Abstract
Myotonic dystrophy type 1 (DM1) is a triplet repeating disorder caused by expanded CTG repeats in the 3′ untranslated region of the dystrophia myotonica protein kinase (DMPK) gene. The transcribed repeats fold into an RNA hairpin with multiple copies of a 5′CUG/3′GUC motif that binds the RNA splicing regulator muscleblind-like 1 protein (MBNL1). Sequestration of MBNL1 by expanded r(CUG) repeats causes splicing defects in a subset of pre-mRNAs including the insulin receptor, the muscle-specific chloride ion channel, Sarco(endo)plasmic reticulum Ca2+ ATPase 1 (Serca1/Atp2a1), and cardiac troponin T (cTNT). Based on these observations, the development of small molecule ligands that target specifically expanded DM1 repeats could serve as therapeutics. In the present study, computational screening was employed to improve the efficacy of pentamidine and Hoechst 33258 ligands that have been shown previously to target the DM1 triplet repeat. A series of inhibitors of the RNA-protein complex with low micromolar IC50’s, which are >20-fold more potent than the query compounds, were identified. Importantly, a bis-benzimidazole identified from the Hoechst query improves DM1-associated pre-mRNA splicing defects in cell and mouse models of DM1 (when dosed with 1 mM and 100 mg/kg, respectively). Since Hoechst 33258 was identified as a DM1 binder through analysis of an RNA motif-ligand database, these studies suggest that lead ligands targeting RNA with improved biological activity can be identified by using a synergistic approach that combines analysis of known RNA-ligand interactions with virtual screening.
INTRODUCTION
RNA plays important roles in biological processes. For example, RNA splicing patterns control the protein isoforms derived from a single primary transcript, and microRNAs can regulate processes such as developmental timing by controlling the lifetime of an mRNA.1–5 RNA-mediated diseases 6–11 can be caused by an RNA loss-of-function or a gain-of-function. RNA gain-of-function causes or contributes to triplet and tetra-repeating disorders such as the myotonic dystrophies,12–14 Huntington’s disease,15 spinocerebellar ataxia,16,17 and fragile X syndrome.18 In myotonic dystrophy type 1 (DM1), an expanded rCUG repeat in the 3′untranslated region (UTR) of the dystrophia myotonica protein kinase (DMPK) mRNA binds and inactivates muscleblind-like 1 protein (MBNL1).19–22 Sequestration of MBNL1 by the expanded repeats results in the mis-splicing of a subset of pre-mRNAs including the muscle-specific chloride ion channel 23 and the insulin receptor 24–26 among others.27,28 Splicing defects in the chloride ion channel and the insulin receptor pre-mRNAs explain the myotonia and insulin insensitivity, respectively, associated with myotonic dystrophy. The presence of expanded rCUG repeats also causes reduced nucleocytoplasmic transport of the DMPK mRNA and thus the expression level of DMPK.21,29 Reduced transport occurs because the expanded repeats bind to various proteins and form nuclear foci.22,30–35
The disease model in which expanded rCUG repeats bind and inactivate MBNL1 points to a therapeutic strategy in which a small molecule or oligonucleotide binds the repeats and frees MBNL1. This therapeutic avenue is supported by several studies 36–39 including the correction of splicing defects by over-expression of MBNL1.20 Several groups have identified small molecules that inhibit the rCUG-MBNL1 interaction including a Hoechst derivative,40 an aminoglycoside derivative,41 an acridine-triazole conjugate,42 pentamidine,38 and peptides and derivatives thereof.36,43 Pentamidine 38 and oligonucleotides 37,39 improve splicing defects in cell culture and animal models.
It is likely that the small molecules that inhibit the rCUG-MBNL1 interaction represent privileged scaffolds for binding rCUG repeats. Therefore, we used Hoechst 33258 and pentamidine as query molecules to identify potential lead ligands in the NCI and eMolecules databases with improved potencies and biological activity; that is, we identified the small molecules that are similar in shape and/or the positioning of functional groups, rings, hydrophobic groups, charge, etc. Once identified, the potencies of the compounds were tested in an in vitro assay. Many ligands are more potent than the parent pentamidine and Hoechst 33258 compounds; the five most potent compounds showed a >20- to >100-fold improvement for disruption of the rCUG-MBNL1 complex. A bis-benzimidazole, which was identified by the Hoechst query, improves DM1-associated pre-mRNA splicing defects in cellular and animal models. Since Hoechst 33258 was identified to bind the repeating 5′CUG/3′GUC motif in the DM1 RNA using an RNA motif-ligand database, these studies suggest that the database approach can be useful to identify lead ligands targeting RNA that can be further improved by using virtual screening.
RESULTS & DISCUSSION
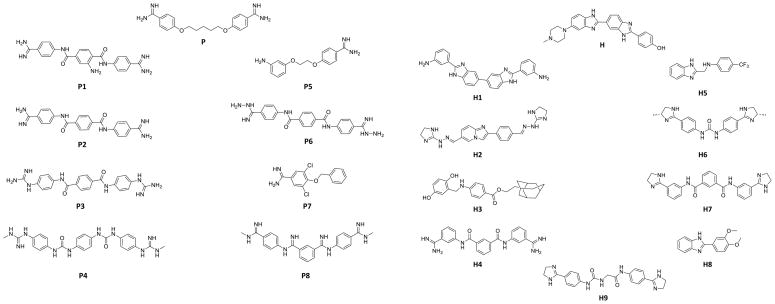
Previous reports have shown that small molecules 40–43 and oligonucleotides 39 can disrupt formation of the toxic RNA-protein interaction that causes myotonic dystrophy. Two of these small molecules, Hoechst 33258 (H) and pentamidine (P) (Figure 1), are interesting lead ligands that are amendable to virtual screening. Hoechst 33258 was identified as a lead ligand by analysis of an RNA motif-ligand database 40,44 while pentamidine was identified from a previous report.38 The goal of virtual screening is to identify compounds with similar shapes or positioning of functional groups that have improved biological activity and/or improved pharmacodynamic or pharmacokinetic properties.
Figure 1.
Structures of the most potent lead compounds that inhibit formation of the r(CUG)12-MBNL1 complex. The compounds were identified from computational screening in which pentamidine (P) and Hoechst 33258 (H) were used as query molecules. Left, the most potent pentamidine-like compounds; right, the most potent Hoechst 33258-like compounds.
Virtual screening methodology
Hoechst 33258 (H) and pentamidine (P) (Figure 1) were used as query molecules to identify compounds in the NCI and eMolecules databases that are similar in shape and/or structure. The NCI database was used because it is an easily accessible, medium sized database (250,000 compounds). It contains the most complete collection of compounds that represents broad chemical space, and the compounds are available free of charge for academic purposes. All compounds used for virtual screening in the NCI database have complete stereochemistry specification, allowing reliable 3D coordinates of stereoisomers to be calculated. The eMolecules database (8,000,000 compounds) was used as it contains the most comprehensive, commercially available collection of small molecules for virtual screening.
Pentamidine (P) and Hoechst 33258 (H) (Figure 1) are attractive lead compound for virtual screening because of their relative structural simplicity, which leads to computationally less demanding 3D shape comparison. Three-dimensional computational shape comparison was completed using the 3D alignment software Rapid Overlay of Chemical Structure (ROCS) because of its fast and efficient implementation of a 3D-shape algorithm.45 ROCS treats atoms as a Gaussian function and thus the overlap between two (or more) atoms are also Gaussian functions. This algorithm implemented in ROCS speeds up the 3-dimensional shape-based calculations, as it involves calculating the maximal intersection of the volumes of two molecules.
For both compounds, 100 conformations were generated with Omega 2.3.2 (v 2.02) software from OpenEye Scientific Software and were compared to 100 conformations of each compound in the NCI and eMolecules database.46 Comparisons were made in two different ways: shape similarities as quantified by the shape Tanimato coefficient 47 and chemistry alignment, or color score.48. Chemistry alignment is specified by the color force field (Implicit Mills Dean48) and accounts for the positioning of hydrogen bond donors, hydrogen bond acceptors, anions, cations, hydrophobic groups and rings. Complete overlap for either the shape or color (chemistry alignment) is assigned a score of “1” while no overlap is assigned a score of “0”. The combo score reported in Table 1 is the sum of the shape Tanimato coefficient and the color score (range = 0–2).
Table 1.
IC50’s for displacement of MBNL1 from rCUG repeats and the degree of similarity to the query molecule (Combo Score).a
| Pentamidine-like | Hoechst-Like | ||||
|---|---|---|---|---|---|
| Small Molecule | Combo Score | IC50 (μM) | Small Molecule | Combo Score | IC50 (μM) |
| Pentamidine | - | >1000 | Hoechst 33258 | - | >1000 |
| P1 | 1.45 | 10 | H1 | 1.63 | 50 |
| P2 | 1.47 | 50 | H2 | 1.21 | 60 |
| P3 | 1.30 | 60 | H3 | 1.13 | 125 |
| P4 | 1.13 | 200 | H4 | 1.12 | 200 |
| P5 | 1.36 | 250 | H5 | 1.04 | 300 |
| P6 | 1.33 | 375 | H6 | 1.13 | 375 |
| P7 | 1.07 | 375 | H7 | 1.15 | 500 |
| P8 | 1.55 | 1000 | H8 | 1.03 | 500 |
| H9 | 1.08 | 500 | |||
The error in the IC50 measurements is ± 10%
Since the compounds are similar to either Hoechst 33258 or pentamidine, it is likely that the compounds have similar binding preferences for RNA internal loops.49 A subset of 40 compounds was chosen for small molecules identified to be most similar to pentamidine; another 40 were chosen for small molecules identified to be similar to Hoechst 33258 (Table S-3). These subsets were selected by visual inspection of the top 500 compounds to allow for maximal chemical diversity and if they are available from the NCI or eMolecules repositories.
Identification of Inhibitors Using Quantitative Time Resolved-Fluorescence Resonance Energy Transfer (qTR-FRET)
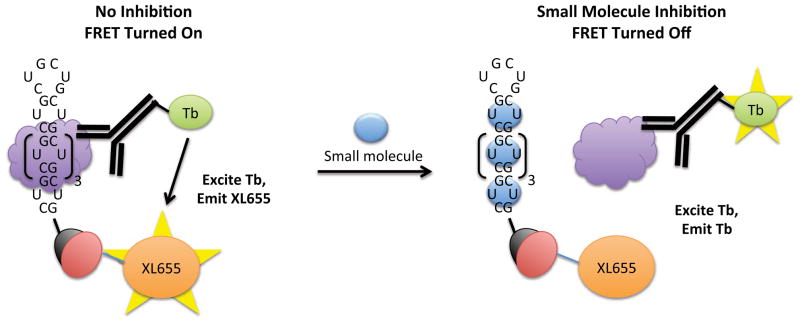
In order to screen the ligands identified via virtual screening for disrupting the r(CUG)6-MBNL1 complex, a qTR-FRET assay was used.50–52 A schematic of the assay is shown in Figure 2. Briefly, 5′-biotinylated r(CUG)6 and MBNL1-His6 are pre-equilibrated, and then the compound of interest is added. Next, streptavidin-XL665 (binds to the biotinylated RNA oligonucleotide) and Tb-Anti-His6 (binds to MBNL1) are added. If the compound does not disrupt the r(CUG)12-MBNL1 interaction, then XL665 and Tb are within close enough proximity to form a FRET pair, and the TR-FRET can be measured. TR-FRET is not observed if the compound inhibits formation of the r(CUG)12-MBNL1 complex. It should be noted that the RNA-protein complex is pre-formed prior to addition of the compound of interest. Therefore, in order to be scored as an active compound, the small molecule must displace MBNL1 from the RNA.
Figure 2.
A schematic of the qTR-FRET assay used to determine the potencies of compounds for disrupting the r(CUG)12-MBNL1 interaction. The r(CUG)12 oligonucleotide is labeled with a 5′-biotin while MBNL1 contains a His6 tag. Left, in the absence of inhibitor, MBNL1 binds to r(CUG)12. Binding is quantified by using two antibodies that form a FRET pair—an anti-His6 antibody labeled with Tb that binds to MBNL1 and streptavidin labeled with XL665 that binds to r(CUG)12. The two fluorophores are within close enough proximity to form a FRET pair. Tb is excited at 345 nm; the resulting emission (~545 nm) excites XL665, which emits at 665 nm. Right, in the presence of inhibitor, the r(CUG)12-MBNL1 interaction is disrupted, and two fluorophores are not within close enough proximity to form a FRET pair. Therefore, emission is only observed at 545 nm (due to Tb). XL665 emission is not observed.
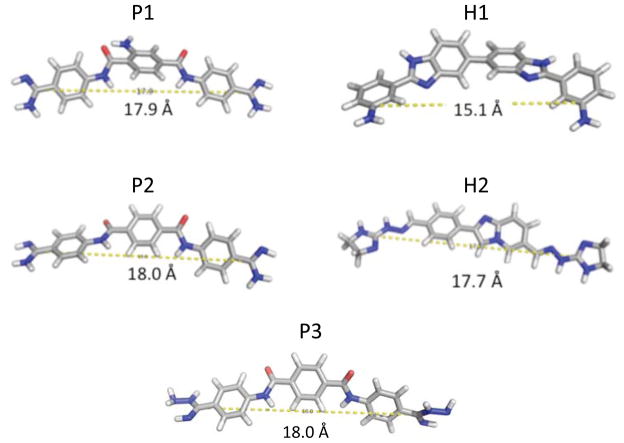
We identified 17 compounds from the qTR-FRET screening assay that inhibited ≥85% of r(CUG)12-MBNL1 complex formation at 1 mM (Figure 1 and Table S-3). Interestingly, pentamidine and Hoechst only inhibit ~10% of r(CUG)12-MBNL1 complex formation at 1 mM. The active compounds were then purified by HPLC, and the IC50’s were measured (Table 1). The IC50’s for the pentamidine-like compounds range from 10 μM to 1000 μM while the IC50’s for the Hoechst-like compounds range from 50 μM to 500 μM. Note that in 2D shape comparison the compounds are chemically quite distinct but they show excellent shape similarity and functional group similarity as quantified by the combo score (Table 1). The best hits (P1, P2, P3, H1, and H2), which have IC50’s ≤60 μM, were structurally analyzed to determine an active chemotype. Each structure has two basic nitrogen atoms that are separated by similar distances, 15.1–18.0 Å (Figure 3).
Figure 3.
Structural analysis of the five most potent compounds that formation of the r(CUG)6-MBNL1 complex using the TR-FRET assay depicted in Figure 2. Each structure has a similar distance between two carbon atoms that display a basic nitrogen.
Specificity of the Small Molecules for the DM1 Motif
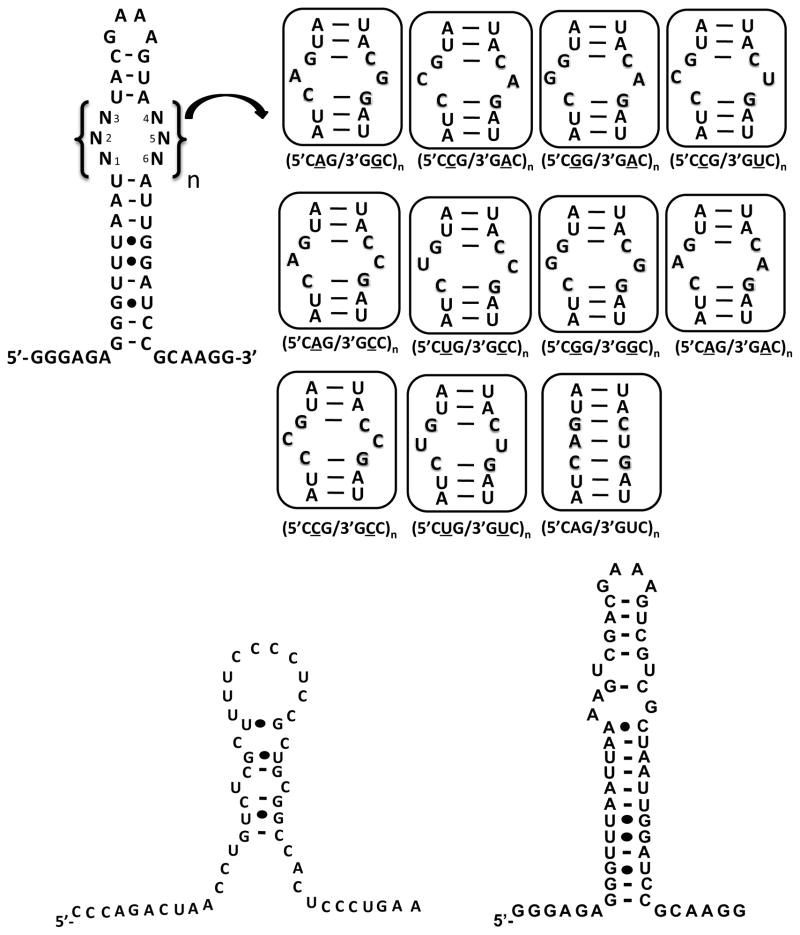
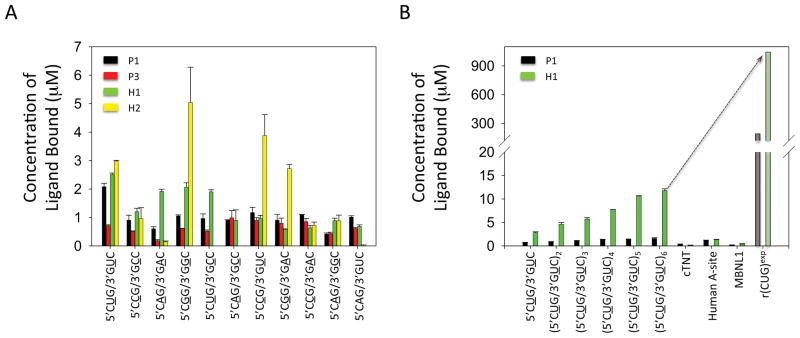
It is important that the small molecules that inhibit the rCUG-MBNL1 interaction show some level of specificity for the DM1 motif, 5′CUG/3′GUC. To estimate specificity, competition dialysis was employed. In these experiment, RNAs displaying the DM1 motif, a fully paired RNA, RNAs displaying other 1×1 nucleotide internal loops, an oligonucleotide mimic of the human A-site, and an oligonucleotide mimic of the cardiac troponin T (cTNT) pre-mRNA were used (Figure 4). The human A-site mimic was used because it is a likely cellular bystander RNA in mammalian cells and because it contains a 1×1 nucleotide UU internal loop motif. Furthermore, the human A-site is accessible for small molecule binding in the context of the ribosome,53–55 and it is a highly abundant cellular RNA.56 cTNT pre-mRNA is a natural substrate for MBNL1.21 It was used in order to determine if the small molecules might inhibit splicing of endogenous genes due to promiscuous binding. Compounds P1, P3, H1, and H2 were investigated; the spectral properties of P2 prevented study by competition dialysis as the wavelengths at which it absorbs overlap with RNA.
Figure 4.
Secondary structures of the RNAs used to determine the specificities of P1, P3, H1, and H2, which are four of the most potent inhibitors of the r(CUG)6-MBNL1 complex. RNAs containing only one copy of a motif are referred to as 5′CXG/3′GXC.
First, the specificities of the compounds for the DM1 motif over a fully paired RNA and RNAs displaying other 1×1 nucleotide internal loops were investigated (Figure 5A). The most selective compound is P1 as it binds the DM1 motif preferentially over all other RNAs investigated. In contrast, P3 binds similarly to RNAs containing 5′CCG/3′GAC, 5′CGG/3′GAC, 5′CCG/3′GUC, 5′CAG/3′GCC, and the DM1 motif. Although the greatest amount of binding for H1 is observed to the DM1 motif, significant binding is also observed to 5′CUG/3′GCC, 5′CGG/3′GGC, and 5′CAG/3′GAC. H2 binding preferences are: 5′CGG/3′GCC > 5′CCG/3′GUC ≈ 5′CUG/3′GUC ≈ 5′CGG/3′GAC.
Figure 5.
Results of competition dialysis experiments used to determine the specificities of a subset of the most potent compounds identified from the qTR-FRET assay. Not all of the compounds could be further investigated due to their spectroscopic properties; that is their absorbance spectra overlapped with the absorbance from the RNAs or protein. A, the specificities of four compounds were investigated by competition dialysis. Each compound was incubated with the 11 RNAs shown in the top panel of Figure 4. The concentration of bound ligand was determined using UV spectroscopy. P1 and H1 were the most promising ligands as they bind an RNA containing 5′CUG/3′GUC more than the other RNAs. B, The specificities of P1 and H1 were further investigated by competition dialysis with RNAs containing one to six copies of the DM1 motif (5′CUG/3′GUC), oligonucleotide mimics of the Human A-site and the cTNT pre-mRNA, and MBNL1. The Human A-site RNA and the cTNT pre-mRNA are representative bystander RNAs. The binding of H1 and P1 to r(CUG)exp (shaded bars) was extrapolated based on the presence of 479 copies of the 5′CUG/3′GUC motif in the DM1 cellular model system used in these studies.
Next, we investigated whether the amount of binding of P1 and H1 increased as a function of the number of DM1 motifs present in an RNA (Figure 5B). Thus, competition dialysis experiments were completed for RNAs containing 1–6 copies of 5′CUG/3′GUC, an oligonucleotide mimic of the human A-site, an oligonucleotide mimic of the cTNT pre-mRNA, and MBNL1 (Figure 4). MBNL1 was included in competition dialysis experiments since significant binding of the small molecules to MBNL1 could inhibit its normal function. In both cases, increased binding of the ligand is observed as the number of 5′CUG/3′GUC motifs present in the RNA increases (Figure 5B). The binding of H1 appears to be approximately stoichiometric, in good agreement with a previous report for another Hoechst derivative.40 Moreover, H1 binds preferentially to RNAs with multiple DM1 motifs over MBNL1, and oligonucleotide mimics of the human A-site and cTNT pre-RNA. In contrast, P1 does not bind stoichiometrically as the number of DM1 motifs increase from one to six. P1 also binds similarly to the human A-site and RNAs containing three or more DM1 motifs, and significant binding is observed to the mimic of the cTNT pre-mRNA (Figure 5B).
The affinity of H1 for an RNA containing one 5′CUG/3′GUC (DM1) motif and a fully paired RNA (Figure 4) were determined by fluorescence anisotropy. In good agreement with our competition dialysis results (Figure 5), H1 binds approximately 2-fold more tightly to the RNA with the DM1 motif (70 ± 32 nM) than to the fully paired RNA (190 ± 61 nM) (Supporting Information, Figure S-25).
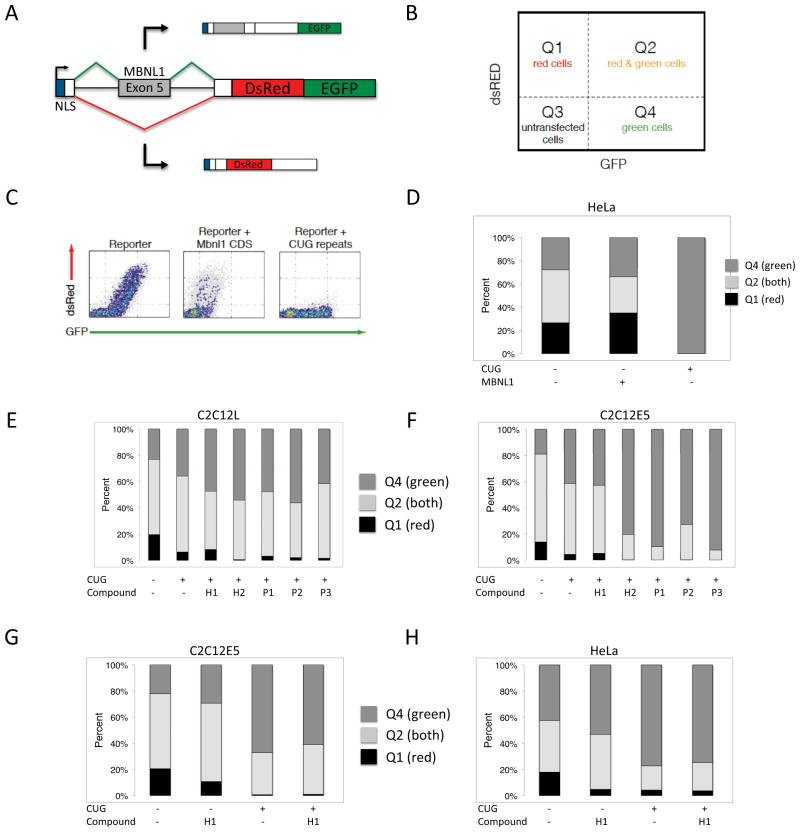
A Bichromatic Reporter System and Flow Cytometry Analysis to Assess Bioactivity
To investigate whether P1, P2, P3, H1, or H2 are biologically active, we performed an initial cell-based screen using a modified bichromatic reporter 57 of alternative splicing (Figure 6A). The reporter contains human Muscleblind exon 5 and adjacent introns. The splicing of this reporter is controlled by MBNL1 itself, through binding and repression close to the 3′ splice site. The reporter was designed such that exon 5 exclusion leads to translation of the DsRed reading frame, and exon 5 inclusion leads to a read-through of the +2 reading frame of DsRed, followed by subsequent translation of the EGFP reading frame. Splicing behavior was assessed by flow cytometry, where the number of cells expressing DsRed and EGFP at various levels can be divided into several quadrants (Figure 6B).
Figure 6.
A) Schematic of bichromatic splicing reporter. The test region is human Muscleblind exon 5 and adjacent introns, and replaces test exons in previous versions of the reporter.57 Inclusion of the exon leads to GFP, and exclusion leads to dsRed. B) A simple method for quantitation of reporter behavior uses quadrants thresholded on GFP and dsRed expression in cells. The fraction of cells in Q1, Q2, and Q4 relative to transfected cells (Q1 + Q2 + Q4) can be used as a metric for reporter behavior. C) Representative flow cytometry plot of the reporter transfected alone, with Mbnl1 coding sequence, or with 960 CUG repeats. The dsRed/GFP ratio is responsive to MBNL1 activity. D) Representative quantitation of reporter behavior in response to Mbnl1 coding sequence or 960 CUG repeats, of which the former leads to increased dsRed, and the latter leads to increased GFP. E–H) Quantitation of reporter response to CUG repeats and compounds in combination. As illustrated in panels E and F, compound H1 exhibits the highest dsRed/GFP ratio relative to all other compounds in the presence of CUG repeats, in two different C2C12 cell subclones (C2C12L and C2C12E5). Panels G and H show instances in which compound H1 can partially rescue the effect of CUG repeats on reporter splicing patterns in both C2C12 and Hela cells. Note that compound H1 alone, in the absence of CUG repeats, leads to a slight increase in GFP relative to dsRed.
The behavior of the splicing reporter is consistent with that of endogenous MBNL1 exon 5, as observed upon transfection into HeLa cells. When transfected alone, ~55% of cells express DsRed or a combination of DsRed and EGFP. Upon co-transfection with a plasmid encoding mouse MBNL1 coding sequence, the percentage of cells that express DsRed or DsRed and EGFP increases to ~70% (Figures 6C and 6D). This is consistent with increased levels of MBNL1 blocking inclusion of the test exon, leading to increased DsRed production relative to EGFP. Likewise, when the reporter is co-transfected with a mini-gene containing 960 CUG repeats in the DMPK context (DT960),21 ~0% of cells express DsRed (Figures 6C and 6D). This is consistent with sequestration of endogenous MBNL1 by CUG repeats, leading to exon inclusion and increased EGFP production relative to DsRed production. Taken together, these results confirm that this bichromatic splicing reporter, which contains MBNL1 exon 5 as a test exon, can be used as a sensitive method for assessing cellular perturbations that affect MBNL1 activity.
To assess the biological activity of our compounds, we co-transfected our bichromatic splicing reporter, along with DT960, into three different cell lines. These lines include two different C2C12 mouse myoblast lines; one is a subclone of the original line (obtained from ATCC), and the other is the same subclone containing a stably integrated control shRNA hairpin that has been designed to not target any endogenous transcripts. After co-transfection of 250 ng of the reporter and 25 ng of DT960, the cells were treated with 20 μM P1, P2, P3, H1, or H2. Two days later, cells were analyzed by flow cytometry.
Similar results were observed in both C2C12 lines (Figures 6E and 6F). As expected, co-transfection with DT960 reduces the fraction of cells that express DsRed. In the presence of DT960, compounds P1, P2, P3, and H2 led to a further reduction in the fraction of cells that express DsRed as compared to untreated cells containing DT960. In contrast, H1 led to a modest increase in the fraction of cells expressing DsRed, suggesting it can block or rescue the effects of CUG repeats on endogenous MBNL1 activity. As an additional control, the effect of H1 on the bichromatic reporter in the absence of DT960 was also tested in C2C12 and HeLa (Figures 6G and 6H). Addition of 20 μM H1 to cells in the absence of DT960 led to a slight decrease in the fraction of cells expressing DsRed. However, addition of 20 μM H1 in the presence of DT960 led to a modest rescue relative to untreated cells in the presence of DT960, as evidenced by increased fractions of C2C12 and HeLa cells expressing DsRed (Figures 6G and 6H). These results suggest that of all the compounds, H1 exhibits biological activity in a manner favorable to counteracting the downstream effects of CUG repeats in the context of DM1.
H1 Improves Splicing Defects in a DM1 Cell Culture Model
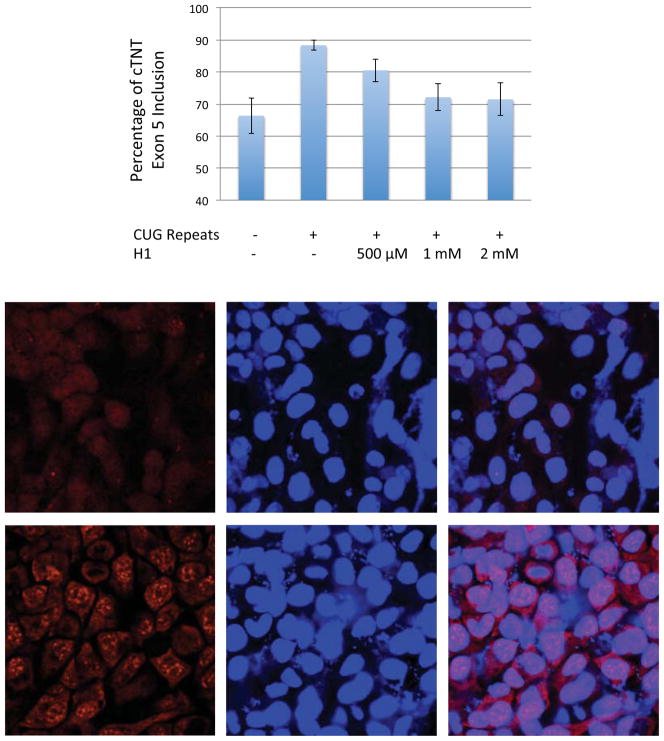
Next, we determined if the five most potent compounds could improve splicing defects in a DM1 cellular model system 38 using a cTNT mini-gene.21 cTNT pre-mRNA is mis-spliced in DM patients.21,58,59 In normal cells, MBNL1 binds upstream of exon 5 in the cTNT pre-mRNA and represses its inclusion.58,60 In the absence of rCUG repeats, approximately 65% of exon 5 is included in cTNT mRNA while in the presence of rCUG repeats, approximately 90% of exon 5 is included (Figure 7A). HeLa cells were co-transfected with plasmids containing a DM1 mini-gene that encodes 960 CTG repeats in a 3′UTR 21 and the cTNT pre-mRNA mini-gene,21 and then the compound of interest was added at a final concentration of 500 μM in growth medium. No improvement in splicing was observed for P1, P2, P3, or H2 (Supporting Information, Figure S-23), in good agreement with a bichromatic splicing reporter (Figure 6).57
Figure 7.
H1 improves splicing defects and disrupt nuclear foci in a DM1 cell culture model. A, H1 improves defects in cTNT pre-mRNA splicing nearly to wild type levels. The error bars indicate standard error. The difference in inclusion percentage for untreated cells that express r(CUG)exp and treated cells (1 mM and 2 mM of H1) is statistically significant. The two tailed p-values are 0.0149 and 0.0192 for cells treated with 1 mM and 2 mM H1, respectively. B, top, H1 disrupts formation of nuclear foci as determined by fluorescence in situ hybridization; bottom, untreated cells with multiple nuclear foci. The microscopic images from left to right are: DY-547 fluorescence indicating the presence of CUG repeats; DAPI; overlay of DY-547 and DAPI.
In contrast, modest improvement of splicing is observed when the cells were treated with 500 μM H1 (Figure 7A). More significant improvement is observed when cells are treated with 1 mM and 2 mM H1, improving splicing patterns almost to wild type. (The difference in inclusion percentage for wild type cells (absence of r(CUG)exp) and r(CUG)exp-expressing cells treated with 1 mM or 2 mM of H1 is not statistically significant.) The difference in inclusion percentage for r(CUG)exp-expressing cells that are untreated (Figure 7A, column 2) and treated with 1 mM or 2 mM of H1 is statistically significant. The two tailed p-values are 0.0149 and 0.0192 for cells treated with 1 mM and 2 mM H1, respectively. Interestingly, Hoechst 33258 does not correct splicing defects in cTNT processing at these concentrations (Supporting Information, Figure S-24). It is important to note that H1 is non-toxic at the concentrations tested.
Control experiments were also completed in order to determine if H1 globally affects splicing. First, HeLa cells were co-transfected with plasmids containing a mini-gene with only five CTG repeats in the 3′UTR and the cTNT mini-gene.21 H1 does not affect cTNT splicing in the absence of CTG repeats (Supporting Information, Figure S-21). We also investigated whether H1 affects the splicing of pre-mRNAs whose splicing is not regulated by MBNL1 (Supporting Information, Figure S-22). A PLEKHH2 mini-gene 38 was co-transfected with the DM1 mini-gene, and the cells were treated with H1. H1 does not affect the splicing of a PLEKHH2 mini-gene at 2 mM. The splicing of endogenous genes TTC8 and CAMKK2 was also not affected. Thus, H1 does not globally affect RNA splicing; rather, it affects the splicing of pre-mRNAs controlled by MBNL1.
The toxicities of the compounds were assessed by flow cytometry using standard forward and side scatter metrics, and propidium iodide (PI) staining (Supporting Information, Figure S-26). PI stains DNA in cells that are still grossly intact but permeabilized; treatment of cells with DMSO results in a slight increase in the percentage of cells stained with PI. Briefly, HeLa cells were treated with 500 μM P1, P2, P3, or H1 and their scatter metrics and PI staining compared to those treated with 2% DMSO and those grown in media alone. Notably, treatment with P1 and P2 yield a large number of observed events with low forward scatter area and height values, indicating that most observed events represent cell debris. Treatment of cells with P3 also leads to loss of cell integrity but to a lesser extent. Following H1 treatment, over half of cells still remain within each gate, indicating that cell integrity is largely preserved. In summary, H1 is by far the least cytotoxic compound as compared to P1, P2, and P3 (Supporting Information, Figure S-26).
H1 Disrupts Formation of Nuclear Foci in a Cell Culture Model of DM1
Another hallmark of myotonic dystrophy is the formation of nuclear foci, which consist of expanded rCUG repeats and proteins including MBNL1.30,34,35,61 In order to determine if H1 is able to disrupt or prevent formation of nuclear foci, a fluorescence in situ hybridization (FISH) assay was used. HeLa cells were co-transfected with a plasmid encoding the DM1 mini-gene. After incubation with H1, the cells were fixed, permeabilized, and incubated with a DY547-labeled, 2′OMe oligonucleotide complementary to the rCUG repeats.38 H1 decreases the percentage of cells with nuclear foci when the cells are dosed with 2 mM and 1 mM compound (Figure 7B) but not with 500 μM.
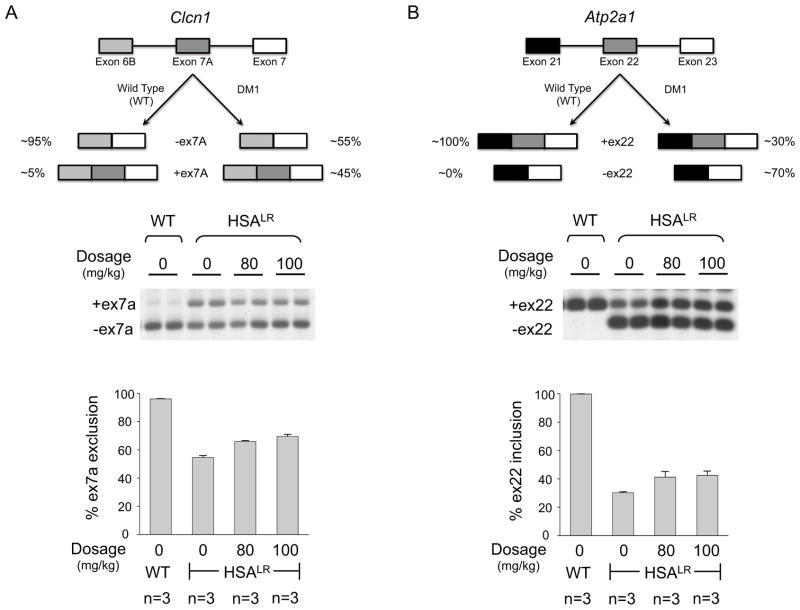
H1 Improves Splicing Defects in a Mouse Model of DM1
A mouse model of DM1 has been reported in which 250 rCUG repeats are expressed using an actin promoter (HSALR).22 The presence of the repeats causes the mis-splicing of the chloride ion channel (Clcn1) and the Serca1 (Atp2a1) pre-mRNAs. Normal adult mice have a Clcn1 exon 7a exclusion rate of 96 ± 0.2%; DM1 mice have an exclusion rate of 54 ± 1.4%. When DM1 mice are dosed with 80 mg/kg and 100 mg/kg, the exclusion rate is partially rescued to 66 ± 0.6% and 69 ± 1.4%, respectively (Figure 8). These improvements in splicing are statistically significant as determined by a t-test. (The two-tailed p-values are 0.0066 for treatment with 80 mg/kg of H1 and 0.0016 for treatment with 100 mg/kg of H1.) Serca1 mis-splicing is also partially rescued. In normal adult mice, the inclusion rate for exon 22 is 99.8 ± 0.1% while the inclusion rate in the HSALR line is only 30 ± 0.6%. Both dosages of H1, 80 mg/kg and 100 mg/kg, rescue splicing similarly with inclusion rates of 41 ± 4.0% and 42 ± 3.0%, respectively (Figure 8). The improvement in splicing is statistically significant when the mice were treated with 100 mg/kg of H1. (The two-tailed p-values are 0.0522 for treatment with 80 mg/kg of H1 and 0.0176 for treatment with 100 mg/kg of H1.)
Figure 8.
H1 improves splicing defects in the muscle-specific chloride ion channel (Clcn1) and Serca1 (Atp2a1) pre-mRNAs in a DM1 mouse model. The DM1 mouse model expresses the human skeletal actin (HSA) transgene containing 250 CTG repeats (HSALR; where LR indicates “long repeats”). Wild type mice contain the HSA transgene with a small number of CTG repeats that do not lead to DM1 (HSASR; where SR indicates “short repeats”).22 A, top: schematic of Clcn1 alternative splicing in wild type and DM1 mice. A, bottom: analysis of Clcn1 alternative splicing by RT-PCR when mice are treated with H1 including a representative gel image and a plot of the corresponding data. The two-tailed p-values are 0.0066 for treatment with 80 mg/kg of H1 and 0.0016 for treatment with 100 mg/kg of H1. B, top: schematic of Atp2a1 (Serca1) alternative splicing in wild type and DM1 mice. B, bottom: analysis of Atp2A1 alternative splicing by RT-PCR when mice are treated with H1 including a representative gel image and a plot of the corresponding data. The two-tailed p-values are 0.0522 for treatment with 80 mg/kg of H1 and 0.0176 for treatment with 100 mg/kg of H1.
Selectivity of H1 Binding to DM1 RNA
Because H1 improves splicing defects in cellular and animal models of DM1 and exhibits little toxicity, it must have at least modest selectivity in vivo. There is little difference in binding affinity or selectivity of H1 for an RNA with a single copy of the DM1 motif and a fully paired RNA or a mimic of the Human A-site (~2-fold). However, the selectivity of H1 for RNAs containing 5′CUG/3′GUC improves as the number of repeats increases (Figure 5). That is, there is only a small difference in the binding of H1 to an RNA with a single motif and a fully paired RNA (Figure 5A) while an ~12-fold difference in binding is observed between an RNA with six DM1 motifs ((5′CUG/3′GUC)6) and a fully paired RNA (Figure 5A and B). Thus, H1 could exhibit some level of specificity for r(CUG)exp in vivo despite the presence of highly expressed bystander RNAs that also contain the DM1 motif or a similar one. For example, the Human A-site (Figure 4) is a highly abundant transcript that contains a single 5′GUC/3′CUG motif (two CUG repeats). At a first consideration, it would be assumed that it would be the most occupied binding site in vivo. However, DM1 is caused by hundreds of copies of the 5′CUG/3′GUC motif. In our cellular model system, r(CUG)exp contains 479 copies of the 5′CUG/3′GUC motif ((CUG)960). Thus, the selectivity of H1 for r(CUG)exp over the A-site could be extrapolated from competition dialysis experiments by normalizing for the number of repeats present in vivo, or 479 times greater than the selectivity of the monomer (~2-fold) (shaded bars in Figure 5B).
Implications
Most RNAs that contribute to disease are unexploited as drug targets. This is generally thought to be due to a lack of information about the privileged scaffolds that specifically target RNA. In this study, a lead molecule was identified by analysis of an RNA motif-ligand database. Previous studies have shown that Hoechst 33258 44 and a derivative thereof 40 bind the regularly repeating 5′CUG/3′GUC motif that is present in DM1 RNA. By using virtual screening, a series of related compounds that have improved in vitro potencies were identified. Importantly, one compound, H1, is specific for RNAs containing multiple 5′CUG/3′GUC motifs over mimics of a highly abundant, accessible RNA target (the human A-site) and a natural MBNL1 substrate (cTNT pre-mRNA). The compound improves splicing defects in cell and animal models and disrupts formation of nuclear foci in cells. Fortuitously, H1 was identified amongst the ~80 compounds that were hand-selected from ~500 virtual screening hits with known activities. Additional compounds could have been screened if required.
Perhaps bioactive small molecules that target other RNAs could be discovered using a similar approach to the one described herein. Lead small molecules that are not bioactive due to poor cellular permeability, toxicity, etc., could be optimized via virtual screening and then tested in vitro or in vivo. Two key developments bolster the likelihood of the applicability of the method: (i) a clearer understanding of the RNA motifs that bind small molecules and the chemical scaffolds that bind RNA and (ii) annotation of the RNA motifs present in RNAs that cause disease, including the HIV genome. By leveraging information in these two areas with compound optimization as described herein, bioactive small molecules that target other RNAs could be identified.
SUMMARY & CONCLUSIONS
A computational approach was used to identify compounds similar to known in vitro inhibitors of a toxic RNA-protein interaction that have improved potencies and bioactivity. A bio-active, non-toxic ligand was identified that is similar to the Hoechst query molecule. Since Hoechst 33258 was identified as a DM1 RNA binder through analysis of an RNA motif-ligand database, these studies suggest that the database can provide lead ligands targeting RNA that can be further improved via computational screening. Furthermore, the H1 ligand module can be further improved for targeting the DM1 RNA by using a modular assembly strategy, which has been shown to improve the in vitro potency of compounds targeting the DM1 RNA by orders of magnitude.40 This approach could be generally applicable to any RNA of interest.
MATERIALS & METHODS
Virtual Screening
Virtual screening was performed on Dell P4 (64-bit) Linux clusters with 32 Xeon processors and Omega 2.3.2 (v 2.02) software from OpenEye Scientific Software. As a starting point for the virtual screen, Chem3D (Cambridge software) was used to energy-minimize (MMFF94force field) a 3-dimensional conformer of pentamidine and Hoechst 33258. Omega 2.3.2 (v 2.02) was used to generate 100 conformers of pentamidine and Hoechst 33258, which were submitted as neutral molecules without any explicit charge. Then, up to 100 conformers of each molecule in the NCI and eMolecules database were generated. (The number of conformers generated for some rigid molecules in the database was less than 100.) The 500 compounds that were the most similar to pentamidine and the 500 compounds that were most similar to Hoechst 33258, as determined by the Tanimato and color scores, were output in rank order as potential hits.45–47 Of these compounds, 88 were obtained (80 from NCI and 8 from ChemDiv (identified from eMolecules database)), and 14 were eliminated from screening due to their insolubility in DMSO or H2O. A complete list of the compounds screened is available in the Supporting Information (Table S-3).
3D-shape comparisons (shape Tanimoto coefficient) were completed using Rapid Overlay of Chemical Structure (ROCS, version 3.0.0) while the chemistry alignment overlap (color score) was completed by using the color force field (Implicit Mills Dean 48) from the OpenEye software package. The combo score is the sum of the shape Tanimoto coefficient and the color score.
Mass Spectroscopy and HPLC Purification
Mass spectra were collected on a Varian 500 MS spectrometer equipped with Varian Prostar Autosampler 410 and/or on an ABI 4800 MALDI-TOF spectrometer.
HPLC purifications were completed on a Waters 1525 Binary HPLC Pump equipped with a Waters 2487 Dual Absorbance Detector system. Compounds were purified using a gradient of 5mL/min, and a linear gradient of 0% to 100% B in A over 55, 50, or 46 min. (A: water + 0.1% trifluoroacetic acid (TFA) (v/v); B: MeOH + 0.1%TFA (v/v).)
The purities of compounds were determined by analytical HPLC using a Waters 1525 Binary HPLC Pump equipped with Waters 2487 Dual λ Absorbance Detector system and the following conditions: a Waters Symmetry® C8 5μm 4.6×150 mm column, room temperature, a flow rate of 2.4 mL/min, and a linear gradient of 0% to 100% B in A over 45 min. (A: water + 0.1% trifluoroacetic acid (TFA) (v/v); B: MeOH + 0.1% TFA (v/v).)
Oligonucleotide Preparation and Purification
The RNA used in qTR-FRET assays (5′biotinylated (CUG)12) was purchased from Dharmacon. The ACE protecting groups were cleaved using Dharmacon’s deprotection buffer. The sample was lyophilized, dissolved in milliQ water and desalted using a PD-10 gel filtration column (GE Healthcare). RNAs used in competition dialysis experiments were prepared via run off-transcription using synthetic DNA templates (Integrated DNA Technologies, Inc.) and a Stratagene RNAMaxx transcription kit. They were purified by denaturing polyacrylamide gel electrophoresis as previously described 62 and dissolved in 1X Competition Dialysis Buffer (8 mM NaH2PO4, 185 mM NaCl and 1 mM EDTA; pH 7.2).
Oligonucleotide concentrations were determined by absorbance at 260 nm (90 °C for the RNA used in qTR-FRET assays; room temperature for RNAs used in competition dialysis) using a Beckman Coutler DU800 UV-Vis spectrophotometer equipped with a Peltier temperature controller unit. Oligonucleotide extinction coefficients were determined using HyTher version 1.0 (Nicolas Peyret and John SantaLucia Jr., Wayne State University, Detroit, MI).(19, 20) The parameters used by HyTher were calculated using information on the extinction coefficients of nearest neighbors in RNA.(21)
Quantitative Time-Resolved Fluorescence Resonance Energy Transfer (qTR-FRET) Assay
The qTR-FRET assay used to identify lead inhibitors of the r(CUG)12-MBNL1 complex is based on PubChem BioAssay AID 2675 (Figure 2). Briefly, 5′-biotinylated r(CUG)12 was folded in 1X Folding Buffer (20 mM HEPES, pH 7.5, 110 mM KCl, and 10 mM NaCl) by heating at 60 °C followed by slowly cooling to room temperature on the bench top. The buffer was adjusted to 1X Assay Buffer (20 mM HEPES, pH 7.5, 110 mM KCl, 10 mM NaCl, 2 mM MgCl2, 2 mM CaCl2, 5 mM DTT, 0.1% BSA, and 0.5% Tween-20) and MBNL1-His6 was added. The final concentrations of RNA and MBNL1 were 80 nM and 60 nM, respectively. The sample was allowed to equilibrate at room temperature for 5 min, and then the compound of interest was added. After 15 min, strepatividin-XL665 (cisbio Bioassays) and anti-His6-Tb (cisbio Bioassays) were added to final concentrations of 40 nM and 0.44 ng/μL, respectively, in a total of 10 μL. The samples were incubated for 1 h at room temperature and then transferred to a well of a black 96-well plate. Time-resolved fluorescence was measured on a Molecular Devices SpectraMax M5 plate reader. Fluorescence was first measured using an excitation wavelength of 345 nm and an emission wavelength of 545 nm (fluorescence due to Tb). TR-FRET was then measured by using an excitation wavelength of 345 nm, an emission wavelength of 665 nm, a 200 μs evolution time, and a 1500 μs integration time. The ratio of fluorescence intensity of 545 nm and 665 nm as compared to the ratios in the absence of ligand and in the absence of RNA were used to determine IC50’s.
Competition Dialysis
Pierce Slide-A-Lyzer MINI dialyzer units (Pierce Biotechnology, Inc.) were dialyzed against water for 24 h in order to ensure that the units did not leak. Competition dialysis was completed as previously described by Chaires.(22) Briefly, RNAs were folded by heating at 60 °C for 5 min followed by slowly cooling to room temperature on the bench top. Dialysis units containing 0.1 mL of 1.6 μM RNA or MBNL1 were placed in 200 mL of 1 μM ligand in 1X Competition Dialysis Buffer. The samples were allowed to equilibrate with the dialysate by stirring the dialysate at 200 rpm for 24 h at room temperature (20–22 °C). Previous studies have shown that this is sufficient time for the sample to reach equilibrium.(19)
At the end of the equilibration period, 67.5 μL of each sample was carefully removed from the dialyzer unit and transferred to microcentrifuge tube. To each sample, 7.5 μL of 10% (w/v) sodium dodecyl sulfate (SDS) was added to give a final concentration of 1% (w/v) SDS, which is sufficient to dissociate the ligand. The SDS step was completed to ensure accurate determination of the concentration of bound ligand as the spectroscopic properties of the bound ligand could be different from that of unbound. The total ligand concentration (Ct) within each dialysis unit was determined spectrophotometrically using an appropriate absorbance wavelength and extinction coefficient for each compound. Appropriate corrections were made for the small dilution resulting from the addition of the 10 μL of SDS.
The free ligand concentration (Cf) was determined from an aliquot of the dialysate solution, which did not vary appreciable from the initial 1 μM. The bound ligand concentration (Cb) was then determined using Equation 1:
| (eq. 1) |
where Cb, Ct, and Cf are concentrations of bound, total, and free ligand, respectively.
Fluorescence Binding Assays
Dissociation constants were determined using an in-solution, fluorescence-based assay. RNA was annealed in DNA buffer (8 mM Na2PO4, pH 7.0, 185 mM NaCl, 0.1 mM EDTA) at 60 °C for 5 min and allowed to slowly cool to room temperature. Then H1 was added to final concentration of 1000 nM. Serial dilutions of the RNA (1:2) were then completed in DNA buffer containing 1000 nM H1. The samples were incubated for 15 min at room temperature and then transferred to a well of a black 384-well plate. Anisotropy intensity was measured using a Molecular Devices SpectraMax M5 plate reader. The change in anisotropy as a function of RNA concentration was fit to standard one site saturation ligand binding equation (eq. 2):
| (eq. 2) |
where y is the observed anisotropy intensity, Bmax is the maximum anisotropy observed, x is the concentration of RNA and Kd is the dissociation constant.
A Bichromatic Reporter System and Flow Cytometry Analysis to Assess Bioactivity
C2C12 and HeLa cell lines were maintained as monolayers in 1X DMEM supplemented with 20% or 10% FBS, respectively. Cells were trypsinized from the surface and plated in 12-well plates in growth medium. While the cells were adhering to the surface, they were co-transfected with 25 ng of a plasmid encoding the DM1 mini-gene 21 and 250 ng of a plasmid encoding a modified bichromatic fluorescent reporter of alternative splicing 57 using Transit (Mirus). (The bichromatic fluorescent reporter contains human MBNL1 exon 5 and adjacent introns in place of cTNT and its adjacent introns (Figure 21A).) The compound of interest was then added (20 μM). After 48 h, the cells were harvested by trypsinzation. The trypsin was inactivated using growth medium, and the cells were immediately subjected to flow cytometry analysis using a BD LSR II.
Improvement of Splicing Defects in a Cell Culture Model Using RT-PCR
In order to determine if H1 improves splicing defects in vivo, a previously reported method was employed.38 Briefly, HeLa cells were grown as monolayers in 12-well plates in growth medium (1X DMEM, 10% FBS, and 1X GlutaMax (Invitrogen)). After the cells reached 90–95% confluency, they were transfected with 800 ng of total plasmid using Lipofectamine 2000 reagent (Invitrogen) per the manufacturer’s standard protocol. Equal amounts of a plasmid expressing a DM1 mini-gene with 960 CTG repeats 21 and a mini-gene of interest (cTNT21 or PLEKHH2 60) were used. Approximately 5 h post-transfection, the transfection cocktail was removed and replaced with growth medium containing the compound of interest. After 16–24 h, the cells were trypsinized from the surface, and total RNA was harvested with a Qiagen RNAEasy kit. An on-column DNA digestion was completed per the manufacturer’s recommended protocol.
A sample of RNA was subjected to reverse transcription-polymerase chain reaction (RT-PCR) as previously described 60 except 5 units of AMV Reverse Transcriptase from Life Sciences were used. Approximately 300 ng were reverse transcribed, and 150 ng were subjected to PCR using a radioactively labeled forward primer. RT-PCR products were observed after 25–30 cycles of: 95 °C for 1 min; 55 °C for 1 min; 72 °C for 2 min and a final extension at 72 °C for 10 min. The products were separated on a denaturing 5% polyacrylamide gel and imaged using a Typhoon phosphorimager.
Control experiments were also completed in which HeLa cells were transfected with a plasmid encoding a mini-gene with five CTG repeats in the 3′ UTR or with a mini-gene that encodes a pre-mRNA whose splicing is not controlled by MBNL1 (PLEKHH2).60 The effect of the compound on the splicing of endogenous mRNAs not under the control of MBNL1 (TTC8 and CAMKK2) was also determined as previously described.38 Differences in alternative splicing were evaluated by a t-test.
Disruption of Nuclear Foci Using Fluorescence In Situ Hybridization (FISH).38
HeLa cells were grown as monolayers in Mat-Tak glass-bottomed, 96-well plates. After the cells reached 90–95% confluency, they were transfected with 100 ng of a plasmid encoding a DM1 mini-gene 21 using Lipfoectamine 2000 per the manufacturer’s standard protocol. The transfection cocktail was removed 5 h post-transfection, and the compound of interest was added in growth medium.
After 16–24 h, the cells were washed with 1X DPBS and fixed with 2% paraformaldehyde in 1X DPBS for 5 min at room temperature. After washing with 1X DPBS, the cells were permeabilized with 1X DPBS + 0.1% Triton X-100 for 5 min at room temperature. The cells were washed with 1X DPBS + 0.1% Triton X-100 three times and then with 30% formamide in 2X SSC Buffer (30 mM sodium citrate, pH 7.0, 300 mM NaCl).
The cells were incubated in 1X FISH Buffer (30% formamide, 2X SSC Buffer, 66 μg/mL bulk yeast tRNA, 2 μg/mL BSA, 2 mM vanadyl complex (New England Bio Labs) and 1 ng/μL DY547-2′OMe-(CAGCAGCAGCAGCAGCAGC)) for 1.5 h at 37 °C. They were then washed with 30% formamide in 2X SSC for 30 min at 37 °C, 1X SSC for 30 min at room temperature, and 1X DPBS + 0.1% Triton X-100 for 5 min at room temperature. Finally, nuclei were stained by incubating the cells with 1 μg/mL DAPI for 5 min at room temperature. The cells were washed with 1X DPBS + 0.1% Triton X-100, and 100 μL of 1X DPBS were added to each well. The cells were imaged using an Olympus FluoView 1000 Confocal Microscope at 100X magnification.
Treatment in mice
All experimental procedures, mouse handling, and husbandry were completed in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care. A mouse model for DM1, HSALR in line 20b,22 was used to investigate if H1 improves splicing defects in animals. HSALR mice express human skeletal actin RNA with 250 CUG repeats in the 3′ UTR. Age- and gender-matched HSALR mice were injected intraperitoneally with 80 or 100 mg/kg H1 in 5% glucose or 5% glucose alone once per day for 7 days. Mice were sacrificed one day after the last injection, and the vastus muscle was obtained. RNA was extracted from the vastus tissue, and cDNA was synthesized as previously described.63 PCR amplification was carried out for 22–24 cycles with the following primer pairs: Clcn1 forward: 5′-TGAAGGAATACCTCACACTCAAGG and reverse: 5′-CACGGAACACAAAGGCACTG; Atp2a1 forward: 5′-GCTCATGGTCCTCAAGATCTCAC and reverse: 5′-GGGTCAGTGCCTCAGCTTTG. The PCR products were separated by agarose gel electrophoresis, and the gel was stained with SYBR Green I (Invitrogen). The gel was scanned with a laser fluorimager (Typhoon, GE Healthcare) and the products quantified using ImageQuant. Differences between two groups were evaluated by a t-test.
Supplementary Material
Acknowledgments
We dedicate this work to Douglas H. Turner on the occasion of his 65th birthday. We thank Sandro Matosevic for assistance with confocal microscopy, the URMC Center for RNA Biology, and the Rochester Wellstone Muscular Dystrophy Cooperative Research Center. This work was funded by the National Institutes of Health (3R01GM079235-02S1 and 1R01GM079235-01A2 to MDD; and AR049077 to CAT), and by The Scripps Research Institute. MDD is a Camille & Henry Dreyfus New Faculty Awardee, a Camille & Henry Dreyfus Teacher-Scholar, and a Research Corporation Cottrell Scholar.
ABBREVIATIONS
- ACE
2-O-[bis[2-(acetyloxy)ethoxy]methyl]
- A-site
aminoacyl tRNA site
- Atp2A1
Sarcoplasmic/endoplasmic reticulum calcium ATPase 1
- CAMKK2
Calcium/calmodulin-dependent protein kinase kinase 2
- Clcn1
chloride channel 1, skeletal muscle
- CUGBP1
CUG binding protein 1
- cTNT
cardiac troponin T
- DAPI
4′,6-diamidino-2-phenylindole
- DM1
myotonic dystrophy type 1
- DMPK
dystrophia myotonica protein kinase
- DPBS
Dulbecco’s Phosphate-Buffered Saline
- DPBS
Dulbecco’s Phosphate-Buffered Saline
- DTT
dithiothreitol
- FDA
Food and Drug Administration
- FISH
fluorescence in situ hybridization
- FRET
fluorescence resonance energy transfer
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- HSALR
human skeletal actin long repeat
- IR
insulin receptor
- MBNL1
muscleblind-like protein 1
- NCI
National Cancer Institute
- PLEKHH2
Pleckstrin-2
- qTR
quantitative time-resolved
- ROCS
Rapid Overlay of Chemical Structures
- RT-PCR
reverse transcription-polymerase chain reaction
- SDS
sodium dodecyl sulfate
- Serca1
Sarcoplasmic/endoplasmic reticulum calcium ATPase 1
- SSC
saline-sodium citrate
- TTC8
tetratricopeptide repeat domain 8
- UTR
untranslated region
Footnotes
Author Contributions: RP and JCL-D contributed equally to this work. All authors contributed to the work.
Supporting Information Available: Supporting Information including full citation for reference 14; a complete list of all compounds tested in the original qTR-FRET screen (Table S-3); the results of mass spectroscopy for all compounds identified from the initial qTR-FRET screen (Tables S-1 and S-2); HPLC retention times and analytical HPLC chromatograms for all compounds identified from the initial qTR-FRET screen (Tables S-1 and S-2 and Figures S-4 – S-20); details of the characterization and synthesis of H1 (Scheme S1, Figures S-1 – S-3); representative autoradiograms of splicing outcomes as determined by RT-PCR (Figures S-21 – S-24); representative binding curves (Figure S-25); assessment of toxicity using flow cytometry (Figure S-26). This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Guthrie C. Science. 1991;253:157. doi: 10.1126/science.1853200. [DOI] [PubMed] [Google Scholar]
- 2.Lee RC, Ambros V. Science. 2001;294:862. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 3.Lau NC, Lim LP, Weinstein EG, Bartel DP. Science. 2001;294:858. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 4.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Science. 2001;294:853. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 5.Chabot B. Trends Genet. 1996;12:472. doi: 10.1016/0168-9525(96)10037-8. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Croce CM. Oncogene. 2006;25:6202. doi: 10.1038/sj.onc.1209910. [DOI] [PubMed] [Google Scholar]
- 7.Caskey CT, Pizzuti A, Fu YH, Fenwick RG, Jr, Nelson DL, Kuhl DP. Science. 1992;256:784. doi: 10.1126/science.1589758. [DOI] [PubMed] [Google Scholar]
- 8.Faustino NA, Cooper TA. Genes Dev. 2003;17:419. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 9.Kalnina Z, Zayakin P, Silina K, Line A. Genes Chromosome Canc. 2005;42:342. doi: 10.1002/gcc.20156. [DOI] [PubMed] [Google Scholar]
- 10.Caskey CT, Pizzuti A, Fu YH, Fenwick RG, Jr, Nelson DL. Science. 1992;256:784. doi: 10.1126/science.1589758. [DOI] [PubMed] [Google Scholar]
- 11.Ranum LP, Cooper TA. Annu Rev Neurosci. 2006;29:259. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- 12.Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, Naylor SL, Day JW, Ranum LP. Science. 2001;293:864. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- 13.Fu YH, Pizzuti A, Fenwick RG, Jr, King J, Rajnarayan S, Dunne PW, Dubel J, Nasser GA, Ashizawa T, de Jong P, Wieringa B, Korneluk R, Perryman MB, Epstein HF, Caskey CT. Science. 1992;255:1256. doi: 10.1126/science.1546326. [DOI] [PubMed] [Google Scholar]
- 14.Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, Hunter K, Stanton VP, Thirion JP, Hudson T, et al. Cell. 1992;68:799. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 15.Mykowska A, Sobczak K, Wojciechowska M, Kozlowski P, Krzyzosiak WJ. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li LB, Yu Z, Teng X, Bonini NM. Nature. 2008;453:1107. doi: 10.1038/nature06909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michlewski G, Krzyzosiak WJ. J Mol Biol. 2004;340:665. doi: 10.1016/j.jmb.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 18.Sellier C, Rau F, Liu Y, Tassone F, Hukema RK, Gattoni R, Schneider A, Richard S, Willemsen R, Elliott DJ, Hagerman PJ, Charlet-Berguerand N. EMBO J. 2010;29:1248. doi: 10.1038/emboj.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS. Science. 2003;302:1978. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 20.Kanadia RN, Shin J, Yuan Y, Beattie SG, Wheeler TM, Thornton CA, Swanson MS. Proc Natl Acad Sci U S A. 2006;103:11748. doi: 10.1073/pnas.0604970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Philips AV, Timchenko LT, Cooper TA. Science. 1998;280:737. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 22.Mankodi A, Logigian E, Callahan L, McClain C, White R, Henderson D, Krym M, Thornton CA. Science. 2000;289:1769. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 23.Mankodi A, Takahashi MP, Jiang H, Beck CL, Bowers WJ, Moxley RT, Cannon SC, Thornton CA. Mol Cell. 2002;10:35. doi: 10.1016/s1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- 24.Paul S, Dansithong W, Kim D, Rossi J, Webster NJ, Comai L, Reddy S. EMBO J. 2006;25:4271. doi: 10.1038/sj.emboj.7601296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dansithong W, Paul S, Comai L, Reddy S. J Biol Chem. 2005;280:5773. doi: 10.1074/jbc.M410781200. [DOI] [PubMed] [Google Scholar]
- 26.Savkur RS, Philips AV, Cooper TA. Nat Genet. 2001;29:40. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 27.Hino S, Kondo S, Sekiya H, Saito A, Kanemoto S, Murakami T, Chihara K, Aoki Y, Nakamori M, Takahashi MP, Imaizumi K. Hum Mol Genet. 2007;16:2834. doi: 10.1093/hmg/ddm239. [DOI] [PubMed] [Google Scholar]
- 28.Kimura T, Nakamori M, Lueck JD, Pouliquin P, Aoike F, Fujimura H, Dirksen RT, Takahashi MP, Dulhunty AF, Sakoda S. Hum Mol Genet. 2005;14:2189. doi: 10.1093/hmg/ddi223. [DOI] [PubMed] [Google Scholar]
- 29.Timchenko NA, Cai ZJ, Welm AL, Reddy S, Ashizawa T, Timchenko LT. J Biol Chem. 2001;276:7820. doi: 10.1074/jbc.M005960200. [DOI] [PubMed] [Google Scholar]
- 30.Fardaei M, Rogers MT, Thorpe HM, Larkin K, Hamshere MG, Harper PS, Brook JD. Hum Mol Genet. 2002;11:805. doi: 10.1093/hmg/11.7.805. [DOI] [PubMed] [Google Scholar]
- 31.Ho TH, Savkur RS, Poulos MG, Mancini MA, Swanson MS, Cooper TA. J Cell Sci. 2005;118:2923. doi: 10.1242/jcs.02404. [DOI] [PubMed] [Google Scholar]
- 32.Jiang H, Mankodi A, Swanson MS, Moxley RT, Thornton CA. Hum Mol Genet. 2004;13:3079. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- 33.Mankodi A, Lin X, Blaxall BC, Swanson MS, Thornton CA. Circ Res. 2005;97:1152. doi: 10.1161/01.RES.0000193598.89753.e3. [DOI] [PubMed] [Google Scholar]
- 34.Mankodi A, Urbinati CR, Yuan QP, Moxley RT, Sansone V, Krym M, Henderson D, Schalling M, Swanson MS, Thornton CA. Hum Mol Genet. 2001;10:2165. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- 35.Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, Swanson MS. EMBO J. 2000;19:4439. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia-Lopez A, Llamusi B, Orzaez M, Perez-Paya E, Artero RD. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1018213108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mulders SA, van den Broek WJ, Wheeler TM, Croes HJ, van Kuik-Romeijn P, de Kimpe SJ, Furling D, Platenburg GJ, Gourdon G, Thornton CA, Wieringa B, Wansink DG. Proc Natl Acad Sci U S A. 2009;106:13915. doi: 10.1073/pnas.0905780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warf MB, Nakamori M, Matthys CM, Thornton CA, Berglund JA. Proc Natl Acad Sci U S A. 2009;106:18551. doi: 10.1073/pnas.0903234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wheeler TM, Sobczak K, Lueck JD, Osborne RJ, Lin X, Dirksen RT, Thornton CA. Science. 2009;325:336. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pushechnikov A, Lee MM, Childs-Disney JL, Sobczak K, French JM, Thornton CA, Disney MD. J Am Chem Soc. 2009;131:9767. doi: 10.1021/ja9020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee MM, Childs-Disney JL, Pushechnikov A, French JM, Sobczak K, Thornton CA, Disney MD. J Am Chem Soc. 2009;131:17464. doi: 10.1021/ja906877y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arambula JF, Ramisetty SR, Baranger AM, Zimmerman SC. Proc Natl Acad Sci U S A. 2009;106:16068. doi: 10.1073/pnas.0901824106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gareiss PC, Sobczak K, McNaughton BR, Palde PB, Thornton CA, Miller BL. J Am Chem Soc. 2008;130:16254. doi: 10.1021/ja804398y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho J, Hamasaki K, Rando RR. Biochemistry. 1998;37:4985. doi: 10.1021/bi972757h. [DOI] [PubMed] [Google Scholar]
- 45.Grant JA, Gallardo MA, Pickup BT. J Comput Chem. 1996;17:1653. [Google Scholar]
- 46.Bostrom J, Greenwood JR, Gottfries J. J Mol Graph Model. 2003;21:449. doi: 10.1016/s1093-3263(02)00204-8. [DOI] [PubMed] [Google Scholar]
- 47.Haigh JA, Pickup BT, Grant JA, Nicholls AJ. Chem Inf Model. 2005;45:673. doi: 10.1021/ci049651v. [DOI] [PubMed] [Google Scholar]
- 48.Mills JE, Dean PM. J Comput Aided Mol Des. 1996;10:607. doi: 10.1007/BF00134183. [DOI] [PubMed] [Google Scholar]
- 49.Bostrom J, Hogner A, Schmitt SJ. Med Chem. 2006;49:6716. doi: 10.1021/jm060167o. [DOI] [PubMed] [Google Scholar]
- 50.Boger DL, Fink BE, Brunette SR, Tse WC, Hedrick MP. J Am Chem Soc. 2001;123:5878. doi: 10.1021/ja010041a. [DOI] [PubMed] [Google Scholar]
- 51.Tse WC, Boger DL. Acc Chem Res. 2004;37:61. doi: 10.1021/ar030113y. [DOI] [PubMed] [Google Scholar]
- 52.Krishnamurthy M, Schirle NT, Beal PA. Bioorg Med Chem. 2008;16:8914. doi: 10.1016/j.bmc.2008.08.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Diop D, Chauvin C, Jean-Jean O. C R Biol. 2007;330:71. doi: 10.1016/j.crvi.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 54.Manuvakhova M, Keeling K, Bedwell DM. RNA. 2000;6:1044. doi: 10.1017/s1355838200000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nudelman I, Rebibo-Sabbah A, Cherniavsky M, Belakhov V, Hainrichson M, Chen F, Schacht J, Pilch DS, Ben-Yosef T, Baasov T. J Med Chem. 2009;52:2836. doi: 10.1021/jm801640k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berg JM, Tymoczko JL, Stryer L. Biochemistry. 6. W. H. Freeman and Company; New York: 2007. [Google Scholar]
- 57.Orengo JP, Bundman D, Cooper TA. Nucleic Acids Res. 2006;34:e148. doi: 10.1093/nar/gkl967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ho TH, Charlet BN, Poulos MG, Singh G, Swanson MS, Cooper TA. EMBO J. 2004;23:3103. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nezu Y, Kino Y, Sasagawa N, Nishino I, Ishiura S. Neuromuscul Disord. 2007;17:306. doi: 10.1016/j.nmd.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 60.Warf MB, Berglund JA. RNA. 2007;13:2238. doi: 10.1261/rna.610607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mankodi A, Teng-Umnuay P, Krym M, Henderson D, Swanson M, Thornton CA. Ann Neurol. 2003;54:760. doi: 10.1002/ana.10763. [DOI] [PubMed] [Google Scholar]
- 62.Disney MD, Childs-Disney JL. Chembiochem. 2007;8:649. doi: 10.1002/cbic.200600569. [DOI] [PubMed] [Google Scholar]
- 63.Lin X, Miller JW, Mankodi A, Kanadia RN, Yuan Y, Moxley RT, Swanson MS, Thornton CA. Hum Mol Genet. 2006;15:2087. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.