiA) to alanine and found that the resulting protein lacked tRNase activity in vitro even in the presence of CysK-His6 (Fig. 3A,B). The loss of nuclease activity was not the result of gross structural perturbation, because the mutant CdiA-CT(H178A) still bound tightly to the cognate CdiI-His6 immunity protein (data not shown). Taken together, these results demonstrate that tRNase activity is intrinsic to the C-terminal 145 residues of UPEC536 CdiA.
Figure 3.
CdiA-CT is an intrinsic tRNase. (A) Multiple sequence alignment suggests that UPEC536 CdiA-CT is comprised of two domains. Homologous CdiA-CT regions from various proteobacteria were aligned with UPEC536 CdiA-CT using ClustalW. The N-terminal region of UPEC536 CdiA-CT (Val 1–Tyr 82) shares sequence identity with the corresponding regions in CdiA proteins from Y. pestis (Uniprot Q74T84), P. luminescens (Q7MB60), and E. cloacae (D5CBA0). The C-terminal region (Asn 89–Ile 227) is homologous to CdiA-related proteins from E. coli MS 153-1 (E6AEZ0), Enterobacter sp. 638 (A4W4W8), and Pseudomonas syringae (F3FGM0). UPEC536 CdiA-CT residues Gly 83 and His 178 (numbered with respect to Val 1 of the VENN motif) are highlighted in red. The alignment was rendered with 50% conservation visibility using Jalview 2.7. (B) The C-terminal domain of CdiA-CT has intrinsic tRNase activity. GITC-phenol-extracted tRNA was treated with the C-terminal domain of CdiA-CT (residues Gly 83–Ile 227) and CdiI-His6 in the presence and absence (mock control) of CysK-His6, and reactions were analyzed by denaturing gel electrophoresis. The migration positions of full-length and cleaved tRNAs are indicated. (C) The His178Ala missense mutation ablates CdiA-CT tRNase activity. GITC-phenol-extracted tRNA was treated with purified CdiA-CT(H178A) and CdiI-His6 in the presence and absence (mock control) of CysK-His6. Reactions were analyzed by denaturing gel electrophoresis.
CysK and CdiA-CT form a stable complex in vitro
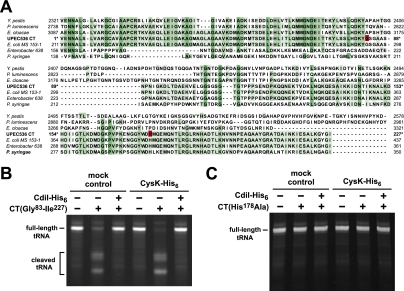
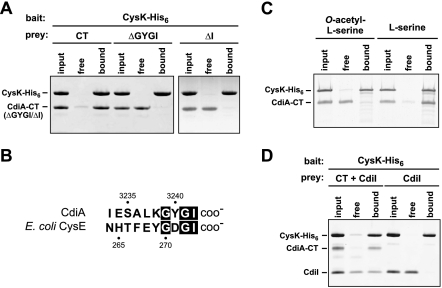
Because the biosynthetic function of CysK is not required for tRNase activation, we asked whether CysK and CdiA-CT form a stable complex. We incubated CdiA-CT with CysK-His6 and then subjected the mixture to Ni2+-affinity chromatography. SDS-PAGE analysis of the bound and free fractions revealed that untagged CdiA-CT was retained with CysK-His6 on the Ni2+-NTA column, suggesting a physical interaction between the proteins (Fig. 4A). Copurification assays also showed that CdiA-CT forms a stable complex with mutant CysK(K42A)-His6 protein but not CysM-His6 (Supplemental Fig. S3), consistent with the results from the in vitro tRNase assays. The only known binding partner of CysK is CysE, the serine O-acetyltransferase that catalyzes the first step in cysteine biosynthesis. CysK and CysE form the cysteine synthase complex in many bacteria and higher plants (Kredich et al. 1969; Droux et al. 1998; Zhu et al. 1998). Structural and biochemical studies of the cysteine synthase complex indicate that the C terminus of CysE binds in the active site cleft of CysK (Mino et al. 2000a; Campanini et al. 2005; Huang et al. 2005; Francois et al. 2006; Zhao et al. 2006; Kumaran et al. 2009). Notably, the C terminus of UPEC536 CdiA shares some sequence identity with the C terminus of CysE (Fig. 4B), raising the possibility that CysK binds the C-terminal Gly–Tyr–Gly–Ile (GYGI) peptide of CdiA-CT. In accord with this hypothesis, CdiA-CT proteins lacking the GYGI sequence (ΔGYGI) or the C-terminal isoleucine residue (ΔI) failed to copurify with CysK-His6 (Fig. 4A) and exhibited no in vitro tRNase activity in either the presence or absence of CysK (Supplemental Fig. S4). The cysteine synthase complex dissociates in the presence of the CysK substrate O-acetyl-L-serine (Kredich et al. 1969), presumably because this small molecule displaces the C-terminal tail of CysE upon binding the CysK active site. Similarly, we found that preincubation of CysK-His6 with O-acetyl-L-serine blocked CdiA-CT/CysK-His6 complex formation, whereas pretreatment with L-serine had little effect on the binding interaction (Fig. 4C). Taken together, these results suggest that the CdiA-CT GYGI motif mimics the C terminus of CysE to bind the CysK active site.
Figure 4.
CdiA-CT and CysK form a stable complex in vitro. (A) CdiA-CT and CysK form a stable complex in vitro. His6-tagged CysK was used as bait protein to test for copurification of untagged CdiA-CT variants. CysK-His6/CdiA-CT mixtures (input) were incubated with Ni2+-NTA agarose, and the free and resin-bound fractions were isolated for analysis by SDS-PAGE. The ΔGYGI and ΔI variants are truncated CdiA-CT proteins lacking the indicated C-terminal residues. The gel migration positions for each protein are indicated. (B) Comparison of the C-terminal residues of UPEC536 CdiA-CT and E. coli CysE. Residue numbers are indicated for each protein. (C) O-acetyl-L-serine blocks formation of the CdiA-CT/CysK complex. CysK-His6 was incubated with either O-acetyl-L-serine or L-serine prior to addition of CdiA-CT. Samples were then processed as described in A. The gel migration positions for each protein are indicated. (D) CdiA-CT/CdiI forms a ternary complex with CysK. Untagged CdiA-CT and/or CdiI proteins were incubated with CysK-His6, and the mixtures were subjected to Ni2+-NTA chromatography. All fractions were analyzed by SDS-PAGE. The gel migration positions for each protein are indicated.
We next asked whether the CdiI immunity protein blocks tRNase activity by disrupting the CysK/CdiA-CT complex. We incubated untagged CdiI with CdiA-CT and CysK-His6 and found that all three proteins copurified as an apparent ternary complex during Ni2+-affinity chromatography (Fig. 4D). CdiI does not appear to interact directly with CysK because these proteins do not copurify in the absence of CdiA-CT (Fig. 4D). These results indicate that CysK and CdiI bind to nonoverlapping sites on CdiA-CT and that immunity function does not entail dissociation of the tRNase complex.
CysK is required for CdiA-CT toxicity in vivo
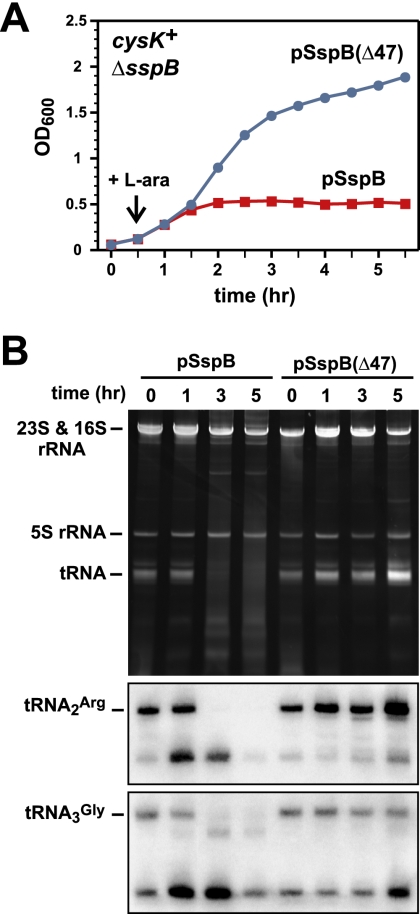
The cleavage of tRNA should block protein synthesis and is presumably the molecular basis of CdiA-CT-mediated growth inhibition. To induce CdiA-CT tRNase activity in vivo, we used the controlled proteolysis system of McGinness et al. (2006) to specifically degrade the CdiI immunity protein. Briefly, CdiI was tagged at its C terminus with the ssrA(DAS) peptide, which targets tagged proteins for degradation by the ClpXP protease only in the presence of the SspB adaptor protein (McGinness et al. 2006). Coexpression of CdiA-CT and CdiI-DAS did not inhibit E. coli ΔsspB cell growth, but production of SspB from a plasmid-borne PBAD promoter resulted in growth inhibition after ∼2 h of induction (Fig. 5A). No growth inhibition was seen in cells expressing SspB(Δ47), which binds the ssrA(DAS) peptide but is unable to deliver tagged proteins to ClpXP (Wah et al. 2002, 2003; McGinness et al. 2006). These results show that growth inhibition is dependent on CdiI degradation. Gel analysis of RNA from the inhibited cells revealed that their tRNAs were degraded (Fig. 5B). Northern blot hybridization and S1 nuclease protection assays showed that tRNA was cleaved in the anti-codon loop upon activation of CdiA-CT (Fig. 5B; Supplemental Fig. S5). The appearance of cleaved tRNA in vivo was coincident with growth inhibition, strongly suggesting that tRNase activity is cytotoxic. This conclusion was supported by a similar controlled degradation experiment using the inactive H178A variant of CdiA-CT, which did not inhibit cell growth (data not shown).
Figure 5.
Activation of CdiA-CT in E. coli cells results in growth arrest and tRNA degradation. (A) E. coli cysK+ ΔsspB cells producing the CdiA-CT/CdiI-DAS complex were treated with L-arabinose (L-ara) to induce synthesis of either SspB or SspB(Δ47) from plasmid vectors. Wild-type SspB delivers DAS-tagged CdiI immunity protein to the ClpXP protease, whereas the SspB(Δ47) variant does so less efficiently. Cell growth was monitored by measuring the OD600 as a function of time. (B) Cells from the experiment in A were removed at various times, and total RNA was isolated for analysis by denaturing gel electrophoresis and Northern blot hybridization. The top panel shows total cellular RNA visualized by ethidium bromide staining. The bottom panels are Northern blots using probes specific for E. coli tRNA2Arg and tRNA3Gly. The gel migration positions of ribosomal RNAs (rRNAs) and tRNAs are indicated.
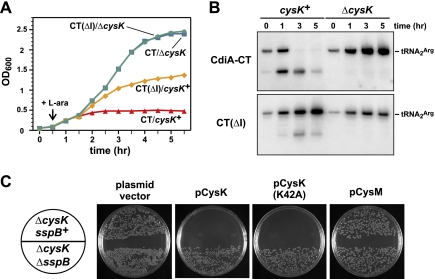
Because tRNase activity arrests cell growth, we next asked whether CysK is required to activate the CdiA-CT toxin in vivo. We repeated the CdiI-DAS degradation experiment in ΔcysK cells and found that SspB induction had no effect on cell growth in this background (Fig. 6A). Northern blot analysis showed no evidence of tRNase activity in ΔcysK cells, whereas tRNA2Arg was cleaved and ultimately degraded in the cysK+ background (Fig. 6B). We also tested CdiA-CT proteins lacking C-terminal residues for growth inhibition activity. The ΔGYGI version of CdiA-CT had no effect on cysK+ and ΔcysK cell growth (data not shown), consistent with its lack of tRNase activity in vitro (Supplemental Fig. S4). Although the ΔI version of CdiA-CT did not form a stable complex with CysK-His6 and had no detectable tRNase activity in vitro (Supplemental Fig. S4), this protein inhibited cell growth in a cysK-dependent manner (Fig. 6A). This growth inhibition was associated with tRNase activity, although at a significantly lower level than that observed with full-length CdiA-CT (Fig. 6B). Together, these results suggest that CdiA-CT(ΔI) retains tRNase activity, but its reduced affinity for CysK limits growth inhibition to an intermediate level.
Figure 6.
CysK is required for CdiA-CT-mediated growth inhibition and tRNase activity in vivo. (A) The DAS-tagged CdiI immunity protein was targeted for proteolysis by SspB production (induced by L-arabinose at the indicated time point) to liberate either CdiA-CT or CdiA-CT(ΔI) proteins in cysK+ and ΔcysK cells. Cell growth was monitored by measuring the OD600 as a function of time. (B) Cells from the experiment shown in A were removed at various times, and total RNA was isolated for Northern blot analysis. The gel migration positions of full-length and cleaved tRNA2Arg are indicated. (C) Complementation of the ΔcysK mutation. E. coli ΔcysK cells expressing the CdiA-CT/CdiI-DAS complex were transformed with plasmids that produce CysK, CysK(K42A), or CysM under control of the arabinose-inducible PBAD promoter. Transformants were selected on LB agar plates supplemented with 0.2% L-arabinose and the appropriate antibiotics. Transformation of E. coli ΔcysK ΔsspB cells serves as a control to demonstrate that CdiI-DAS degradation is required for growth inhibition.
We next sought to complement the ΔcysK mutation with a plasmid-borne copy of the gene. We introduced plasmid pCH450-CysK (which expresses cysK under the control of an arabinose-inducible promoter) into ΔcysK sspB+ and ΔcysK ΔsspB backgrounds and then tested whether the resulting cells could be transformed with a compatible plasmid expressing the CdiA-CT/CdiI-DAS toxin/immunity pair. Control cells carrying the pCH450 vector plasmid without cysK were readily transformed with the CdiA-CT/CdiI-DAS plasmid (Fig. 6C). In contrast, no stable transformants were obtained with ΔcysK sspB+ pCH450-CysK cells when plated on selective medium supplemented with L-arabinose to induce CysK synthesis (Fig. 6C). However, the ΔcysK ΔsspB pCH450-CysK cells could be readily transformed with the CdiA-CT/CdiI-DAS plasmid (Fig. 6C), reconfirming that SspB-dependent degradation of the DAS-tagged CdiI immunity protein is required for growth inhibition. This transformation assay was also used to examine growth inhibition in cells carrying plasmid-borne copies of the cysK(K42A) and cysM genes. Consistent with the in vitro tRNase experiments, we found that cysK(K42A) supported growth inhibition, whereas cysM did not (Fig. 6C). Together, these results demonstrate that CysK is required for CdiA-CT-mediated growth inhibition.
CysK is required for UPEC536 CdiA-mediated CDI
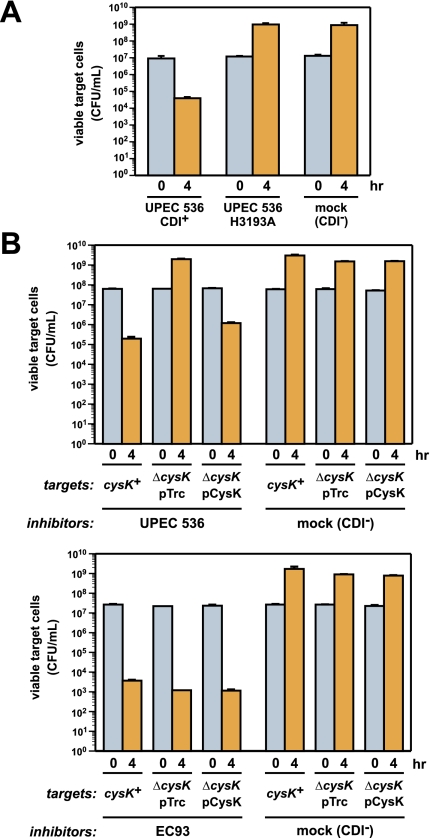
The toxicity of the CdiA-CT tRNase suggests that this nuclease activity is responsible for growth inhibition during cell-mediated CDIUPEC536. However, the UPEC536 CdiA protein is very large (∼331 kDa) and could possess multiple toxic activities that act in concert to inhibit target cell growth. Therefore, we examined the role of tRNase activity by performing growth competitions using inhibitor cells that express full-length CdiA proteins from cosmid-borne CDIUPEC536 systems. Viable target cell counts were reduced ∼100-fold after coculture with inhibitor cell expressing wild-type CdiA, whereas target cell growth was unaffected by inhibitor cells that express the His3193Ala version of CdiA lacking tRNase activity (Fig. 7A). Immunofluorescence staining using antibodies specific for the CdiA-CT confirmed that CdiA(H3193A) protein was expressed on the cell surface at approximately the same level as wild-type CdiA (Supplemental Fig. S6). These data strongly suggest the CdiA tRNase activity is solely responsible for growth inhibition during CDIUPEC536.
Figure 7.
CysK is required for growth inhibition during CDIUPEC536. (A) tRNase activity is required for the inhibition of target cell growth during CDIUPEC536. Target cells were cocultured with inhibitor cells expressing either wild-type CdiA or a variant carrying the His3193Ala missense mutation, which ablates tRNase activity. Growth inhibition was assessed by quantifying the number of viable target cells as colony-forming units per milliliter (CFU/mL) at 0 and 4 h of coculture. The CDI− competition contained mock inhibitor cells carrying a cosmid vector with no CDI system. (B) CysK is required for growth inhibition during CDIUPEC536. Inhibitor cells expressing either the UPEC536 or EC93 CDI system were cocultured with cysK+, ΔcysK, and pCysK-complemented ΔcysK target cells. Viable target cells were determined (CFU/mL) at 0 and 4 h of coculture.
We next tested whether CysK is required to activate CdiA tRNase activity during cell-mediated CDI. Target cells carrying the ΔcysK mutation were completely resistant to inhibitor cells expressing UPEC536 CdiA (Fig. 7B), consistent with the toxin-activating function of CysK. Susceptibility to CDIUPEC536 was almost completely restored by providing ΔcysK target cells with a plasmid-borne copy of the cysK gene (Fig. 7B). In contrast, the ΔcysK mutation provided no protection against the E. coli EC93 CDI system (Fig. 7B), which delivers a different CdiA-CT toxin that is thought to inhibit cell growth by forming a pore in the inner membrane of target cells (Aoki et al. 2009, 2010). Thus, CysK plays a critical and specific role in CDI mediated by the CDIUPEC536 system.
Discussion
The results presented here demonstrate that the CdiA-CT from UPEC536 is an anti-codon nuclease responsible for the inhibition of target cell growth during CDI. Remarkably, the cytotoxic tRNase activity of CdiA-CT is latent and requires activation by the biosynthetic enzyme CysK. CdiA-CT and CysK bind stably to one another, and this complex appears to be required for tRNase activity in vitro and in vivo. CdiA-CT variants lacking one to four C-terminal residues have a much lower affinity for CysK and exhibit no tRNase activity in vitro. Of course, the C-terminal deletions could also interfere with CdiA-CT structure and ablate tRNase activity independently of the CysK-binding defect. However, three observations argue against this latter possibility. First, both C-terminal deletion variants copurify with His6-tagged CdiI immunity protein during Ni2+-affinity chromatography, indicating that CdiA-CT structure is not grossly perturbed by these changes. Second, although CdiA-CT(ΔI) (which lacks the C-terminal Ile residue) has no detectable tRNase in vitro, it significantly interferes with cell growth when expressed in E. coli. This growth inhibition is dependent on CysK and is associated with the partial degradation of cellular tRNAs. These findings suggest that CdiA-CT(ΔI) remains an intrinsic tRNase, but its activity is limited by a lower binding affinity for CysK. Last, appending a Flag peptide epitope onto the C terminus of CdiA-CT also blocks CysK binding and abrogates tRNase activity (EJ Diner and CS Hayes, unpubl.). As with the other CdiA-CT variants, the Flag-tagged domain has high affinity for CdiI, again suggesting that it retains the wild-type fold. Thus, the CysK/CdiA-CT complex is likely to represent the functional tRNase. Importantly, these findings are relevant to cell-mediated CDI. Target cells deleted for the cysK gene are completely resistant to inhibitor cells expressing UPEC536 CdiA, demonstrating that CysK is critical for growth inhibition during CDIUPEC536. This function is unique and distinct from the roles played by other target cell proteins that act as receptors and/or mediators of CdiA-CT translocation. Thus, CysK acts as a permissive factor during CDIUPEC536, allowing the toxic tRNase activity of CdiA-CT to be manifest in the target cell cytoplasm.
Although CysK-binding is required to activate the CdiA-CT tRNase domain, the precise mechanism of activation is not clear. Initially, we hypothesized that the N-terminal domain of CdiA-CT suppressed the activity of its C-terminal tRNase domain and CysK binding altered the orientation of the two domains to relieve inhibition. This model is consistent with the observation that the C-terminal 145 residues of CdiA exhibit tRNase activity in the absence of CysK. However, deletion of the C-terminal Ile residue in the context of the CysK-independent tRNase domain significantly reduces activity in vitro (EJ Diner and CS Hayes, unpubl.), indicating that the GYGI peptide does not function simply as a toggle switch for nuclease activity. Perhaps CysK binding induces a rearrangement of active site residues to promote catalysis. Presumably, these conformational changes in the tRNase domain are subtle because CysK-bound CdiA-CT retains its affinity for the CdiI immunity protein. Alternatively, CysK could facilitate substrate binding, perhaps even making direct contacts with tRNA. Finally, it is possible that the oligomerization of CdiA-CT is required for activation. CysK is a dimeric protein, and the proposed CdiA-CT-binding sites are presented on the same side of the homodimer (Rabeh and Cook 2004; Francois et al. 2006). Therefore, binding to CysK would tether two CdiA-CT proteins together in close approximation to one another. It is not immediately clear why an anti-codon nuclease should require dimerization for activity, but this mechanism may explain why the truncated tRNase domain (Gly 83–Ile 227) requires 10-fold higher concentrations to attain the same activity of the CysK/CdiA-CT complex. We are currently testing these models using biochemical approaches and high-resolution crystallography.
In many bacteria and higher plants, CysK also binds stably to the CysE protein to form the cysteine synthase complex (Kredich et al. 1969; Droux et al. 1998; Zhu et al. 1998; Mino et al. 2000b; Kumaran et al. 2009). The molecular basis of the CysK/CysE interaction has been examined in detail, and these studies have demonstrated that the C terminus of CysE binds in the active site cleft of CysK (Bogdanova and Hell 1997; Mino et al. 2000a; Campanini et al. 2005; Huang et al. 2005; Kumaran et al. 2009). The C-terminal Ile residue of CysE is particularly critical for complex formation and mediates many of the contacts with CysK (Francois et al. 2006; Salsi et al. 2010). CdiA-CT contains a C-terminal GYGI peptide motif that is similar to the GDGI sequence in CysE, and like the cysteine synthase complex, the CdiA-CT/CysK interaction requires the C-terminal Ile residue of CdiA-CT and can be disrupted by O-acetyl-L-serine. These observations suggest that CdiA-CT and CysE both bind CysK in the same manner. The evolutionary significance of this molecular mimicry is unclear, but perhaps the CdiA-CT has coopted a well-conserved protein–protein interaction to ensure activation in a wide variety of bacteria. The CysK proteins from other bacterial species are capable of promoting CdiA-CT nuclease activity, but CDI growth inhibitory activities appear to be restricted to the same or closely related species (EJ Diner, ZC Ruhe, and CS Hayes, unpubl.). Intriguingly, the CdiA-CT tRNase domain shares significant sequence identity with a handful of predicted proteins from the Gram-positive species Ruminococcus lactaris, Clostridium acetobutylicum, Listeria innocua, and Geobacillus sp. Y412MC10 (see Supplemental Fig. S2). In Ruminococcus, Geobacillus, and C. acetobutylicum, the genes encoding the CdiA-CT homologs are immediately followed by genes for proteins that are clearly related to UPEC536 CdiI, suggesting that these genes encode toxin/immunity pairs. The function of these gene pairs has not been examined, but they may represent a growth competition system analogous to CDI. The CdiA-CT homolog in Geobacillus contains a WXG100 domain (Pfam PF06013) in its N terminus, suggesting it is exported through an ESX/type VII secretion pathway (Simeone et al. 2009; Sutcliffe 2011). It is unclear whether these Gram-positive tRNase domains also require activation by CysK. Although each Gram-positive homolog contains the GYGI motif, these proteins are predicted to have additional C-terminal residues, which should prevent CysK binding. However, CysK from Bacillus subtilis activates CdiA-CT in vitro and in E. coli cells (EJ Diner and CS Hayes, unpubl.), suggesting that the same activation mechanism could operate in Gram-positive bacteria, provided the C terminus of the tRNase domain is processed to expose the GYGI motif.
If growth inhibition is the sole function of the CDIUPEC536 system, then the benefit conferred by CysK-mediated toxin activation is not immediately obvious. In fact, toxin activation provides an additional opportunity for target cells to evolve resistance to CDIUPEC536 because cysK is not an essential gene. In contrast, soluble bacteriocins also deliver toxic domains into bacteria to inhibit growth, but typically only require cell envelope proteins for recognition and translocation of the toxin (Cascales et al. 2007; Chauleau et al. 2011). One exception appears to be colicin M (an inhibitor of peptidoglycan synthesis), which is remodeled into its active conformation by a peptidyl-prolyl cis-trans isomerase (FkpA) in the periplasm of target cells (Schaller et al. 1982; El Ghachi et al. 2006; Hullmann et al. 2008; Helbig et al. 2011). CysK has no known chaperone activity, but could perhaps stabilize the CdiA-CT in target cells. The hydrophobic C-terminal GYGI sequence resembles known recognition determinants for the ClpXP and ClpAP proteases (Sauer et al. 2004; Sauer and Baker 2011). Therefore, CysK could accentuate growth inhibition by protecting the tRNase domain from proteolysis. However, E. coli ΔclpX and ΔclpP mutants are no more sensitive to CDIUPEC536 than wild-type cells (CM Beck, JS Webb, DA Low, and CS Hayes, unpubl.). Perhaps the CysK/CdiA-CT complex serves other functions in addition to its characterized role in growth inhibition. For example, the interaction with CysK provides a possible mechanism to modulate sulfur metabolism and cysteine biosynthesis, which influence stress responses, antibiotic resistance, and biofilm formation (Ren et al. 2005; Ackerley et al. 2006; Fuentes et al. 2007). According to this model, the delivery of CdiA-CT between immune CDI+ cells would serve a contact-dependent signaling function to coordinate cellular activities in the same manner as quorum-sensing systems. Prior work with E. coli EC93 indicates that there is no exclusion mechanism to prevent CdiA-CT delivery between CDI+ bacteria (Aoki et al. 2010), suggesting that toxin exchange between isogenic cells is common. Intriguingly, we also found that UPEC536 carrying the His3193Ala missense mutation in cdiA exhibits a defect in biofilm formation compared with wild-type CDI+ cells (JS Webb, T Smith, and DA Low, unpubl.). We are currently exploring how tRNase activity impinges on the regulatory pathways that control biofilm formation and whether CdiA-CT exchange between UPEC536 serves an intercellular signaling function.
Materials and methods
Bacterial strains and plasmids
All bacterial strains used in this study are listed in Supplemental Table S2. The ΔcysK::kan, ΔsucC::kan, ΔahpC::kan, ΔtreC::kan, and ΔsucD::kan gene disruptions were obtained from the Keio collection (Baba et al. 2006) and transduced into E. coli strains CH1944 (Garza-Sánchez et al. 2006) or JCM158 wzb::kan (Aoki et al. 2008) using bacteriophage P1-mediated generalized transduction. Kanamycin resistance cassettes were excised using FLP recombinase as described (Datsenko and Wanner 2000). All plasmids and cosmids are listed in Supplemental Table S2, and construction details are outlined in the Supplemental Material.
Protein purification
All CdiA-CT/CdiI-His6 complexes were overproduced and purified under nondenaturing conditions in reaction buffer (20 mM sodium phosphate at pH 7.0, 150 mM NaCl, 10 mM β-mercaptoethanol) as described previously (Aoki et al. 2010). Each complex was denatured in reaction buffer containing 6 M guanidine-HCl, and the CdiA-CT variants were isolated from CdiI-His6 by Ni2+-affinity chromatography. Purified CdiA-CT and CdiI-His6 proteins were refolded by dialysis into reaction buffer. Untagged CdiI was purified in the same manner from a complex with His6-tagged CdiA-CT. The various CysK-His6 and E. coli CysM-His6 proteins were purified by Ni2+-affinity chromatography under nondenaturing conditions. All His6-tagged proteins were eluted from Ni2+-NTA resin using reaction buffer supplemented with 250 mM imidazole. Imidazole was subsequently removed by dialysis against reaction buffer, and the purified proteins were quantified by absorbance at 280 nm.
Protein–protein interaction assessed by Ni2+-affinity chromatography
Binding interactions between CdiA-CT and His6-tagged CysK proteins were assayed by copurification on Ni2+-NTA resin. Briefly, His6-tagged “bait” (i.e., CysK or CysM) and untagged “prey” CdiA-CT proteins were mixed together at a final concentration of 10 μM in reaction buffer. An aliquot of the protein mixture was removed (for subsequent SDS-PAGE analysis), and then Ni2+-NTA resin was added and the mixture was incubated at room temperature for 1 h The reactions were centrifuged to collect the resin, and the supernatant was removed and analyzed as the “free” fraction. The resin was then washed three times with reaction buffer supplemented with 20 mM imidazole. Resin-bound proteins were eluted with reaction buffer supplemented with 250 mM imidazole. For the CdiA-CT/CysK complex-blocking experiments, CysK-His6 was preincubated for 15 min with 50 mM O-acetyl-L-serine (or 50 mM L-serine) in 100 mM Tris-HCl (pH 6.5) prior to the addition of CdiA-CT. All fractions (input, free, and bound) were analyzed by SDS-PAGE and staining with Coomassie brilliant blue.
In vitro tRNase assays
Dialyzed S100 fractions were used for tRNase activity assays, as described previously (Aoki et al. 2010). Purified tRNA substrates were prepared by either GITC-phenol extraction (Garza-Sánchez et al. 2006) or DEAE-cellulose chromatography of E. coli S100 lysates (as described below). Reactions contained 0.1 μM CdiA-CT (and equimolar CdiI-His6, where indicated) and 130 ng/μL tRNA in reaction buffer supplemented with 10 mM MgCl2. The truncated CdiA-CT(G83-I227) domain was used at a higher concentration (1 μM final) for in vitro tRNase assays. Where indicated, CysM-His6 and CysK-His6 protein variants were included at a final concentration of 0.1 μM. Reactions were extracted with GITC-phenol extraction, and the isolated tRNAs were analyzed by gel electrophoresis on 8.3 M urea/6% polyacrylamide gels in 1× Tris-borate-EDTA (TBE) buffer. Gels were stained with ethidium bromide to visualize nucleic acids.
Purification of the CdiA-CT-activating factor
E. coli strain X90 was grown to OD600 ∼1.0 in LB medium at 37°C and harvested over crushed ice. Cells were broken by French press, and the S100 fraction was prepared by ultracentrifugation at 100,000g for 4 h at 4°C as described (Powers and Noller 1991). Following ultracentrifugation, the S100 fraction was dialyzed against 20 mM Tris-HCl (pH 6.0) and 250 mM NaCl. The dialyzed S100 was passed over a DEAE-cellulose 52 column pre-equilibrated in 20 mM Tris-HCl (pH 6.0) and 250 mM NaCl. The flowthrough fraction (fraction A) contained most of the proteins present in the original S100 fraction (as determined by SDS-PAGE). Fraction B (which contained tRNAs and some proteins) was eluted from the DEAE-cellulose column with 20 mM Tris-HCl (pH 6.0) and 500 mM NaCl and dialyzed against reaction buffer for use as substrate tRNA for in vitro tRNase assays. Fraction A was dialyzed against 20 mM sodium phosphate (pH 7.0) and 75 mM NaCl and further fractionated by DEAE-cellulose chromatography to enrich for the CdiA-CT-activating factor. Proteins were eluted using a linear gradient of 75–150 mM NaCl in 20 mM sodium phosphate (pH 7.0), and individual fractions from the gradient were tested for the ability to stimulate the tRNase activity. The activating fractions were pooled, adjusted to 0.8 M (NH4)2SO4, and applied to a HiTrap butyl-Sepharose hydrophobic interaction column (GE Healthcare). Proteins were eluted by decreasing the (NH4)2SO4 concentration using a step gradient. Each fraction was dialyzed against reaction buffer and assayed for tRNase activity with 0.1 μM purified CdiA-CT. The fraction eluting at 0.6 M (NH4)2SO4 activated CdiA-CT and was analyzed by SDS-PAGE. This protein fraction (35 μg) was dialyzed into 50 mM ammonium bicarbonate and lyophilized for trypsin digestion and liquid chromatography–mass spectrometry analysis.
Growth inhibition activity of CdiA-CT
E. coli strains X90 ΔsspB and X90 ΔsspB ΔcysK carrying plasmids pCH410::sspB or pCH410::sspB(Δ47) (Poole et al. 2011) were transformed with plasmid pTrc99a::CdiA-CT/CdiI-DAS, and the resulting cells were grown to mid-log phase in LB supplemented with 150 μg/mL ampicillin and 10 μg/mL tetracycline and then diluted to OD600 of 0.05 in fresh LB medium supplemented with ampicillin and tetracycline. Cells were grown for 30 min and then adjusted to 0.4% L-arabinose to induce the synthesis of SspB or SspB(Δ47). The OD600 was measured every 30 min following induction to monitor cell growth. Cell samples from each culture were removed and fixed with an equal volume of ice-cold methanol. Whole-cell RNA was then extracted by GITC-phenol for analysis by denaturing polyacrylamide gel electrophoresis. Specific tRNAs were also analyzed by Northern blot hybridization using end-labeled oligonucleotide probes as described (Hayes and Sauer 2003; Garza-Sánchez et al. 2006). Complementation experiments were performed with E. coli X90 ΔcysK and X90 ΔsspB ΔcysK strains carrying plasmid pTrc99a::CdiA-CT/CdiI-DAS. These cells were made chemically competent and then transformed with plasmid constructs that express cysM and cysK variants under control of the L-arabinose-inducible PBAD promoter. Transformed cells were selected on LB agar plates supplemented with L-arabinose (0.2%), ampicillin (150 μg/mL), and tetracycline (10 μg/mL).
Growth competition assays
Inhibitor and target E. coli strains were grown separately to mid-log phase (OD600 ∼0.5) at 37°C in LB medium without antibiotics. Inhibitor and target cells were then diluted to ∼3 × 107 colony-forming units (CFU) per milliliter and mixed at a 1:1 ratio in 10 mL of fresh LB medium. The coculture was then incubated with shaking in a 125-mL baffled flask at 37°C. Samples were taken at the 0-h and 4-h time points for serial dilution and quantification of viable cells (CFUs per milliliter) on LB agar plates supplemented with the appropriate antibiotics to select for either inhibitor or target cells. All inhibitor cells were based on E. coli strain EPI100 carrying cosmids pWEB-TNC (CDI−), pDAL866 (CdiAUPEC536), pDAL660Δ1-39 (CdiAEC93), and pDAL893 [CdiA(H3193A)UPEC536]. All target cells were based on the JCM158 Δwzb background. For the ΔcysK complementation studies, target cells carrying pTrc99A::cysK (or pTrc99A vector control) were grown in 1 mM IPTG prior to coculture with inhibitor cells.
Cell surface immunofluorescence
Cells expressing UPEC536 CdiA were grown to OD600 ∼0.8 in LB medium at 37°C with aeration. Cells were harvested by centrifugation and resuspended in 0.2 mL of 4% formaldehyde in phosphate-buffered saline (PBS). After fixation for 15 min at room temperature, the cells were washed twice with 0.2 mL of PBS and resuspended in 1% bovine serum albumin in PBS. Rabbit polyclonal antisera specific for the CdiA-CT (1:50 dilution) were added and incubated at room temperature for 30 min. Cells were washed twice with 0.2 mL of PBS and resuspended in 1% bovine serum albumin in PBS. Anti-rabbit AlexaFluor 488 antibody was added and incubated on ice for 30 min. Cells were washed, resuspended in PBS supplemented with 0.02% sodium azide, and pipetted onto poly-D-lysine slides. After drying for 5 min, slides were washed three times with 50 μL of Nanopure water and dried once more. One drop of FluoroGel in TES buffer (Electron Microscopy Sciences) was applied, followed by the cover slip. Slides were allowed to dry overnight at 4°C prior to fluorescence microscopy.
Acknowledgments
We thank Travis Smith and Aubrey MacMillan for performing preliminary growth competition experiments. Mass spectrometry and peptide identification were conducted at the University of California Davis Proteomics core facility. This work was supported by grants GM078634 (to C.S.H.) and U54AI065359 (to D.A.L.) from the National Institutes of Health, and grant 0642052 (to D.A.L.) from the National Science Foundation.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.182345.111.
References
- Ackerley DF, Barak Y, Lynch SV, Curtin J, Matin A 2006. Effect of chromate stress on Escherichia coli K-12. J Bacteriol 188: 3371–3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki SK, Pamma R, Hernday AD, Bickham JE, Braaten BA, Low DA 2005. Contact-dependent inhibition of growth in Escherichia coli. Science 309: 1245–1248 [DOI] [PubMed] [Google Scholar]
- Aoki SK, Malinverni JC, Jacoby K, Thomas B, Pamma R, Trinh BN, Remers S, Webb J, Braaten BA, Silhavy TJ, et al. 2008. Contact-dependent growth inhibition requires the essential outer membrane protein BamA (YaeT) as the receptor and the inner membrane transport protein AcrB. Mol Microbiol 70: 323–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki SK, Webb JS, Braaten BA, Low DA 2009. Contact-dependent growth inhibition causes reversible metabolic downregulation in Escherichia coli. J Bacteriol 191: 1777–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki SK, Diner EJ, de Roodenbeke CT, Burgess BR, Poole SJ, Braaten BA, Jones AM, Webb JS, Hayes CS, Cotter PA, et al. 2010. A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature 468: 439–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol 2: 2006.0008 doi: 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanova N, Hell R 1997. Cysteine synthesis in plants: Protein–protein interactions of serine acetyltransferase from Arabidopsis thaliana. Plant J 11: 251–262 [DOI] [PubMed] [Google Scholar]
- Campanini B, Speroni F, Salsi E, Cook PF, Roderick SL, Huang B, Bettati S, Mozzarelli A 2005. Interaction of serine acetyltransferase with O-acetylserine sulfhydrylase active site: Evidence from fluorescence spectroscopy. Protein Sci 14: 2115–212415987896 [Google Scholar]
- Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, Postle K, Riley M, Slatin S, Cavard D 2007. Colicin biology. Microbiol Mol Biol Rev 71: 158–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauleau M, Mora L, Serba J, de Zamaroczy M 2011. FtsH-dependent processing of RNase colicins D and E3 means that only the cytotoxic domains are imported into the cytoplasm. J Biol Chem 286: 29397–29407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droux M, Ruffet ML, Douce R, Job D 1998. Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants–structural and kinetic properties of the free and bound enzymes. Eur J Biochem 255: 235–245 [DOI] [PubMed] [Google Scholar]
- El Ghachi M, Bouhss A, Barreteau H, Touze T, Auger G, Blanot D, Mengin-Lecreulx D 2006. Colicin M exerts its bacteriolytic effect via enzymatic degradation of undecaprenyl phosphate-linked peptidoglycan precursors. J Biol Chem 281: 22761–22772 [DOI] [PubMed] [Google Scholar]
- Francois JA, Kumaran S, Jez JM 2006. Structural basis for interaction of O-acetylserine sulfhydrylase and serine acetyltransferase in the Arabidopsis cysteine synthase complex. Plant Cell 18: 3647–3655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes DE, Fuentes EL, Castro ME, Perez JM, Araya MA, Chasteen TG, Pichuantes SE, Vasquez CC 2007. Cysteine metabolism-related genes and bacterial resistance to potassium tellurite. J Bacteriol 189: 8953–8960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza-Sánchez F, Janssen BD, Hayes CS 2006. Prolyl-tRNA(Pro) in the A-site of SecM-arrested ribosomes inhibits the recruitment of transfer-messenger RNA. J Biol Chem 281: 34258–34268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes CS, Sauer RT 2003. Cleavage of the A site mRNA codon during ribosome pausing provides a mechanism for translational quality control. Mol Cell 12: 903–911 [DOI] [PubMed] [Google Scholar]
- Hayes CS, Aoki SK, Low DA 2010. Bacterial contact-dependent delivery systems. Annu Rev Genet 44: 71–90 [DOI] [PubMed] [Google Scholar]
- Helbig S, Patzer SI, Schiene-Fischer C, Zeth K, Braun V 2011. Activation of colicin M by the FkpA prolyl cis-trans isomerase/chaperone. J Biol Chem 286: 6280–6290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Vetting MW, Roderick SL 2005. The active site of O-acetylserine sulfhydrylase is the anchor point for bienzyme complex formation with serine acetyltransferase. J Bacteriol 187: 3201–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hullmann J, Patzer SI, Romer C, Hantke K, Braun V 2008. Periplasmic chaperone FkpA is essential for imported colicin M toxicity. Mol Microbiol 69: 926–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajava AV, Cheng N, Cleaver R, Kessel M, Simon MN, Willery E, Jacob-Dubuisson F, Locht C, Steven AC 2001. β-Helix model for the filamentous haemagglutinin adhesin of Bordetella pertussis and related bacterial secretory proteins. Mol Microbiol 42: 279–292 [DOI] [PubMed] [Google Scholar]
- Kredich NM, Becker MA, Tomkins GM 1969. Purification and characterization of cysteine synthetase, a bifunctional protein complex, from Salmonella typhimurium. J Biol Chem 244: 2428–2439 [PubMed] [Google Scholar]
- Kumaran S, Jez JM 2007. Thermodynamics of the interaction between O-acetylserine sulfhydrylase and the C-terminus of serine acetyltransferase. Biochemistry 46: 5586–5594 [DOI] [PubMed] [Google Scholar]
- Kumaran S, Yi H, Krishnan HB, Jez JM 2009. Assembly of the cysteine synthase complex and the regulatory role of protein-protein interactions. J Biol Chem 284: 10268–10275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinness KE, Baker TA, Sauer RT 2006. Engineering controllable protein degradation. Mol Cell 22: 701–707 [DOI] [PubMed] [Google Scholar]
- Mino K, Yamanoue T, Sakiyama T, Eisaki N, Matsuyama A, Nakanishi K 1999. Purification and characterization of serine acetyltransferase from Escherichia coli partially truncated at the C-terminal region. Biosci Biotechnol Biochem 63: 168–179 [DOI] [PubMed] [Google Scholar]
- Mino K, Hiraoka K, Imamura K, Sakiyama T, Eisaki N, Matsuyama A, Nakanishi K 2000a. Characteristics of serine acetyltransferase from Escherichia coli deleting different lengths of amino acid residues from the C-terminus. Biosci Biotechnol Biochem 64: 1874–1880 [DOI] [PubMed] [Google Scholar]
- Mino K, Yamanoue T, Sakiyama T, Eisaki N, Matsuyama A, Nakanishi K 2000b. Effects of bienzyme complex formation of cysteine synthetase from Escherichia coli on some properties and kinetics. Biosci Biotechnol Biochem 64: 1628–1640 [DOI] [PubMed] [Google Scholar]
- Poole SJ, Diner EJ, Aoki SK, Braaten BA, t'Kint de Roodenbeke C, Low DA, Hayes CS 2011. Identification of functional toxin/immunity genes linked to contact-dependent growth inhibition (CDI) and rearrangement hotspot (Rhs) systems. PLoS Genet 7: e1002217 doi: 10.1371/journal.pgen.1002217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T, Noller HF 1991. A functional pseudoknot in 16S ribosomal RNA. EMBO J 10: 2203–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabeh WM, Cook PF 2004. Structure and mechanism of O-acetylserine sulfhydrylase. J Biol Chem 279: 26803–26806 [DOI] [PubMed] [Google Scholar]
- Ren D, Zuo R, Gonzalez Barrios AF, Bedzyk LA, Eldridge GR, Pasmore ME, Wood TK 2005. Differential gene expression for investigation of Escherichia coli biofilm inhibition by plant extract ursolic acid. Appl Environ Microbiol 71: 4022–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salsi E, Bayden AS, Spyrakis F, Amadasi A, Campanini B, Bettati S, Dodatko T, Cozzini P, Kellogg GE, Cook PF, et al. 2010. Design of O-acetylserine sulfhydrylase inhibitors by mimicking nature. J Med Chem 53: 345–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer RT, Baker TA 2011. AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem 80: 587–612 [DOI] [PubMed] [Google Scholar]
- Sauer RT, Bolon DN, Burton BM, Burton RE, Flynn JM, Grant RA, Hersch GL, Joshi SA, Kenniston JA, Levchenko I, et al. 2004. Sculpting the proteome with AAA+ proteases and disassembly machines. Cell 119: 9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller K, Holtje JV, Braun V 1982. Colicin M is an inhibitor of murein biosynthesis. J Bacteriol 152: 994–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone R, Bottai D, Brosch R 2009. ESX/type VII secretion systems and their role in host-pathogen interaction. Curr Opin Microbiol 12: 4–10 [DOI] [PubMed] [Google Scholar]
- Sutcliffe IC 2011. New insights into the distribution of WXG100 protein secretion systems. Antonie van Leeuwenhoek 99: 127–131 [DOI] [PubMed] [Google Scholar]
- Tai CH, Nalabolu SR, Jacobson TM, Minter DE, Cook PF 1993. Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 using the natural and alternative reactants. Biochemistry 32: 6433–6442 [DOI] [PubMed] [Google Scholar]
- Wah DA, Levchenko I, Baker TA, Sauer RT 2002. Characterization of a specificity factor for an AAA+ ATPase: Assembly of SspB dimers with ssrA-tagged proteins and the ClpX hexamer. Chem Biol 9: 1237–1245 [DOI] [PubMed] [Google Scholar]
- Wah DA, Levchenko I, Rieckhof GE, Bolon DN, Baker TA, Sauer RT 2003. Flexible linkers leash the substrate binding domain of SspB to a peptide module that stabilizes delivery complexes with the AAA+ ClpXP protease. Mol Cell 12: 355–363 [DOI] [PubMed] [Google Scholar]
- Zhao C, Moriga Y, Feng B, Kumada Y, Imanaka H, Imamura K, Nakanishi K 2006. On the interaction site of serine acetyltransferase in the cysteine synthase complex from Escherichia coli. Biochem Biophys Res Commun 341: 911–916 [DOI] [PubMed] [Google Scholar]
- Zhu X, Yamaguchi T, Masada M 1998. Complexes of serine acetyltransferase and isozymes of cysteine synthase in spinach leaves. Biosci Biotechnol Biochem 62: 947–952 [DOI] [PubMed] [Google Scholar]