Abstract
We describe a conditional in vivo protein trap mutagenesis system that reveals spatio-temporal protein expression dynamics and assesses gene function in the vertebrate Danio rerio. Integration of pGBT-RP2 (RP2), a gene-breaking transposon containing a protein trap, efficiently disrupts gene expression with >97% knockdown of normal transcript levels while simultaneously reporting protein expression of each locus. The mutant alleles are revertible in somatic tissues via Cre recombinase or splice-site blocking morpholinos, thus representing the first systematic conditional mutant alleles outside the mouse model. We report a collection of 350 zebrafish lines including a diverse array of molecular loci. RP2 integrations reveal the complexity of genomic architecture and gene function in a living organism and can provide information on protein subcellular localization. The RP2 mutagenesis system is a step towards a unified codex of protein expression and direct functional annotation of the vertebrate genome.
Innovation and evolution have led to diverse biological systems and biochemical pathways in vertebrates1–3. Understanding which genes are necessary, important, or advantageous for survival in complex, multi-cellular organisms requires an examination of gene expression and function. The cumulative information on a single gene can be thought of as a codex, which contains multiple types of data including sequence, expression domains, and function, with each data type further containing multiple observations (e.g. sequence variations, dynamic expression over time or in response to genetic or environmental cues). Here we present an in vivo mutagenesis tool that bridges the gap between sequence data and gene function. The RP2 protein trap system recapitulates endogenous gene expression, disrupts gene splicing with nearly all tested lines displaying >99% knockdown of native transcript, and provides the first systematic collection of conditional mutants in the zebrafish.
Genomic Assessment Tool RP2
We developed a gene-break transposon (GBT) mutagenesis system for the vertebrate model system Danio rerio to facilitate genome annotation and understanding of gene function. The GBT-RP2.1 (RP2) vector has several features that efficiently cooperate to report gene sequence, expression, and function (Figure 1). The mutagenic core of RP2 contains two “trap” domains that are used to “capture” genomic information by affecting transcription, resulting in efficient knockdown of endogenous loci by a trapping vector in zebrafish for the first time (Table 1). These two key components, a protein trap and a 3’ exon trap, are placed within inverted terminal repeats of the Tol2 transposon to permit efficient genome-wide delivery4,5.
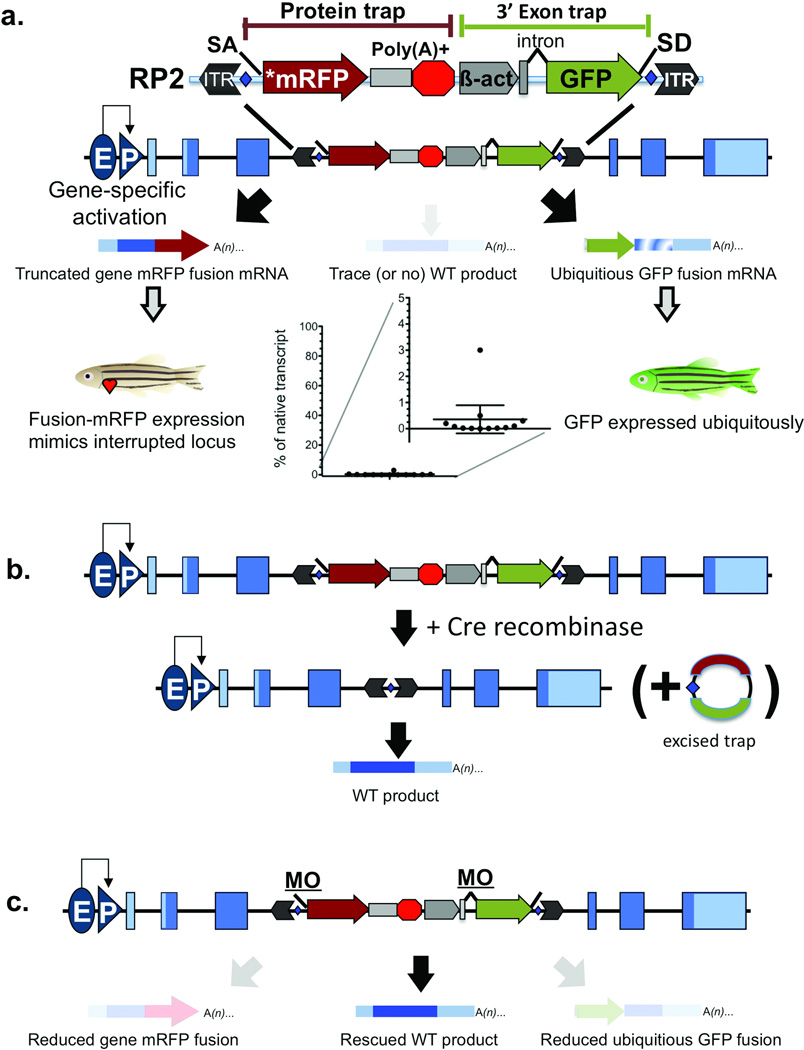
Figure 1. RP2 Gene-Break Transposon and Reversion Systems.
(a) The schematic shows the RP2 gene-break transposon system composed of a protein trap cassette with a transcriptional stop and 3’ exon trap cassette. The interrupted wild-type mRNA is either not produced or is produced at trace levels. (b) The schematic shows RP2 reversion with Cre recombinase. Blue diamonds show loxP sites flanking the mutagenic cassettes. By supplying Cre, the protein and 3’ exon traps are excised, resulting in normal levels of the nascent mRNA. (c) The schematic shows RP2 reversion with splice acceptor masking morpholinos. Both splice acceptors are derived from carp beta actin intron 1. ITR (inverted terminal repeat), SA (splice acceptor), *mRFP (AUG-less monomeric red fluorescent protein), Poly(A)+ (polyadenylation signal with extra transcriptional terminator and putative border element), β-act (carp beta actin enhancer, promoter, non-coding exon, and intron 1 sequences), GFP (green fluorescent protein), SD (splice donor), E (enhancer), P (promoter).
Table 1. Gene disruptions of GBTs.
The column categories present data for each molecularly characterized GBT line. The putative human ortholog (GBT0137 lists the putative mouse ortholog) and putative zebrafish (zf) co-ortholog are listed. % mRNA denotes the relative mRNA remaining in homozygous GBT larvae; in some cases, the transposon inserts into an exon directly disrupting the transcript. Fusion tag lists the seven proximal amino acids at the fusion site with mRFP. Pos indicates the relative position of the truncation in the predicted wild-type protein; for a description of special fusions a, b, and c please refer to the methods. LG, linkage group; DPF, days post-fertilization; HV, homozygous viable as adults; sih, silent heart; nic, nicotine response; swim, reduced swimming; fin, fin malformation; ND, no data.
| Vector | Line | LG | Gene ID | Ortholog | zf Co-Ortholog | % mRNA | Phenotype | Fusion Tag | Pos |
|---|---|---|---|---|---|---|---|---|---|
| R14 | GBT0019 | 17 | k2p10.1 | TREK2 | LOC563228 | 20.7 | None (HV) | QVNWDPE | 17 |
| R14 | GBT0031 | 23 | tnnt2a | TNNT2 | 5.6 | * sih | EETTQEH | 50 | |
| R14 | GBT0046 | 2 | epha4b | EPHA5 | ek1 | ND | None (HV) | RNYPENE | 42 |
| R15 | GBT0001 | 23 | casz1 | CASZ1 | 13 | None (5 DPF) | M | 1 | |
| R15 | GBT0002 | 14 | si:ch211-51g4.4 | SORBS2 | 24.3 | None (5 DPF) | MPSFK | 5(a) | |
| R15 | GBT0005 | 2 | itgb1b | ITGB1 | itgb1a | 33.3 | None (HV) | GLSRAQQ | 19 |
| R15 | GBT0010 | 7 | cdh11 | CDH11 | ND | None (5 DPF) | FSLKDNR | 551 | |
| R15 | GBT0039 | 19 | gabbr1.2 | GABBR1 | gabbr1a | <5 | * nic (HV) | HYDRHYT | 161 |
| R15 | GBT0043 | 7 | CD99l2 | CD99L2 | 14.4 | None (HV) | GKDSGKG | 108 | |
| R16 | GBT0007 | 17 | LOC794348 | C14ORF102 | 0.1 | None (HV) | TALQKVK | ~20 | |
| R16 | GBT0016 | 6 | pbx1a | PBX1 | zgc:15882 | ND | None (HV) | CEIKEKT | 87 |
| R16 | GBT0021 | 22 | si:ch73-150k18.1 | CNTNAP5 | LOC569185 | exon | None (HV) | VHGEGQR | 285 |
| RP2 | GBT0033 | 7 | si:dkeyp-73c8.2 | LRCH4 | ND | None (5 DPF) | LSDITHA | 57 | |
| RP2 | GBT0035 | 13 | LOC559134 | PARG | si:dkey-259k14.2 | ND | None (5 DPF) | NECLIIT | 578 |
| RP2 | GBT0040 | 19 | hoxA5a-hoxA3a | HOXA cluster | HoxAb cluster | 0.1 | None (HV) | RGPALVQ | 20(b) |
| RP2 | GBT0060 | 9 | crfb4 | IL10RB | ND | None (5 DPF) | NIIGVIT | 19 | |
| RP2 | GBT0067 | 16 | myom3 | MYOM3 | LOC100330205 | 0.2 | None (HV) | LQLSKQC | ~263 |
| RP2 | GBT0078 | 4 | grip1 | GRIP1 | ND | * Reduced Adult Viab. | SLLKNVG | 151 | |
| RP2 | GBT0091 | 9 | enox1 | ENOX1 | ND | ND | TGQQLVS | 49 | |
| RP2 | GBT0101 | 11 | dido1 | DIDO1 | exon | * lethal 10–16 DPF | LLHHIHS | 38(c) | |
| RP2 | GBT0125 | Un | LOC100333685 | CD302 | 0.02 | None (HV) | ARGEDDN | 36 | |
| RP2 | GBT0126 | 9 | nrp2b | NRP2 | nrp2a | 0.04 | None (HV) | LYGCQIT | 427 |
| RP2 | GBT0133 | 9 | zic2a | ZIC2 | zic2b | exon | None (5 DPF) | DSAHMGA | 47 |
| RP2 | GBT0137 | 1 | zgc:110335 | eef1a1 | zgc:73138 | 0.5 | None (HV) | QDVYKIG | 257 |
| RP2 | GBT0141 | 1 | gpm6ba | GPM6B | gpm6bb | 0.02 | None (HV) | DERDESK | 21 |
| RP2 | GBT0145 | 3 | LOC560542 | EPN2 | ND | None (5 DPF) | SPASYHG | 198 | |
| RP2 | GBT0154 | 8 | si:ch211-163121.8 | KIAA1324 | ND | None (5 DPF) | GFYSNGT | 351 | |
| RP2 | GBT0156 | 5 | fras1 | FRAS1 | ND | * fin | SRAGHCH | 652 | |
| RP2 | GBT0166 | 23 | atp1b2a | ATP1B2 | atp1b2b | ND | None (5 DPF) | GRTASSW | 33 |
| RP2 | GBT0168 | 14 | fgf13 (1y+1v) | FGF13 | fgf13l | ND | None (5 DPF) | KENSEPE | 72 |
| RP2 | GBT0175 | 6 | arhgef25b | GEFT | ND | None (5 DPF) | VCCFNQK | 89 | |
| RP2 | GBT0231 | 7 | neo1 | NEO1 | ND | None (5 DPF) | AITRPQS | 810 | |
| RP2 | GBT0242 | 7 | zgc:110022 | TEX261 | ND | ND | CFVTLAI | 23 | |
| RP2 | GBT0283 | 24 | LOC793623 | SH3KBP1 | ND | ND | MDNEAEK | 209 | |
| RP2 | GBT0325 | 11 | LOC557764 | MEGF6 | megf6a | ND | ND | TGVVCNE | 446 |
| RP2 | GBT0340 | 7 | NFATC3 | NFATC3 | LOC566869 | ND | ND | QPPLGPA | 30 |
| RP2 | GBT0348 | 18 | ryr1b | RYR1 | ryr1a | 3 | * swim | QMISACK | 1444 |
| RP2 | GBT0363 | 8 | XU0032 | 0 | * lethal | ||||
| RP2 | GBT0364 | 10 | XU0135 | 0.3 | * lethal | ||||
| RP2 | GBT0365 | 12 | XU0231 | 0.1 | None (5 DPF) |
The protein trap domain in RP2 generates the expression profile, including subsequent protein localization and accumulation, while mutating the gene. In cases where RP2 integrates in the sense orientation of a transcription unit, the protein trap’s splice acceptor (SA) overrides normal splicing of the transcription unit, creating a fusion between endogenous upstream exons and the monomeric red fluorescent protein (mRFP) reporter sequences. An in-frame fusion between the reporter and the tagged protein is required to produce red fluorescence due to removal of the start codon from mRFP. Since the trapped gene’s promoter produces the mRFP fusion transcript, it is made when and where the endogenous gene’s mRNA is transcribed (Supplementary Figure 1). We note that the localization of the mRFP fusion protein can be dependent on protein trafficking signals within the trapped protein fragment. The efficient mutagenesis observed in RP2-disrupted loci occurs by termination of the fusion transcript through signals derived from the ocean pout antifreeze gene, consisting of a strong polyadenylation signal (poly(A)), transcriptional terminator, and putative border element6.
The second key domain of RP2 includes the previously described 3’ exon trap or poly(A) trap used in zebrafish forward genetic applications6,7. GFP activation indicates a high likelihood of transposon insertion in the sense orientation of a transcription unit and does not require concomitant endogenous gene expression. Indeed, all identified mRFP-expressing loci also exhibit linked GFP expression (Fig. 2a). Variability of GFP expression is generally limited to ubiquitous changes in GFP intensity. Extensive sequencing of molecular tags created by 3’ exon trap activation in zebrafish somatic tissue demonstrated the ability of this vector to identify native transcripts, including novel RNA species (Supplementary Note, Supplementary Figure 2).
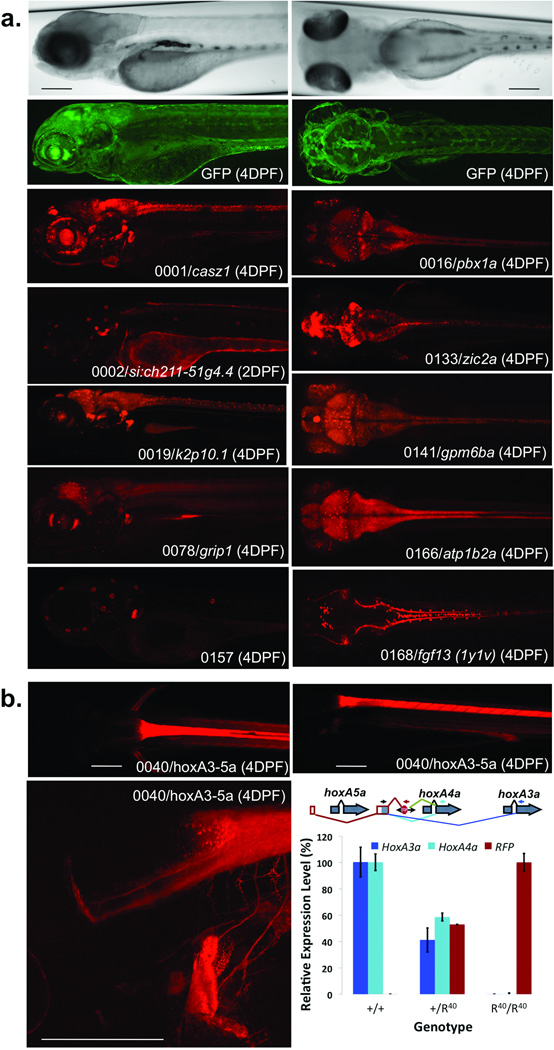
Figure 2. Protein Expression Codex.
(a) The images show examples of protein trap expression patterns; they are maximal image projections of z-stacks in sagittal (left) and coronal (right) planes. Shown at the top are brightfield and corresponding GFP images of 4 DPF larvae. For each mRFP image, the identifier, gene name and age of the larval fish are indicated. Images are scaled similarly with the scale bar representing 200 µM. (b) The mRFP expression pattern in GBT0040, an integration within the HoxAa cluster between hoxA5a and hoxA4a. Scale bars represent 100 µM. The schematic demonstrates the annotated genes of this region of the hoxAa cluster. The red framed exons and red splicing lines show exons spliced to RP2. The green splice lines show the primary splice event from the 3’ exon trap and the dark and light blue splicing lines show alternative splicing identified by RT-PCR. The graph shows relative transcript abundance containing both the shared exon and indicated downstream cassette (hoxA3a, hoxA4a, or RFP) within the given genotypes (95% Confidence Interval, n = 4).
An RP2 protein-trap mutagenized gene produces mRFP expression that mimics endogenous expression while quantitatively removing the normal full-length protein. To permit selective correction of the mutagenized locus, we flanked RP2 with Cre recombinase recognition sites (loxP) to excise the mutagenic core, leaving behind only the Tol2 inverted terminal repeats (ITR) (Fig 1b) (for review of recombinase systems8). In contrast to common mouse conditional mutagenesis methods, RP2 mutagenic core excision results in conditional rescue when the integration occurs within an intron (Figs. 3a and 4d). To revert the GBT mutation in the entire embryo, Cre recombinase mRNA is delivered at the single cell stage. Alternatively, inducible or tissue-specific expression of Cre can create conditional rescue, permitting investigation of the spatio-temporal requirements for the mutagenized locus.
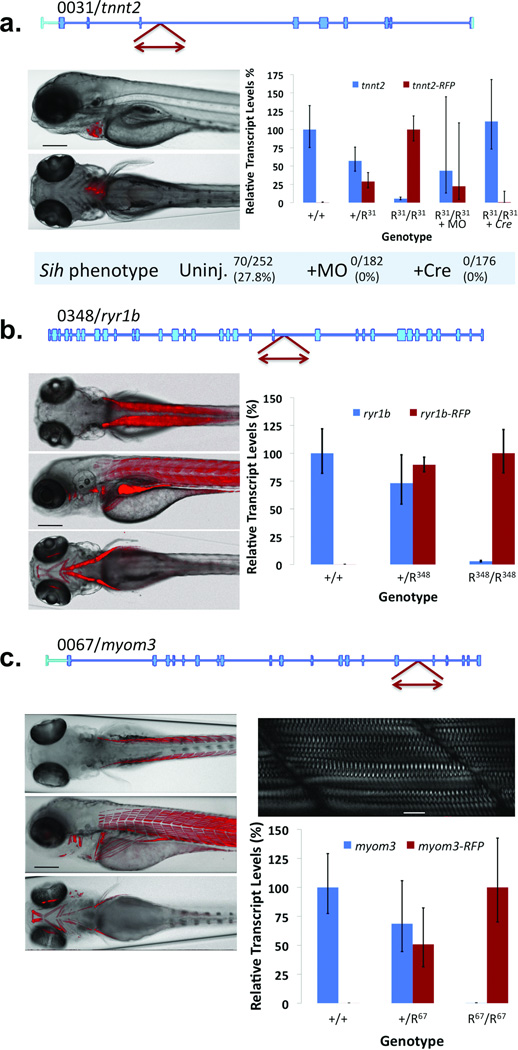
Figure 3. Protein Trap Integrations into muscle-specific genes.
(a) Integration into the 5th intron of troponin T2 (GBT0031/tnnt2) results in mRFP expression in heart tissue. The plot shows relative transcript levels in fish of the indicated genotypes (n = 11) and after addition of splice acceptor masking morpholino (+MO, n = 8) or Cre recombinase (+Cre, n = 4) to homozygous mutant fish R31/R31 (tnnt2amn0031Gt/mn0031Gt). Error bars represent 95% confidence interval. Scale bar represents 200 µM. (b) Integration into the 37th intron of ryanodine receptor 1b (GBT0348/ryr1b) results in mRFP expression in fast-twitch muscle. The plot shows relative transcript levels in the indicated genotypes (95% confidence interval, n = 4). Scale bar represents 200 µM. (c) Integration into the 17th intron of myomesin 3 (GBT0067/myom3) results in mRFP expression in slow-twitch muscle in heterozygous (+/R67) or homozygous larvae (R67/R67 = myom3mn0067Gt/mn0067Gt). Scale bar in the bright-field fluorescent overlay, 200 µM; inset scale bar, 20 µM. The plot shows the relative transcript levels in the indicated genotypes (95% confidence interval, n = 4).
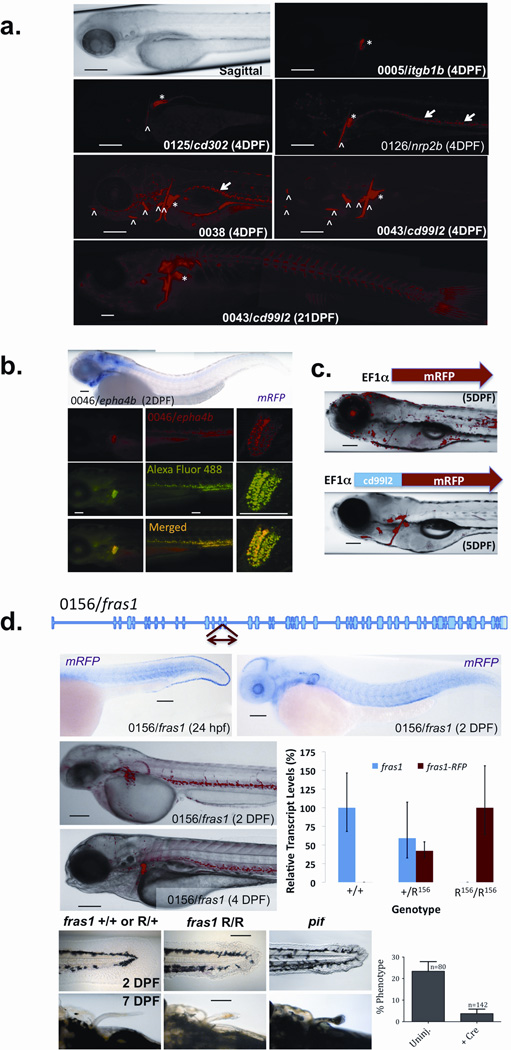
Figure 4. Secretion of mRFP fusions.
(a) Representative images showing common patterns of mRFP accumulation of secretory lines. mRFP accumulation is marked by arrows in vessels or white blood cells, asterisks in kidney tubules, and carrots in bone. Scale bars represent 200 µM. (b) The top panel shows an in situ hybridization of an mRFP probe in a GBT0046/ephaA4b larva. Fluorescence images of the head, tail, and kidney tubules (also apparent in the head image) show mRFP in GBT0046/epha4b fish (top), an Alexa-Fluor 488 dextran-injected fish (middle), and a merged image (bottom). Scale bars represent 200µM. (c) The images demonstrate the effect of the cd99l2 N-terminal fusion on mRFP protein distribution within injected embryos. Scale bars represent 200 µM. (d) The schematic shows GBT integration into fras1 (GBT0156/fras1). Images show in situ hybridization of an mRFP probe (top) and mRFP expression in GBT0046/ fras1 larvae (middle). The plot shows the relative transcript levels in the indicated genotypes, (95% confidence interval, n = 4). The bottom panel of images show developing fins in larvae of the indicated genotypes (R156/R156 = fras1mn0156Gt/mn0156Gt, pif = fras1te262/te262). The plot (bottom right) shows the fraction of fras1mn0156Gt/mn0156Gt fish showing the fin phenotype with (+Cre) and without (uninj.) Cre mRNA (s.e.m., n = 3 families). Scale bars represent 200 µM.
Cre-mediated excision results in genetically irreversible deletion of the mutagenic core. However, many genes have additional functions later in development or have adult functions that differ from their early embryonic roles. In these cases, a transient rescue strategy that bypasses the early embryonic roles or functions may be preferable. GBT splice acceptor (SA)-targeted antisense morpholino oligonucleotides can be used to transiently rescue a GBT mutation (Fig. 1c) for 3 or 4 days post-fertilization (DPF), permitting the study of a gene’s function later in development. Both the primary and secondary splice acceptors, in front of the protein trap and 3’ exon trap GFP, respectively, are derived from the same source. Therefore, antisense morpholinos targeted against the common SA sequence effectively mask both SAs from the spliceosome (Fig. 1c), resulting in a transient boost of the wild-type product from the GBT-mutated locus. This strategy can provide rapid proof that rescued phenotypes are caused by the protein trap affecting splicing of the mutant locus.
Expression profiling with RP2
Unlike prior generation zebrafish vectors (i.e. enhancer traps)9–11, the requirement in RP2 of an in-frame translational fusion means that the expression pattern obtained from a GBT protein trap is directly associated with the tagged gene product (Fig. 1a). We prioritized and catalogued lines with robust expression at two and four DPF. GBT strain generation is an ongoing process, with updated results available through an interactive online database, http://zfishbook.org. All lines are freely available now through zfishbook, and, once the collection is synchronized, will be accessible from the Zebrafish International Resource Center (http://zebrafish.org).
Of our initial collection of 350 protein trap lines with mRFP expression, about 1/3rd exhibit neuronal expression, with instances of both diverse and redundant expression domains (Fig. 2a). For instance, multiple lines show mRFP expression in sensory ganglia (for example, GBT0001 and GBT0019) or neuromasts (for example, GBT0002 and GBT0157). Despite having shared expression domains in sensory ganglia or neuromasts, mRFP expression in these lines differs in other tissues. [AU: previous sentence OK as edited?] In many cases, fluorescent puncta appear within a general neural background of mRFP, with distinct differences in the number, location, or coverage of mRFP-positive spots (for example, GBT0016 and GBT0141). In some cases, a GBT insertion only affects a single protein isoform. For example, due to the integration site within fgf13, GBT0168 only traps the 1y+1v isoform of fgf1312. As the complexity of the protein trap expression library increases, each line will contribute to its own codex as well as providing morphological, developmental, and molecular annotation of the zebrafish.
Annotation of transcriptional organization with RP2
Hox clusters are highly conserved among animals, but the mechanism underlying this strict conservation remains largely unclear13. GBT0040 is an insertion in the zebrafish HoxAa cluster, between the annotated exons of the hoxA5a and hoxA4a genes resulting in an mRFP pattern that resembles hoxA4a expression (Fig. 2b). Upstream exons fused to the mRFP transcript in GBT0040 contained two exons, one non-coding exon 5’ of hox5Aa and one protein-coding exon between hox5Aa and hox4Aa (Fig. 2b). Transcriptional assessment demonstrated that the protein-coding exon splices to downstream exons from both hoxA4a and hoxA3a to produce protein-coding sequences with novel N-terminal sequences (Fig. 2b). Notably, this single insertion results in the effective loss (>99% knockdown) of both of these alternate mRNA transcripts (Fig. 2b). Exon sharing and alternate transcripts within Hox clusters has been previously observed in the transcriptome14,15. The exon sharing and alternate transcripts apparent within the GBT0040 allele suggest a mechanism underlying the conserved retention of hox genes within a single genomic cluster.
Annotating gene function with RP2
The GBT protein trap system provides in vivo functional annotation beyond reporting the dynamic expression patterns of vertebrate genes and promoter or splicing variations at interrupted loci. In Figure 3, we show three examples of genes expressed within distinct domains of the zebrafish musculature: tnnt2 (GBT0031, Fig. 3a), ryr1b (GBT0348, Fig 3b), and myom3 (GBT0067, Fig. 3c).
GBT0031/tnnt2 (Fig. 3a) is a first generation protein trap insertion (R14, Supplementary Figure 3) into troponinT2 (tnnt2) with strong cardiac muscle-specific mRFP expression. Protein trap disruption of tnnt2 results in recessive loss of heartbeat, that phenocopies a previously documented tnnt2 mutation, silent heart16,17. To test the somatic reversibility of this protein trap system, we injected Cre recombinase mRNA or splice acceptor masking morpholino into embryos from a GBT0031 incross. Whereas 28% (n = 252) of uninjected embryos developed the silent heart phenotype, injecting either Cre recombinase or masking morpholino reversed the silent heart phenotype (Fig. 3a). We also measured the knockdown of intact tnnt2 mRNA via quantitative PCR across the exons flanking the protein trap insertion. Homozygous GBT0031 (tnnt2amn31-gt−/mn31gt−) embryos expressed 6% of the intact mRNA as compared to wild-type siblings. Injection of either the gene-break masking morpholino or Cre recombinase restored the level of intact mRNA to near heterozygotic (tnnt2a+/mn31gt−) or wild-type (tnnt2a+/+) levels, respectively (Figure 3A). Note that the GBT0031/tnnt2 locus was made from an early generation protein trap vector, pGBT-R14 (R14) (Supplementary Figure 3). Although these first successful protein trap vectors including R14, pGBT-R15 (R15)7, and pGBT-R16 (R16) often demonstrate a stronger knockdown than earlier published gene trap vectors in zebrafish11, the overall knockdown can be incomplete (Supplementary Figure 3; Table 1). Addition of the strong transcriptional terminator or “stop” cassette and an additional splice acceptor in RP2 results in a notably stronger knockdown effect (Fig. 1; Table 1; Supplementary Figure 3).
An RP2 insertion into the ryanodine receptor 1b gene (ryr1b/GT0348) results in only 3% of its mRNA intact in homozygous (ryr1bmn348gt−/mn348gt−) offspring. This is the weakest knockdown we have measured in an RP2 line to date (Table 1), but represents a sharp improvement over the early protein trap vectors R14, R15, and R16. GBT0348/ryr1b produces mRFP expression in the fast-twitch muscle of the zebrafish (Fig. 3b). The disruption of ryr1b expression in homozygous larvae results in a slow swimming phenotype that leads to growth impairment and lethality by 14 DPF (data not shown). The GBT0348 phenotype is similar to a previously reported allele of ryr1b that impacts swimming called relatively relaxed18 (personal communication James Dowling, University of Michigan).
The third muscle-specific gene insertion is in myomesin 3 (GBT0067/myom3; Fig. 3c), a structural component of the M-band of intermediate fibers of skeletal muscle19. Myom3 is expressed in the slow or intermediate muscle fibers of the larval zebrafish. Homozygous fish can survive to fertility in a laboratory environment despite having less than 1% of intact myom3 mRNA.
RP2 annotation of protein localization and trafficking
Protein trap fusions can reveal protein subcellular localization; for example, the myom3-mRFP fusion is found in the M-band of the sarcomere (Fig. 3c). They can also be used to study subsets of proteins with common trafficking properties. The signal most frequently observed in this collection of protein trap lines was the N-terminal signal sequence, found in members of the secretome20. Understanding the function of the secretome is particularly crucial because of the extensive roles that cellular context and cell-cell signaling play in vertebrate biology and physiology. Key subsets of the secretome such as G-protein coupled receptors represent critical drug targets.
About 25% of the 350 lines we generated have some mRFP accumulation in the kidney tubules, white blood cells or developing bone (Fig. 4a). Based on the presence of signal sequences at the N-terminus of genes identified from fish exclusively with these mRFP patterns, we hypothesized that the kidney and white blood cells were filtering or in other ways accumulating the mRFP fusion proteins. To test the ability of these cell types to remove fluorescent particles from the blood, we injected Alexa Fluor 488 conjugated dextran into the bloodstream of GBT0046 embryos at 3 DPF. Within eight hours the dextran particles had accumulated in both the kidney ducts and white blood cells where the mRFP fusion protein was also localized (Fig. 4b). The presumed trafficking of secreted mRFP-protein fusions results in diverse expression outcomes. In some lines, mRFP fusion proteins demonstrate some local retention indicating where the mRFP fusion protein originated7. Other lines show no mRFP fusion protein locally, an effect that is readily documented using an antisense mRFP probe for in situ expression data (Fig. 4b,d).
Strong mRFP accumulation in developing bone occurs in a few protein trap lines and is dependent on the fusion protein (Fig. 4a), suggesting an additional protein trafficking mechanism. For instance, we observed strong mRFP accumulation in developing bone upon RP2 integration into cd99 antigen like-2 (GBT0043/cd99l2); the cd99l2-mRFP fusion protein accumulates in or near developing bone (Fig. 4a), even though cd99l2 mRNA is not expressed at or near this tissue (data not shown). We hypothesized that the amino acids present in the cd99l2 protein fusion of GBT0043 direct secretion and subsequent accumulation of mRFP to the bone matrix. We tested this by injecting DNA that would produce mRFP with and without the cd99l2 fusion sequence in a ubiquitous, albeit mosaic, manner in the zebrafish. When mRFP alone is expressed from a ubiquitous promoter, we see generalized, mosaic expression of mRFP. However, when the cd99l2 sequences are fused to the mRFP, fluorescence localized to developing bone (Fig. 4c).
Protein trap integrations into secretome genes are efficient mutagens as illustrated by integration into the Fraser syndrome 1 gene (GBT0156/ fras1) (Fig. 4d). The RP2 insertion is in the 15th intron of fras1, creating a fusion protein with the extracellular domain of fras1. The secreted fras1-mRFP fusion protein in GBT0156 accumulates in both the kidney and white blood cells. However the mRFP-fusion mRNA is located within the brain, lens, muscle, and developing fins and skin, similar to endogenous expression patterns for fras121,22. GBT0156 fish homozygous for the insertion (fras1mn0156gt−/mn0156gt−) express only 0.01% of intact fras1 transcript and display a phenotype mimicking pinfin, a previously described mutation in fras121,23. Microinjecting Cre mRNA reverts the pinfin phenotype in GBT0156 embryos. If further prioritization of secreted lines based on mRNA expression is desired, in situ hybridization using a single antisense mRFP probe documents the expression pattern of tagged lines.
Discussion
This set of GBTs represents the first in vivo protein trap alleles in a vertebrate with reporter production and line selection occurring within the animal rather than using in vitro cells for production or molecular characterization. The approach deployed here is based in part on the extensive gene-trapping work in mouse embryonic stem (ES) cells, including vectors that produce protein fusions– one form of protein traps (for review24). It also complements the rich history of in vivo protein trapping in Drosophila, where the protein traps are not designed to terminate the interrupted protein and therefore are less mutagenic25–27. The zebrafish model is well suited for high-throughput genome mutagenesis and real-time expression analysis using protein trap technology. Moreover, the RP2 transposon system is not restricted to use in zebrafish and can be applied to other model organisms.
Near-random integration of transposons into the vertebrate genome permits unbiased identification of genes regardless of whether they are completely novel, predicted, or well characterized in other species. In addition, the visible expression pattern of the trapped locus can be used to intentionally bias a genetic screen by preselecting mutations in genes expressed in a tissue of interest. Furthermore, the dominant reporter in GBT mutants opens the door to an array of genetic screens difficult or impossible to perform with chemical mutagenesis, including behavioural genetics. The ability to sort a population based on the presence of a single gene mutation allows quantitative assessment of the role of a single gene in modifying complex, multigenic phenotypes. Localized reversion of the mutation by the use of tissue-specific Cre will permit mapping of tissues or neural networks that are required for proper function.
The combination of conditional mutants in a vertebrate with in vivo protein trap technology enables a direct link between DNA sequence, expression, and function for each genetic locus, a concept we term a gene ‘codex’. When combined with high-resolution analyses of sequence variation, the zebrafish is a key model for assembling a full codex for the genetic complement of the vertebrate genome and can provide new insights into genomic complexity that have been difficult to directly study and functionally annotate.
The RP2 mutagenesis system presented here is a first step. To complete the codex, multiple alleles for each protein-coding gene will be needed to understand expression, trafficking, and function depending on insert location. For example, integration at N-terminal locations is more likely to produce functional null alleles since large portions of the protein are truncated. In contrast, integration into more C-terminal locations is more likely to produce proteins with at least some function, but that are informative about subcellular protein localization and trafficking. Comprehensive mutagenesis of a vertebrate genome with insertional mutagens, like RP2, could require millions of insertional events28. Therefore to maximize genome coverage, it is recommended to utilize complementary vector systems to take advantage of differences in integration preferences28,29. A first step will be to produce all three reading frames of the RP2 vector. Subsequent complementary transposon systems and yet-to-be established reverse genetic approaches will increase genome coverage to achieve the goal of a comprehensive codex of the vertebrate genome.
Supplementary Material
Acknowledgments
The National Institute of Drug Abuse (DA14546), National Institute of General Medical Sciences (GM63904) and the Mayo Foundation provided funding for this research. S.S. and V.S. acknowledge funding support from CSIR (Grant FAC002), India. We thank the Center for Genome Engineering at the University of Minnesota for providing collaborative discussion and resources. We thank the staff of the Zebrafish Core Facility at the Mayo Clinic for providing zebrafish care. We thank Dr. Eric Klee at the Mayo Clinic for bioinformatic analyses of the zebrafish transcriptome used in theoretical design and testing of the RP2 system. We thank Dr. Anthony Person for assistance in injection of fluorescently labelled dextrans. We thank InSciEd Out (www.insciedout.org) participants and Summer Undergraduate Research Fellow, Nicole Boczek, for imaging RFP-positive fish.
Footnotes
Author Contributions. K.J.C., D.B., S.S., and S.C.E. designed the experiments. D.B. built R14, R15, R16, RP2, and the pEF1a-cd99l2-RFP vectors. S.S. built pX. J.N. made the pEF1a-RFP vector. D.B. piloted the production of R14, R15, and R16 lines. K.J.C. trained and managed the team that produced >300 RP2 lines and molecularly characterized lines other than GBT0031, GBT0039, and GBT0043 (D.B.). M.D.U., T.M.G., A.N., K.J.S., and K.J.C. indentified and propagated mRFP-positive lines. A.M.P., M.D.U., and K.J.C. photographed mRFP expression patterns. K.J.S. and K.J.C. molecularly characterized lines by 5’RACE, inverse PCR, and QRT. T.M.G., K.J.C., and D.B. characterized the reversion of GBT0031 by Cre mRNA and morpholino injection. D.B. injected fluorescently conjugated dextran into GBT0046 and imaged its uptake. J.N. imaged GBT0043 fish and together with K.J.C. injected fish with EF1a-RFP and EF1a-cd99l2-RFP vectors- K.J.C. imaged the injected fish. S.E.W. analyzed the GBT0156/frasI phenotype and reversion. V.M.B. conducted in situ hybridizations. Y.D. initially identified GBT0348 in a screen performed in X.X.’s lab and contributed QRT data of three additional RP2 lines. H-M.P. tested the GBT protocol in M.H.’s lab and produced several independent lines, including the initial assessment and imaging of GBT0040. A.P., V.S., and S.S. produced and analyzed 3’ exon trap data for Supplementary Figure 2. K.J.C. and S.C.E. were the primary authors of the text.
References
- 1.Stevens CW. The evolution of vertebrate opioid receptors. Front Biosci. 2009;14:1247–1269. doi: 10.2741/3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huxley-Jones J, Robertson DL, Boot-Handford RP. On the origins of the extracellular matrix in vertebrates. Matrix Biol. 2007;26:2–11. doi: 10.1016/j.matbio.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 3.Sauka-Spengler T, Bronner-Fraser M. Evolution of the neural crest viewed from a gene regulatory perspective. Genesis. 2008;46:673–682. doi: 10.1002/dvg.20436. [DOI] [PubMed] [Google Scholar]
- 4.Balciunas D, et al. Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLoS Genet. 2006;2:e169. doi: 10.1371/journal.pgen.0020169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Urasaki A, Morvan G, Kawakami K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics. 2006;174:639–649. doi: 10.1534/genetics.106.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sivasubbu S, et al. Gene-breaking transposon mutagenesis reveals an essential role for histone H2afza in zebrafish larval development. Mech Dev. 2006;123:513–529. doi: 10.1016/j.mod.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Petzold AM, et al. Nicotine response genetics in the zebrafish. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0908247106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Branda CS, Dymecki SM. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- 9.Parinov S, Kondrichin I, Korzh V, Emelyanov A. Tol2 transposon-mediated enhancer trap to identify developmentally regulated zebrafish genes in vivo. Dev Dyn. 2004;231:449–459. doi: 10.1002/dvdy.20157. [DOI] [PubMed] [Google Scholar]
- 10.Balciunas D, et al. Enhancer trapping in zebrafish using the Sleeping Beauty transposon. BMC genomics. 2004;5:62. doi: 10.1186/1471-2164-5-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawakami K, et al. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 12.Munoz-Sanjuan I, Simandl BK, Fallon JF, Nathans J. Expression of chicken fibroblast growth factor homologous factor (FHF)-1 and of differentially spliced isoforms of FHF-2 in chicken limb development. Development. 1999;126:409–421. doi: 10.1242/dev.126.2.409. [DOI] [PubMed] [Google Scholar]
- 13.Holland PW. Beyond the Hox: how widespread is homeobox gene clustering? J Anat. 2001;199:13–23. doi: 10.1046/j.1469-7580.2001.19910013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coulombe Y, et al. Multiple promoters and alternative splicing: Hoxa5 transcriptional complexity in the mouse embryo. PLoS ONE. 2010;5:e10600. doi: 10.1371/journal.pone.0010600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hadrys T, Prince V, Hunter M, Baker R, Rinkwitz S. Comparative genomic analysis of vertebrate Hox3 and Hox4 genes. J Exp Zool B Mol Dev Evol. 2004;302:147–164. doi: 10.1002/jez.b.20012. [DOI] [PubMed] [Google Scholar]
- 16.Chen JN, et al. Mutations affecting the cardiovascular system and other internal organs in zebrafish. Development. 1996;123:293–302. doi: 10.1242/dev.123.1.293. [DOI] [PubMed] [Google Scholar]
- 17.Sehnert AJ. Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat Genet. 2002;31:106–110. doi: 10.1038/ng875. [DOI] [PubMed] [Google Scholar]
- 18.Hirata H. Zebrafish relatively relaxed mutants have a ryanodine receptor defect, show slow swimming and provide a model of multi-minicore disease. Development. 2007;134:2771–2781. doi: 10.1242/dev.004531. [DOI] [PubMed] [Google Scholar]
- 19.Schoenauer R. Myomesin 3, a novel structural component of the M-band in striated muscle. J Mol Biol. 2008;376:338–351. doi: 10.1016/j.jmb.2007.11.048. [DOI] [PubMed] [Google Scholar]
- 20.Klee EW. The zebrafish secretome. Zebrafish. 2008;5:131–138. doi: 10.1089/zeb.2008.0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carney TJ. Genetic analysis of fin development in zebrafish identifies furin and hemicentin1 as potential novel fraser syndrome disease genes. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1000907. e1000907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gautier P, Naranjo-Golborne C, Taylor MS, Jackson IJ, Smyth I. Expression of the fras1/frem gene family during zebrafish development and fin morphogenesis. Dev Dyn. 2008;237:3295–3304. doi: 10.1002/dvdy.21729. [DOI] [PubMed] [Google Scholar]
- 23.van Eeden FJ, et al. Genetic analysis of fin formation in the zebrafish, Danio rerio. Development. 1996;123:255–262. doi: 10.1242/dev.123.1.255. [DOI] [PubMed] [Google Scholar]
- 24.Friedel RH, Soriano P. In: Methods in Enzymology. Wassarman Paul M, Soriano Philippe M, editors. Vol. Volume 477. Academic Press; 2010. pp. 243–269. [Google Scholar]
- 25.Aleksic J, Lazic R, Müller I, Russell SR, Adryan B. Biases in Drosophila melanogaster protein trap screens. BMC genomics. 2009;10:249. doi: 10.1186/1471-2164-10-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buszczak M, et al. The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics. 2007;175:1505–1531. doi: 10.1534/genetics.106.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morin X, Daneman R, Zavortink M, Chia W. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc Natl Acad Sci USA. 2001;98:15050–15055. doi: 10.1073/pnas.261408198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nord AS, et al. Modeling insertional mutagenesis using gene length and expression in murine embryonic stem cells. PLoS ONE. 2007;2:e617. doi: 10.1371/journal.pone.0000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skarnes WC, et al. A public gene trap resource for mouse functional genomics. Nat Genet. 2004;36:543–544. doi: 10.1038/ng0604-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 31.Gibbs P, Schmale M. GFP as a genetic marker scorable throughout the life cycle of transgenic zebra fish. Marine Biotechnology. 2000;2:107–125. doi: 10.1007/s101269900014. [DOI] [PubMed] [Google Scholar]
- 32.Campbell RE. A monomeric red fluorescent protein. Proc Natl Acad Sci USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petzold AM. SCORE imaging: specimen in a corrected optical rotational enclosure. Zebrafish. 2010;7:149–154. doi: 10.1089/zeb.2010.0660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clark KJ, Geurts AM, Bell JB, Hackett PB. Transposon vectors for gene-trap insertional mutagenesis in vertebrates. Genesis. 2004;39:225–233. doi: 10.1002/gene.20049. [DOI] [PubMed] [Google Scholar]
- 35.Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- 36.Bill BR, Petzold AM, Clark KJ, Schimmenti LA, Ekker SC. A primer for morpholino use in zebrafish. Zebrafish. 2009;6:69–77. doi: 10.1089/zeb.2008.0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.