Abstract
Isocitrate dehydrogenase (IDH) mutations are frequent in blast-phase myeloproliferative neoplasms and might therefore contribute to leukemic transformation. We examined this possibility in 301 consecutive patients with chronic-phase primary myelofibrosis (PMF). The mutant IDH was detected in 12 patients (4%): 7 IDH2 (5 R140Q, 1 R140W and 1 R172G) and 5 IDH1 (3 R132S and 2 R132C). In all, 6 (50%) of the 12 IDH-mutated patients also expressed JAK2V617F. Overall, 18 (6%) patients displayed only MPL and 164 (54.3%) only JAK2 mutations. Multivariable analysis that accounted for conventional risk factors disclosed inferior overall survival (OS; P=0.03) and leukemia-free survival (LFS; P=0.003) in IDH-mutated patients: OS hazard ratio (HR) was 0.39 (95% confidence interval (95% CI) 0.2–0.75), 0.50 (95% CI 0.27–0.95) and 0.53 (95% CI 0.23–1.2) for patients with no, JAK2 or MPL mutations, respectively. Further analysis disclosed a more pronounced effect for the mutant IDH on OS and LFS in the presence (P=0.0002 and P<0.0001, respectively) as opposed to the absence (P=0.34 and P=0.64) of concomitant JAK2V617F. Analysis of paired samples obtained during chronic- and blast-phase disease revealed the presence of both IDH and JAK2 mutations at both time points. Our observations suggest that IDH mutations in PMF are independent predictors of leukemic transformation and raise the possibility of leukemogenic collaboration with JAK2V617F.
Keywords: IDH1, IDH2, myeloproliferative, prognosis, cytogenetics
Introduction
Among the three BCR-ABL1-negative myeloproliferative neoplasms (MPNs), including polycythemia vera, essential thrombocythemia and primary myelofibrosis (PMF), the latter is by far the worst in terms of both survival and quality of life.1, 2 The more aggressive disease biology in PMF is also manifest by the higher prevalence of cytogenetic abnormalities3 and somatic mutations.4 The latter involve JAK2, MPL, TET2, ASXL1, CBL, isocitrate dehydrogenase (IDH)1, IDH2, IKZF1, LNK, EZH2 and DNMT3A.4, 5 Recent studies have reported higher frequencies of IDH1/IDH2 and LNK mutations in blast-phase MPN,6, 7 suggesting a pathogenetic contribution to disease progression.
Isocitrate dehydrogenase-1 is located on chromosome 2q33.3 and IDH2 on chromosome 15q26.1. Both genes encode enzymes that catalyze oxidative decarboxylation of isocitrate to α-ketoglutarate. IDH mutations involve exon 4 and affect three specific arginine residues: R132 (IDH1), R172 (IDH2) and R140 (IDH2).8 The mutant IDH has decreased affinity for isocitrate but displays catalytic activity in converting α-ketoglutarate to 2-hydroxyglutarate.9, 10, 11, 12 Decreased supply of α-ketoglutarate or accumulation of 2-hydroxyglutarate is believed to underlie the oncogenic properties of the mutant IDH.9, 13
IDH mutations are prevalent in low-grade gliomas and secondary glioblastomas (mutational frequency ∼70%)14 and they have also been described, although at a much lower frequency, in myeloid malignancies including acute myeloid leukemia (AML; 10–20%),15, 16, 17, 18 myelodysplastic syndrome (MDS; 3–5%),19, 20 MPN (1–4%),8, 18 MDS/MPN including chronic myelomonocytic leukemia (∼9%),20 post-MDS AML (∼15%),19 post-MPN AML (∼22%),8 post-MDS/MPN AML (∼10%),20 del(5q)-associated high-risk MDS or AML (∼22%)21 and blast-phase chronic myelogenous leukemia (∼4%).22 Single case reports also included angioimmunoblastic lymphoma23 and acute lymphoblastic leukemia.18
Several studies have examined the phenotypic and prognostic effects of both IDH1 and IDH2 mutations in AML, and most have shown a consistent association with normal or intermediate-risk karyotype, sole trisomy 8 and NPM1 mutations.16, 17, 23, 24, 25, 26, 27 The mutant IDH1 was associated with worse prognosis in cytogenetically normal AML with NPM1+/FLT3− molecular profile17, 28, 29 and better prognosis in FLT3+ AML.27 In some17 but not other30 studies, the mutant IDH2 was associated with unfavorable prognosis in cytogenetically normal AML,17 whereas a more recent study suggested that the mutant IDH2R140 was prognostically more favorable than the mutant IDH2R172.31
Unlike the case with AML, there is limited information on the prognostic impact of IDH mutations in chronic myeloid neoplasms, including MDS19 and MPN.8 We recently reported on IDH1 and IDH2 mutational frequencies among 1473 patients with BCR-ABL1-negative MPN:8 0.8% in essential thrombocythemia, 1.9% in polycythemia vera, 4.1% in PMF, 1% in post-essential thrombocythemia/polycythemia vera MF, 0% in blast-phase essential thrombocythemia, 25% in blast-phase polycythemia vera and 25% in blast-phase PMF. The particular study included only 111 patients with chronic-phase PMF and complete clinical information; therefore, detailed prognostic analysis was limited, especially in terms of clinically relevant mutation interactions. In the current study, we examined the phenotypic and prognostic effects of IDH1 and IDH2 mutations among 301 patients with chronic-phase PMF, in the context of other MPN-associated mutations.
Materials and methods
This study was approved by the Mayo Clinic Institutional Review Board. All patients provided informed written consent for study sample collection and permission for use in research. Study eligibility criteria included availability of bone marrow histology and cytogenetic information at the time of referral to the Mayo Clinic. The diagnoses of PMF and leukemic transformation were according to the World Health Organization criteria.32 Patients with blast-phase disease at the time of their referral to the Mayo Clinic were excluded from the study because one of the objectives of the study was to assess mutation impact on leukemic transformation. Unfavorable karyotype designation and DIPSS-plus (Dynamic International Prognostic Scoring System-plus) risk categorization were as described previously.33, 34 All study patients were fully characterized for karyotype, JAK2 and MPL mutational status and DIPSS-plus risk category.
DNA from bone marrow or peripheral blood was extracted using conventional methods. MPL and JAK2 mutation analyses were performed according to previously published methods.35, 36, 37, 38 IDH1 and IDH2 mutations were analyzed by direct sequencing and/or high-resolution melting assay. Direct sequencing for IDH1 exon 4 mutations was performed using the following primer sequences: sense, 5′-CGGTCTTCAGAGAAGCCATT-3′ and anti-sense, 5′-CACATTATTGCCAACATGAC-3′.18 IDH2 exon 4 was amplified using sense, 5′-CCACTATTATCTCTGTCCTC-3′ and anti-sense, 5′-GCTAGGCGAGGAGCTCCAGT-3′.19 Both reactions were performed in 25 μl volume containing 100 ng of DNA, 0.25 Units Taq polymerase, 0.3 m each of dATP, dCTP, dGTP and dTTP, 5 μl of a 10 × PCR Buffer (Roche Diagnostics, Indianapolis, IN, USA) and 0.2 μ each of sense and anti-sense primers. The reaction was denatured at 94 °C for 3 min, followed by 35 cycles of denaturing at 94 °C for 30 s, annealing at 57 °C for 30 s and extension at 72 °C for 40 s. After a final extension at 72 °C for 2 min, the products were confirmed by 1.3% agarose gel and purified using Qiagen's PCR quick purification kit (Qiagen, Santa Clarita, CA, USA). The product was sequenced using the ABI PRISM 3730xl analyzer (Applied Biosystems Inc., Foster City, CA, USA) to screen for the presence of mutations.
High-resolution melting was performed using the LightCycler 480 Real-Time PCR system (Roche Diagnostics), using the above-mentioned primers for IDH1 mutations (R130) and the following primers for IDH2 mutations (R140 and R172): R140 sense, 5′-GCTGAAGAAGATGTGGAA-3′ and anti-sense, 5′-TGATGGGCTCCCGGAAGA-3′ R172 sense, 5′-CCAAGCCCATCACCATTG-3′ and anti-sense, 5′-CCCAGGTCAGTGGATCCC-3′.
All statistical analyses considered clinical and laboratory parameters obtained at the time of first referral to the Mayo Clinic, which in most instances coincided with the time of bone marrow biopsy at the Mayo Clinic and study sample collection. Differences in the distribution of continuous variables between categories were analyzed by either the Mann–Whitney (for comparison of two groups) or the Kruskal–Wallis (comparison of three or more groups) test. Patient groups with nominal variables were compared by the χ2-test. Overall survival (OS) was calculated from the date of first referral to the date of death (uncensored) or last contact (censored). Leukemia-free survival (LFS) was calculated from the date of first referral to the date of leukemic transformation (uncensored) or death/last contact (censored). OS and LFS curves were prepared by the Kaplan–Meier method and compared by the log-rank test. Cox's proportional hazard regression model was used for multivariable analysis. P-values <l0.05 were considered significant. The Stat View (SAS Institute, Cary, NC, USA) statistical package was used for all calculations.
Results
A total of 301 consecutive patients with PMF were included in this study. The median age at the time of study was 63 years (range, 14–82) and 65% were males. DIPSS-plus risk distribution was 11% low, 16% intermediate-1, 36% intermediate-2 and 37% high. Other clinical and laboratory characteristics at the time of Mayo Clinic referral are outlined in Table 1; 40 (13%) patients had received cytoreductive therapy at the time of their first referral at our institution. The study population included 178 patients who were evaluated at or near the time of their diagnosis and their presenting characteristics are separately outlined in Table 2.
Table 1. Comparison of clinical characteristics of patients with primary myelofibrosis stratified by the presence or absence of IDH, MPL and JAK2 mutations.
| Variables | All patients (n=301) | IDH mutated (n=12) | MPL mutated (n=18) | JAK2 mutated (n=164) | IDH/MPL/JAK2 unmutated (n=107) | P-value |
|---|---|---|---|---|---|---|
| Age (years); median (range) | 63 (14–82) | 66 (50–74) | 62 (35–82) | 65 (28–81) | 58 (14–79) | 0.0002 |
| Age >65 years; n (%) | 96 (32%) | 6 (50%) | 6 (33%) | 77 (47%) | 30 (28%) | 0.02 |
| Males (%) | 197 (65%) | 7 (58%) | 11 (61%) | 105 (64%) | 74 (69%) | 0.75 |
| Hemoglobin, g/dl; median (range) | 10 (6–15) | 11 (7–15) | 10 (6–14) | 10 (7–15) | 10 (6–14) | 0.97 |
| Leukocyte count, × 109/l; median (range) | 9 (1–176) | 9 (4–48) | 11 (4–50) | 10 (1–176) | 7 (1–147) | 0.11 |
| Platelet count, × 109/l; median (range) | 222 (11–1493) | 141 (66–410) | 128 (14–662) | 218 (11–984) | 257 (13–1493) | 0.06 |
| DIPSS-plus risk group (%) | ||||||
| Low | 34 (11%) | 0 | 4 (22%) | 18 (11%) | 12 (11%) | |
| Intermediate-1 | 47 (16%) | 2 (16%) | 2 (11%) | 19 (12%) | 24 (22%) | |
| Intermediate-2 | 109 (36%) | 5 (42%) | 3 (17%) | 60 (37%) | 41 (38%) | 0.11 |
| High | 111 (37%) | 5 (42%) | 9 (50%) | 67 (41%) | 30 (28%) | |
| Constitutional symptoms; n (%) | 115 (38%) | 5 (42%) | 4 (22%) | 69 (42%) | 37 (35%) | 0.30 |
| Circulating blasts ⩾1% n (%) | 190 (63%) | 10 (83%) | 11 (61%) | 101 (62%) | 68 (64%) | 0.50 |
| Hemoglobin <10 g/dl; n (%) | 151(50%) | 5 (42%) | 10 (56%) | 88 (54%) | 48 (45%) | 0.46 |
| Leukocytes >25 × 109/l; n (%) | 52 (17%) | 2 (17%) | 4 (22%) | 33 (20%) | 13 (12%) | 0.36 |
| Platelets <100 × 109/l; n (%) | 73 (24%) | 3 (25%) | 8 (44%) | 35 (21%) | 27 (25%) | 0.19 |
| Leukocytes <4 × 109/l; n (%) | 46 (15%) | 1 (8%) | 1 (6%) | 24 (15%) | 20 (19%) | 0.43 |
| Palpable spleen >10 cm; n (%) | 109 (36%) | 2 (17%) | 5 (28%) | 67 (41%) | 35 (33%) | 0.20 |
| Splenectomy; n (%) | 53 (18%) | 2 (17%) | 4 (22%) | 28 (17%) | 19 (18%) | 0.96 |
| Cytogenetic categories | ||||||
| Normal | 181 (60%) | 9 (75%) | 12 (67%) | 92 (56%) | 68 (64%) | |
| Favorable | 89 (30%) | 2 (17%) | 6 (33%) | 53 (32%) | 28 (26%) | 0.55 |
| Unfavorable | 31 (10%) | 1 (8%) | 0 (0%) | 19 (12%) | 11 (10%) | |
| Transplanted; n (%) | 24 (8%) | 0 (0%) | 2 (11%) | 10 (6%) | 13 (12%) | 0.31 |
| Deaths; n (%) | 192 (64%) | 11 (92%) | 12 (67%) | 109 (66%) | 58 (54%) | 0.04 |
| Leukemic transformations; n (%) | 36 (12%) | 5 (42%) | 1 (6%) | 17 (11%) | 13 (12%) | 0.01 |
Abbreviation: DIPSS-plus, Dynamic International Prognostic Scoring System-Plus.
Bold values indicate significant differences.
Table 2. Comparison of clinical characteristics of patients with primary myelofibrosis, who were evaluated within 1 year of diagnosis, stratified by the presence or absence of IDH, MPL and JAK2 mutations.
| Variables | All patients (n=178) | IDH mutated (n=10) | MPL mutated (n=9) | JAK2 mutated (n=96) | IDH/MPL/JAK2 un-mutated (n=63) | P-value |
|---|---|---|---|---|---|---|
| Age (years); median (range) | 63 (14–81) | 66 (50–74) | 60 (35–66) | 65 (28–81) | 58 (14–79) | 0.005 |
| Age >65 years; n (%) | 70 (39%) | 5 (50%) | 1 (11%) | 48 (50%) | 16 (25%) | 0.004 |
| Males (%) | 116 (65%) | 7 (70%) | 7 (80%) | 63 (66%) | 39 (62%) | 0.79 |
| Hemoglobin, g/dl; median (range) | 10 (6–15) | 11 (7–15) | 10 (6–14) | 10 (7–15) | 11 (6–14) | 0.50 |
| Leukocyte count, × 109/l; median (range) | 8 (1–147) | 10 (4–48) | 11 (4–50) | 9 (1–99) | 7 (2–147) | 0.15 |
| Platelet count, × 109/l; median (range) | 253 (12–1493) | 159 (79–410) | 146 (31–662) | 245 (12–984) | 316 (14–1493) | 0.13 |
| DIPSS-plus risk group (%) | ||||||
| Low | 25 (14%) | 0 | 2 (22%) | 13 (14%) | 10 (16%) | 0.60 |
| Intermediate-1 | 36 (20%) | 2 (20%) | 1 (11%) | 15 (16%) | 18 (29%) | |
| Intermediate-2 | 62 (35%) | 4 (40%) | 3 (33%) | 35 (36%) | 20 (32%) | |
| High | 55(31%) | 4 (40%) | 3 (33%) | 33 (34%) | 15 (24%) | |
| Constitutional symptoms; n (%) | 65 (37%) | 4 (40%) | 2 (22%) | 38 (40%) | 21 (33%) | 0.68 |
| Circulating blasts ⩾1% n (%) | 101 (57%) | 8 (80%) | 5 (56%) | 54 (56%) | 34(53%) | 0.49 |
| Hemoglobin <10 g/dl; n (%) | 77 (43%) | 4 (40%) | 6 (67%) | 47 (49%) | 20 (32%) | 0.08 |
| Leukocytes >25 × 109/l; n (%) | 26 (15%) | 2 (20%) | 2 (22%) | 14 (15%) | 8 (13%) | 0.84 |
| Platelets <100 × 109/l; n (%) | 36 (20%) | 1 (10%) | 4 (44%) | 17 (18%) | 14 (22%) | 0.22 |
| Leukocytes <4 × 109/l; n (%) | 27 (15%) | 1 (10%) | 1 (11%) | 12 (13%) | 13 (21%) | 0.51 |
| Palpable spleen >10 cm; n (%) | 44 (25%) | 2 (20%) | 2 (22%) | 28 (29%) | 12 (19%) | 0.45 |
| Splenectomy; n (%) | 24 (13%) | 1 (10%) | 3 (33%) | 12 (13%) | 8 (13%) | 0.35 |
| Cytogenetic categories | ||||||
| Normal | 116 (65%) | 7 (70%) | 6 (67%) | 58 (60%) | 45 (71%) | 0.75 |
| Favorable | 48 (27%) | 2 (20%) | 3 (33%) | 30 (31%) | 13 (21%) | |
| Unfavorable | 14 (8%) | 1 (10%) | 0 (0%) | 8 (8%) | 5 (8%) | |
| Transplanted; n (%) | 15 (8%) | 0 (0%) | 2 (22%) | 5 (5%) | 8 (13%) | 0.12 |
| Deaths; n (%) | 107 (60%) | 9 (90%) | 6 (67%) | 59 (61%) | 32 (51%) | 0.09 |
| Leukemic transformations; n (%) | 22 (12%) | 4 (40%) | 1 (11%) | 13 (14%) | 4 (6%) | 0.03 |
Abbreviation: DIPSS-plus, Dynamic International Prognostic Scoring System-Plus.
Bold values indicate significant differences.
The mutant IDH was detected in 12 patients (4%): 7 IDH2 (5 R140Q, 1 R140W and 1 R172G) and 5 IDH1 (3 R132S and 2 R132C). MPL exon 10 was mutated in 18 patients (6.3%) and constituted W515L in 14 patients, W515K in 3 and a novel frameshift mutation in 1 patient. JAK2V617F was detected in 169 (56%) patients. Six patients displayed both JAK2V617F and IDH mutations (IDH2R140Q in two patients, IDH2R140W in one and IDH1R132S in three); JAK2V617F allele burdens in these six patients with concomitant mutant IDH were 1, 7, 22, 27, 30 and 96%, respectively. One patient displayed both IDHR140Q and MPLW515R. In all, 107 (36%) patients were negative for all three mutations (that is, JAK2V617F, MPL exon 10 and IDH1/2).
The 12 IDH-mutated patients, with or without concomitant JAK2V617F, were clinically compared with patients belonging to the 3 other molecular subgroups: mutated for JAK2 only (n=164), mutated for MPL only (n=18) and unmutated for all three (n=107). As can be seen in Tables 1 and 2, the four molecular subgroups were remarkably similar in their phenotype with few exceptions; IDH-mutated patients were significantly older than those with no mutations (P=0.04), whereas age distribution was similar between patients with mutant IDH, MPL or JAK2. At the time of this writing, 192 (64%) deaths and 36 (12%) leukemic transformations were documented. The median follow-up time for living patients was 68 months (range 12–296). Treatment over the course of the disease was primarily with conventional drugs, and a total of 53 therapeutic splenectomies and 24 transplants were documented.
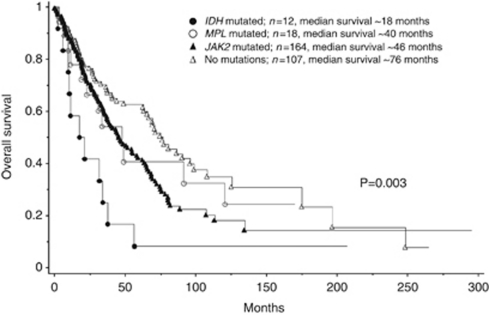
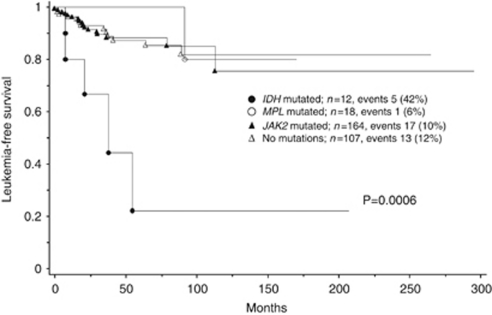
In univariate analysis, IDH-mutated patients lived shorter than did those with JAK2 (P=0.03), MPL (P=0.047) or no mutations (P=0.0009). The OS data for the four molecular subgroups are shown in Figure 1. IDH-mutated patients also showed significantly shorter LFS, compared with those with JAK2 (P=0.0008), MPL (P=0.02) or no mutations (P=0.001), as shown in Figure 2. LFS was similar between patients with no mutations and those with either MPL (P=0.47) or JAK2 (P=0.99). The OS of patients with no mutations was significantly longer than those with JAK2V617F (P=0.01), but not than those with MPL mutations (P=0.41). After accounting for age, the OS difference between patients with JAK2V617F and no mutations became insignificant (P=0.40), whereas the presence of the mutant IDH remained a significant disadvantage for both OS (P=0.04) and LFS (P=0.005).
Figure 1.
Overall survival data for 301 patients with primary myelofibrosis stratified by the presence or absence of IDH, MPL and JAK2 mutations.
Figure 2.
Leukemia-free survival data for 301 patients with primary myelofibrosis stratified by the presence or absence of IDH, MPL and JAK2 mutations.
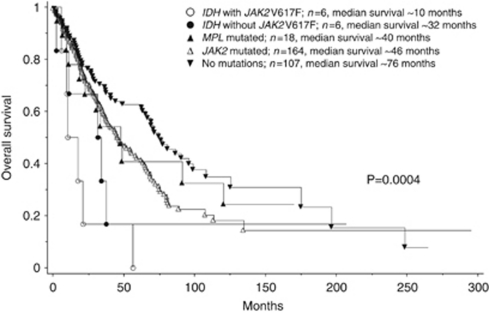
Multivariable analysis of OS that included risk categorization per DIPSS-plus33 confirmed the independent prognostic relevance of the mutant IDH (P=0.03): hazard ratio (HR) for patients with no mutations=0.39, 95% confidence interval (95% CI) 0.2–0.75; HR for JAK2-mutated patients=0.50, 95% CI, 0.27–0.95; HR for MPL-mutated patients=0.53, 95% CI, 0.23–1.2. A similar analysis for LFS that included risk factors for leukemic transformation (that is, unfavorable karyotype and platelet count <100 × 109/l) as covariates also confirmed the prognostic relevance of the mutant IDH (P=0.003): HR for patients with no mutations=0.16; 95% CI, 0.06–0.46; HR for JAK2-mutated patients=0.18; 95% CI, 0.06–0.48; HR for MPL-mutated patients=0.09; 95% CI, 0.01–0.76).33 Further analysis disclosed that the negative OS (Figure 3) and LFS (Figure 4) effect of the mutant IDH was most pronounced in the presence (P=0.0002 and P<0.0001, respectively) as opposed to the absence (P=0.34 and P=0.64, respectively) of concomitant JAK2V617F expression. Analysis of paired samples obtained during the chronic and blast phases of the disease was possible in two IDH-mutated patients and showed the presence of both IDH and JAK2 mutations at both time points.
Figure 3.
Overall survival data for 301 patients with primary myelofibrosis stratified by the presence or absence of MPL, JAK2 or IDH mutations with or without concomitant JAK2V617F expression.
Figure 4.
Leukemia-free survival data for 301 patients with primary myelofibrosis stratified by the presence or absence of MPL, JAK2 or IDH mutations with or without concomitant JAK2V617F expression.
Discussion
The most feared disease complication in MPN is leukemic transformation.39 In PMF, risk factors for leukemic progression include unfavorable karyotype,3 thrombocytopenia (platelet count <100 × 109/l) and ⩾3% circulating blasts;33, 40 the 10-year incidence of AML was estimated at 12% in the absence of unfavorable karyotype and thrombocytopenia and 31% in the presence of either one of the two risk factors.33 Prognosis in post-PMF AML is dismal with a median survival of <3 months and is not favorably affected by conventional chemotherapy.39 The discovery of JAK2V617F in the majority of patients with PMF raised hopes of better outcome with effective molecularly targeted therapy.41 However, it has since been realized that the presence or absence of JAK2V617F in PMF did not affect leukemic transformation35 and that leukemic blasts in JAK2-mutated patients who develop AML did not necessarily express the mutation.42 These observations suggest that JAK2V617F is neither necessary nor sufficient for leukemic progression in PMF.
This study suggests that the presence of the mutant IDH signifies an increased risk of leukemic transformation in PMF and also raises the intriguing possibility of leukemogenic collaboration between the mutant IDH and JAK2V617F; 4 (67%) of 6 patients with concomitant IDH and JAK2 mutations developed AML as opposed to only 1 (17%) of 6 IDH-mutated patients without concomitant JAK2V617F, 1 (6%) of 18 with MPL mutations, 17 (10%) of 164 with only JAK2 mutation and 13 (12%) of 107 patients with no mutations (P<0.0001; Figure 4). A similar clinical observation was made in a recent report that showed an inferior LFS in IDH-mutated myeloid malignancies with isolated del(5q);43 in the particular study, two of six patients with IDH mutations also carried JAK2V617F and both had transformed into AML, whereas only two of the remaining four IDH-mutated patients without concomitant JAK2V617F had transformed into AML.43 The possibility that the mutant IDH collaborates with other oncogenes is further supported by a recent report in which the mutant IDH enhanced growth and mitogen-activated protein kinase and signal transducer and activator of transcription-3 signaling in BRAF-mutated melanoma cells.44
Currently known mutations in PMF are believed to represent late genetic events derived from an ancestral abnormal clone the genetic make up of which remains elusive. The fact that many of these mutations are infrequent and lack disease specificity further undermines their pathogenetic contribution to disease initiation.4 On the other hand, the absence of mutual exclusivity and the higher prevalence of some MPN-associated mutations (for example, IDH,6 LNK,7 IKZF145 and TP5346 mutations) in blast-phase, as opposed to chronic-phase, disease suggests possible pathogenetic contribution to leukemic transformation. The observations from this study suggest one possibility in which mutations with non-redundant functional consequences collaborate to amplify the development of AML. Alternatively, the presence of mutations of interest (such as the mutant IDH) in at-risk patients might simply constitute a marker of genomic instability associated with impending leukemic transformation. A third possibility considers the distribution of specific mutations in independent clones that arise from a common ancestral clone that is susceptible to both emergence of mutations of interest and leukemic transformation.47
The prognostic impact of IDH mutations in AML has been studied extensively.16, 17, 23, 24, 25, 26, 27, 28, 29, 30 In contrast, very few studies have looked into this matter in chronic myeloid malignancies. Both IDH1 and IDH2 mutations occur in MDS, although some studies19 have reported a preponderance of IDH1 mutations, whereas others have shown the opposite.20 In the current PMF study, 7 of the 12 IDH mutations involved IDH2. In MDS and other myeloid neoplasms associated with sole del(5q), the presence of mutant IDH has been associated with inferior OS and LFS.19, 21, 43 It is noteworthy that the MDS study showing a detrimental prognostic effect of the mutant IDH involved only IDH1 mutations.19 In this study, there was no evidence to suggest that IDH1 and IDH2 mutations were prognostically different (data not shown). Regardless, the number of cases with IDH mutations in this study (n=12) was too small to accurately determine the individual prognostic contribution of IDH1 vs IDH2 mutations in PMF.
JAK2V617F in PMF and other MPN is associated with advanced age.48 This study suggests that IDH mutations in PMF also cluster with older age. A similar observation has been made in AML as well28 and underscores the importance of accounting for age in evaluating the prognostic significance of IDH mutations. Another characteristic feature of IDH-mutated PMF in this study was the relative paucity of abnormal or unfavorable karyotype. This particular observation has also been noted in the context of AML, MDS, MDS/MPN and post-MDS/MPN AML8, 18, 20 and suggests that IDH mutations are not simply markers of genomic instability.
Our clinical observations underscore the potential relevance of looking for other mutations or epigenetic abnormalities that functionally mimic the mutant IDH, in JAK2-mutated PMF.13 It would also be interesting to examine the mutant IDH-induced phenotypic modifications of JAK2V617F mouse models. Whether therapeutic targeting of the mutant IDH or interfering with the production/function of its ‘oncogenic' metabolite (that is, 2-hydroxyglutarate) would favorably affect leukemic progression in PMF remains to be seen.
Acknowledgments
This study is supported in part by grants from the ‘Myeloproliferative Disorders Foundation, Chicago, IL, USA' and ‘The Henry J Predolin Foundation for Research in Leukemia, Mayo Clinic, Rochester, MN, USA'.
The authors declare no conflict of interest.
References
- Tefferi A, Vainchenker W. Myeloproliferative neoplasms: molecular pathophysiology, essential clinical understanding, and treatment strategies. J Clin Oncol. 2011;29:573–582. doi: 10.1200/JCO.2010.29.8711. [DOI] [PubMed] [Google Scholar]
- Mesa RA, Niblack J, Wadleigh M, Verstovsek S, Camoriano J, Barnes S, et al. The burden of fatigue and quality of life in myeloproliferative disorders (MPDs): an international Internet-based survey of 1179 MPD patients. Cancer. 2007;109:68–76. doi: 10.1002/cncr.22365. [DOI] [PubMed] [Google Scholar]
- Caramazza D, Begna KH, Gangat N, Vaidya R, Siragusa S, Van Dyke DL, et al. Refined cytogenetic-risk categorization for overall and leukemia-free survival in primary myelofibrosis: a single center study of 433 patients. Leukemia. 2011;25:82–88. doi: 10.1038/leu.2010.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefferi A. Novel mutations and their functional and clinical relevance in myeloproliferative neoplasms: JAK2, MPL, TET2, ASXL1, CBL, IDH and IKZF1. Leukemia. 2010;24:1128–1138. doi: 10.1038/leu.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Wahab O, Pardanani A, Rampal R, Lasho TL, Levine RL, Tefferi A. DNMT3A mutational analysis in primary myelofibrosis, chronic myelomonocytic leukemia and advanced phases of myeloproliferative neoplasms. Leukemia. 2011;25:1219–1220. doi: 10.1038/leu.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardanani A, Lasho TL, Finke CM, Mai M, McClure RF, Tefferi A. IDH1 and IDH2 mutation analysis in chronic- and blast-phase myeloproliferative neoplasms. Leukemia. 2010;24:1146–1151. doi: 10.1038/leu.2010.77. [DOI] [PubMed] [Google Scholar]
- Pardanani A, Lasho T, Finke C, Oh ST, Gotlib J, Tefferi A. LNK mutation studies in blast-phase myeloproliferative neoplasms, and in chronic-phase disease with TET2, IDH, JAK2 or MPL mutations. Leukemia. 2010;24:1713–1718. doi: 10.1038/leu.2010.163. [DOI] [PubMed] [Google Scholar]
- Tefferi A, Lasho TL, Abdel-Wahab O, Guglielmelli P, Patel J, Caramazza D, et al. IDH1 and IDH2 mutation studies in 1473 patients with chronic-, fibrotic- or blast-phase essential thrombocythemia, polycythemia vera or myelofibrosis. Leukemia. 2010;24:1302–1309. doi: 10.1038/leu.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324:261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med. 2010;207:339–344. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou WC, Lei WC, Ko BS, Hou HA, Chen CY, Tang JL, et al. The prognostic impact and stability of isocitrate dehydrogenase 2 mutation in adult patients with acute myeloid leukemia. Leukemia. 2011;25:246–253. doi: 10.1038/leu.2010.267. [DOI] [PubMed] [Google Scholar]
- Boissel N, Nibourel O, Renneville A, Gardin C, Reman O, Contentin N, et al. Prognostic impact of isocitrate dehydrogenase enzyme isoforms 1 and 2 mutations in acute myeloid leukemia: a study by the Acute Leukemia French Association group. J Clin Oncol. 2010;28:3717–3723. doi: 10.1200/JCO.2010.28.2285. [DOI] [PubMed] [Google Scholar]
- Abbas S, Lugthart S, Kavelaars FG, Schelen A, Koenders JE, Zeilemaker A, et al. Acquired mutations in the genes encoding IDH1 and IDH2 both are recurrent aberrations in acute myeloid leukemia: prevalence and prognostic value. Blood. 2010;116:2122–2126. doi: 10.1182/blood-2009-11-250878. [DOI] [PubMed] [Google Scholar]
- Thol F, Weissinger EM, Krauter J, Wagner K, Damm F, Wichmann M, et al. IDH1 mutations in patients with myelodysplastic syndromes are associated with an unfavorable prognosis. Haematologica. 2010;95:1668–1674. doi: 10.3324/haematol.2010.025494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmider O, Gelsi-Boyer V, Slama L, Dreyfus F, Beyne-Rauzy O, Quesnel B, et al. Mutations of IDH1 and IDH2 genes in early and accelerated phases of myelodysplastic syndromes and MDS/myeloproliferative neoplasms. Leukemia. 2010;24:1094–1096. doi: 10.1038/leu.2010.52. [DOI] [PubMed] [Google Scholar]
- Pardanani A, Patnaik MM, Lasho TL, Mai M, Knudson RA, Finke C, et al. Recurrent IDH mutations in high-risk myelodysplastic syndrome or acute myeloid leukemia with isolated del(5q) Leukemia. 2010;24:1370–1372. doi: 10.1038/leu.2010.98. [DOI] [PubMed] [Google Scholar]
- Soverini S, Score J, Iacobucci I, Poerio A, Lonetti A, Gnani A, et al. IDH2 somatic mutations in chronic myeloid leukemia patients in blast crisis. Leukemia. 2011;25:178–181. doi: 10.1038/leu.2010.236. [DOI] [PubMed] [Google Scholar]
- Caramazza D, Lasho TL, Finke CM, Gangat N, Dingli D, Knudson RA, et al. IDH mutations and trisomy 8 in myelodysplastic syndromes and acute myeloid leukemia. Leukemia. 2010;24:2120–2122. doi: 10.1038/leu.2010.213. [DOI] [PubMed] [Google Scholar]
- Thol F, Damm F, Wagner K, Gohring G, Schlegelberger B, Hoelzer D, et al. Prognostic impact of IDH2 mutations in cytogenetically normal acute myeloid leukemia. Blood. 2010;116:614–616. doi: 10.1182/blood-2010-03-272146. [DOI] [PubMed] [Google Scholar]
- Chou WC, Lei WC, Ko BS, Hou HA, Chen CY, Tang JL, et al. The prognostic impact and stability of isocitrate dehydrogenase 2 mutation in adult patients with acute myeloid leukemia. Leukemia. 2011;25:246–253. doi: 10.1038/leu.2010.267. [DOI] [PubMed] [Google Scholar]
- Schnittger S, Haferlach C, Ulke M, Alpermann T, Kern W, Haferlach T. IDH1 mutations are detected in 6.6% of 1414 AML patients and are associated with intermediate risk karyotype and unfavorable prognosis in adults younger than 60 years and unmutated NPM1 status. Blood. 2010;116:5486–5496. doi: 10.1182/blood-2010-02-267955. [DOI] [PubMed] [Google Scholar]
- Green CL, Evans CM, Hills RK, Burnett AK, Linch DC, Gale RE. The prognostic significance of IDH1 mutations in younger adult patients with acute myeloid leukemia is dependent on FLT3/ITD status. Blood. 2010;116:2779–2782. doi: 10.1182/blood-2010-02-270926. [DOI] [PubMed] [Google Scholar]
- Paschka P, Schlenk RF, Gaidzik VI, Habdank M, Kronke J, Bullinger L, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28:3636–3643. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- Marcucci G, Maharry K, Wu YZ, Radmacher MD, Mrozek K, Margeson D, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:2348–2355. doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner K, Damm F, Gohring G, Gorlich K, Heuser M, Schafer I, et al. Impact of IDH1 R132 mutations and an IDH1 single nucleotide polymorphism in cytogenetically normal acute myeloid leukemia: SNP rs11554137 is an adverse prognostic factor. J Clin Oncol. 2010;28:2356–2364. doi: 10.1200/JCO.2009.27.6899. [DOI] [PubMed] [Google Scholar]
- Green CL, Evans CM, Zhao L, Hills RK, Burnett AK, Linch DC, et al. The prognostic significance of IDH2 mutations depends on the location of the mutation. Blood. 2011;118:409–412. doi: 10.1182/blood-2010-12-322479. [DOI] [PubMed] [Google Scholar]
- Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114:937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- Gangat N, Caramazza D, Vaidya R, George G, Begna K, Schwager S, et al. DIPSS plus: a refined Dynamic International Prognostic Scoring System for primary myelofibrosis that incorporates prognostic information from karyotype, platelet count, and transfusion status. J Clin Oncol. 2011;29:392–397. doi: 10.1200/JCO.2010.32.2446. [DOI] [PubMed] [Google Scholar]
- Passamonti F, Cervantes F, Vannucchi AM, Morra E, Rumi E, Pereira A, et al. A dynamic prognostic model to predict survival in primary myelofibrosis: a study by the IWG-MRT (International Working Group for Myeloproliferative Neoplasms Research and Treatment) Blood. 2010;115:1703–1708. doi: 10.1182/blood-2009-09-245837. [DOI] [PubMed] [Google Scholar]
- Tefferi A, Lasho TL, Huang J, Finke C, Mesa RA, Li CY, et al. Low JAK2V617F allele burden in primary myelofibrosis, compared to either a higher allele burden or unmutated status, is associated with inferior overall and leukemia-free survival. Leukemia. 2008;22:756–761. doi: 10.1038/sj.leu.2405097. [DOI] [PubMed] [Google Scholar]
- Pardanani AD, Levine RL, Lasho T, Pikman Y, Mesa RA, Wadleigh M, et al. MPL515 mutations in myeloproliferative and other myeloid disorders: a study of 1182 patients. Blood. 2006;108:3472–3476. doi: 10.1182/blood-2006-04-018879. [DOI] [PubMed] [Google Scholar]
- Tefferi A, Levine RL, Lim KH, Abdel-Wahab O, Lasho TL, Patel J, et al. Frequent TET2 mutations in systemic mastocytosis: clinical, KITD816V and FIP1L1-PDGFRA correlates. Leukemia. 2009;23:900–904. doi: 10.1038/leu.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefferi A, Lasho TL, Patnaik MM, Finke CM, Hussein K, Hogan WJ, et al. JAK2 germline genetic variation affects disease susceptibility in primary myelofibrosis regardless of V617F mutational status: nullizygosity for the JAK2 46/1 haplotype is associated with inferior survival. Leukemia. 2010;24:105–109. doi: 10.1038/leu.2009.225. [DOI] [PubMed] [Google Scholar]
- Mesa RA, Li CY, Ketterling RP, Schroeder GS, Knudson RA, Tefferi A. Leukemic transformation in myelofibrosis with myeloid metaplasia: a single-institution experience with 91 cases. Blood. 2005;105:973–977. doi: 10.1182/blood-2004-07-2864. [DOI] [PubMed] [Google Scholar]
- Huang J, Li CY, Mesa RA, Wu W, Hanson CA, Pardanani A, et al. Risk factors for leukemic transformation in patients with primary myelofibrosis. Cancer. 2008;112:2726–2732. doi: 10.1002/cncr.23505. [DOI] [PubMed] [Google Scholar]
- James C, Ugo V, Le Couedic JP, Staerk J, Delhommeau F, Lacout C, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- Theocharides A, Boissinot M, Girodon F, Garand R, Teo SS, Lippert E, et al. Leukemic blasts in transformed JAK2-V617F-positive myeloproliferative disorders are frequently negative for the JAK2-V617F mutation. Blood. 2007;110:375–379. doi: 10.1182/blood-2006-12-062125. [DOI] [PubMed] [Google Scholar]
- Patnaik MM, Lasho TL, Finke CM, Knudson RA, Ketterling RP, Chen D, et al. Isolated del(5q) in myeloid malignancies: clinicopathologic and molecular features in 143 consecutive patients. Am J Hematol. 2011;86:393–398. doi: 10.1002/ajh.21984. [DOI] [PubMed] [Google Scholar]
- Shibata T, Kokubu A, Miyamoto M, Sasajima Y, Yamazaki N. Mutant IDH1 confers an in vivo growth in a melanoma cell line with BRAF mutation. Am J Pathol. 2011;178:1395–1402. doi: 10.1016/j.ajpath.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager R, Gisslinger H, Passamonti F, Rumi E, Berg T, Gisslinger B, et al. Deletions of the transcription factor Ikaros in myeloproliferative neoplasms. Leukemia. 2010;24:1290–1298. doi: 10.1038/leu.2010.99. [DOI] [PubMed] [Google Scholar]
- Harutyunyan A, Klampfl T, Cazzola M, Kralovics R. p53 lesions in leukemic transformation. N Engl J Med. 2011;364:488–490. doi: 10.1056/NEJMc1012718. [DOI] [PubMed] [Google Scholar]
- Green A, Beer P. Somatic mutations of IDH1 and IDH2 in the leukemic transformation of myeloproliferative neoplasms. N Engl J Med. 2010;362:369–370. doi: 10.1056/NEJMc0910063. [DOI] [PubMed] [Google Scholar]
- Vannucchi AM, Antonioli E, Guglielmelli P, Pardanani A, Tefferi A. Clinical correlates of JAK2V617F presence or allele burden in myeloproliferative neoplasms: a critical reappraisal. Leukemia. 2008;22:1299–1307. doi: 10.1038/leu.2008.113. [DOI] [PubMed] [Google Scholar]