The question of how traits and behaviors pass from one generation to the next has been the subject of intense interest throughout the history of science. Simple parent-child correlations are open to multiple interpretations, as parents transmit both environment and genome to their children. Until recently, genotyping – or the direct measurement of variation in an individual’s DNA sequence through biological assays – was exorbitantly expensive; distinguishing the roles of genetics and environment was the realm of behavioral genetics, in which samples of twin, adoption or other pedigree data were analyzed. However, with the completion of the Human Genome Project in the early 2000s (Venter et al., 2001; Lander et al., 2001) and the advent of inexpensive, genome-wide scans of variation, it is now increasingly feasible to directly examine specific genetic variants that predict individual differences.
In fact, the costs of comprehensively genotyping human subjects have fallen to the point where major funding bodies, even in the social sciences, are beginning to incorporate genetic and biological markers into major social surveys. The National Longitudinal Study of Adolescent Health, the Wisconsin Longitudinal Study, and the Health and Retirement Survey have launched, or are in the process of launching, datasets with comprehensively genotyped subjects. Similar efforts are also underway in Europe, for example with the Biobank Project in the United Kingdom (Ollier, Sprosen and Peakman, 2005) and the large scale genotyping of subjects at several European twin registries. These samples contain, or will soon contain, data on hundreds of thousands of genetic markers for each individual in the sample as well as, in most cases, basic economic variables. How, if at all, should economists use and combine molecular genetic and economic data? What challenges arise when analyzing genetically informative data?
In this article, we lay out the terrain for such questions. We use the term “genoeconomics,” originally proposed by Benjamin et al. (2007), to describe the use of molecular genetic information in economics. To illustrate some of the challenges that researchers in this field are likely to encounter, we present results from a “genome-wide association study” of educational attainment, one of the first of its kind in economics.1 This type of study involves analyzing hundreds of thousands of genetic markers and seeking to understand their association with some trait of interest. We use a sample of 7,500 individuals from the Framingham Heart Study. After quality controls, our dataset contains over 360,000 genetic markers per person. Despite some initially promising results, the main findings from this dataset fail to replicate in a second large replication sample of 9,500 people from the Rotterdam Study, suggesting that the original results were probably spurious. These findings are unfortunately typical in molecular genetics, and therefore also cautionary.
The frequent replication failures in the molecular genetics literature are likely a result of several forces, the most important of which is probably that the samples used in research are too small to ensure that there is adequate power to detect true associations (Ioannidis, 2005; Ioannidis, 2007). When true effect sizes are small, the power to detect true associations will of course be poor and the ratio of true to false signals will hence be low. Indeed, an important implication of the genome-wide association study results reported in this paper is that they confirm that common genetic variants with large main effects are likely to be extremely rare for economic variables, which tend to be far removed from the molecular genetic variant in the chain of causation. We perform power analyses to demonstrate this point and show that under plausible assumptions about the effect sizes of a specific type of common variation in the human genome, samples in the tens of thousands, perhaps more, may be necessary to detect genetic influences on most complex economic variables in a robust manner. This insight suggests that most existing genoeconomic studies, which are based on samples in the hundreds, are dramatically underpowered and that we should expect a high false discovery rate until this is remedied. Our choice of educational attainment as the outcome variable of study was determined by the widespread availability of this characteristic in cohorts that have already been genotyped. An important next step of a successful genoeconomic research agenda is to start measuring more biologically proximate variables – such as preferences– in large samples. Variables which are less distant from the genome in the chain of causation are more likely to require smaller samples in order for genetic associations to be reliably detected, and any detected associations are more likely to have a biologically meaningful interpretation and economically meaningful implications.
The empirical results in this paper are also used to discuss several other methodological issues that arise in the analysis of molecular genetic data. Our overall assessment is cautiously optimistic: this new data source has the potential not only to complement traditional behavioral genetic studies but also to add a new dimension to our understanding of heterogeneity in economic behaviors and outcomes, especially when it comes to traits that are close to the underlying biology. But for this ample potential to be realized, researchers and consumers of this literature should be wary of the pitfalls that lie ahead (Benjamin et al., 2007). The most urgent of these challenges is the difficulty of doing reliable inference when faced with multiple hypothesis problems, which are on a scale that has never before been encountered in social science.
Behavioral Genetics
Over the past few decades, behavioral geneticists have produced a compelling array of evidence that genetic variance does affect in economic behaviors, outcomes, and preferences. The general approach in these studies is to make assumptions about the extent to which the different sibling types share genetic and environmental conditions and infer the fraction of variance that can be statistically accounted for by genetic variation (heritability, denoted h2), rearing conditions (common environment, denoted c2), and idiosyncratic factors (unique environment, denoted e2). These studies often compare the resemblance of adoptees reared in the same family to that of biological siblings reared in the same family, or the resemblance of identical (monozygotic) twins, who share their entire genomes, to that of fraternal (dizygotic) twins, who share approximately half their genomes. Sacerdote (2010) provides an accessible introduction for economists. The standard textbook is Plomin et al. (2008).
The simplest behavioral genetic model is based on a host of strong assumptions, including the independence of genetic and family effects, functional form assumptions, and fails to take assortative mating and non-linear genetic effects into account. In the 1970s, when the debate between environmentalists and hereditarians reached its peak, there was much controversy over whether the high heritability estimates, especially for IQ, were artifacts of the simplistic behavioral genetic framework that would go away in more elaborate designs and with better data. In response, behavioral geneticists have built much richer datasets and expanded their models, relaxing the various problematic assumptions. They have consistently found that personality, IQ and most other traits remain highly, or at least moderately, correlated with genetic endowments (Bouchard and McGue, 2003). In fact, the consensus in behavioral genetics that there is genetic variance in virtually all human traits is so strong that it has been elevated to the status of a “law” (Turkheimer, 2000).2
Economic behaviors, preferences and outcomes are no exception. Behavioral genetic methods originally made limited inroads into economics through the work of Taubman and coauthors (for example, Taubman, 1976), who demonstrated that genetically identical (monozygotic) twins exhibit greater similarity than fraternal (dizygotic) twins in both educational attainment and income. Since then, a number of papers have followed suit in applying behavioral genetic research designs to the study of economic outcomes. Many of these studies rely on quasi-experiments such as adoption (Sacerdote, 2007; Björklund, Lindahl and Plug, 2006; Björklund, Jäntti and Solon, 2005), twinning (Taubman, 1976; Lichtenstein, Pedersen and McClearn, 1992) or comparisons of multiple sibling types (Björklund, Jäntti and Solon, 2005). More recent work has also demonstrated that economic preferences elicited from incentivized experiments or surveys are heritable, with estimates typically in the 20–30 percent range (Wallace, Cesarini, Johannesson and Lichtenstein, 2007; Cesarini, Dawes, Johannesson, Lichtenstein and Wallace, 2009a, 2009b). These estimates are biased downward because they do not take into account measurement error in the preference elicitation.3 Two other studies of portfolio choice data found heritability estimates of about 0.25–0.60 for various financial decision-making variables (Barnea, Cronqvist and Siegel, 2010; Cesarini, Johannesson, Lichtenstein, Sandewall and Wallace, 2010).
In interpreting heritability estimates, it is crucial to appreciate the possibility that genetic effects may operate via environmental effects, because genotypes may either evoke environmental responses or cause an individual to select a particular environment endogenously (Becker and Tomes, 1979; Dickens and Flynn, 2001; Fowler, Settle and Christakis, 2010; Jencks, 1980; Ridley, 2003). This possibility has given rise to the expression “nature via nurture” – as opposed to “nature versus nurture”. Estimates of the behavioral genetic model can therefore be thought of as reduced form coefficients from a more general model in which some environments are endogenous to genotype (Dickens and Flynn, 2001; Jencks and Brown, 1977; Jencks, 1980; Lizzeri and Siniscalchi, 2008).
As pointed out by Jencks (1980), a common mistake is to equate “genetic” with “immutable”: the fact that a person’s DNA sequence is in some sense fixed does not mean that the effects of that sequence are fixed. Goldberger (1979) provides several examples of how the implications of heritability estimates have been misstated and notes that high estimates do not imply that interventions are doomed to failure. While genetic variation can statistically account for a moderate to large share of income in contemporary Western societies, this does not mean that it would be infeasible to use redistributive policies or policies that encourage human capital formation to change the distribution of income. Heritability is a population parameter which depends on both the environmental effects operating in a specific population at a certain point in time and on the genetic variation in that population. It says little about what would happen to the mean and variance of the trait were the environment to change. Therefore, there is no contradiction between observing a high heritability for height, say, and secular increases in height over time as the environment changes. Heritability estimates do not tell us how the genetic effects operate, of course, nor do they tell us much about whether the mechanisms are easy or hard to modify. But far from being useless, as has sometimes been asserted, heritability estimates tell us that for most traits, a sizeable fraction of the within-family resemblance can ultimately be traced to shared DNA. We suspect that were it not for the impressive cumulative progress in behavioral genetics over the last couple of decades, the issue would still be contentious. Figuring out how and why genetic factors matter is an interesting scientific activity, and molecular genetic methods are an exciting tool to bring to bear on these questions.
Elementary Molecular Genetic Concepts
Molecular genetics is the branch of genetics that studies the structure and function of DNA at its most basic level. Recent decades have seen major advances, allowing researchers to better understand the numerous ways in which genomes vary between individuals. The human genome consists of 23 pairs of chromosomes that package DNA. One member of each pair of chromosomes is inherited from the mother and the other from the father. DNA itself consists of two strands of elementary building blocks that together form a double helix structure. The elementary building blocks, called nucleotides, each contain one of four bases: A (adenine), C (cytosine), T (thymine), or G (guanine), resulting in four distinct nucleotides. Due to a property of DNA called complementarity, a nucleotide with the base A is always paired with a nucleotide with the base T and a nucleotide with the base C is always paired with a nucleotide with the base G, forming so-called base pairs and holding the two strands of DNA together.
A locus is a specific position of a DNA sequence on a (pair of) chromosome(s). A locus thus refers to a pair of base pairs (or nucleotide pairs), one base pair coming from the paternal chromosome and the other base pair coming from the maternal chromosome. The human genome consists of approximately three billion such pairs of base pairs arranged into the 23 (pairs of) chromosomes. Because of complementarity, the second base of a base pair can be directly identified from knowledge of the first one, and so it is common practice to refer to a locus as consisting of two single bases rather than of a pair of base pairs. For example, the genotype AT-AT would be referred to as AA or as TT.4
Genes are sequences of nucleotide base pairs that code for some types of RNA products, many of which in turn code for proteins. These RNA products and proteins begin cascades of interactions that regulate bodily structures and functions. Only a small portion of the genome actually consists of genes, and both genetic variation in the genes and in the remaining portion of the genome can account for variation in behaviors and traits. However, because of genes’ functional importance, many researchers have focused their attention on genetic variation in the genes; also, it is often said, loosely, that “a gene causes” a behavior or trait even though what is meant is that genetic variation at a given locus – often not even in a gene – accounts for some of the variation in the behavior or trait.
Humans share most, but not all, of their genetic material: approximately 99.6 percent of base pairs are the same when comparing any two unrelated individuals (Kidd et al., 2008). Genetic variation comes in many forms, but most can be traced to one of two types of mutation events. The simplest mutation event is a base substitution, in which the base pair of a nucleotide pair is substituted for another. Whenever a nucleotide varies at a specific locus across individuals in the population, it is said to be a single nucleotide polymorphism, or SNP (pronounced “snip”), with the different genetic variants of a SNP called “alleles”. Most other forms of genetic variation are due to repeated segments of DNA. In variable number of tandem repeat (VNTR) polymorphisms, there are differences across individuals in the number of times that particular short segments of DNA are repeated. In copy number variation (CNV) polymorphisms, there are differences in the number of repetitions of long segment of DNA – of at least 1,000 base pairs and often many more.
Genotyping SNPs and other genetic variants is performed with technology that allows high-throughput typing of hundreds of thousands of genetic variants per individual. Current technologies type around 500,000 SNPs, but versions with over one million SNPs and other variants are already available and this number is expected to increase in the very near future. Within a decade, it will be possible to genotype entire genomes at relatively low cost. Because SNPs in the vicinity of each other are often highly correlated, it is generally possible to impute unobserved SNPs with high accuracy if a neighboring set of SNPs has been genotyped5; for that reason, even though most arrays type only a minute fraction of the three billion base pairs in the haploid human genome, they can in principle capture a large part of the relevant genetic variation.
In some rare cases, a difference at a specific locus on a chromosome can single-handedly lead to a disease: Huntington’s disease is an example. However, the vast majority of physical characteristics are “polygenic,” meaning they are influenced by multiple genetic polymorphisms (Mackay, 2001). The fact that there are so many potentially etiologically relevant SNPs in the genome leads to challenges of inference in the face of multiple hypothesis testing of a magnitude that social scientists have never before faced. Should gene-gene interactions also turn out to be important, the combinatorics would simply be staggering. However, as we later argue, the weight of the evidence suggests that a significant portion of the genetic variance in complex traits is “additive.”
Many traits of interest to economists are several steps removed from the original genotype in the chain of causation. Therefore, it is unlikely that genetic variants with a proximal effect on socioeconomic status exist and will ever be found. However, simple association models between candidate SNPs and various outcomes are still useful, because the main effects discovered could suggest areas for further exploration and point to the more proximate biological mechanism (for example, pointing to influences on the production of the neurotransmitter dopamine in the brain).
Candidate Gene Studies
Genetic association studies are becoming increasingly common in the social sciences, including economics. Many of the studies carried out to date have examined the relationship between some economic characteristics – typically an experimentally elicited preference parameter – and a relatively small number of genetic markers that are selected based on some a priori hypotheses derived from information about their biological function. The studies that follow this research design are therefore known as “candidate-gene” studies.
The first candidate-gene study in genoeconomics of which we are aware was by Knafo et al. (2008), who reported that polymorphisms of the AVPR1a gene were associated with the level of generosity exhibited by a sample of 203 college students playing a standard dictator game (Forsythe, Horowitz, Savin and Sefton, 1994). There have subsequently been many other reported associations between experimentally elicited preferences and genetic variants (Apicella et al., 2010; Crisan et al., 2009; Dreber et al., 2009; Israel et al., 2009; Kuhnen and Chiao, 2009; McDermott, Tingley, Cowden, Frazzetto and Johnson, 2009; Roe et al., 2009; Zhong et al., 2009, Zhong, Israel, Xue, Ebstein and Chew, 2009). For a review of the research to date, see Ebstein, Israel, Chew, Zhong and Knafo (2010).
The genetic markers used in these studies were typically selected based on evidence about their neurochemical function, and the studies test the hypothesis that these genetic markers are correlated with behavior. As an example, because the dopamine receptor D4 (DRD4) gene affects dopamine receptors in the brain and because dopamine plays an important role in learning and the processing of reward (Schultz, Dayan and Montague, 1997), many researchers initially suspected that variation in the DRD4 gene is associated with variation in risk preferences. Two early genoeconomics studies, Dreber et al. (2009) and Kuhnen and Chiao (2009), both reported an association between a particular variant of DRD4 and experimentally elicited risk preferences. However, Carpenter, Garcia and Lum (2009) subsequently failed to replicate this finding, and in fact reported a borderline significant association in the opposite direction.
Yet another set of studies drew on pharmacological randomized control studies which demonstrated that exogenous administration of the neuropeptide oxytocin increases trust in humans relative to a placebo (Kosfeld, Heinrichs, Zak, Fischbacher and Fehr, 2005). Such findings suggest that variation in the gene OXTR, which encodes the oxytocin receptor, may account for the heritable variation in prosocial behavior (Cesarini, Dawes, Johannesson, Lichtenstein and Wallace, 2009a). Israel et al. (2009) reported an association between an OXTR variant and giving in the dictator game, but Apicella et al. (2010) failed to replicate this association. Similarly, Nicolaou, Shane, Adi, Mangino and Harris (2011) reported an association between a variant of the dopamine receptor D3 gene and self-employment but the association did not replicate in a significantly larger sample (van der Loos et al., 2011).
An Illustration of Genoeconomic Research
Though genoeconomics research has so far mainly focused on candidate-gene studies, another important research design is the genome-wide association study. In this approach, tens or hundreds of thousands of genetic markers are individually tested for association with a trait of interest. The molecular genetic approach is fundamentally different from the behavioral genetic approach in that the effect of a genotype is estimated directly, rather than inferred by contrasting the resemblance of different types of relatives. During the last decade, genome-wide association studies have emerged as a standard research tool in medical genetics. The approach has been somewhat successful in uncovering associations between common variation and complex diseases and traits, with over 700 published studies for more than 140 traits (Hindorff, Junkins, Hall, Mehta, and Manolio, 2010), a figure that is rapidly growing. The advantages of genome-wide association studies are that they require no prior knowledge about the pathways between genotype and outcome or behavior of interest and that they scan all the available genome data, rather than study a particular variant. This may lead to unanticipated findings. However, a potential downside is limited power, as proper adjustment for multiple hypothesis testing requires extremely stringent significance thresholds.
In this section, we describe a project in which we conducted a genome-wide association study seeking to identify SNPs associated with educational attainment. In the process, we describe the main steps generally required to work with genotypic data as well as some of the challenges that arise in this type of research. What follows is an overview of the main steps; additional technical details are available in an Appendix available online with this paper at <http://e-jep.org>. In the first stage of this study, we analyzed data on about 7,500 individuals who have been genotyped at over half a million SNPs, to search for specific SNPs associated with educational attainment. In the replication stage of our genome-wide association study, we attempted to replicate the 20 most significant associations from the first stage in an independent dataset with more than 9,500 genotyped individuals
The First Stage: Data from the Framingham Heart Study
The data for the first stage of the genome-wide association study comes from the Framingham Heart Study (Dawber, Meadors and Moore, 1951; Feinleib, Kannel, Garrison, McNamara and Castelli, 1975), a longitudinal study which was started six decades ago by the U.S. Public Health Service to study cardiovascular diseases and which has played an important role in the medical literature (Levy and Brink, 2005). The study was initiated in 1948, when two-thirds of all adults domiciled in the town of Framingham were enrolled. The study was expanded in 1971 and 2002, when first and second generation descendants were enrolled, respectively. The spouses of first-generation descendants were also enrolled. Out of a total of 14,531 individuals included in the study, 8,496 had complete genotypic, education, and basic demographic data. Genotyping was conducted using the Affymetrix 500K chip, an array which contains 500,568 SNPs (Affymetrix, 2009). More details on the data used here, on the construction of the educational attainment variables, and on the genotyping are provided in the online Appendix.
As genome-wide association studies have become increasingly popular in medical genetics in the last few years, a number of methodological conventions have been developed, both to deal with the complexities of genotypic data and also with the issue of testing hypotheses in this context (Pearson and Manolio, 2008; Sullivan and Purcell, 2008). We followed these conventions here. We first applied a number of quality control measures to the Framingham sample comprising all 9,237 individuals with genetic data. We dropped 499 individuals from the sample because more than 5 percent of their genotypic data were missing; a high “missingness” can be suggestive that problems occurred in the genotyping procedure for the individual. Next, we only kept individual SNPs which satisfied each of three additional quality controls: (i) a missing data frequency less than 2.5 percent; (ii) a test that the observed genotype frequencies are equal to their theoretical expectations under random mating (a test of “Hardy-Weinberg equilibrium”); (iii) the frequency of the minor allele (the least common allele) greater than 1 percent. Failure to meet (i) and (ii) may indicate genotyping errors for the SNP; failure to meet (iii) generally leads to imprecisely estimated coefficients. From the 500,568 SNPs on our Affymetrix 500k array, 363,776 satisfied all three criteria.
Another important step for the researcher working with genotypic data is to control for population stratification – the differences in allele frequencies across subpopulations. Such differences can occur as a consequence of founder effects, genetic drift, and differences in natural selection pressures (Hartl, 1988)6. When both the frequencies of alleles and environmental factors affecting a trait of interest differ across subpopulations, failure to control properly for these differences can lead to omitted-variables bias and to spurious associations between those alleles and the trait. An illustration of population stratification was provided by Hamer and Sirota (2000), who asked their readers to entertain the thought experiment of looking for genetic markers for chopstick use in a sample comprising Caucasian and East Asian individuals. Without population stratification controls, markers which differ significantly in frequency between the Caucasian and Asian subpopulations will be found to be associated with chopstick use, but those associations will of course be due to cultural, not genetic, differences. Although the individuals in the Framingham Heart Study are almost all of European ancestry, population stratification has been shown to be a concern even in samples of European Americans (Campbell et al., 2005).
Several approaches have been proposed in the literature to control for population stratification. The most convincing solution to this problem is to only use within-family variation in genotype, using sibling fixed effects models. However, this comes at the cost of not using any of the between-family variation and thus of much reduced power. We employed a standard approach (the EIGENSTRAT method) developed by Price et al. (2006), which applies “principal component analysis” to the genotypic data. The first principal component of a set of variables is the linear combination of the variables with the coefficients chosen to capture as much of the sample variation as possible. The second principal component is obtained analogously, but subject to the constraint that it seek to capture whatever variation is remaining after the first principal component has been applied—and so on. We used the scores of each individual on each of the 10 first principal components as control variables in the main regression specification; in effect, these 10 values capture common variation across the population structure, and thus offer at least a partial control for population stratification. Consistent with standard practice, we also dropped individuals who were outliers from the sample, where outliers are defined as individuals whose score is at least six standard deviations from the mean on one of the top ten principal components. As a result, 531 outliers were eliminated, leaving 8,207 individuals with satisfactory genotypic data. The final sample used for the genome-wide association study comprised 7,574 individuals with satisfactory data on genotype and on observed characteristics. The sample size for each regression was generally a bit smaller than that, because for each SNP there were some individuals with missing genotypic information.
Association Analysis
For each individual SNP that passed our quality control standards, we specified the following standard regression model,
where Edu is years of education, SNPS is the number of copies of the minor allele (0, 1, or 2) an individual has at SNP s, PC is a vector of the individual’s principal component scores on the 10 top principal components of the genome of the sample, and X is a vector of controls.
A number of questions immediately arise in looking at this framework. For example, is the relationship between the genetic differences and educational attainment linear? (In genetic parlance, the model assumes that all genetic variation is “additive.”) The model is misspecified if, in expectation, the educational attainment of the heterozygotes (those with one copy of the minor allele) is in fact not the midpoint of the two homozygotes (those with zero or two copies of the minor allele). The main justification for specifying an additive model, besides parsimony, is that theory as well as converging evidence from behavioral genetics and animal breeding (Hill, Goddard and Visscher, 2008) suggest that an additive, polygenic, model of inheritance fits the data surprisingly well for most complex traits. Among other observations, if higher order interactions between genetic polymorphisms were important, one would observe a much more rapid decay of resemblance when comparing, say, monozygotic twins to full siblings and cousins.
Here, the coefficient estimate of β1 cannot be regarded as an estimate of the causal effect of SNPs on education; in particular, SNPs can be correlated with neighboring genetic polymorphisms that causally affect education but are not included in the regression. Because the Framingham sample is family-based and related individuals share parts of their environments and large portions of their genomes, it is necessary to account for the non-independence of the error terms. The interested reader is referred to the online Appendix for additional details of the necessary calculations.
Inference under Multiple Hypothesis
A challenging issue that arises in genome-wide association studies is the huge number of potentially relevant genetic variables – that is, a very large number of hypotheses are being tested. Because of this, many SNPs will inevitably turn out to have a statistically significant correlation with the dependent variables, at least at conventional levels of statistical significance, due to sampling variation and other chance events. Several methods have been proposed to draw statistical inferences in this context. The most commonly used – and the most stringent – solution is to use an approach named after the Italian mathematician Carlo Bonferroni, in which the conventional significance threshold is divided by the number of tests performed to obtain a Bonferroni-corrected significance threshold (or equivalently, all p-values are multiplied by the number of tests performed to obtain Bonferroni-corrected p-values). In the study with the Framingham data, 363,776 tests were performed (one for each SNP that passed the quality-control filters), thus yielding a Bonferroni-corrected significance threshold of 0.05 / 363,776 = 1.37×10−7.
The Bonferroni approach is generally agreed to be overly conservative because SNPs that are close to one another are generally correlated, and thus not statistically independent; thus, the Bonferroni correction divides by too large a number. With this consideration in mind, one commonly used threshold in the early literature for genome-wide association studies based on 500,000-SNP array data was set by the Wellcome Trust Case Control Consortium at 5×10−7 (Wellcome Trust Case Control Consortium, 2007). The standard threshold is now 5·10−8 (McCarthy et al., 2008). For simplicity, we only report below the Bonferroni-corrected p-values, but several more sophisticated methods have been developed to deal with this situation.7 However, as we discuss below, despite the theoretical soundness of these methods, previous experience with false positives in the field of medical genetics has led researchers to be cautious in interpreting results that have not been replicated in an independent sample.
The Replication Stage: Data from the Rotterdam Study
The data for the replication stage of the genome-wide association study comes from the Rotterdam Study (Hofman et al., 2009), a study that currently consists of three cohorts. Recruitment for the first, second, and third cohorts begun in 1990, 2000, and 2006, respectively. Together, these three cohorts contain data on 14,926 individuals from the well-defined Ommoord district in Rotterdam, all of whom were 45 years old or more when the third cohort was recruited. Of these, 9,535 individuals have complete genotypic, education and basic demographic data. Genotyping was done with the Illumina 550K and 610K arrays.
In the replication stage, we attempted to replicate the 20 most significant associations from the first stage using the Rotterdam data as an independent sample.8 The procedures we followed were very similar to those outlined above for the first stage, but many of the 20 SNPs were not directly available in the Rotterdam data and so had to be imputed, since the Framingham Heart Study and the Rotterdam Study used different genotyping platforms. Imputation is performed by using the correlation structure of an independent, more densely genotyped reference sample to infer the expected genotypes at the SNPs that have not been genotyped in the sample of interest. Though this procedure inevitably results in measurement error for the imputed SNPs (and thus to downward bias in their coefficient estimates), this effect is relatively small here as all 20 SNPs were imputed with a R2 greater than 0.92. We used the HapMap CEU sample (the International HapMap Consortium, 2003) as reference sample. Again, the online Appendix provides more detail on this procedure.
Results
Table 1 reports results for the 20 SNPs that attained the highest statistical significance in the first stage analysis of the Framingham dataset. The first column gives the rs number – the most widely used SNP identifier – of each SNP with the chromosome on which it is located in parentheses. The second column shows a letter of the base pair corresponding to the minor allele of the SNP; this can be useful to compare the direction of the estimated coefficients across studies. The third column indicates whether the SNP is close (less than 100,000 base pairs away) to any known genes (as mentioned above, SNPs need not be within genes).
TABLE I.
20 Most Significant Associations from First Stage and Results from Replication Stage
| SNP (Chromosome) |
Minor Allele |
Nearby Genes? |
First Stage | Replication Stage | |||
|---|---|---|---|---|---|---|---|
| β | p-value | Bonferroni | β | p-value | |||
| rs11758688† (6) | T | Yes | −0.253 | 2.97 · 10−7 | 0.107 | −0.0674 | 0.1209 |
| rs12527415† (6) | T | Yes | −0.253 | 3.03 · 10−7 | 0.109 | −0.0689 | 0.1138 |
| rs17365411 (2) | C | No | 0.26 | 3.73 · 10−7 | 0.134 | −0.0314 | 0.4553 |
| rs7655595 (4) | G | No | −0.266 | 3.99 · 10−7 | 0.144 | 0.0007 | 0.9877 |
| rs17350845 (1) | C | Yes | −0.291 | 6.22 · 10−7 | 0.224 | 0.0175 | 0.7162 |
| rs12691894 (2) | G | No | −0.246 | 6.67 · 10−7 | 0.24 | 0.0698 | 0.0998 |
| rs9646799 (2) | T | Yes | 0.271 | 7.41 · 10−7 | 0.267 | −0.0139 | 0.7579 |
| rs11722767 (4) | C | No | −0.257 | 7.77 · 10−7 | 0.28 | 0.0007 | 0.9877 |
| rs10947091† (6) | T | Yes | −0.245 | 9.03 · 10−7 | 0.325 | −0.0674 | 0.1229 |
| rs6536456 (4) | C | No | 0.23 | 1.32 · 10−6 | 0.474 | 0.0272 | 0.4917 |
| rs1580882 (4) | T | No | 0.229 | 1.43 · 10−6 | 0.516 | 0.026 | 0.5105 |
| rs6536463 (4) | G | No | 0.229 | 1.48 · 10−6 | 0.533 | 0.0209 | 0.5962 |
| rs1502720 (4) | C | No | 0.228 | 1.66 · 10−6 | 0.56 | 0.026 | 0.5213 |
| rs10436961 (1) | A | Yes | −0.268 | 1.82 · 10−6 | 0.657 | 0.0173 | 0.7196 |
| rs4845129 (1) | G | Yes | −0.265 | 2.07 · 10−6 | 0.745 | 0.0175 | 0.7164 |
| rs17365432 (2) | G | No | 0.257 | 2.32 · 10−6 | 0.836 | −0.0326 | 0.4675 |
| rs11225388 (11) | G | Yes | 0.261 | 2.51 · 10−6 | 0.904 | −0.0648 | 0.1476 |
| rs7743593 (6) | C | Yes | 0.301 | 2.68 · 10−6 | 0.965 | 0.0172 | 0.7397 |
| rs10028331 (4) | G | No | −0.259 | 2.93 · 10−6 | 1 | −0.0226 | 0.6317 |
| rs11964691 (6) | T | Yes | 0.307 | 3.29 · 10−6 | 1 | −0.0441 | 0.4137 |
NOTES: This panel reports the 20 SNPs with the highest statistical significance from the first stage of the GWAS as well as results for those SNPs from both the first and the replication stages. Column 3 indicates whether the SNP is less than 100,000 base pairs away from any known genes. Columns 5 and 8 report the (uncorrected) p-values from the first and from the replication stages, respectively. Columns 6 reports the Bonferroni-corrected p-values for the first stage, with corrections for 363,776 tests.
These three SNPs are the least likely to be false positives, and future work in still other replication datasets may clarify what role, if any, they play.
The fourth column shows the estimated regression coefficients; these are clustered around 0.25 for most SNPs, meaning that in our sample, the difference between the two homozygotes is about 0.5 years in educational attainment. However, these estimates are subject to upward bias because of a “winner's curse” type of selection bias (Zhong and Prentice, 2008). Put simply, the significance of a SNP depends in part on the size of the regression coefficient in the sample studied, so the most significant SNPs are likely to have actual regression coefficients that are inflated relative to actual population parameter. In fact, the similarity in the magnitude of our reported coefficient estimates may suggest that the bias may count for much more than the underlying true coefficients. The estimated effect sizes are usually smaller in follow-up studies than in the original study, even when replication attempts are successful (Ioannidis, Ntzani, Trikalinos, and Contopoulos-Ioannidis, 2001).
The fifth column reports the raw p-value of each SNP. Four of the SNPs are significant at the 5·10−7 level and none of them at the 5·10−8 level. As shown in the sixth column, none of the SNPs survive a Bonferroni correction at the 10 percent level, the two lowest Bonferroni-corrected p-values being 0.11. The top two hits are in the vicinity of several known genes, the closest being the IER2 gene, which is located a little over 40,000 base pairs away from the two SNPs.9 As a robustness check, we also computed standard errors by clustering at the level of the family. In general, the clustered standard errors were considerably smaller than the standard errors used to compute the p-values reported in Table 1, and in this calculation eight of the top 20 hits survived the Bonferroni correction at the 10 percent level.
The last two columns of Table I report the results of the replication attempt, with the independent Rotterdam data, of the 20 most significant associations from the first stage. As shown, none of the top 20 SNPs has a statistically significant association with educational attainment in the Rotterdam data. In fact, the signs of the estimated beta coefficients from the first stage and the replication stage are identical for only nine of the 20 SNPs and the correlation between the 20 regression coefficients in the two samples is 0.01. In other words, although our first-stage results might have seemed somewhat promising, the attempt to replicate them in an independent sample was a complete failure. Although this may be a false non-replication of true effects, we argue below that it is more likely that these initial results were spurious.
Multiple Testing, False Positives, and Power Considerations
Our examination of the Framingham data revealed a number of markers with “suggestive” associations with educational attainment, but an attempt to replicate these results in the independent Rotterdam sample failed. This experience points to a number of valuable lessons for economists interested in studying the molecular genetic basis of economic behaviors and outcomes. As noted earlier, a number of studies using the “candidate gene” approach have also found interesting associations, which then failed to replicate in independent samples.
This pattern of failed replications is not unique to economics or the social sciences. After the decoding of the Human Genome Project in the early 2000s, there was a rush to find the molecular genetic correlates of important diseases; hundreds of studies, many with strikingly small samples and often not properly controlling for multiple testing, were published to report genetic associations with some traits and diseases. However, later meta-analyses and review studies revealed that most of these associations failed to replicate (Ioannidis, 2005; Ioannidis, 2007). Though it is possible that some of the published associations were true only in the particular populations in which they were originally found because of treatment effect heterogeneity or that they were falsely non-replicated (Ioannidis, 2007), the most plausible explanation is that the initial findings were spurious. We are not suggesting that replications are always necessary: one can imagine cases where the corrected p-values are so small that the results can be judged convincing on the basis of one sample alone. However, as a practical matter, replication has emerged as an important method for evaluating work in this area, and we think it is advisable that economists adopt this as a convention, at least in the sense that the onus is on those who wish to depart from the default to provide a convincing motivation.
More generally, it would greatly benefit the genoeconomics enterprise if economists and other social scientists learned from the mistakes that have been made in genetic research in the past decade, rather than repeat them in our own discipline. In our judgment, the plethora of false positives can be attributed to two main factors: studies that are statistically underpowered because sample sizes are too small; and bias toward publishing results that seem to show positive and significant correlations. We will discuss these in turn.
Most published association studies, especially in economics and the social sciences, are seriously underpowered given what we now know about the plausible effect sizes of common genetic variants. Consequently, the probability that an association study will detect a true signal is vanishingly small. In other words, with the small samples that are typical of many studies, the probability that an association in the data is due to a true signal is low relative to the probability that it is due to noise (in Bayesian language, the posterior odds of a true association are low). This point is now close to universally accepted in the molecular genetics community and is illustrated by our above results from our genome-wide association study of educational attainment. Accumulating evidence from the genetics literature suggests that for most complex traits, the genetic variance is highly diffuse throughout the genome, that most true causal variants have very small effect sizes, and that genetic markers with an R2 greater than 1 percent are rare (Visscher, 2008). In the case of height, which is one of the most studied traits with a very high heritability of around 80 percent (Visscher, Hill and Wray, 2008), the 180 loci identified by the largest study to date, with more than 180,000 subjects, only explain a total of about 10 percent of the variance in this characteristic (Allen et al., 2011).
Several non-replicated published results notwithstanding, there is no reason to expect traits of interest to economists to be different from medical traits with respect to the distribution of the effect sizes of the genetic markers. In fact, the situation is likely to be even more difficult for economists, because socioeconomic traits are generally very distal from the genome in the chain of causation and because of the low reliability of many measured economic traits. For example, it is not clear how well behavior in economic laboratory experiments captures attitudes toward altruism, cooperation or risk.
For most traits, thus, the genetic markers that have been found so far generally explain only a small fraction of the population variance. In contrast, estimates of the effect of heritability from behavioral genetics often hover around 30 to 50 percent or more. This gap has given rise to a debate in the genetics community about the causes of this “missing heritability” (Eichler et al., 2010; Manolio et al., 2009). Many explanations have been proposed, the main ones being about structural variants such as CNVs and VNTRs (discussed earlier), which until recently were not typed by the main platforms; low power to detect multi-gene interactions; very rare causal variants that are not captured by the main genotyping platforms; and – perhaps the most plausible to us – very large numbers of causal variants, each with very small effect sizes. A recent paper provided support for the latter explanation as well as some hope to the genetic community: Yang et al. (2010) showed that considering all the SNPs on some common genotyping chips simultaneously – as opposed to the usual approach in a genome-wide association study of testing each SNP individually with some stringent significance threshold – could account for 45 percent of the variance in human height. The 45 percent figure is bound to rise with more comprehensive genotyping platforms and the advent of whole genome sequencing. Similar results were recently documented for IQ (Davies, Tenesa, Payton, Yang and Harris, 2011).
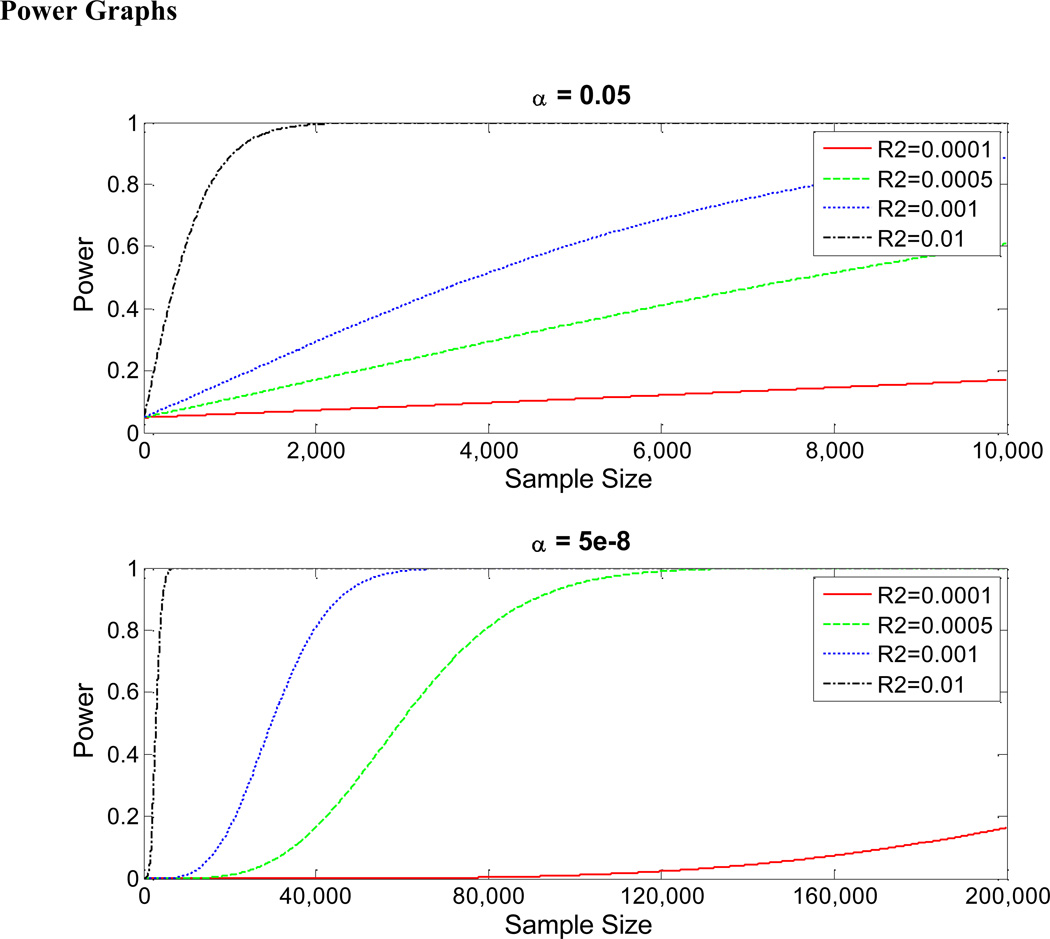
With the newest genotyping platforms now integrating structural variants and increasing their coverage to include rarer SNPs, very large samples will be needed to have adequate statistical power to make progress in mapping the genetic architecture of complex traits. To illustrate this point more formally, Figure 1 shows power graphs for the conventional 5 percent level of significance often used in the social sciences and for the much more stringent 5·10−8 level of statistical significance. Statistical power, measured on the vertical axis, is the probability that the null hypothesis of no association will be rejected at a given significance threshold. This probability of rejecting the null hypothesis varies with the sample size, shown on the horizontal axis. The four different curves in each panel correspond to markers that truly explain (that is, their true population R2 are) 0.01, 0.05, 0.1, and 1 percent of the variance of the trait. Thus, these graphs show the statistical power to detect a true marker-trait association as a function of the sample size. In the first panel, for the α=0.05 level of statistical significance – a significance threshold that is much higher than what is generally relevant in association studies given the multiple testing involved –, a sample of about 4,000 subjects is required to have power of 50 percent to detect a marker with a true R2 of 0.1 percent (with no correction for multiple testing). For a marker with a true R2 of 0.01 percent and with a sample of 10,000 individuals, the power of detecting the association is less than 20 percent. Even for a marker with a very large true R2 of 1 percent – larger than the R2's of all the markers that have been found to predict height – a sample of almost 800 subjects is needed to have power of 80 percent. As can be seen in the second panel, for the α=5·10−8 level of statistical significance, sample sizes in the tens of thousands are needed to have confidence in statistical estimates if the marker has an R2 of 0.05 percent, 0.1 percent, or 1 percent. If the marker has an R2 of 0.01 percent, even sample sizes of 200,000 will have power of less than 20 percent.
FIGURE I.
NOTES: Each curve in these graphs shows the statistical power to detect a true marker-trait association as a function of the sample size. Statistical power is the probability that the null hypothesis of no association will be rejected at a given significance threshold. The four different curves in each panel correspond to markers that truly explain (that is, their true population R2 are) 0.01%, 0.05%, 0.1%, and 1% of the variance of the trait. The two panels correspond to two different significance thresholds (α = 0.05 and α = 5e-8). These power graphs are for single tests – that is, without correction for multiple hypothesis testing.
As a result of such considerations, large multi-sample consortia are rapidly becoming the norm in medical genetics, with total sample sizes in the tens of thousands and sometimes exceeding 50,000; examples are the Wellcome Trust Case Control Consortium (2007), CHARGE (Psaty et al., 2009), and GEFOS (Rivadeneira et al., 2009). The Gentrepreneur Consortium (van der Loos, Koellinger, Groenen and Thurik, 2010), aimed at finding genetic predictors of entrepreneurship in a sample of over 60,000 individuals, is one of the first such consortia in economics; we and some collaborators are also in the process of forming a similar consortium for educational attainment. We believe consortia of this type to be the appropriate constructive response to the otherwise quite dismaying findings reported here.
The second main factor behind finding so many false positives in the literature is publication bias. It is much easier to publish “positive results” than properly powered negative findings, and this creates incentives for “data mining” and selective reporting. Researchers invest significant effort in assembling datasets with rich genotypic and phenotypic information. With little theory to discipline the empirical work, and with a still-scant biological background regarding the genetics of socio-economic traits, the temptation is great to experiment with different specifications on different traits and markers before settling on the “right” tests to run. Unless the p-values in the subsequently published manuscript fully account for the multiple testing and the uncertainty stemming from the specification search, the resulting p-values will be too low. For example, if additive, non-additive, and sex-specific models are estimated, but only findings from the model with lowest p-values are reported, the resulting inference will of course be incorrect. It is difficult to assess the magnitude of these problems in genoeconomics, but the reported p-values in many published papers are likely to be grossly misleading. The lack of convincing replications and the known failed replications in the field are consistent with this interpretation. Journal editors and referees also have a responsibility to avoid publishing headline-grabbing results, at least when experience suggests that such results are likely to evaporate under later replication attempts. And there needs to be some reward to carefully executed and well-powered studies with negative findings.
In our view, the problem is not the fact that the approach of the genome-wide association study is a largely atheoretical exercise. Exploratory data mining is a perfectly legitimate scientific endeavor when it is identified and treated as such. Many of the replicable findings that thus far have emerged from genome-wide association studies of medical outcomes were unexpected and unlikely to have been uncovered from candidate-based approaches. But data mining must be done with discipline, meaning a complete reporting of and correction for the number of hypotheses tested, including any pre-testing of the model specification.
The nature of molecular genetic data makes the multiple hypothesis testing problem particularly acute, but there may also be a silver lining. In particular, it is generally relatively easy to attempt a replication of a genoeconomic finding in independent samples. Moreover, awareness of these issues is growing among genoeconomists. Indeed, we believe that economists have much to learn from the different scientific culture that prevails in the medical sciences in general and in genetics in particular. Though the problems of multiple hypothesis testing and publication bias have received some attention in economics (for example, Anderson, 2008; Card and Krueger, 1995; De Long and Lang, 1992; Kling, Liebman and Katz, 2007), they are generally overlooked. A culture that encourages – when this is technically feasible – collaboration across groups of authors to pool data and to replicate results across datasets would doubtless contribute to this desirable goal.
Molecular Genetic Data Providers
Most available genotyped samples have been collected for medical research purposes. These studies vary in sample size from a few hundred to several thousand observations. In addition to medical outcomes, the studies usually also collect some background information about their participants that are interesting for economists, such as education, profession, labor market experience, and income. Many of the large data-providers have formed research consortia, the most important of which are the CHARGE and GIANT consortia. As the costs of genotyping technologies are falling rapidly, social science data providers are expressing greater interest in adding molecular genetic information to their lists of variables The first major U.S. data provider to include such information was the Longitudinal Study of Adolescent Health which, as part of its third wave of data collection between 1999 and 2001, collected saliva samples and genotyped approximately 2,500 individuals for six genetic markers on or near genes linked to functions of the neurotransmitters dopamine and serotonin. A major provider of genoeconomic data in the coming years will be the U.S. Health and Retirement Survey, which is presently being expanded to include genome-wide association data – over one million genetic markers – in approximately 13,000 subjects. Given the large sample, and the rich set of phenotypic data on health, psychological well-being and economic and financial behavior, we anticipate that this data will emerge as a valuable resource for researchers in the social sciences interested in studying human genetic variation.
We anticipate that a steady stream of new datasets with economic variables and comprehensively genotyped subjects will be made available to researchers over time. One promising approach for the near future is to focus on the collection of additional, standardized socioeconomic variables in samples that already contain genotyped data. Over time, adding genotype information to existing socioeconomic datasets should provide an important new source of data to economists interested in studying the molecular genetic associates of economic behaviors and outcomes. Socioeconomic datasets already contain a wealth of relevant variables and are unlikely to be matched by data collected for other purposes in terms of measurement accuracy and breadth or duration of longitudinal follow-up. We further anticipate the formation of genoeconomic research consortia for the social sciences. An obvious challenge with this approach is that for research consortia to be effective, everyone will have to measure the same set of traits using reliable batteries which are uniform across surveys. The National Human Genome Research Institute is funding the PhenX project to develop consensus measures for phenotypes and exposures to be used in genome-wide association studies (https://www.phenx.org/).
Potential Benefits of Molecular Genetic Data
We have thus far discussed a number of methodological challenges that face researchers who use molecular genetics to reliably identify genetic associates of economic traits. In addition, the possibility of genome-based information and interventions raise a host of privacy and ethical issues that we have not pursued here. Despite those difficulties, we believe there are several reasons, beyond sheer intellectual curiosity, why the use of molecular genetic data will ultimately benefit the economic sciences.
First, knowledge of the biological mechanisms might suggest additional policies or interventions that had previously not been anticipated. There are already examples in the medical literature of genome-wide association studies leading to unanticipated findings that in turn have helped inspire new therapies (Hirschhorn, 2009). It is not a necessary condition that the genetic markers identified have large effects on the outcome, only that the biological pathway they implicate provides useful cues about possible prevention mechanisms. For some behavioral outcomes, genetic markers may also be of diagnostic and predictive utility, regardless of whether or not they shed direct light on biological mechanisms. For example, Benjamin (2010) points out that if dyslexia can eventually be predicted sufficiently well by genetic screening, children with dyslexia-susceptibility markers could be taught differently how to read from a very young age. More generally, if molecular genetic data can be used to predict which individuals are at high “genetic risk” for adverse outcomes, then it would in principle be possible to use such information to evaluate the costs and benefits of taking preventive measures targeted at helping at-risk individuals.
Given the current state of knowledge, the predictive utility of the genetic markers which have been identified thus far (with very few, but important, exceptions) is too weak for these hopes to be realized any time soon for the overwhelming majority of potential outcomes. The initial promises that genomics would revolutionize “personalized medicine” have not yet been realized. If anything, the main use of genotyping technology has proven to be prognostic, not therapeutic, as anticipated by some (Christakis, 1999, p.196). Nonetheless, in many applications where the purpose is risk prediction, several statistical techniques may yield informative predictions without having precise estimates of the effect of individual markers, by making efficient use of all the available genotypic information, using so called polygenic risk scores (Purcell et al., 2009). As increasingly rich genetic information becomes available and as new statistical techniques are developed, the predictive utility of the genetic data will doubtless rise.
Second, analysis of molecular genetic data may also help empirical economists to estimate causal effects. As genetic endowments become increasingly easy to measure, it will be possible to control for genetic determinants of the dependent variable, thereby reducing omitted variable bias (Bernheim, 2009). For example, a successful genome-wide association study of educational attainment might be able to identify genetic markers that influence the general likelihood of high intellectual performance, delayed gratification, and persistence. Such markers would be a valuable control variable in empirical studies on a number of economic questions, including income, labor supply, occupational choice, or savings decisions (though the absence of these variables is not a source of bias in a properly controlled randomized evaluation).
Third, a more speculative empirical use of genetic data is to use markers as instruments in empirical settings where reverse causality or omitted variable bias is otherwise a concern. This use of genetic markers was anticipated by Davey-Smith and Ebrahim (2003) and was pioneered in economic analyses by Ding, Lehrer, Rosequist and Audrain-McGovern (2006), who used molecular genetic data to instrument for health in a regression of academic outcomes on health. To serve as valid instrument, the marker Z needs to impact the outcome Y only through its impact on the intervening variable X. This chain of causation will be exceedingly, perhaps impossibly, difficult to establish in many cases that interest economists (Conley, 2009; Cawley, Han and Norton, 2011). There are two major challenges. First, many markers are pleiotropic, meaning that they influence multiple outcomes directly. If the marker Z also affects Y through other unobserved channels for which the researcher cannot control, then the so-called exclusion restriction fails. Second, even if the marker does not have pleiotropic effects, it may be correlated (in linkage disequilibrium) with a genetic marker that affects Y through the unobservables. Economists will likely be in a better position to assess the plausibility that a given marker satisfies the exclusion restriction once its biological function is intricately understood. Conley (2009) argues that it is very unlikely that any markers which satisfy the requirements of a valid instrument will ever be found.
Finally, and perhaps most importantly, molecular genetic data may prove helpful in understanding variation in policy response across individuals. Plausibly uncovering such treatment-effect heterogeneity will require a detailed research design where plausibly exogenous environmental variation is used to estimate the gene-environment interactions (Conley, 2009). The most frequently cited example of a gene-environment interaction is the work of Caspi et al. (2002). These authors studied the determinants of a composite index of anti-social behavior in a sample of 1,037 male children in New Zealand. In a regression of their index on a particular polymorphism in a gene called MAOA, childhood maltreatment, and the interaction of these two variables, they found a significant interaction and concluded that the MAOA gene modifies the influence of the childhood maltreatment. While this paper is a useful illustration of the possibilities in this area, it is important to note that childhood maltreatment is not necessarily exogenous but may be correlated with other unobserved determinants of poor outcomes. Also, a recent meta-study (Risch et al., 2009) was unable to corroborate the Caspi et al. (2002) findings. This is not too surprising, because the study is vulnerable to exactly the same criticisms that we have made against candidate genes studies in economics. Economists have a powerful toolkit for the estimation and identification of causal effects which has the potential to contribute to the current gene-environment interaction literature. Presently, this literature suffers from the limitation that the environmental variables used may be endogenous, as existing studies are not based on experimentally or quasi-experimentally generated exogenous variation in environment (Conley, 2009).
The cost of genotyping has now fallen to a level where it is feasible to directly study molecular genetic variation and many social science surveys are in the process of incorporating this information. We believe that such data will add a valuable new understanding to our dimension of heterogeneity in economic behavior. If researchers adhere to high empirical standards, we are confident that genoeconomics will generate a number of durable scientific insights that will ultimately lead to a richer and more comprehensive economic science.
Supplementary Material
Acknowledgements
The authors wish to thank the JEP editorial team, along with Anders Björklund, Markus Jäntti, Sandy Jencks and Peter Visscher, for comments on an earlier version of this paper. We are also grateful to Sarah Medland and Peter Visscher for the intellectual generosity they have shown us in responding to a number of methodological queries. Given that the list of authors is a lengthy one for an economics paper, it we spell out contributions. Beauchamp and Cesarini were the primary authors for the paper, while Koellinger and van der Loos contributed to the writing. Beauchamp designed the research protocol and analyzed the Framingham data. Van der Loos analyzed the Rotterdam data. Beauchamp, Cesarini, Christakis, Johannesson conceived the project and provided impetus. Christakis, Johannesson, Fowler, Groenen, Rosenquist, and Thurik critically reviewed the manuscript. As always, any views expressed here are those of the authors alone and we retain responsibility for any errors.
Footnotes
Preliminary results from a genome-wide association study of educational attainment have previously been reported by Posthuma et al. (2008) and Beauchamp et al. (2010). A genome-wide association study of self-employment has been initiated by van der Loos et al. (2010).
These estimates have also recently been corroborated by techniques which utilize molecular genetic data in ingenuous ways to estimate heritability (Visscher et al., 2006; Yang et al., 2010).
Adjusting for noise appears to approximately double the heritability estimates (Beauchamp, Cesarini, Rosenquist, Fowler and Christakis, 2011). Test retest data (unpublished) has been collected for a sample of about 100 twins that participated in the experiments reported in Wallace, Cesarini, Johannesson and Lichtenstein (2007) and Cesarini, Dawes, Johannesson, Lichtenstein and Wallace (2009a, b). These data suggest a test retest correlation of about 0.5. Adjusting for measurement noise would thus approximately double the heritability estimates.
For an accessible introduction to the basic concepts in molecular genetics, see Strachan and Read (2003) or Carey (2003).
When two SNPs are correlated, geneticists say that they are in "linkage disequilibrium." Even though the human genome is about 3 billion base pairs, half a million well chosen markers are sufficient to cover (“tag”) much of the genetic variation.
In population genetics, a founder effect is defined as the reduction in genetic variance that arises when a new population is established by a low number of isolated individuals. Genetic drift refers to changes in the allele frequencies in a population due to chance events.
Some possibilities include permutation-based approaches (Churchill and Doerge, 1994), Bayesian approaches that incorporate prior biological knowledge and SNP characteristics (Stephens and Balding, 2009), estimation and control of the false discovery rate (the expected proportion of significant associations that are false positives) (Hochberg and Benjamini, 1990; Sabatti, Service and Freimer, 2003), and approaches that calculate the effective number of independent tests (Gao, Becker, Becker, Starmer and Province, 2010).
An alternative approach would have been to pool all the coefficient estimates from the two samples and then meta-analyze the results. This approach is typical in consortium studies.
In Table V of the online Appendix, we report the SNPs (from the above set of 20) which are in or near any known genes, along with the distances in base pairs.
Contributor Information
Jonathan P. Beauchamp, Harvard University, Cambridge, Massachusetts..
David Cesarini, Center for Experimental Social Science, New York University, New York City, New York, and an Affiliated Researcher, Institute for Industrial Economics (IFN), Stockholm, Sweden..
Magnus Johannesson, Stockholm School of Economics, Stockholm, Sweden..
Matthijs J. H. M. van der Loos, Erasmus School of Economics, Rotterdam, Netherlands..
Philipp D. Koellinger, Erasmus School of Economics, Rotterdam, Netherlands..
Patrick J. F. Groenen, Erasmus School of Economics, Rotterdam, Netherlands..
James H. Fowler, University of California at San Diego, La Jolla, California..
J. Niels Rosenquist, Harvard Medical School and the Massachusetts General Hospital's Psychiatric and Neurodevelopmental Genetics Unit and a Research Fellow at the Institute for Quantitative Social Science at Harvard, Cambridge, Massachusetts..
A. Roy Thurik, Erasmus School of Economics, Rotterdam, Netherlands..
Nicholas A. Christakis, Harvard Medical School, Cambridge, Massachusetts..
References
- Affymetrix. Affymetrix® Genome-Wide Human SNP Array 5.0. 2009 Available at http://www.affymetrix.com/support/technical/datasheets/genomewide_snp5_datasheet.pdf.
- Allen Hana Lango, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson Michael L. Multiple Inference and Gender Differences in the Effects of Early Intervention: A Reevaluation of the Abecedarian, Perry Preschool, and Early Training Projects. Journal of the American Statistical Association. 2008;103(484):1481–1495. [Google Scholar]
- Apicella Coren L, Cesarini David, Johanneson Magnus, Dawes Christopher T, Lichtenstein Paul, Wallace Björn, Beauchamp Jonathan, Westberg Lars. No Association Between Oxytocin Receptor (OXTR) Gene Polymorphisms and Experimentally Elicited Social Preferences. PLoS ONE. 2010;5(6):e11153. doi: 10.1371/journal.pone.0011153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnea Amir, Cronqvist Henrik, Siegel Stephan. Nature or nurture: What determines investor behavior? Journal of Financial Economics. 2010;98(3):583–604. [Google Scholar]
- Beauchamp Jonathan, Cesarini David J, Rosenquist Niels, Fowler James H, Christakis Nicholas A. A Genome-Wide Association Study of Educational Attainment. Genes, Brains and the Labor Market; Working Paper, Presented at IZA conference.2010. [Google Scholar]
- Becker Gary S, Tomes Nigel. An Equilibrium Theory of the Distribution of Income and Intergenerational Mobility. The Journal of Political Economy. 1979;87(6):1153–1189. [Google Scholar]
- Benjamin Daniel J, Chabris Christopher F, Glaeser Edward L, Gudnason Vilmundur, Harris Tamara B, Laibson David I, Launer Lenore J, Purcell Shaun. Genoeconomics. In: Maxine Weinstein, Vaupel James W, Wachter Kenneth W., editors. Biosocial Surveys. Washington: The National Academies Press; 2007. pp. 192–289. [Google Scholar]
- Benjamin Daniel J. White Paper for NSF Workshop on Genes, Cognition, and Social Behavior. 2010 Manuscript. [Google Scholar]
- Bernheim Douglas B. On the Potential of Neuroeconomics: A Critical (But Hopeful) Appraisal. American Economic Journal: Microeconomics. 2009;1(2):1–41. [Google Scholar]
- Björklund Anders, Jäntti Markus, Solon Gary. Influences of Nature and Nurture on Earnings Variation: A Report on a Study of Various Sibling Types in Sweden. In: Samuel Bowles, Gintis Herbert, Graves Melissa Osborne., editors. Unequal Chances: Family Background and Economic Success. Princeton: Princeton University Press; 2005. [Google Scholar]
- Björklund Anders, Lindahl Mikael, Plug Erik. The Origins of Intergenerational Associations: Lessons from Swedish Adoption Data. Quarterly Journal of Economics. 2006;121(3):999–1028. [Google Scholar]
- Campbell Catarina D, Ogburn Elizabeth L, Lunetta Kathryn L, Lyon Helen N, Freedman Matthew L, et al. Demonstrating Stratification in a European American Population. Nature Genetics. 2005;37(8):868–872. doi: 10.1038/ng1607. [DOI] [PubMed] [Google Scholar]
- Card David, Krueger Alan B. Time-Series Minimum-Wage Studies: A Meta Analysis. The American Economic Review. 1995;85(2):238–243. [Google Scholar]
- Carey Gregory. Human Genetics for the Social Sciences. Thousand Oaks: Sage Publications; 2003. [Google Scholar]
- Carpenter Jeffrey, Garcia Justin, Lum J. Koji. Can Dopamine Receptor Genes Explain Economic Preferences and Outcomes? 2009 Manuscript. [Google Scholar]
- Caspi Avshalom, McClay Joseph, Moffitt Terrie E, Mill Jonathan, Martin Judy, Craig Ian W, Taylor Alan, Poulton Richie. Role of Genotype in the Cycle of Violence in Maltreated Children. Science. 2002;297(5582):851–854. doi: 10.1126/science.1072290. [DOI] [PubMed] [Google Scholar]
- Cawley John, Han Euna, Norton Edward C. The validity of genes related to neurotransmitters as instrumental variables. Health Economics. 2011;20(8):884–888. doi: 10.1002/hec.1744. [DOI] [PubMed] [Google Scholar]
- Cesarini David, Dawes Christopher T, Johannesson Magnus, Lichtenstein Paul, Wallace Björn. Genetic Variation in Preferences for Giving and Risk-Taking. Quarterly Journal of Economics. 2009a;124(2):809–841. [Google Scholar]
- Cesarini David, Dawes Christopher T, Johannesson Magnus, Lichtenstein Paul, Wallace Björn. Heritability of overconfidence. Journal of the European Economic Association. 2009b;7(2–3):617–627. [Google Scholar]
- Cesarini David, Johannesson Magnus, Lichtenstein Paul, Sandewall Örjan, Wallace Björn. Genetic Variation in Financial Decision-Making. Journal of Finance. 2010;65(5):1725–1754. [Google Scholar]
- Christakis Nicholas A. Death Foretold: Prophecy and Prognosis in Medical Care. Chicago: University of Chicago Press; 1999. [Google Scholar]
- Churchill Gary A, Doerge Rebecca W. Empirical Threshold Values for Quantitative Trait Mapping. Genetics. 1994:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley Dalton. The Promise and Challenges of Incorporating Genetic Data into Longitudinal Social Science Surveys and Research. Biodemography and Social Biology. 2009;55(2):238–251. doi: 10.1080/19485560903415807. [DOI] [PubMed] [Google Scholar]
- Crisan Liviu G, Pana Simona, Vulturar Romana, Heilman Renata M, Szekely Raluca, Druga Bogdan, Dragos Nicolae, Miu Andrei C. Genetic Contributions of the Serotonin Transporter to Social Learning of Fear and Economic Decision Making. Social Cognitive and Affective Neuroscience. 2009;4(4):399–408. doi: 10.1093/scan/nsp019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey Smith George, Ebrahim Shah. Mandelian Randomization: Can Genetic Epidemiology Contribute to Understanding Environmental Determinants of Disease. International Journal of Epidemiology. 2003;32(1):1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- Davies Gail, Tenesa Albert, Payton Tony, Yang Jian, Harris Sarah E. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Molecular Psychiatry. 2011 doi: 10.1038/mp.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawber Thomas R, Meadors Gilcin F, Moore Felix E. Epidemiological Approaches to Heart Disease: The Framingham Study. American Journal Public Health. 1951;41:279–286. doi: 10.2105/ajph.41.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Long Bradford J, Lang Kevin. Are All Economic Hypotheses False? Journal of Political Economy. 1992;100(6):1257–1272. [Google Scholar]
- Dickens William T, Flynn James R. Heritability Estimates versus Large Environmental Effects: The IQ Paradox Resolved. Psychological Review. 2001;108(2):346–369. doi: 10.1037/0033-295x.108.2.346. [DOI] [PubMed] [Google Scholar]
- Ding Weili, Lehrer Steven F, Rosenquist Niels J, Audrain-McGovern Janet. The Impact of Poor Health on Education: New Evidence Using Genetic Markers. 2006 doi: 10.1016/j.jhealeco.2008.11.006. NBER Working Paper w12304. [DOI] [PubMed] [Google Scholar]
- Dreber Anna, Apicella Coren L, Eisenberg Daniel TA, Garcia Justin R, Zamore Richard J, Lum Koji, Campbell Benjamin. The 7R Polymorphism in the Dopamine Receptor D4 Gene (DRD4) Is Associated with Financial Risk-Taking in Men. Evolution and Human Behavior. 2009;30(2):85–92. [Google Scholar]
- Ebstein Richard P, Israel Salomon, Hong Chew Soo, Zhong Songfa, Knafo Ariel. Genetics of Human Social Behavior. Neuron. 2010;65:831–844. doi: 10.1016/j.neuron.2010.02.020. [DOI] [PubMed] [Google Scholar]
- Eichler Evan E, Flint Jonathan, Gibson Greg, Kong Augustine, Leal Suzanne M, Moore Jason H, Nadeau Joseph H. Missing Heritability and Strategies for Finding the Underlying Causes of Complex Disease. Nature Reviews Genetics. 2010;11(6):446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinleib Manning, Kannel William B, Garrison Robert J, McNamara Patricia M, Castelli William P. The Framingham Offspring Study: Design and Preliminary Data. Preventive Medicine. 1975;4:518–552. doi: 10.1016/0091-7435(75)90037-7. [DOI] [PubMed] [Google Scholar]
- Fowler James H, Settle Jaime, Christakis Nicholas A. Correlated genotypes in friendship networks. Proceedings of the National Academy of Sciences. 2011;108(5):1993–1997. doi: 10.1073/pnas.1011687108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsythe Robert, Horowitz Joel L, Savin NE, Sefton Martin. Fairness in Simple Bargaining Experiments. Games and Economic Behavior. 1994;6:347–369. [Google Scholar]
- Gao Xiaoyi, Becker Lewis C, Becker Diane M, Starmer Joshua D, Province Michael A. Avoiding the High Bonferroni Penalty in Genome-Wide Association Studies. Genetic Epidemiology. 2010;34(1):100–105. doi: 10.1002/gepi.20430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberger Arthur S. Heritability. Economica. 1979;46(184):327–347. [Google Scholar]
- Hamer Dean H, Sirota Leo. Beware the Chopstick Gene. Molecular Psychiatry. 2000;(1):11–13. doi: 10.1038/sj.mp.4000662. [DOI] [PubMed] [Google Scholar]
- Hartl Daniel L. A Primer of Population Genetics. Sunderland: Sinauer Associates; 1988. [Google Scholar]
- Hochberg Yosef, Benjamini Yoav. More Powerful Procedures for Multiple Significance Testing. Statistics in Medicine. 1990;9(7):811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- Hofman Albert, Breteler Monique MB, van Duijn Cornelia M, Janssen Harry LA, Krestin Gabriel P, et al. The Rotterdam Study: 2010 Objectives and Design Update. European Journal of Epidemiology. 2009;24(9):553–572. doi: 10.1007/s10654-009-9386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill William G, Goddard Michael E, Visscher Peter M. Data and Theory Point to Mainly Additive Genetic Variance for Complex Traits. Plos Genetics. 2008;4(2):e1000008. doi: 10.1371/journal.pgen.1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff Lucia A, Junkins Heather A, Hall PN, Mehta JP, Manolio TA. A Catalog of Published Genome-Wide Association Studies. [Accessed 24 august 2010]; Available at: www.genome.gov/gwastudies.
- Hirschhorn JN. Genomewide Association Studies--Illuminating Biologic Pathways. New England Journal of Medicine. 2009;360(17):1699–1701. doi: 10.1056/NEJMp0808934. [DOI] [PubMed] [Google Scholar]
- Ioannidis John PA, Ntzani Evangelia E, Trikalinos Thomas A, Contopoulos-Ioannidis Despina G. Replication Validity of Genetic Association Studies. Nature Genetics. 2001;29:306–309. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- Ioannidis John PA. Why Most Published Research Findings are False. PLoS Medicine. 2005;2(8):e124. doi: 10.1371/journal.pmed.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis John PA. Non-replication and Inconsistency in the Genome-Wide Association Setting. Human Heredity. 2007;64(4):203–213. doi: 10.1159/000103512. [DOI] [PubMed] [Google Scholar]
- Israel Salomon, Lerer Elad, Shalev Idan, Uzefovsky Florina, Riebold Mathias, et al. The Oxytocin Receptor (OXTR) Contributes to Prosocial Fund Allocations in the Dictator Game and the Social Value Orientations Task. PLoS ONE. 2009;4(5):e5535. doi: 10.1371/journal.pone.0005535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jencks Christopher. Heredity, Environment, and Public Policy Reconsidered. American Sociological Review. 1980 October;45:723–736. [PubMed] [Google Scholar]
- Jencks Christopher, Brown Marsha. Genes and Social Stratification. In: Paul Taubman., editor. Kinometrics. Amsterdam: North-Holland; 1977. pp. 169–233. [Google Scholar]
- Kidd Jeffrey M, Cooper Gregory M, Donahue William F, Hayden Hillary S, Sampas Nick, et al. Mapping and Sequencing of Structural Variation from Eight Human Genomes. Nature. 2008;453(7191):56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kling Jeffrey R, Liebman Jeffrey B, Katz Lawrence F. Experimental Analysis of Neighborhood Effects. Econometrica. 2007;75(1):83–119. [Google Scholar]
- Kosfeld Michael, Heinrichs Marcus, Zak Paul J, Fischbacher Urs, Fehr Ernst. Oxytocin Increases Trust in Humans. Nature. 2005;435(7042):673–676. doi: 10.1038/nature03701. [DOI] [PubMed] [Google Scholar]
- Knafo Ariel, Israel Salomon, Darvasi Ariel, Bachner-Melman Rachel, Uzefovsky Florina, et al. Individual Differences in Allocation of Funds in the Dictator Game Associated with Length of the Arginine Vasopressin 1a Receptor (AVPR1a) RS3 Promoter Region and Correlation between RS3 Length and Hippocampal mRNA. Genes, Brain and Behavior. 2008;7(3):266–275. doi: 10.1111/j.1601-183X.2007.00341.x. [DOI] [PubMed] [Google Scholar]
- Kuhnen Camelia M, Chiao Joan Y. Genetic Determinants of Financial Risk Taking. PLoS ONE. 2009;4(2):e4362. doi: 10.1371/journal.pone.0004362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander Eric S, Linton Lauren M, Birren Bruce, Nusbaum Chad, Zody Michael C, Baldwin Jennifer, et al. Initial Sequencing and Analysis of the Human Genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Levy Daniel, Brink Susan. A Change of Heart: How the People of Framingham, Massachusetts Helped Unravel the Mysteries of Cardiovascular Disease. New York: Knopf; 2005. [PubMed] [Google Scholar]
- Lichtenstein Paul, Pedersen Nancy L, McClearn Gerald E. The Origins of Individual Differences in Occupational Status and Educational Level. Acta Sociologica. 1992;35(1):13–31. [Google Scholar]
- Lizzeri Alessandro, Siniscalchi Marciano. Parental Guidance and Supervised Learning. Quarterly Journal of Economics. 2008;123(3):1161–1195. [Google Scholar]
- Mackay Trudy. The Genetic Architecture of Quantitative Traits. Annual Review of Genetics. 2001;35:303–339. doi: 10.1146/annurev.genet.35.102401.090633. [DOI] [PubMed] [Google Scholar]
- Manolio Teri A, Collins Francis S, Cox Nancy J, Goldstein David B, Hindorff Lucia A, et al. Finding the Missing Heritability of Complex Diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark I. McCarthy, Abecasis Goncalo R, Cardon Lon R, Goldstein David B, Little Julian, Ioannidis John PA, Hirschhorn Joel N. Genome-wide association studies for complex traits: Consensus, uncertainty and challenges. Nature Review Genetics. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- McDermott Rose, Tingley Dustin, Cowden Jonathan, Frazzetto Giovanni, Johnson Dominic DP. Monoamine Oxidase A Gene (MAOA) Predicts Behavioral Aggression Following Provocation. Proceedings of the National Academy of Sciences. 2009;106(7):2118–2123. doi: 10.1073/pnas.0808376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaou Nicos, Shane Scott, Adi Georgina, Mangino Massimo, Harris Juliette. A polymorphism associated with entrepreneurship: Evidence from dopamine receptor candidate genes. Small Business Economics. 2011;36(2):151–155. [Google Scholar]
- Ollier William, Sprosen Tim, Peakman Tim. UK Biobank: From Concept to Reality. Pharmacogenomics. 2005;6(6):639–646. doi: 10.2217/14622416.6.6.639. [DOI] [PubMed] [Google Scholar]
- Pearson Thomas A, Manolio Teri A. How to Interpret a Genome-Wide Association Study. Journal of the American Medical Association. 2008;299(11):335–1344. doi: 10.1001/jama.299.11.1335. [DOI] [PubMed] [Google Scholar]
- Plomin Robert, DeFries John C, McClearn Gerald E, McGuffin Peter. Behavioral Genetics. 5th Edition. New York: Worth Publishers; 2008. [Google Scholar]
- Posthuma Danielle, Willemsen Gonneke, van der Sluis S, Sullivan Patrick, Smit JH, et al. A Genome-Wide Association Study for Educational Attainment. Behavior Genetics. 2008;38:643–644. [Google Scholar]
- Price Alkes L, Patterson Nick J, Plenge Robert M, Weinblatt Michael E, Shadick Nancy A, Reich David. Principal Components Analysis Corrects for Stratification in Genome-Wide Association Studies. Nature Genetics. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Psaty Bruce M, O'Donnell Christopher J, Gudnason Vilmundur, Lunetta Kathryn L, Folsom Aaron R, et al. on behalf of the CHARGE Consortium. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of Prospective Meta-Analyses of Genome-Wide Association Studies from Five Cohorts. Circulation: Cardiovascular Genetics. 2009;2:73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell Shaun M, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley Matt. Nature via Nurture: Genes, Experience, and What Makes Us Human. New York: Harper Collins Publishers; 2003. [Google Scholar]
- Rivadeneira F, Styrkársdottir U, Estrada K, Halldórsson BV, Hsu YH, et al. Twenty Bone-Mineral-Density Loci Identified by Large-Scale Meta-Analysis of Genome-Wide Association Studies. Nature Genetics. 2009;41(11):1199–1206. doi: 10.1038/ng.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch Neil, Herrell Richard, Lehner Thomas, Liang Kung-Yee, Eaves Lindon, et al. Interaction Between the Serotonin Transporter Gene (5-HTTLPR), Stressful Life Events, and Risk of Depression: A Meta-analysis. Journal of the American Medical Association. 2009;301(23):2462–2471. doi: 10.1001/jama.2009.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe Brian E, Tilley Michael R, Gu Howard H, Beversdorf David Q, Sadee Wolfgang, et al. Financial and Psychological Risk Attitudes Associated with Two Single Nucleotide Polymorphisms in the Nicotine Receptor (CHRNA4) Gene. PLoS ONE. 2009;4(8):e6704. doi: 10.1371/journal.pone.0006704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatti Chiara, Service Susan, Freimer Nelson. False Discovery Rate in Linkage and Association Genome Screens for Complex Disorders. Genetics. 2003;164(2):829–833. doi: 10.1093/genetics/164.2.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacerdote Bruce. How Large Are the Effects from Changes in Family Environment. A Study of Korean American Adoptees. Quarterly Journal of Economics. 2007;122(1):119–157. [Google Scholar]
- Sacerdote Bruce. Nature And Nurture Effects On Children's Outcomes: What Have We Learned From Studies Of Twins And Adoptees? In: Jess Benhabib, Jackson Matthew, Bisin Alberto., editors. Handbook of Social Economics. Amsterdam: North Holland; 2010. [Google Scholar]
- Schultz Wolfram, Dayan Peter, Montague Read P. A Neural Substrate of Prediction and Reward. Science. 1997;275(5306):1593. doi: 10.1126/science.275.5306.1593. [DOI] [PubMed] [Google Scholar]
- Stephens Matthew, Balding David J. Bayesian Statistical Methods for Genetic Association Studies. Nature Reviews Genetics. 2009;10(10):681–690. doi: 10.1038/nrg2615. [DOI] [PubMed] [Google Scholar]
- Strachan Tom, Read Andrew. Human Molecular Genetics. 3rd edition. London: Garland Science/Taylor & Francis Group; 2003. [Google Scholar]
- Sullivan Patrick F, Purcell Shaun. Analyzing Genome-Wide Association Study Data: A Tutorial Using PLINK. In: Benjamin M Neale, Ferreira Manuel AR, Medland Sarah, Posthuma Danielle., editors. Statistical Genetics; Gene Mapping Through Linkage and Association. New York: Taylor & Francis Group; 2008. pp. 355–394. [Google Scholar]
- Taubman Paul. The Determinants of Earnings: Genetics, Family and Other Environments: A Study of White Male Twins. American Economic Review. 1976;66(5):858–870. [Google Scholar]
- Turkheimer Eric. Three Laws of Behavior Genetics and What They Mean. Current Directions in Psychological Science. 2000;9(5):160–164. [Google Scholar]
- The International HapMap Consortium. The International HapMap Project. Nature. 2003:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Van der Loos Matthijs JHM, Koellinger Philipp D, Groenen Patrick JF, Thurik Roy A. Genome-Wide Association Studies and the Genetics of Entrepreneurship. European Journal of Epidemiology. 2010;25:1–3. doi: 10.1007/s10654-009-9418-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Loos Matthijs JHM, Koellinger Philipp D, Groenen Patrick JF, Fernando Rivadeneira Cornelius A, van Rooij Frank JA, Uitterlinden André G, Hofman Albert, Thurik A Roy. Candidate gene studies and the quest for the entrepreneurial gene. Small Business Economics. 2012;38(1) forthcoming. [Google Scholar]
- Venter J. Craig, Adams Mark D, Myers Eugene W, Li Peter W, Mural Richard J, et al. The Sequence of the Human Genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Visscher PM, Hill William G, Wray Naomi R. Heritability in the Genomics Era Concepts and Misconceptions. Nature Reviews Genetics. 2008;9(4):255–266. doi: 10.1038/nrg2322. [DOI] [PubMed] [Google Scholar]
- Visscher Peter M. Sizing up Human Height Variation. Nature Genetics. 2008;40(5):489–490. doi: 10.1038/ng0508-489. [DOI] [PubMed] [Google Scholar]
- Visscher Peter M, Medland Sarah E, Ferreira Manuel AR, Morley Katherine I, Zhu Gu, Cornes Belinda K, Montgomery Grant W, Martin Nicholas G. Assumption-Free Estimation of Heritability from Genome-Wide Identity-by-Descent Sharing between Full Siblings. Plos Genetics. 2006;2(3):e41. doi: 10.1371/journal.pgen.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace Björn, Cesarini David, Johannesson Magnus, Lichtenstein Paul. Heritability of Ultimatum Game Responder Behavior. Proceedings of the National Academy of Sciences. 2007;104(40):15631–15634. doi: 10.1073/pnas.0706642104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium. Genome-Wide Association Study of 14,000 Cases of Seven Common Diseases and 3,000 Shared Controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Jian, Benyamin Beben, McEvoy Brian P, Gordon Scott, Henders Anjali K, et al. Common SNPs Explain a Large Proportion of the Heritability for Human Height. Nature Genetics. 2010;42(7):565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Hua, Prentice Ross L. Bias-Reduced Estimators and Confidence Intervals for Odds Ratios in Genome-Wide Association Studies. Biostatistics. 2008;9(4):621–634. doi: 10.1093/biostatistics/kxn001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Songfa, Israel Salomon, Xue Hong, Sham Pak C, Ebstein Richard P, Chew Soo Hong. A Neurochemical Approach to Valuation Sensitivity over Gains and Losses. Proceedings of the Royal Society B: Biological Sciences. 2009a;276(1676):4181–4188. doi: 10.1098/rspb.2009.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Songfa, Israel Salomon, Xue Hong, Ebstein Richard P, Chew Soo Hong. Monoamine Oxidase A Gene (MAOA) Associated with Attitude Towards Longshot Risks. PlosONE. 2009b;4(12):e8516. doi: 10.1371/journal.pone.0008516. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.