Abstract
The expression of the renoprotective antiaging gene Klotho is decreased in uremia. Recent studies suggest that Klotho may be a tumor suppressor, and its expression may be repressed by DNA hypermethylation in cancer cells. Here we investigated the effects and possible mechanisms by which Klotho expression is regulated during uremia in uninephrectomized B-6 mice given the uremic toxins indoxyl sulfate or p-cresyl sulfate. Cultured human renal tubular HK2 cells treated with these toxins were used as an in vitro model. Injections of indoxyl sulfate or p-cresyl sulfate increased their serum concentrations, kidney fibrosis, CpG hypermethylation of the Klotho gene, and decreased Klotho expression in renal tubules of these mice. The expression of DNA methyltransferases 1, 3a, and 3b isoforms in HK2 cells treated with indoxyl sulfate or p-cresyl sulfate was significantly increased. Specific inhibition of DNA methyltransferase isoform 1 by 5-aza-2′-deoxycytidine caused demethylation of the Klotho gene and increased Klotho expression in vitro. Thus, inhibition of Klotho gene expression by uremic toxins correlates with gene hypermethylation, suggesting that epigenetic modification of specific genes by uremic toxins may be an important pathological mechanism of disease.
Keywords: DNA methylation, DNA methyltransferase, Klotho, uremic toxin
Uremic patients have higher risks of cardiovascular disease and mortality than the normal population.1 The antiaging gene, Klotho, encodes a single-pass transmembrane protein that forms a complex with multiple fibroblast growth factor receptors, and is most abundant in the renal tubules.2 A defect in Klotho gene expression in mice results in shortened lifespan and aging-like phenotypes.3, 4, 5, 6 Recent studies also showed that Klotho functions as a renoprotective factor.7, 8
Klotho expression is decreased in uremic patients.9 Increased oxidative stress and inflammation, which are common in uremia status, cause depressed Klotho expression.4, 10 Previous studies indicated that uremic toxins increased the oxidative stress and reduced cell viability.11, 12 An animal study showed that treatment with the uremic toxin sorbent, AST-120, increased Klotho expression and inhibited cell senescence in the kidneys of uremic rats.13
Epigenetic modification of specific genes by uremic toxins might be an important pathological mechanism in uremic milieu. The expression of the Klotho gene is regulated by epigenetics. A recent study showed that CpG hypermethylation of the Klotho gene was associated with decreased Klotho expression in cervical cancer cells.14 The putative role of DNA methylation on regulation of Klotho expression in the uremia is currently still not clear.
Indoxyl sulfate (IS) and p-cresyl sulfate (PCS), which are protein-bound uremic toxins, increase significantly during kidney injury.15 Previous studies have shown that IS and PCS cause cellular toxicity by inducing oxidative stress.11, 16 It is known that oxidative stress affects DNA methyltransferase (DNMT) expression and modulates epigenetic regulation of gene expression during carcinogenesis.17, 18 Recent studies have demonstrated that IS and PCS significantly associated with mortality and kidney disease progression in patients with chronic kidney disease (CKD).19, 20, 21 We hypothesized that protein-bound uremic toxins, IS and PCS, suppressed Klotho expression by DNA hypermethylation via increased DNMT expression. An animal model of CKD and an in vitro study with human real epithelial cells treated with IS and PCS were used to test this hypothesis.
RESULTS
Chronic IS and PCS injection increased kidney fibrosis and serum IS and PCS concentrations in experimental mice
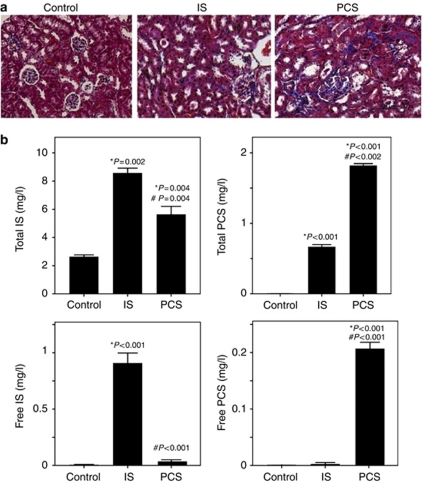
Histological staining results showed that IS- and PCS-injected mice had significantly increased kidney fibrosis than the control mice (Figure 1a). The serum IS and PCS concentrations of study animals are plotted in Figure 1b. The serum total IS concentrations in IS-injected (8.55±0.37 mg/l; P=0.002) and PCS-injected (5.61±0.60 mg/l; P=0.004) mice were significantly higher than control mice (2.39±0.15 mg/l). The serum total PCS concentrations in IS-injected (0.66±0.04 mg/l; P<0.001) and PCS-injected (1.82±0.03 mg/l; P<0.001) mice were also significantly higher than control mice (<0.225 mg/l). The free IS levels of IS-injected mice were significantly higher than control mice (0.87±0.14 vs. <0.5 mg/l, P<0.001). The free PCS levels of PCS-injected mice were significantly higher than control mice (0.21±0.01 vs. <0.15 mg/l, P<0.001).
Figure 1.
Results of Masson's trichrome staining and the serum levels of indoxyl sulfate (IS) and p-cresyl sulfate (PCS) in experimental mice. (a) Masson's trichrome staining showed that both IS- and PCS-injected mice had significantly increased kidney fibrosis than control mice. The total IS concentrations of control, IS, and PCS mice were 2.39±0.15, 8.55±0.37, and 5.61±0.60 mg/l, respectively. The total PCS concentrations of control, IS, and PCS study groups were <0.225, 0.66±0.04, and 1.82±0.03 mg/l, respectively. (b) The total IS and PCS concentrations of IS- and PCS-injected mice were significantly higher than those of control mice. The free IS levels of control, IS, and PCS study groups were 0.87±0.14, <0.50, and <0.50 mg/l, respectively. The free IS levels of IS-injected mice were significantly higher than control mice (P<0.001). The free PCS levels of control, IS, and PCS study groups were <0.15, <0.15, and 0.21±0.01 mg/l, respectively. (b) The free PCS levels of PCS-injected mice were significantly higher than control mice (P<0.001). *P<0.05 vs. control; #P<0.05 vs. IS.
The serum blood urea nitrogen levels of control, IS-, and PCS-injected mice were 33.87±1.87, 34.50±1.71, and 36.36±1.12 mg/dl, respectively. The serum creatinine levels of control, IS-, and PCS-injected mice were 0.34±0.01, 0.35±0.02, and 0.37±0.04 mg/dl, respectively. The blood urea nitrogen and creatinine levels of IS- and PCS-injected mice were not significantly different from those of the control mice (P>0.05). At the end of study, the body weights of control, IS-, and PCS-injected mice were 25.75±1.04, 20.73±2.16, and 21.15±2.21 g, respectively. The body weights of IS- and PCS-injected mice were significantly lower than the control mice (P>0.05).
IS- and PCS-injected mice had increased DNMT 1 expression and DNA hypermethylation of the Klotho gene
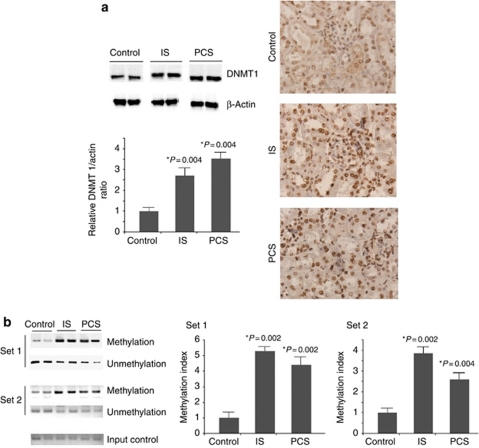
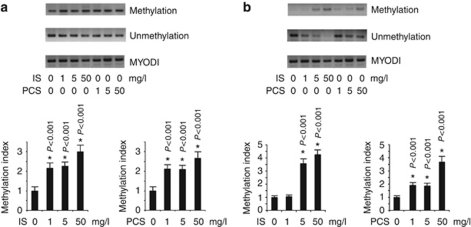
The western blot results revealed that the expression of DNMT 1 in IS- and PCS-injected mice significantly increased compared with the control mice. It was also noted by immunohistochemistry staining that IS and PCS significantly increased the DNMT 1 expression in the nuclei of the glomerular and tubular cells (Figure 2a). Methylation-specific PCR (MSP) analysis with primer sets 1 and 2 showed that both IS- and PCS-injected mice had significantly higher methylation indexes of the Klotho gene than control mice (Figure 2b). The relative methylation indexes of IS- and PCS-injected mice vs. control mice were 5.3:1 and 4.4:1, respectively, in the position of primer set 1. The relative methylation indexes of IS- and PCS-injected mice vs. control mice were 3.8:1 and 2.7:1, respectively, in the position of primer set 2.
Figure 2.
Indoxyl sulfate (IS)- and p-cresyl sulfate (PCS)-injected mice had increased DNA methyltransferase 1 (DNMT 1) expression and DNA hypermethylation of the Klotho gene. (a) The results of western blot analysis and immunostaining showed that IS and PCS significantly increased the DNMT 1 expression. (b) Methylation-specific PCR (MSP) analysis with primer sets 1 and 2 showed that both IS- and PCS-injected mice had significantly higher methylation indexes of Klotho genes than control mice. *P<0.05 vs. control.
IS- and PCS-injected mice had reduced Klotho expression in renal tubules
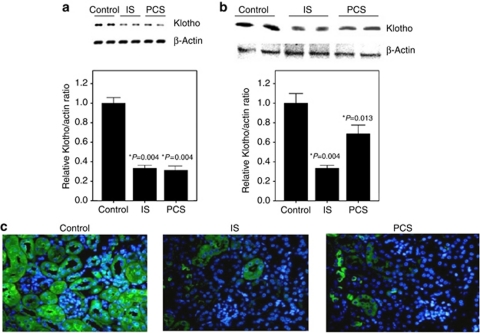
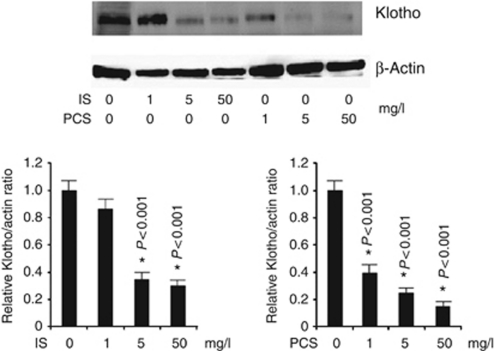
Real-time PCR and western blotting analysis showed that mRNA and protein expression of the Klotho gene were significantly decreased in the renal cortex of IS- and PCS-injected mice when compared with the ½-nephrectomized mice without IS and PCS injection (Figure 3a and b). Immunofluorescence staining also showed that the intensity of positive Klotho staining in renal tubules was significantly decreased in IS and PCS mice (Figure 3c). The results of western blotting showed that the expression of α-glutathione S-transferases increased significantly in IS- and PCS-injected mice when compared with the control mice (Supplementary Figure S1a online). The results of immunofluorescence double staining with Klotho and α-glutathione S-transferases showed that IS and PCS significantly decreased Klotho expression and increased α-glutathione S-transferase expression in the renal tubule cells in vivo (Supplementary Figure S1b online).
Figure 3.
Indoxyl sulfate (IS) and p-cresyl sulfate (PCS) injection reduced the Klotho expression in renal tubules. (a) Real-time PCR and (b) western blotting revealed that Klotho mRNA and protein expression were significantly lower in kidneys treated with IS or PCS. (c) Immunofluorescence staining indicated that the intensity of positive Klotho staining in renal tubules was significantly decreased in mice treated with IS or PCS. *P<0.05 vs. control.
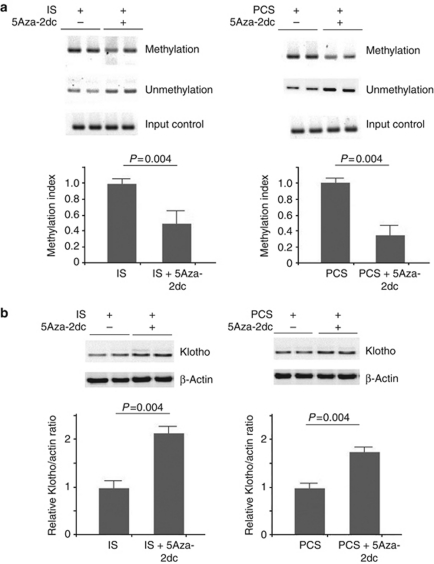
5-Aza-2′-deoxycytidine (5Aza-2dc) decreased the DNA hypermethylation of the Klotho gene and increased Klotho expression in vivo
MSP analysis showed that inhibiting DNMT 1 by using 5Aza-2dc significantly decreased the DNA methylation of Klotho gene in IS- and PCS-injected mice when compared with the mice treated with IS or PCS alone (Figure 4a). In addition, the results of western blotting revealed that treatment with 5Aza-2dc significantly increased Klotho expression in IS- and PCS-injected mice when compared with the mice treated with IS or PCS alone (Figure 4b).
Figure 4.
DNA methyltransferase 1 (DNMT 1) inhibitor demethylated the Klotho gene and increased Klotho expression in vivo. (a) Methylation-specific PCR (MSP) with mouse Klotho set 1 primers showed that simultaneous treatment with 5-aza-2′-deoxycytidine (5Aza-2dc) in indoxyl sulfate (IS)- and p-cresyl sulfate (PCS)-injected mice significantly demethylated the Klotho gene. (b) Western blotting showed that 5Aza-2dc significantly increased the Klotho expression in IS- and PCS-injected mice.
IS and PCS increased DNA methylation of the Klotho gene and decreased Klotho protein expression in vitro
In the animal study, both the serum IS and PCS concentrations increased significantly in IS- and PCS-injected mice. For discriminating the individual effects of IS and PCS on the DNA methylation and gene expression of Klotho, we conducted an in vitro study with cultured human renal tubular cells treated with IS and PCS, respectively. MSP analysis indicated that HK2 cells treated with IS had significantly increased CpG methylation of the Klotho gene in the positions of primer set 1 (at concentrations of 1, 5, and 50 mg/l) and 2 (at concentrations of 5 and 50 mg/l). HK2 cells treated with PCS (1, 5, and 50 mg/l) had significantly increased CpG hypermethylation of the Klotho gene in the positions of primer set 1 (Figure 5a) and 2 (Figure 5b).
Figure 5.
Indoxyl sulfate (IS) and p-cresyl sulfate (PCS) increased DNA methylation of the Klotho gene in vitro. Chromosome DNA samples from cultured HK2 cells treated with IS or PCS for 72 h were analyzed. Methylation-specific PCR (MSP) with human Klotho primer sets (a) 1 and (b) 2 revealed that the methylation indexes of HK2 cells treated with IS or PCS were significantly increased. *P<0.05 vs. lane 1.
Western blotting analysis showed that IS significantly decreased Klotho protein expression in cultured HK2 cells, at concentrations of 5 and 50 mg/l. PCS (1, 5, and 50 mg/l) also decreased Klotho protein expression significantly in vitro when compared with the cells without PCS treatment (Figure 6).
Figure 6.
Indoxyl sulfate (IS) and p-cresyl sulfate (PCS) reduced Klotho expression in vitro. Western blotting showed that the Klotho protein expression was significantly decreased in HK2 cells treated with IS or PCS. *P<0.05 vs. lane 1.
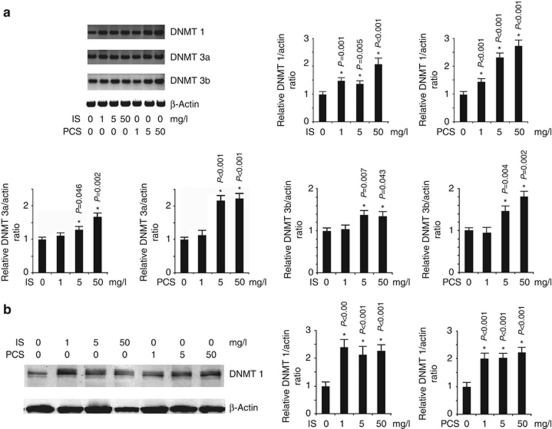
IS and PCS increased DNMT 1, 3a, and 3b expression in vitro
The results of real-time PCR indicated that IS and PCS could significantly increase DNMT 1 mRNA expression in cultured HK2 cells at a concentration of 1, 5, and 50 mg/l. The mRNA expressions of DNMT 3a and 3b were significantly increased in HK2 cells treated with IS or PCS at concentrations of 5 and 50 mg/l (Figure 7a). Western blotting for DNMT 1 revealed that IS and PCS could significantly increase DNMT 1 protein expression in vitro (Figure 7b).
Figure 7.
Indoxyl sulfate (IS) and p-cresyl sulfate (PCS) increased DNA methyltransferase (DNMT)1, 3a, and 3b expression in vitro. (a) Compared with HK2 cells without IS/PCS treatment, real-time PCR analysis showed that IS and PCS significantly increased DNMT 1, 3a, and 3b expression. (b) Results of western blotting for DNMT 1 showed that IS and PCS at concentrations of 1, 5, and 50 mg/l significantly increased DNMT 1 protein expression in cultured HK2 cells. *P<0.05 vs. lane 1.
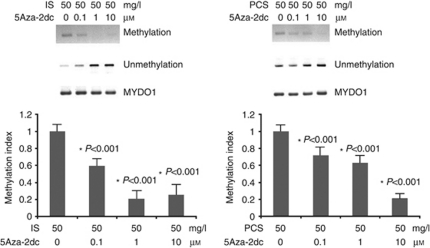
5Aza-2dc decreased the DNA hypermethylation of the Klotho gene and increased Klotho expression in HK2 cells treated with IS or PCS
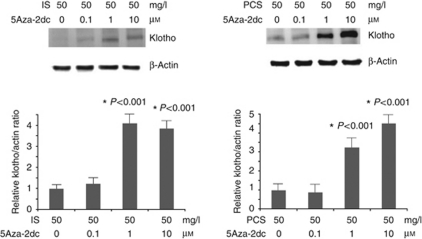
MSP analysis with set 1 primers demonstrated that the DNMT 1 inhibitor, 5Aza-2dc, could significantly decrease the CpG hypermethylation of the Klotho gene in cultured HK2 cells treated with 50 mg/l IS or PCS (Figure 8). The results of western blotting showed that 5Aza-2dc, at concentrations of 1 and 10 μmol/l, significantly increased the Klotho protein expression in HK2 cells treated with 50 mg/l IS or PCS (Figure 9).
Figure 8.
DNA methyltransferase 1 (DNMT 1) inhibitor demethylated the Klotho gene. Methylation-specific PCR (MSP) with human Klotho set 1 primers showed that 5-Aza-2′-deoxycytidine (5Aza-2dc) inhibited Klotho gene hypermethylation in cultured HK2 cells treated with 50 mg/l indoxyl sulfate (IS) or p-cresyl sulfate (PCS). *P<0.05 vs. lane 1.
Figure 9.
DNA methyltransferase 1 (DNMT 1) inhibitor increased Klotho expression in vitro. Western blotting showed that 5-aza-2′-deoxycytidine (5Aza-2dc) at concentrations of 1 and 10 μmol/l increased the Klotho protein expression significantly in HK2 cells treated with 50 mg/l indoxyl sulfate (IS) or p-cresyl sulfate (PCS). *P<0.05 vs. lane 1.
DISCUSSION
Uremic status increases oxidative stress, chronic inflammation, and malnutrition in patients with CKD.22 The unphysiological uremic environment and the epigenotype are considered to be important for the clinical outcomes in the CKD patient group.23 However, current clinical studies with normal subjects and CKD patients have not shown a significant correlation between the changes of global DNA methylation and renal function.24, 25 The content of uremic milieu is very complex;22 therefore, studies focusing on specific uremic toxins and site-specific DNA methylation might be critical. In this study, we explored the epigenetic regulation of the Klotho gene by undialyzable protein-bound uremic toxins, IS and PCS.15 A previous study showed that administration of IS to hypertensive rats reduced renal expression of Klotho, promoted cell senescence, and increased renal fibrosis.26 Our study further demonstrated that both IS and PCS decreased Klotho expression in renal tubules in vitro and in vivo. We also showed that epigenetic silencing of the Klotho gene by IS and PCS might be the possible regulation mechanism.
DNMTs are the key enzymes for the regulation of DNA methylation.27 DNMT 1 is the most abundant DNMT, and is considered to be the key maintenance methyltransferase in mammals.27, 28 Accumulated evidence reveals that DNA methylation regulated by DNMTs is associated with the development and progression of diseases such as malignancies and autoimmune diseases.29, 30 Epigenetic inactivation of tumor-suppressor genes by DNMTs is a critical pathological mechanism for malignancies.31, 32 Several studies have identified an association between Klotho and cancer in humans. Recent evidence suggested that Klotho had tumor-suppressor activities, but decreased Klotho expression was noted during the carcinogenesis process.14, 33, 34 Our study with cultured human renal tubular cells suggested that administration of IS and PCS increased DNMT 1, 3a, and 3b expressions. Inhibiting DNMT 1 activity with 5Aza-2dc demethylated CpG of the Klotho gene, and increased Klotho expression in HK2 cells treated with IS or PCS.
Oxidative stress has an important role for the tissue injury caused by IS and PCS.11, 12, 16 Previous studies have revealed that oxidative stress activates the Ras-MEK pathway in renal cells.35, 36 Oxidative stress increases the DNMT expression during carcinogenesis.17 It has been shown that the expression of DNMTs is upregulated by activated Ras, and DNA methylation may be regulated by the Ras signaling.17, 37 On the basis of this evidence, we speculate that oxidative stress induced by IS and PCS might increase DNMT expression via the Ras-MEK pathway. The increased expression of DNMTs might further hypermethylate the Klotho gene.
Recent studies have demonstrated that IS downregulates renal expression of Klotho through production of reactive oxygen species and activation of nuclear factor-κB, and promotes cell senescence with expression of senescence-related proteins such as p16, p21, p53, and retinoblastoma protein in the kidney of hypertensive rats.26, 38 Klotho is considered as a regulator of oxidative stress and senescence.39 Inhibiting Klotho expression by Klotho RNA interference could upregulate the p53/p21 pathway and induce premature senescence of human cells.40 It is considered that regulating Klotho expression via DNA methylation by IS and PCS might have a critical role in the cell senescence process caused by uremic toxins.
There were some limitations in this study. The levels of IS and PCS in CKD patients vary between the disease stages.19, 21, 41 Comparing with the IS and PCS levels of CKD patients, the serum IS and PCS levels of experimental mice in this study were similar to the levels of patients with late-stage CKD. It was also shown that IS and PCS caused significant renal fibrosis. However, IS and PCS did not significantly increase the blood urea nitrogen and creatinine levels of experimental mice in this study. We thought that decreased body weights in IS- and PCS-injected mice might negate the severity of uremia. The animal models used in this study might not present the full-scale uremia.
In conclusion, this study suggests that transcriptional suppression of Klotho by IS and PCS is correlated with CpG hypermethylation of the Klotho gene. Our study also showed that increased DNMT expression caused by IS and PCS might be responsible for CpG hypermethylation of the Klotho gene.
MATERIALS AND METHODS
Study animals
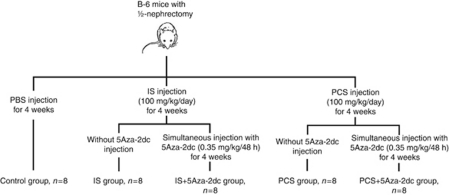
Ten-week-old B-6 mice with ½-nephrectomy were used in this study. The experimental mice received intraperitoneal injection with IS (Sigma, St Louis, MO; n=8) or PCS (Kureha, Tokyo, Japan; n=8) at a dosage of 100 mg/kg/day for 4 weeks. The control mice (n=8) received daily phosphate-buffered saline injection for 4 weeks at the same volume that was administered to the experimental mice. At the end of study, the body weight of study animal was recorded, and the serum levels of blood urea nitrogen and creatinine were analyzed. The renal cortex was microdissected for further analysis. In the 5Aza-2dc treatment study, the IS- and PCS-injected mice received intraperitoneal injection with 5Aza-2dc (Sigma) simultaneously at a dosage of 0.35 mg/kg per 48 h for 4 weeks (n=8 for each group). All animal experiments were approved by the experimental animal ethics committee at the Chang Gung Memorial Hospital. The flow diagram for animal study is summarized in Figure 10.
Figure 10.
The flow diagram for animal study. 5Aza-2dc, 5-aza-2′-deoxycytidine; IS, indoxyl sulfate; PBS, phosphate-buffered saline; PCS, p-cresyl sulfate.
IS and PCS measurement
The method for the measurement of total IS and PCS has been described in our previous report.21 Briefly, serum samples were deproteinized by the addition of three parts methanol to one part serum for determination of total IS and total PCS. All analyses were performed on a Waters ACQUITY Ultra Performance Liquid Chromatography (UPLC) system (Milford, MA). IS and PCS were detected at 280 and 260 nm. Buffer flow was 0.4 ml/min using 10 mmol/l NH4H2PO4 (pH=4.0) (A) and 100% acetonitrile (B) with a gradient from 82.5% A/17.5% B to 55% A/45% B, over 9 min. The limits of detection of this assay were 1.0 mg/l for total IS and 0.225 mg/l for total PCS. The detection limits for free IS and PCS were 0.50 and 0.15 mg/l. Calibration curves were constructed by plotting the peak areas versus the concentrations of each analyte. Quantitative results were obtained and expressed as concentrations (mg/l).
Methylation-specific PCR
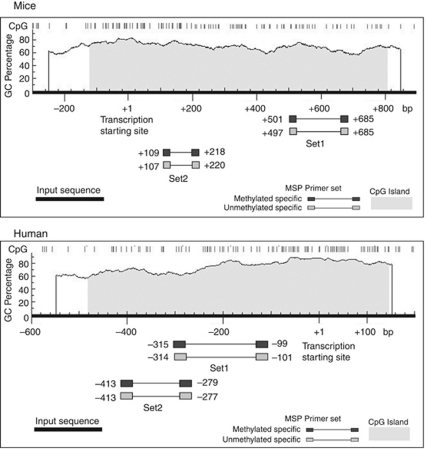
The prediction of CpG islands in the Klotho gene and the designing of MSP primers were performed with the MethPrimer software.42 The genomic regions near the transcription starting site (+1) of the Klotho gene (mouse: −300 to +900 bp; human: −600 to +200 bp) were inputted for CpG island analysis. A previous study has shown that the genomic region ranging from −1830 bp to +7 bp of human Klotho gene has significant promoter activities.43 The criteria for the CpG island prediction were as follows: island size >100 bp, GC percent >50.0, and observed/expected >0.6. Two sets of MSP primers were designed for Klotho of both mice and humans. The predicted CpG islands of the Klotho genes in mice and humans and the locations of MSP primers are plotted in Figure 11. The genomic DNA was extracted (Quick-gDNA MiniPrep, Zymo Research, Irvine, CA) and modified by bisulfate treatment (EZ DNA Methylation-Gold Kit, Zymo Research) for MSP analyses with two sets of gene promoter–specific primer pairs that recognize the methylated and unmethylated CpG sites. The PCR product of genomic DNA without bisulfate treatment, with primers located in the promoter region of the mouse Klotho gene, was used as the inputted control for the MSP. In the MSP for HK2 cells, the human MYOD1 gene was used as an inputted control. The IQ5 real-time PCR detection system (Bio-Rad, Hercules, CA) was used for MSP analysis. The PCR products were visualized by ethidium bromide staining in 2% agarose gels, and the densitometric intensity corresponding to each band was quantified. Each reaction was performed in triplicates. The methylation index was calculated as (band intensity of MSP with methylated primers) / (band intensity of inputted control). The methylation index was plotted after normalization to the control group. The sequence of MSP primers and the amplification program are summarized in Supplementary Table S1 online.
Figure 11.
Plots of CpG islands of the Klotho gene in mice and humans. The genomic region near the transcription start site (+1) of the Klotho gene (mouse: −300 to +900 bp; human: −600 to +200 bp) was inputted for CpG island analysis. The predictive CpG island of the mouse Klotho gene was 931 bp (−123 to +808 bp). The predictive CpG island of the human Klotho gene was 629 bp (−482 to +147 bp). The locations of the methylation-specific PCR (MSP) primers relative to transcription starting site are presented.
Quantitative real-time PCR
Total RNA was isolated with a commercial kit (RNeasy Kit, Qiagen, Valencia, CA) according to the manufacturer's instructions, including DNase treatment. A measure of 5 μg of total RNA was then reverse transcribed by reverse transcriptase (Bio-Rad) with random primers. Real-time PCR was performed in 25 μl SYBR Green PCR Master Mix (Applied Biosystems, Carlsbad, CA) containing 0.6 mol/l primers and 1 μg cDNA using an iQ5 real-time PCR detection system (Bio-Rad). The thermal cycling program and PCR primers for Klotho, DNMTs, and β-actin are listed in Supplementary Table S2 online. Each PCR reaction was performed in triplicates, and the mean Ct value was used for static analysis. Messenger RNA expression was standardized to β-actin expression levels, followed by normalization to the control group.
Western blotting analysis
Total protein was extracted using a commercial kit according to the manufacturer's instructions (Protein Extraction Kit, Millipore, Billerica, MA). A measure of 30 μg of protein from each sample was mixed with a sample-loading buffer and loaded onto separate lanes on 12% sodium dodecyl sulfate-polyacrylamide gel. Proteins were electrotransferred onto polyvinylidene fluoride membranes (0.2 μm: Immun-Blot, Bio-Rad), and then immunoblotted with antibodies against Klotho (Abcam, Cambridge, MA), DNMT 1 (Cell Signaling, Denver, MA), α-glutathione S-transferase (Abcam), and β-actin (Abcam). The intensity of each band was quantified using the NIH Image software (Bethesda, MD), and the densitometric intensity corresponding to each band was normalized against β-actin expression.
Masson's trichrome, immunohistochemistry, and immunofluorescence staining
Tissue sections from a paraffin-embedded block were stained with standard Masson's trichrome method. Paraffin tissue sections were cut, mounted, deparaffinized, rehydrated, and stained with hematoxylin–eosin using standard histological techniques. For immunohistochemical staining, the Ventana Benchmark automated staining system and Ventana reagents were used (Ventana Medical Systems, Tucson, AZ), and primary antibodies against DNMT 1 (1:100 dilution at 4 °C overnight; Cell Signaling) were used. For immunofluorescence staining, serial cryostat sections of renal cortex were incubated with a primary rabbit antibody against Klotho (1:100 dilution at 4 °C overnight; Abcam), followed by incubation with a fluorescein isothiocyanate-conjugated anti-rabbit IgG antibody (1:200 dilution at room temperature for 45 min; Abcam). For detecting the renal tubular injury, a biomarker for proximal renal tubular injury, α-glutathione S-transferase, was stained. Double immunofluorescence staining with Klotho and α-glutathione S-transferases (goat antibody; 1:300 dilution at 4 °C overnight; Abcam) was done. The staining for α-glutathione S-transferases was followed by incubation with an Alexa Fluor 568 dye-labeled anti-goat IgG (1:200 dilution at room temperature for 45 min; Invitrogen, Grand Island, NY). The sections were counterstained with 4′,6-diamidino-2-phenylindole (dilution 1:500; Sigma) to identify cellular nuclei.
Cell culture and treatment
HK2 cells were cultured as reported previously.44 HK2 cell cultures at ∼70% confluence were synchronized under serum-free conditions for 48 h. The HK2 cells under the serum-free condition were then treated with IS or PCS at concentrations of 0, 1, 5, and 50 mg/l for 72 h. For inhibition experiments, 5Aza-2dc at concentrations of 0, 0.1, 1, and 10 μmol/l were added to HK2 cells treated with IS or PCS at a concentration of 50 mg/l for 72 h. Each reaction was repeated in triplicates.
Statistical analysis
All data were expressed as mean±s.e. One-way analysis of variance with Bonferroni corrections was performed for analyzing the data of cell culture study. Data from different study animal groups were compared using the Wilcoxon–Mann–Whitney test, and P-values of <0.05 were considered statistically significant.
Acknowledgments
We thank the staff of Keelung Chang Gung Memorial Hospital Research Center for their assistance with this investigation. We also thank the Kureha Corporation for providing the P-cresyl sulfate. This work was funded by a grant from the CMRPG280091.
All the authors declared no competing interests.
Footnotes
SUPPLEMENTARY MATERIAL
Table S1. The primers for thermocycler program of methylation-specific PCR.
Table S2. The primers for thermocycler program of real-time PCR.
Figure S1. IS and PCS increased the α-glutathione S-transferase expression in vivo.
Supplementary material is linked to the online version of the paper at http://www.nature.com/ki
Supplementary Material
References
- Ayodele OE, Alebiosu CO. Burden of chronic kidney disease: an international perspective. Adv Chronic Kidney Dis. 2010;17:215–224. doi: 10.1053/j.ackd.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Kuro-o M. Klotho and aging. Biochim Biophys Acta. 2009;1790:1049–1058. doi: 10.1016/j.bbagen.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosu H, Yamamoto M, Clark JD, et al. Suppression of aging in mice by the hormone Klotho. Science. 2005;309:1829–1833. doi: 10.1126/science.1112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T, Yamada K, Kim HC, et al. Cognition impairment in the genetic model of aging Klotho gene mutant mice: a role of oxidative stress. FASEB J. 2003;17:50–52. doi: 10.1096/fj.02-0448fje. [DOI] [PubMed] [Google Scholar]
- Kamemori M, Ohyama Y, Kurabayashi M, et al. Expression of Klotho protein in the inner ear. Hear Res. 2002;171:103–110. doi: 10.1016/s0378-5955(02)00483-5. [DOI] [PubMed] [Google Scholar]
- Anamizu Y, Kawaguchi H, Seichi A, et al. Klotho insufficiency causes decrease of ribosomal RNA gene transcription activity, cytoplasmic RNA and rough ER in the spinal anterior horn cells. Acta Neuropathol. 2005;109:457–466. doi: 10.1007/s00401-004-0971-7. [DOI] [PubMed] [Google Scholar]
- Haruna Y, Kashihara N, Satoh M, et al. Amelioration of progressive renal injury by genetic manipulation of Klotho gene. Proc Natl Acad Sci USA. 2007;104:2331–2336. doi: 10.1073/pnas.0611079104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura H, Yoshida T, Tsuchiya K, et al. Klotho reduces apoptosis in experimental ischaemic acute renal failure. Nephrol Dial Transplant. 2005;20:2636–2645. doi: 10.1093/ndt/gfi165. [DOI] [PubMed] [Google Scholar]
- Koh N, Fujimori T, Nishiguchi S, et al. Severely reduced production of Klotho in human chronic renal failure kidney. Biochem Biophys Res Commun. 2001;280:1015–1020. doi: 10.1006/bbrc.2000.4226. [DOI] [PubMed] [Google Scholar]
- Ohyama Y, Kurabayashi M, Masuda H, et al. Molecular cloning of rat klotho cDNA: markedly decreased expression of klotho by acute inflammatory stress. Biochem Biophys Res Commun. 1998;251:920–925. doi: 10.1006/bbrc.1998.9576. [DOI] [PubMed] [Google Scholar]
- Tumur Z, Niwa T. Indoxyl sulfate inhibits nitric oxide production and cell viability by inducing oxidative stress in vascular endothelial cells. Am J Nephrol. 2009;29:551–557. doi: 10.1159/000191468. [DOI] [PubMed] [Google Scholar]
- Palm F, Nangaku M, Fasching A, et al. Uremia induces abnormal oxygen consumption in tubules and aggravates chronic hypoxia of the kidney via oxidative stress. Am J Physiol Renal Physiol. 2010;299:F380–F386. doi: 10.1152/ajprenal.00175.2010. [DOI] [PubMed] [Google Scholar]
- Adijiang A, Niwa T. An oral sorbent, AST-120, increases Klotho expression and inhibits cell senescence in the kidney of uremic rats. Am J Nephrol. 2010;31:160–164. doi: 10.1159/000264634. [DOI] [PubMed] [Google Scholar]
- Lee J, Jeong DJ, Kim J, et al. The anti-aging gene Klotho is a novel target for epigenetic silencing in human cervical carcinoma. Mol Cancer. 2010;9:109–118. doi: 10.1186/1476-4598-9-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff AC, Meyer TW, Hostetter TH. New insights into uremic toxicity. Curr Opin Nephrol Hypertens. 2008;17:560–565. doi: 10.1097/MNH.0b013e32830f45b6. [DOI] [PubMed] [Google Scholar]
- Schepers E, Meert N, Glorieux G, et al. P-cresylsulphate, the main in vivo metabolite of p-cresol, activates leucocyte free radical production. Nephrol Dial Transplant. 2007;22:592–596. doi: 10.1093/ndt/gfl584. [DOI] [PubMed] [Google Scholar]
- Campos AC, Molognoni F, Melo FH, et al. Oxidative stress modulates DNA methylation during melanocyte anchorage blockade associated with malignant transformation. Neoplasia. 2007;9:1111–1121. doi: 10.1593/neo.07712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco R, Schoneveld O, Georgakilas AG, et al. Oxidative stress, DNA methylation and carcinogenesis. Cancer Lett. 2008;266:6–11. doi: 10.1016/j.canlet.2008.02.026. [DOI] [PubMed] [Google Scholar]
- Barreto FC, Barreto DV, Liabeuf S, et al. Serum indoxyl sulfate is associated with vascular disease and mortalityin chronic kidney disease patients. Clin J Am Soc Nephrol. 2009;4:1551–1558. doi: 10.2215/CJN.03980609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bammens B, Evenepoel P, Keuleers H, et al. Free serum concentrations of the protein-bound retention solute p-cresol predict mortality in hemodialysis patients. Kidney Int. 2006;69:1081–1087. doi: 10.1038/sj.ki.5000115. [DOI] [PubMed] [Google Scholar]
- Wu IW, Hsu KH, Lee CC, et al. p-Cresyl sulphate and indoxyl sulphate predict progression of chronic kidney disease. Nephrol Dial Transplant. 2011;26:938–947. doi: 10.1093/ndt/gfq580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yavuz A, Tetta C, Ersoy FF, et al. Uremic toxins: a new focus on an old subject. Semin Dial. 2005;18:203–211. doi: 10.1111/j.1525-139X.2005.18313.x. [DOI] [PubMed] [Google Scholar]
- Dwivedi RS, Herman JG, McCaffrey TA, et al. Beyond genetics: epigenetic code in chronic kidney disease. Kidney Int. 2011;79:23–32. doi: 10.1038/ki.2010.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanayakkara PW, Kiefte-de Jong JC, Stehouwer CD, et al. Association between global leukocyte DNA methylation, renal function, carotid intima-media thickness and plasma homocysteine in patients with stage 2-4 chronic kidney disease. Nephrol Dial Transplant. 2008;23:2586–2592. doi: 10.1093/ndt/gfn040. [DOI] [PubMed] [Google Scholar]
- Stenvinkel P, Karimi M, Johansson S, et al. Impact of inflammation on epigenetic DNA methylation - a novel risk factor for cardiovascular disease. J Intern Med. 2007;261:488–499. doi: 10.1111/j.1365-2796.2007.01777.x. [DOI] [PubMed] [Google Scholar]
- Adijiang A, Shimizu H, Higuchi Y, et al. Indoxyl sulfate reduces klotho expression and promotes senescence in the kidneys of hypertensive rats. J Ren Nutr. 2011;21:105–109. doi: 10.1053/j.jrn.2010.10.020. [DOI] [PubMed] [Google Scholar]
- Turek-Plewa J, odziñski PP. The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell Mol Biol Lett. 2005;10:631–647. [PubMed] [Google Scholar]
- Ting AH, Jair KW, Schuebel KE, et al. Differential requirement for DNA methyltransferase 1 in maintaining human cancer cell gene promoter hypermethylation. Cancer Res. 2006;66:729–735. doi: 10.1158/0008-5472.CAN-05-1537. [DOI] [PubMed] [Google Scholar]
- Fandy TE. Development of DNA methyltransferase inhibitors for the treatment of neoplastic diseases. Curr Med Chem. 2009;16:2075–2085. doi: 10.2174/092986709788612738. [DOI] [PubMed] [Google Scholar]
- Pan Y, Sawalha AH. Epigenetic regulation and the pathogenesis of systemic lupus erythematosus. Transl Res. 2009;153:4–10. doi: 10.1016/j.trsl.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- Robert MF, Morin S, Beaulieu N, et al. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat Genet. 2003;33:61–65. doi: 10.1038/ng1068. [DOI] [PubMed] [Google Scholar]
- Wolf I, Levanon-Cohen S, Bose S, et al. Klotho: a tumor suppressor and a modulator of the IGF-1 and FGF pathways in human breast cancer. Oncogene. 2008;27:7094–7105. doi: 10.1038/onc.2008.292. [DOI] [PubMed] [Google Scholar]
- Camilli TC, Xu M, O'Connell MP, et al. Loss of Klotho during melanoma progression leads to increased filamin cleavage, increased Wnt5A expression, and enhanced melanoma cell motility. Pigment Cell Melanoma Res. 2011;24:175–186. doi: 10.1111/j.1755-148X.2010.00792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arany I, Megyesi JK, Nelkin BD, et al. STAT3 attenuates EGFR-mediated ERK activation and cell survival during oxidant stress in mouse proximal tubular cells. Kidney Int. 2006;70:669–674. doi: 10.1038/sj.ki.5001604. [DOI] [PubMed] [Google Scholar]
- Arany I, Faisal A, Nagamine Y, et al. p66shc inhibits pro-survival epidermal growth factor receptor/ERK signaling during severe oxidative stress in mouse renal proximal tubule cells. J Biol Chem. 2008;283:6110–6117. doi: 10.1074/jbc.M708799200. [DOI] [PubMed] [Google Scholar]
- Chang HC, Cho CY, Hung WC. Silencing of the metastasis suppressor RECK by RAS oncogene is mediated by DNA methyltransferase 3b-induced promoter methylation. Cancer Res. 2006;66:8413–8420. doi: 10.1158/0008-5472.CAN-06-0685. [DOI] [PubMed] [Google Scholar]
- Shimizu H, Bolati D, Adijiang A, et al. Indoxyl sulfate downregulates renal expression of Klotho through production of ROS and activation of nuclear factor-B. Am J Nephrol. 2011;33:319–324. doi: 10.1159/000324885. [DOI] [PubMed] [Google Scholar]
- Kuro-o M. Klotho as a regulator of oxidative stress and senescence. Biol Chem. 2008;389:233–241. doi: 10.1515/BC.2008.028. [DOI] [PubMed] [Google Scholar]
- de Oliveira RM. Klotho RNAi induces premature senescence of human cells via a p53/p21 dependent pathway. FEBS Lett. 2006;580:5753–5758. doi: 10.1016/j.febslet.2006.09.036. [DOI] [PubMed] [Google Scholar]
- Calaf R, Cerini C, Génovésio C, et al. Determination of uremic solutes in biological fluids of chronic kidney disease patients by HPLC assay. J Chromatogr B Analyt Technol Biomed Life Sci. 2011;879:2281–2286. doi: 10.1016/j.jchromb.2011.06.014. [DOI] [PubMed] [Google Scholar]
- Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- Choi BH, Kim CG, Lim Y, et al. Transcriptional activation of the human Klotho gene by epidermal growth factor in HEK293 cells; role of Egr-1. Gene. 2010;450:121–127. doi: 10.1016/j.gene.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Nightingale J, Patel S, Suzuki N, et al. Oncostatin M, a cytokine released by activated mononuclear cells, induces epithelial cell-myofibroblast transdifferentiation via Jak/Stat pathway activation. J Am Soc Nephrol. 2004;15:21–32. doi: 10.1097/01.asn.0000102479.92582.43. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.