Abstract
Gene expression profiling has provided insights into different cancer types and revealed tissue-specific expression signatures. Alterations in microRNA expression contribute to the pathogenesis of many types of human diseases. Few studies have integrated all levels of gene expression, miRNA and methylation to uncover correlations between these data types. We performed an integrated profiling to discover instances of miRNAs associated with a gene expression and DNA methylation signature across multiple cancer types. Using data from The Cancer Genome Atlas (TCGA), we revealed a concordant gene expression and methylation signature associated with the microRNA hsa-miR-142 across the same samples. In all cancer types examined, we found a signature of co-expression of a gene set R and methylated sites M, which correlate positively (M+) or negatively (M−) with the expression of hsa-miR-142. The set R consistently contains many genes, such as TRAF3IP3, NCKAP1L, CD53, LAPTM5, PTPRC, EVI2B, DOCK2, LCP2, CYBB and FYB. The signature is preserved across glioblastoma, ovarian, breast, colon, kidney, lung, uterine and rectum cancer. There is 28% overlap of methylation sites in M between glioblastoma (GBM) and ovarian cancer. There is 60% overlap of genes in R between GBM and ovarian (P = 1.3e−11). Most of the genes in R are known to be expressed in lymphocytes and haematopoietic stem cells, while M reflects membrane proteins involved in cell-cell adhesion functions. We speculate that the hsa-miR-142 associated signature may signal haematopoietic-specific processes and an accumulation of methylation events triggering a progressive loss of cell-cell adhesion. We also observed that GBM samples belonging to the proneural subtype tend to have underexpressed hsa-miR-142 and R genes, hypomethylated M+ and hypermethylated M−, while the mesenchymal samples have the opposite profile.
Keywords: cancer, microRNA, gene expression, methylation, correlation, integrated analysis
Introduction
Gene expression regulation through mechanisms that involve microRNAs and epigenetics (DNA methylation) has attracted much attention recently.1 MiRNAs are a class of small RNA molecules that target mRNAs, causing translation repression. MiRNAs regulate genes associated with different biological processes, such as apoptosis, stress response, or tumorigenesis.2 In the context of cancer, miRNAs have emerged as new molecular players involved in carcinogenesis. Deregulation of miRNAs has been shown in glioblastoma, colon and ovarian cancer.3,4 Recently, it was shown that miR-10b is upregulated in gliomas, even though it is not expressed in healthy human brain tissue.5 Distinguishing cancer subtypes on the basis of miRNA expression may help to personalize therapies; this would enable doctors to better match a patient with the treatment the patient is likely to respond to with fewest side effects.6–11
Expression of genes is affected by miRNA expression and DNA methylation, which are known to regulate each other in both directions.12 MiRNAs are known to be targets of epigenetic regulation, as well as to target the epigenome, allowing for self-regulatory loops. One third of all human miRNA genes have a CpG island in the upstream region, suggesting epigenetic regulation of miRNA expression.13–15 On the other hand, several miRNAs are known to drive methylation signatures via interactions with DNA methyltransferases (DNMTs) and RBL2 proteins16,17 DNMTs are responsible for the methylation of the CpG islands of genes in an RBL2-dependent manner. The over expression of DNA methyltransferases (DNMTs) is often a poor prognostic indicator for cancer, since they induce hypermethylation and underexpression of tumor suppressors.18,19 It still remains largely unknown how, exactly, miRNAs affect genome methylation at the epigenetic level. The precise epigenetic mechanisms underlying the alteration of miRNA expression also remain largely unknown.
Few studies have performed an integrated analysis of gene expression, miRNA expression and methylation using data from the same samples.2,20–24 In this study we explore relations of these three data types, in terms of their correlation to one another. Our goal was to identify particular miRNAs having strong and consistent correlative relationships with gene expression and methylation. We focused on integrated cancer datasets that are available from The Cancer Genome Atlas (TCGA) repository. The TCGA Research Network was established to generate the comprehensive catalog of genomic abnormalities driving tumorigenesis.25,26 We found that hsa-miR-142 is consistently associated with a gene expression and methylation signature in all cancer datasets.
Methods
Our analysis involved a computational method to identify miRNA, methylation sites and genes of potential importance in a set of samples, based on correlations among the different data types across samples.27 We performed an integrated correlation analysis of the miRNA, methylation and gene expression data that is available for glioblastoma and ovarian cancer. Our method’s aim is to evaluate the correlations between methylation-miRNA and mRNA-miRNA. For example, the correlation of miRNA Mi’s and gene Ri’s expression (across the samples for which both miRNA and gene expression are available) would show if there is a positive or negative correlation between Mi and Ri.28
We downloaded methylation, miRNA and gene expression data for glioblastoma and ovarian cancer, which was available on The Cancer Genome Atlas (TCGA) repository in April 2011. We used the level 3 data from TCGA and filtered out the healthy samples, such that we considered only tumor samples. For gene and miRNA expression the level 3 data represent the expression for particular genes or miRNAs per sample. For methylation the level 3 data represent the methylated sites/genes per sample. Level 3 data was derived after the raw signals per probe (level 1) were normalized per probe set (level 2) and then averaged for a gene or miRNA. Supplementary file 6 shows the number of tumor samples that were available on TCGA for each combination of data types. Additionally, for breast, colon, kidney, uterine and rectum cancer we downloaded the sequencing-based RNASeq and miRNASeq level 3 data available on TCGA in November 2011. This data represents the calculated expression calls for genes and miRNAs per sample. It is derived after calculating the expression signal for all reads aligning to a particular gene or miRNA (level 1) and normalizing the reads with TMM normalization (level 2).
Discretization of gene expression, miRNAs and methylation
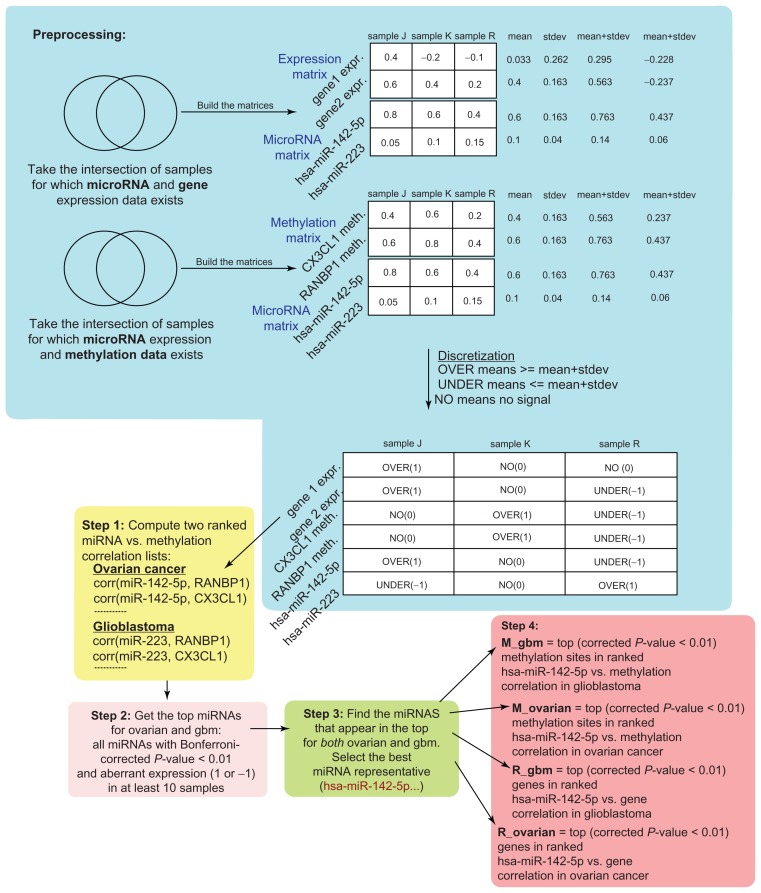
We performed a pair wise correlation analysis between the different data types, as Figure 1 shows. Initially, the algorithm compares the sample-specific methylation of each gene with the expression level of each miRNA, across all samples. To evaluate the pairwise Pearson correlation between different data types (gene expression, miRNA expression and methylation), we discretized all three data types. A gene’s expression (similarly, a methylation site’s or miRNA’s) took a discrete value of −1, 0, or 1 for a sample, representing underexpression (hypomethylation), moderate, or overexpression (hypermethylation) in the sample. The discretization is based on whether the gene’s (or methylation site’s) expression is at least one standard deviation away from the gene’s mean value over all samples. We represented a gene (or miRNA or methylation site) as a vector of discrete values across all samples. Discretization of the values serves the following two purposes: (1) it allows us to compare two data types on a common basis, ie, whether there is aberrant expression or methylation observed in the same samples. (2) It allows us to filter out high correlations if there is moderate (non-extreme) expression/methylation (0) in most or all of the samples.
Figure 1.
Preprocessing: We discretized the expression and methylation data to determine if a gene is under, over, or moderately (non-extremely) expressed/methylated in each sample. To turn a gene (or miRNA or methylation site) into a discrete vector over all tumor samples, we evaluated the gene expression’s mean value and standard deviation over all samples. Then, if the gene had value greater than the mean plus standard deviation in a sample, we represented it as 1 (over-expressed or hypermethylated). If the gene had value lower than the mean minus standard deviation in a sample, we represented it as −1 (under-expressed or hypomethylated). Otherwise, the gene was represented as 0 in the sample. We used the discrete vector representation of each miRNA’s expression and methylation over all samples (either gbm or ovarian) from the preprocessing step. Step 1: We evaluated the Pearson correlation between all pairs of miRNAs and methylation sites in glioblastoma and ovarian cancer. Then, we ranked all pairwise correlations in descending order, as shown. Step 2: We kept the top ranked miRNAs for ovarian and gbm. A condition was that the Bonferroni-corrected P-value, derived from a two-tailed t-test that evaluated the Pearson correlation, should be less than 0.01. Step 3: We found the miRNAs appearing in the top ranks in both ovarian and gbm and we selected the best miRNA as representative. Step 4: Using the best miRNA as representative, we found the top correlated methylation sites and genes in gbm and ovarian. We refer to the resulting sets as M_gbm, M_ovarian (methylation sites) and R_gbm, R_ovarian (gene expression).
Our analysis method differs from that of29 who used z-scores for discretizing the data. The z-scores indicate how many standard deviations a value is above or below the mean; previous studies discretized a gene expression value if z-score >2 or <−2. The study then used Fisher’s exact P-value to evaluate correlations between gene expression and mutations by assigning an exact P-value to a correlation, considering only up or only down regulation, and keeping P < 0.01. Pearson correlation, on the other hand, allows us to consider both up and down regulation for a pair, offering us two benefits. Using the Pearson correlation allows us to find negative correlations, as well as positive ones, such as concordantly over-expressed genes and hypomethylated sites. Moreover, Pearson correlation allows us to find negative correlations and it allows us to find subclasses of samples defined by opposite expression/methylation patterns (such as, the M+ and M− patterns). Since we are not dealing with somatic mutations, we believe Pearson correlation is a more suitable choice than Fisher’s exact P-value for our analysis.
To evaluate the statistical significance of the Pearson correlation between miRNA and gene expression (or methylation) we used a two-tailed t-test. The t-test was based on the Student’s t-distribution with n-2 degrees of freedom, where n is the number of samples. The t-test was two-tailed since either positive or negative correlation may be of interest. We performed Bonferroni-correction by multiplying the P-value with the number of genes or methylation sites. We set a threshold of 0.01 for the Bonferroni-corrected P-value. Additionally, we made one million random permutations of the samples and evaluated the correlation from the permutated data; a correlation higher than the original is a false discovery. We applied a cutoff false discovery rate of 0.01, such that if the calculated FDR was greater than 0.01, the original correlation was rejected as false. In other words, the FDR was derived by counting how often the permutated absolute correlation was at least as high as the original correlation.
Association with gene ontology
Gene ontology enrichment was assessed using the Database for Annotation, Visualization, and Integrated Discovery—DAVID.30 We also used the Gene Set Enrichment Analysis tool by the Broad Institute to evaluate the annotation enrichment of M and R. The full results are given in Supplementary files 12 and 13.31
Association with proneural and mesenchymal GBM subclasses
We used the Suppl. Info from26 to associate R and M genes to different glioblastoma subclasses, as well as tumor samples to proneural, neural, classical or mesenchymal subclasses. We used Suppl. Table S3 to retrieve the genes highly expressed in different subclasses. We used Table S7 to retrieve samples with specific subclass associations.
Results
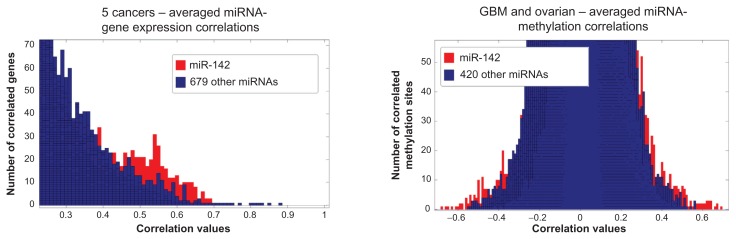
We first examined the relative significance of all miRNAs in the context of all miRNA-gene expression (or miRNA-methylation) Pearson correlation coefficient values. For this purpose, we first matched miRNA with gene expression data (or miRNA with methylation data) on the same samples. Then, we created histograms covering all miRNA-gene expression and miRNA-methylation correlation values, including all 17,814 genes and 27,578 methylation sites, over multiple cancers. We found that hsa-miR-142 stands out significantly among other miRNAs as having higher correlation values with more genes and methylation sites. Figure 2 shows the resulting histograms after averaging the correlation coefficients for miRNA-gene expression and miRNA-methylation across all cancer types available. Highlighted in red color are the occurrences where the histogram involving hsa-miR-142 lies outside the envelope of all other histograms. A significant amount of such histogram “tails” exist at the right side of the gene expression histograms (R), and at both sides of the methylation histograms (M+ and M−). Supplementary files 1–2 give the lists of the highest and lowest correlations for hsa-miR-142- methylation and hsa-miR-142-gene expression in all cancer types (corrected P-value <0.01). Supplementary file 5 shows the miRNA-gene expression correlation histograms for individual cancer types.
Figure 2.
Left: Histogram of the miRNA-gene expression correlations for cancer types that had miRNA and gene expression data (RNASeq and miRNASeq) available. We matched the miRNA-gene expression data on the same samples. Since we did not find a significant negative miRNA-gene correlation, the left graph shows just positive values. Right: the miRNA-methylation correlations for GBM and ovarian cancer (the other TCGA cancer types lacked integrated miRNA-methylation data). We matched the miRNA-methylation data on the same samples. We plotted the miRNAs that appear in all cancer types, which resulted in 680 miRNAs and 421 miRNAs, respectively. For each miRNA we included the correlation values for all 17,814 genes or 27,578 methylation sites. We averaged the correlations over all cancer types to determine if a correlation remains consistently high in all cancers. As shown, the miRNA hsa-miR-142 is highly correlated with a larger set of genes or methylation sites than other miRNAs.
Core of the signature: R and M
After we determined the significance of hsa-miR-142 in terms of high correlation with a set of genes and methylation sites across multiple cancer types, our aim is to define the sets of genes R and methylation sites M with which hsa-miR-142 is most frequently associated.
In glioblastoma and ovarian cancer, we derived the sets R and M by ranking the genes and methylation sites in descending order by their correlation with hsa-miR-142. We derived R for breast, colon, kidney, uterine and rectum cancer by correlating the RNASeq with the miRNASeq sequencing-based data available on TCGA. Then, R and M consisted of the genes or methylation sites with a Bonferroni-corrected P-value less than 0.01.
We derived M for breast, colon, kidney, uterine, rectum and lung cancer by using a metagene approach since sequencing-based methylation data was not available to correlate with the miRNASeq data. We first aggregated the 100 genes that had the highest average correlation with has-miR-142 in GBM and ovarian; the average value of these 100 genes over all samples in a cancer gave the first metagene. In each of 11 cancers, we ranked all genes based on their mutual information with the metagene. Then, we averaged the mutual information of the genes with the metagene across all 11 cancers and took the 15 top genes to create a second metagene. We ranked the correlation of this second metagene to all methylation sites in breast, colon, kidney, lung, rectum and uterine cancer. Supplementary files 7–10 give the metagene and the derived M.
M and R signature and overlaps between cancers
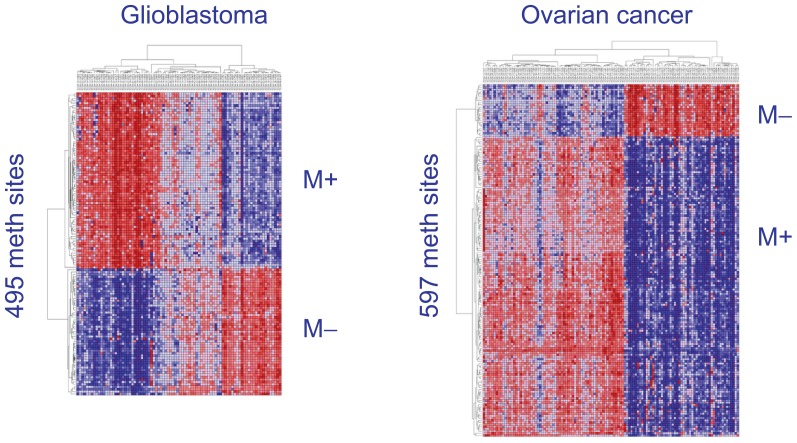
Figure 3 illustrates the M+ and M− signatures in glioblastoma and ovarian cancer. Sites positively correlated with hsa-miR-142 are M+, while negatively correlated sites are M−. The M sets shown consist of the methylation sites that have a Bonferroni-corrected P-value of less than 0.01 with hsa-miR-142 over the samples. This heatmap is the result of step4 of our analysis (see Methods section).
Figure 3.
Results of step4 of our analysis method (see Methods section). Left: The methylation sites that are most correlated with hsa-miR-142, either positively or negatively, in glioblastoma (M_gbm). Right: M_ovarian in ovarian cancer. We distinguish the signature M into the methylation sites having positive correlation with hsa-miR-142 (M+) and those with negative correlation (M−). The overlap of methylation sites between M+_gbm (236) and M+_ovarian (471) is 76, while the overlap between M−_gbm (259) and M−_ovarian (126) is 63.
Tables 1–2 show the overlaps of the R and M signatures between all cancer types considered. As shown, we found a significant overlap of M and R between different cancer types. We matched the precise probes of the methylation sites in M between different cancers.
Table 1.
Overlap of R between cancers.
| R_GBM | R_COAD | R_BRCA | R_UCEC | R_READ | R_KIRC | |
|---|---|---|---|---|---|---|
| R_OV (1338) | 671 | 224 | 279 | 382 | 82 | 629 |
| P = 1.3e-11 | P = 1.7e-11 | P = 4e-11 | P = 5.4e-11 | P = 3.1e-11 | P = 3e-11 | |
| R_GBM (1106) | – | 155 | 194 | 271 | 61 | 453 |
| P = 0 | P = 0 | P = 2e-11 | P = 0 | P = 0 | ||
| R_COAD (289) | – | – | 197 | 224 | 85 | 254 |
| P = 1.8e-11 | P = 3.3e-11 | P = 2.2e-12 | P = 0 | |||
| R_BRCA (404) | – | – | – | 305 | 78 | 368 |
| P = 4e-11 | P = 6.3e-12 | P = 0 | ||||
| R_UCEC (486) | – | – | – | – | 87 | 441 |
| P = 3.7e-11 | P = 3.5e-11 | |||||
| R_READ (101) | – | – | – | – | – | 88 |
| P = 0 | ||||||
| R_KIRC (1325) | – | – | – | – | – | – |
Notes: The parentheses show the number of genes in R for each cancer type. The cells show the R overlap sizes between different cancer types and the p-values of the overlaps using the hypergeometric cumulative distribution function.
Table 2.
Overlap of M signatures between cancers.
| M_GBM | M_COAD | M_BRCA | M_UCEC | M_READ | M_KIRC | M_KIRP | M_LUSC | |
|---|---|---|---|---|---|---|---|---|
| M_OV (597) | 139 | 341 | 375 | 143 | 68 | 268 | 8 | 351 |
| P = 1e-11 | P = 4e-12 | P = 1e-11 | P = 1e-11 | P = 0 | P = 1e-11 | P = 0 | P = 0 | |
| M_GBM (495) | – | 190 | 241 | 55 | 31 | 133 | 4 | 156 |
| P = 1e-11 | P = 3e-12 | P = 3e-11 | P = 0 | P = 3e-12 | P = 1e-5 | P = 0 | ||
| M_COAD (1749) | – | – | 705 | 143 | 131 | 403 | 9 | 628 |
| P = 1e-11 | P = 1e-11 | P = 0 | P = 1e-11 | P = 1e-10 | P = 0 | |||
| M_BRCA (1787) | – | – | – | 155 | 98 | 466 | 7 | 709 |
| P = 2e-11 | P = 0 | P = 1e-11 | P = 4e-7 | P = 0 | ||||
| M_UCEC (184) | – | – | – | – | 39 | 121 | 5 | 149 |
| P = 0 | P = 4e-11 | P = 3e-9 | P = 3e-12 | |||||
| M_READ (131) | – | – | – | – | – | 73 | 4 | 86 |
| P = 0 | P = 9e-8 | P = 0 | ||||||
| M_KIRC (866) | – | – | – | – | – | – | 9 | 361 |
| P = 0 | P = 0 | |||||||
| M_KIRP (10) | – | – | – | – | – | – | – | 7 |
| P = 4e-8 | ||||||||
| M_LUSC (1258) | – | – | – | – | – | – | – | – |
Notes: The parentheses show the number of methylation sites in M for each cancer type. The cells show the M overlap sizes between different cancer types and the P-values of the overlaps using the hypergeometric cumulative distribution function.
We used P-values (matlab’s hygecdf function) to evaluate the significance of the overlap sizes of M and R between different cancer types, getting small P-values near zero, as Tables 1–2 show. We also evaluated the significance of the correlations between miRNA, methylation and gene expression by using permutation-based P-values. This involved repeatedly shuffling the values of one of the data types over all samples and re-computing the correlations over one million trials; the P-values were always near zero, which points to the correlations’ significance.
We further evaluated whether the R and M associations were specific for the particular genes and methylation sites in GBM. For this purpose, we reversed roles by looking for any correlation of expression of M genes with methylation of R genes. When we looked for correlation of CX3CL1 gene expression (rather than methylation) with the methylation of any genes in R_GBM, there was always zero correlation. This supports that the results are specific to M methylation and R gene expression and hsa-miR-142 is a good representative of the signature.
R is enriched in glioblastoma mesenchymal subclasses
We found the signature profile to reflect two genetically distinct subclasses, which in glioblastoma tend to correspond to the previously identified proneural and mesenchymal subclasses. Table 3 shows how the miRNAs and expressed genes (R) are differentially expressed between the different glioblastoma subclasses. The miRNA hsa-miR-142 and genes R were distinctly expressed between proneural-mesenchymal glioblastoma subclasses. Our integrated analysis revealed that the signature differs between glioblastoma subclasses in that the miRNA hsa-miR-142 and genes R are underexpressed in proneural and overexpressed in mesenchymal. The majority of samples with over-expressed R and hsa-miR-142 belong to the mesenchymal subclass and the P-value of the distributions illustrates the significance.
Table 3.
The distribution of glioblastoma samples between proneural, neural, classical and mesenchymal classes according to R gene and miRNA expression.
| R genes and hsa-miR-142 expression | Proneural | Neural | Classical | Mesenchymal | Multinomial probability |
|---|---|---|---|---|---|
| Over-expressed | 2 | 4 | 2 | 25 | 5.49178E-09 |
| Under-expressed | 12 | 2 | 5 | 0 | 0.001937516 |
Notes: We observed a significant difference between the classes: in proneural R is under-expressed, while in mesenchymal R is over-expressed. The multinomial probability in the last column is the total probability under the null hypothesis that at least 25 out of 33 over-expressed samples (or at least 12 out of 19 under-expressed samples) would have been classified in any one of the four classes.
We examined the overlap of the R_GBM genes and the R_ovarian genes with the mesenchymal (216), classical (162), proneural (178) and neural (129) genes from (Verhaak and others 2010).26 182 of the R_GBM genes are found in the mesenchymal genes and 122 of the R_ovarian genes are found in the mesenchymal genes. There was significantly less overlap of R_GBM and R_ovarian with proneural genes (53 and 15, respectively), neural genes (3 and 10) or classical genes (8 and 11). Therefore, the genes in R significantly overlap with the genes in26 that distinguish between proneural vs. mesenchymal subtypes in glioblastoma. The P-values of the overlaps are <0.0001.
Discussion
Table 4 gives the functional annotations of the top-ranked genes in R; we derived these genes as shown in Supplementary files 8–9. The genes R are all expressed in human hematopoietic stem cells or leukocytes. Many of these genes are involved in regulating T-cell activation; T-cell is a type of lymphocyte. The gene set R is particularly enriched in genes known to be preferentially expressed in T-cell differentiation stages.32 Some of the R genes represent membrane proteins involved in cell-cell adhesion in leukocytes. Lymphocytes are known to play a role in cancer, but their role remains quite controversial.33 Lymphocytes are generally known to detect and eliminate cancer cells, but on the other hand infiltration of lymphocytes, such as B cells and T cells, into cancer can be an indication of poor prognosis (recurrence and metastasis). It was shown that lymphocytes that make the RANKL inflammatory protein are critical for metastasis. No metastasis was found in mice without lymphocytes, but adding RANKL in mice restored the potential for metastasis compensating for the function of lymphocytes.34
Table 4.
The known functions of the top-ranked genes in R.
| SASH3 (CXorf9) | Signaling adapter protein in lymphocytes | CD2 | Cell adhesion molecule found on the surface of T cells and natural killer cells |
| CCL5 | Regulated upon activation, normal T-cell expressed | CD37 | Transmembrane protein, leukocyte antigen, may play a role in T-cell-B-cell interactions |
| FLJ21438 | Proteins in B-cell exosomes | DOK3 | Negative regulator of JNK signaling in B-cells |
| Rgr | Membrane protein, retinal G-protein coupled receptor | PSCD4 | Plasma membrane, regulation of cell adhesion |
| TLR2 | Membrane protein, immune system signaling pathway | ITGB2 | Leukocyte cell adhesion molecule |
| APBB1IP | Peripheral membrane protein, mediates Rap1-induced adhesion | BTK | Peripheral membrane protein, plays a crucial role in B-cell development (mature B lymphocytes) |
| DOCK2 | Peripheral membrane protein, haematopoietic and lymphocyte cell-specific protein | CXCR3 | Membrane protein, expressed primarily on activated T lymphocytes and NK cells, regulate leukocyte trafficking |
| ARHGAP9 | Regulates adhesion of hematopoietic cells to the extracellular matrix | FYB | Signaling transduction in T cells, modulates the expression of interleukin-2 |
| LCP2 | Lymphocyte protein promoting T cell development | PTPRC | Required for T-cell activation. Interleukin-12-dependent in activated lymphocytes |
| CD53 | Leukocyte surface antigen, signal transduction in T cells | LAIR1 | Leukocyte-associated receptor, found on NK cells, T cells, and B cells |
| TRAF3IP3 | Gene expressed in t-lyphocytes | URP2 | Cell adhesion in hematopoietic cells. Required for leukocyte adhesion to endothelial cells |
| CYBB | Glycoprotein integral to plasma membrane | AIF1 | Promotes the proliferation of T-lymphocytes. Enhances lymphocyte migration |
| NCKAP1L | Membrane-associated haematopoietic protein | ITGB2 | Leukocyte cell adhesion molecule |
| IL10RA | Interleukin-10 receptor in membrane proteins, expressed in hemopoietic cells and lymphocytes | HAVCR2 | T-cell membrane protein |
| LAPTM5 | Transmembrane protein associated with lysosomes, may play a role in hematopoiesis | CD48 | Ligand for CD2. Might facilitate interaction between activated lymphocytes. Probably involved in regulating T-cell activation |
| PLEK | Hemopoietic progenitor cell differentiation | SLA | Negatively regulates T-cell receptor (TCR) signaling |
| ARHGAP30 | Rho GTPase activating protein 30 | CCR5 | Integral membrane protein, mainly expressed on T cells |
| ARHGAP25 | Actin remodeling, cell migration | EVI2B | Integral membrane protein, bone marrow and blood expression |
Note: All of the genes are expressed in leukocytes, specifically lymphocytes, or haematopoietic stem cells.
Table 5 shows the methylation sites that appear in M for both ovarian cancer and GBM. Similar to R, most of M consists of trans-membrane proteins, which are related to cell-cell adhesion functions. For example, SSTR3, SSTR5, TMEM149, are all trans-membrane proteins with cell-cell adhesion functions. A few of the proteins are also involved in signal transduction. The most prominent correlation was between hsa-miR-142 and the CX3CL1 methylation site, which was shown to control cell invasion in glioma.35 Many of the genes in R are also found in the M− set, which suggests that their undermethylation affects their expression. In recent work, hsa-miR-142 was shown to be expressed in hematopoietic and leukocytic cells, which supports the miRNA’s association with R and M.27,36–39
Table 5.
Functional annotations of the methylation sites that overlap between M_GBM and M_ovarian.
| AFF3 (M−) | Expressed in the lymphoid system, transcription regulation | LAPTM5 (M−) | Transmembrane receptor associated with lysosomes |
| ALDH3A1 (M+) | Metabolism of neurotransmitters | LCP2 (M−) | Lymphocyte protein promoting T cell development |
| ARHGAP25 (M−) | Actin remodeling, cell migration | LRP3 (M+) | Integral membrane protein |
| BIN2 (M−) | Breast cancer-associated | LSM7 (M+) | Ribonucleoprotein complex |
| C10orf27 (M−) | Cell differentiation | LY86 (M−) | Inflammatoy response, apoptosis |
| C16orf54 (M−) | Transmembrane protein | MAMSTR (M+) | Transcription regulation |
| C2orf40 (M−) | Cancer-related augurin precursor | MPHOSPH9 (M−) | Peripheral membrane protein |
| C6orf25 (M−) | Plasma membrane-bound cell surface receptor | MTMR11 (M +) | Protein-tyrosine phosphatase |
| CARD8 (M−) | Apoptotic protein | NCF2 (M−) | Superoxide-generating NADPH oxidase activity |
| CCDC80 (M+) | Promotes cell adhesion and matrix assembly | NCOR2 (M+) | Transcriptional repression |
| CD101 (M−) | Leukocyte surface membrane protein | OGG1 (M+) | DNA repair enzyme |
| CD6 (M−) | Plasma membrane protein involved in T-cell activation | OSM (M−) | Tumor inhibitor |
| CD79B (M−) | B lymphocyte receptor | PAQR6 (M+) | Integral membrane protein |
| CHRM1 (M+) | G protein-coupled receptor membrane protein | PHKG1 (M+) | Protein kinase activity |
| CX3CL1 (M+) | T cell leukocyte adhesion and migration process at the endothelium | PLD4 (M−) | Single-pass membrane protein |
| DAPP1 (M−) | Peripheral membrane protein, B lymphocyte adapter protein | PLEKHA4 (M+) | Peripheral Membrane protein |
| DAPK2 (M+) | Cell apoptosis inducer | POR (M−) | ER membrane oxidoreductase |
| DDAH1 (M+) | Regulator of nitric oxide generation | PPP2R1A (M+) | Protein phosphatase |
| DNAI1 (M+) | Dynein intermediate chain, cytoplasmic | PRELP (M+) | Extracellular matrix, collagen binding in connective tissue |
| FBN3 (M+) | Extracellular matrix structural constituent, fibrillin. | PTGFRN (M+) | Integral membrane protein, Single-pass type I membrane protein |
| FAM113B (M−) | Hydrolase activity | PTPRCAP (M−) | Transmembrane phosphoprotein, plasma membrane, integral membrane protein |
| FAM78A (M−) | Hypothetical protein | RB1 (M−) | Tumor suppressor, negative regulator of the cell cycle |
| FGR (M−) | Cell migration and adhesion | ROBO4 (M−) | External side of plasma membrane |
| FUT3 (M+) | Membrane protein, tumor metastasis and adhesion | RPE65 (M+) | Plasma membrane protein |
| GGT1 (M+) | Membrane protein | RUNX1 (M−) | Acute myeloid leukemia 1 protein |
| GIPC1 (M+) | Regulator cell surface receptor, trafficking | SEMA3B (M−) | Extracellular membrane, Neuronal development, tumor suppression by apoptosis induction |
| GPX2 (M−) | Glutathione peroxidase | SHROOM1 (M−) | Neuronal development |
| PLEK (M−) | Hemopoietic progenitor cell differentiation | SLA (M−) | Negative regulator T-cell receptor (TCR) signaling |
| AFF3 (M−) | Expressed in the lymphoid system, transcription regulation | LAPTM5 (M−) | Transmembrane receptor associated with lysosomes |
| GRIP1 (M+) | Glutamate receptor-interacting protein 1 | SLC44A2 (M+) | Plasma membrane protein |
| HKDC1 (M+) | Hexokinase domain-containing protein 1 | SNCG (M+) | Breast cancer-specific gene 1 protein |
| IL17RE (M−) | Membrane protein, interleukin receptor | SSTR3 (M+) | Plasma membrane protein |
| IL18BP (M−) | Extracellular binding | SSTR5 (M+) | Plasma membrane protein |
| IL22RA1 (M+) | Membrane protein, interleukin receptor | TMEM149 (M−) | Transmembrane protein |
| INCA1 (M+) | Inhibitor of CDK | TNFAIP8L2 (M−) | Tumor necrosis factor, immune homeostasis |
| INPP5J (M+) | Plasma membrane protein | TNKS1BP1 (M+) | Tankyrase-1-binding protein, enzyme binding |
| KCNQ1 (M+) | Potassium voltage-gated channel | TRAF1 (M−) | TNF receptor associated factor |
| KIAA0427 (M+) | Regulation of translational initiation | WFDC2 (M−) | Extracellular region, proteolysis |
| KLHL34 (M−) | Kelch-like protein 34 | ZNF205 (M+) | Zinc finger protein, transcription regulation |
| KLHL6 (M−) | Protein binding, B-lymphocyte antigen receptor signaling | ZNF48 (M+) | Zinc finger protein, transcription regulation |
| LAMB2 (M+) | Basement membrane protein, attachment, migration and organization of cells into tissues during embryonic development | ZNF512B (M+) | Zinc finger protein, transcription regulation |
Notes: Many of the genes in R are also found in the M− set and the names of these genes are highlighted in bold.
We examined the annotation enrichment of the 139 M methylation sites and 671 R genes that overlap between ovarian cancer and GBM, using the Broad Institute’s Gene Set Enrichment Analysis tool. There was an overlap with several modules that were previously found to be implicated in cancer.40 The enriched Gene Ontology annotations included plasma membrane, response to wounding, cell-cell adhesion, extracellular space, cell-surface receptor signal transduction, inflammatory response, oxidoreductase activity. The P-values of the overlaps of M and R with these modules is <1.0e-5. These results suggest the methylation of M+ and M−, as well as miRNA and gene expression, may play a role in progressive disruption of adhesion and junction molecules and loss of cell adhesion components. In several types of cancer, disruption of cell-cell adhesion molecules is a hallmark in phenotypes such as the epithelial-mesenchymal transition and invasion.41
Table 6 shows the top clusters for a functional annotation clustering of the 671 R genes that overlap between ovarian cancer and GBM, as derived with the DAVID tool.30 Inflammatory and defense response are the most significant annotations, which is consistent with the anti-inflammatory functions of the interleukin genes found in the top-ranked R genes (see Table 4). Integral membrane and transmembrane annotations are also among the most enriched annotations. Additionally, we examined the R genes in the Broad Institute’s Gene Set Enrichment Analysis (GSEA) tool, finding that one third of the genes overlap with sets of known membrane proteins. Several subsets of these genes were previously implicated in breast cancer (74 up regulated and 43 down regulated), liver cancer (41), bladder cancer (59 up regulated) and thyroid cancer (38 up regulated). The P-values of the overlaps is low. Supplementary files 12–13 give the complete functional annotation clusters for R as derived using DAVID, as well as the enriched annotations in M using the Broad Institute’s GSEA tool.
Table 6.
The top three functional annotation term clusters associated with the list of 671 R genes that overlap between R_ovarian and R_GBM.
| Gene ontology annotation | Count | P-value |
|---|---|---|
| GO:0006952~defense response | 81 | 3.31E-43 |
| GO:0006954~inflammatory response | 47 | 4.55E-26 |
| GO:0009611~response to wounding | 54 | 1.12E-22 |
| Disulfide bond | 150 | 9.53E-37 |
| Disulfide bond | 146 | 1.09E-35 |
| Topological domain:Extracellular | 135 | 1.61E-30 |
| Glycoprotein | 174 | 1.83E-30 |
| Glycosylation site:N-linked (GlcNAc...) | 168 | 3.25E-29 |
| Topological domain:Cytoplasmic | 147 | 1.20E-27 |
| Signal | 137 | 3.38E-24 |
| Signal peptide | 137 | 6.20E-24 |
| Membrane | 202 | 9.05E-24 |
| Receptor | 88 | 4.19E-22 |
| GO:0005886~plasma membrane | 162 | 1.80E-21 |
| Transmembrane region | 170 | 2.00E-21 |
| Transmembrane | 170 | 4.00E-21 |
| GO:0031224~intrinsic to membrane | 189 | 5.47E-15 |
| GO:0016021~integral to membrane | 184 | 1.11E-14 |
| GO:0005886~plasma membrane | 162 | 1.80E-21 |
| GO:0005887~integral to plasma membrane | 81 | 3.86E-20 |
| GO:0031226~intrinsic to plasma membrane | 81 | 1.52E-19 |
| GO:0044459~plasma membrane part | 113 | 4.82E-19 |
Note: Annotations are clustered together if they have similar gene members; the more common genes annotations share, the higher the chance they will be grouped together. The count shows how frequently the particular annotation occurs in a cluster and the p-value shows the likelihood that such a count or a higher one would be observed in a random cluster. The p-value associated with each annotation term inside a cluster is statistically measured by Fisher Exact in DAVID system.
Several miRNAs are known to suppress methylation via interactions with DNMTs and RBL2 proteins. DNMTs are known to be responsible for the methylation of the CpG islands of tumor suppressors in an RBL2-dependent manner.16,17 Therefore, miRNAs and their DNMT targets are known to cause epigenetic changes (or their reversal) that contribute to malignant transformation in human tumors.19 The aberrant expression of DNA methyltransferases (DNMTs) is often a poor prognostic indicator for cancer.42 We speculate that the M signature may be partially induced because hsa-miR-142 targets the DNA methyltransferases (DNMTs).31,43 Either DNMT3L or DNMT3B was part of our M+ signature in most cancers. DNMT3L was hypermethylated in mesenchymal GBM phenotype together with hsa-miR-142 overexpression. The miRNA hsa- miR-142 is known to target the DNA methyltransferases DNMT1 and DNMT3A, which are also regulated by DNMT3L.44 DNMT3L is a regulatory factor of DNA methyltransferases, which is essential for the function of DNMT3A and DNMT3B.45,46 In previous studies, the coexpression of DNMT3L with DNMT3A resulted in a striking stimulation of de novo methylation by DNMT3A.47 Whether hsa-miR-142 influences the methylation of M via its regulation of DNMTs remains to be determined in lab experiments. Additionally, hsa-miR-142 has CpG islands embedded in its promoter region.15 Genes associated with CpG islands are believed to be targeted by DNA methyltransferases and are known to be prone to methylation in cancer14 suggesting hsa-miR-142 may be regulated by epigenetics (see Supp. File 4).
Conclusion
By performing miRNA vs. gene expression and miRNA vs. methylation analysis, our work identified hsa-miR-142 as the miRNA most strongly and consistently correlated with a set of genes and methylation sites. Our results show that miRNA expression is associated with altered expression profiles of genes in R and methylated sites in M. The hsa-miR-142 miRNA is the best representative of this signature for all cancers (see Supp. Files 5 and 11). When we used hsa-miR-142 as a proxy in several cancers the resulting M and R overlapped significantly between cancer types. The P-value of the resulting overlaps between cancer types was close to 0. This signature appears to be associated with transformation from proneural to mesenchymal subclass in glioblastoma. The results suggest that in the mesenchymal tumor subtypes R genes tend to be over-expressed, M+ is hypermethylated, M− is hypomethylated and hsa-miR-142 is over-expressed. In proneural subtypes, on the other hand, the gene and miRNA expression and methylation are the opposite. Our study is the first that associated this signature with several cancer types, showing distinct methylation and expression patterns between the proneural and mesenchymal subclasses.
All of the R genes are known to be expressed in leukocytes and haematopoietic stem cells. Many of the M genes represent membrane proteins related to cell-cell adhesion functions. We speculate that in cancer the signature may contribute to a progressive loss of cell-cell adhesion. We found either DNA methyltransferase DNMT3L or DNMT3B to be methylated and associated strongly with hsa-miR-142 expression in the cancer types examined. From a previous study, DNA methyltransferases were shown to be associated with the methylation of cell adhesion-related genes in carcinomas.41 Integrated microRNA and gene expression and methylation data for healthy human samples was unavailable at the time of analysis; in the future, a human dataset of healthy samples may provide additional insights.
Supplementary Files
The supplementary material for this manuscript is available from 9037SupplementaryFiles.zip.
Supp 1: The R signature in ovarian cancer, GBM, colon, kidney, uterine, rectum, breast cancer. Includes Bonferroni-corrected P-values, correlations and mutual information. Derived based on correlations with hsa-miR-142.
Supp 2: The M signature in ovarian cancer and GBM. Includes Bonferroni-corrected P-values, correlations and mutual information. Derived based on correlations with hsa-miR-142.
Supp 3: Joint R and M signatures averaged between ovarian cancer and glioblastoma.
Supp 4: The CpG islands located in the promoters of miRNAs on the genome.
Supp 5: The miRNA-gene expression correlation value histograms for individual cancer types.
Supp 6: The number of samples with M and R signature in glioblastoma and ovarian cancer.
Supp 7: The first metagene is the list of R genes that occur in both ovarian cancer and GBM at P-value <0.01, sorted in terms of the joint ovarian/GBM average correlation with hsa-miR-142.
Supp 8: The R signature is verified using the first metagene in 11 cancers (glioblastoma, ovarian, breast, colon, kidney clear/papillary cell, lung squamous cell/adenocarcinoma, lower grade glioma, uterine, rectum). In all cancers we ranked the genes based on their correlation to the first metagene.
Supp 9: The second metagene. Derived by averaging the mutual information of each gene to the first metagene (weighted by the number of samples in each cancer) over 11 cancer types and ranking the genes.
Supp 10: The M signature is verified using the second metagene in glioblastoma, ovarian, breast, colon, uterine, rectum, kidney and lung cancer. In all cancers we ranked the methylation sites based on their correlation to the second metagene.
Supp 11: The significance of the microRNAs is verified using the second metagene in glioblastoma and ovarian cancer. In GBM and ovarian we ranked the microRNAs based on their correlation to the second metagene.
Supp 12: Functional annotation clustering of the R genes using the DAVID tool.
Supp 13: Annotations enriched in the M methylation sites using the Broad Institute’s Gene Set Enrichment Analysis Tool (GSEA).
Acknowledgements
The authors would like to thank Wei Yi Cheng for performing the analysis of RNASeq and miRNASeq data and Dr. Hoon Kim for helpful discussions.
List of Abbreviations Used
- R_GBM
the R signature of gene expression for glioblastoma
- R_OV
the R signature of gene expression for ovarian cancer
- M_GBM
the M signature of methylation for glioblastoma
- M_OV
the M signature of methylation for ovarian cancer
- M_COAD
the M signature of methylation for colon cancer
- M_BRCA
the M signature of methylation for breast cancer
- M_UCEC
the M signature of methylation for uterine cancer
- M_READ
the M signature of methylation for rectum adenocarcinoma
- M_KIRC
the M signature of methylation for kidney renal clear cell carcinoma
- M_KIRP
the M signature of methylation for kidney renal papillary cell carcinoma
- M_LUSC
the M signature of methylation for lung squamous cell carcinoma
- M+
the methylation sites in M that are positively correlated with hsa-miR-142
- M−
the methylation sites in M that are negatively correlated with hsa-miR-142
- miRNA
microRNA
- DNMT
DNA methyltransferase
- GBM
glioblastoma
Footnotes
Author Contributions
Conceived and designed the experiments: BA, DA. Analysed the data: BA, DA. Wrote the first draft of the manuscript: BA. Contributed to the writing of the manuscript: BA, DA. Agree with manuscript results and conclusions: BA, DA. Jointly developed the structure and arguments for the paper: BA, DA. Made critical revisions and approved final version: BA, DA. All authors reviewed and approved of the final manuscript.
Disclosures and Ethics
As a requirement of publication author(s) have provided to the publisher signed confirmation of compliance with legal and ethical obligations including but not limited to the following: authorship and contributorship, conflicts of interest, privacy and confidentiality and (where applicable) protection of human and animal research subjects. The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material. Any disclosures are made in this section. The external blind peer reviewers report no conflicts of interest.
References
- 1.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 2.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bandres E, Agirre X, Bitarte N, et al. Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer. 2009;125:2737–43. doi: 10.1002/ijc.24638. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Gabriely G, Yi M, Narayan RS, et al. Human glioma growth is controlled by microRNA-10b. Cancer Res. 2011;71:3563–72. doi: 10.1158/0008-5472.CAN-10-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bredel M, Scholtens DM, Harsh GR, et al. A network model of a cooperative genetic landscape in brain tumors. JAMA. 2009;302:261–75. doi: 10.1001/jama.2009.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim TM, Huang W, Park R, Park PJ, Johnson MD. A developmental taxonomy of glioblastoma defined and maintained by MicroRNAs. Cancer Res. 2011;71:3387–99. doi: 10.1158/0008-5472.CAN-10-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li A, Walling J, Ahn S, et al. Unsupervised analysis of transcriptomic profiles reveals six glioma subtypes. Cancer Res. 2009;69:2091–9. doi: 10.1158/0008-5472.CAN-08-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer cell. 2006;9:157–73. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 10.Spence T, Nguyen J, Bouffet E, Huang A. MicroRNAs in Brain Tumors MicroRNAs in Cancer Translational Research. Springer Science + Business Media BV; 2011. [Google Scholar]
- 11.Srinivasan S, Patric IR, Somasundaram K. A ten-microRNA expression signature predicts survival in glioblastoma. PLoS One. 2011;6:e17438. doi: 10.1371/journal.pone.0017438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer cell. 2010;17:510–22. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lujambio A, Calin GA, Villanueva A, et al. A microRNA DNA methylation signature for human cancer metastasis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13556–61. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pavicic W, Perkio E, Kaur S, Peltomaki P. Altered methylation at microRNA-associated CpG islands in hereditary and sporadic carcinomas: a methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA)-based approach. Mol Med. 2011;17:726–35. doi: 10.2119/molmed.2010.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou X, Ruan J, Wang G, Zhang W. Characterization and identification of microRNA core promoters in four model species. PLoS Comput Biol. 2007;3:e37. doi: 10.1371/journal.pcbi.0030037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benetti R, Gonzalo S, Jaco I, et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol. 2008;15:998. doi: 10.1038/nsmb0908-998b. [DOI] [PubMed] [Google Scholar]
- 17.Sinkkonen L, Hugenschmidt T, Berninger P, et al. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol. 2008;15:259–67. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- 18.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 19.Croce CM. Nat Rev Genet. Vol. 10. Causes and consequences; 2009. of microRNA dysregulation in cancer; pp. 704–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buffa FM, Camps C, Winchester L, et al. MicroRNA-associated progression pathways and potential therapeutic targets identified by integrated mRNA and microRNA expression profiling in breast cancer. Cancer Res. 2011;71:5635–45. doi: 10.1158/0008-5472.CAN-11-0489. [DOI] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas Research N. Bell D, Berchuck A, et al. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong H, Siu H, Luo L, Fang X, Jin L, Xiong M. Investigation gene and microRNA expression in glioblastoma. BMC Genomics. 2010;11(Suppl 3):S16. doi: 10.1186/1471-2164-11-S3-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Z, Asmann YW, Kalari KR, et al. Integrated analysis of gene expression, CpG island methylation, and gene copy number in breast cancer cells by deep sequencing. PLoS One. 2011;6:e17490. doi: 10.1371/journal.pone.0017490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu K, Cui J, Olman V, Yang Q, Puett D, Xu Y. A comparative analysis of gene-expression data of multiple cancer types. PLoS One. 2010;5:e13696. doi: 10.1371/journal.pone.0013696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Enerly E, Steinfeld I, Kleivi K, et al. miRNA-mRNA integrated analysis reveals roles for miRNAs in primary breast tumors. PLoS One. 2011;6:e16915. doi: 10.1371/journal.pone.0016915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chin L, Hahn WC, Getz G, Meyerson M. Making sense of cancer genomic data. Genes Dev. 2011;25:534–5. doi: 10.1101/gad.2017311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masica DL, Karchin R. Correlation of somatic mutation and expression identifies genes important in human glioblastoma progression and survival. Cancer Res. 2011;71:4550–61. doi: 10.1158/0008-5472.CAN-11-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 31.Dietmann S, Lee W, Wong P, Rodchenkov I, Antonov AV. CCancer: a bird’s eye view on gene lists reported in cancer-related studies. Nucleic Acids Res. 2010;38:W118–23. doi: 10.1093/nar/gkq515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee MS, Hanspers K, Barker CS, Korn AP, McCune JM. Gene expression profiles during human CD4+ T cell differentiation. Int Immunol. 2004;16:1109–24. doi: 10.1093/intimm/dxh112. [DOI] [PubMed] [Google Scholar]
- 33.DeNardo DG, Coussens LM. Inflammation and breast cancer. Balancing immune response: crosstalk between adaptive and innate immune cells during breast cancer progression. Breast Cancer Res. 2007;9:212. doi: 10.1186/bcr1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan W, Zhang W, Strasner A, et al. Tumour-infiltrating regulatory T cells stimulate mammary cancer metastasis through RANKL-RANK signalling. Nature. 2011;470:548–3. doi: 10.1038/nature09707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sciume G, Soriani A, Piccoli M, Frati L, Santoni A, Bernardini G. CX3CR1/CX3CL1 axis negatively controls glioma cell invasion and is modulated by transforming growth factor-beta1. Neuro Oncol. 2010 doi: 10.1093/neuonc/nop076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kasashima K, Nakamura Y, Kozu T. Altered expression profiles of microRNAs during TPA-induced differentiation of HL-60 cells. Biochem Biophys Res Commun. 2004;322:403–10. doi: 10.1016/j.bbrc.2004.07.130. [DOI] [PubMed] [Google Scholar]
- 37.Sun W, Shen W, Yang S, Hu F, Li H, Zhu TH. miR-223 and miR-142 attenuate hematopoietic cell proliferation, and miR-223 positively regulates miR-142 through LMO2 isoforms and CEBP-beta. Cell Res. 2010;20:1158–69. doi: 10.1038/cr.2010.134. [DOI] [PubMed] [Google Scholar]
- 38.Yuan W, Sun W, Yang S, et al. Downregulation of microRNA-142 by protooncogene LMO2 and its co-factors. Leukemia. 2008;22:1067–71. doi: 10.1038/sj.leu.2405001. [DOI] [PubMed] [Google Scholar]
- 39.Zhao S, Wang Y, Liang Y, et al. MicroRNA-126 regulates DNA methylation in CD4+ T cells and contributes to systemic lupus erythematosus by targeting DNA methyltransferase 1. Arthritis Rheum. 2011;63:1376–86. doi: 10.1002/art.30196. [DOI] [PubMed] [Google Scholar]
- 40.Segal E, Friedman N, Koller D, Regev A. A module map showing conditional activity of expression modules in cancer. Nat Genet. 2004;36:1090–8. doi: 10.1038/ng1434. [DOI] [PubMed] [Google Scholar]
- 41.Liu WB, Cui ZH, Ao L, et al. Aberrant methylation accounts for cell adhesion-related gene silencing during 3-methylcholanthrene and diethylnitrosamine induced multistep rat lung carcinogenesis associated with over-expression of DNA methyltransferases 1 and 3a. Toxicol Appl Pharmacol. 2011;251:70–8. doi: 10.1016/j.taap.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 42.McCabe MT, Brandes JC, Vertino PM. Cancer DNA methylation: molecular mechanisms and clinical implications. Clin Cancer Res. 2009;15:3927–7. doi: 10.1158/1078-0432.CCR-08-2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veeck J, Esteller M. Breast cancer epigenetics: from DNA methylation to microRNAs. J Mammary Gland Biol Neoplasia. 2010;15:5–17. doi: 10.1007/s10911-010-9165-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanwal R, Gupta S. Epigenetics and cancer. J Appl Physiol. 2010;109:598–605. doi: 10.1152/japplphysiol.00066.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Emburgh BO, Robertson KD. Modulation of Dnmt3b function in vitro by interactions with Dnmt3L, Dnmt3a and Dnmt3b splice variants. Nucleic Acids Res. 2011;39:4984–5002. doi: 10.1093/nar/gkr116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wienholz BL, Kareta MS, Moarefi AH, Gordon CA, Ginno PA, Chedin F. DNMT3L modulates significant and distinct flanking sequence preference for DNA methylation by DNMT3A and DNMT3B in vivo. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chedin F, Lieber MR, Hsieh CL. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:16916–21. doi: 10.1073/pnas.262443999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The supplementary material for this manuscript is available from 9037SupplementaryFiles.zip.
Supp 1: The R signature in ovarian cancer, GBM, colon, kidney, uterine, rectum, breast cancer. Includes Bonferroni-corrected P-values, correlations and mutual information. Derived based on correlations with hsa-miR-142.
Supp 2: The M signature in ovarian cancer and GBM. Includes Bonferroni-corrected P-values, correlations and mutual information. Derived based on correlations with hsa-miR-142.
Supp 3: Joint R and M signatures averaged between ovarian cancer and glioblastoma.
Supp 4: The CpG islands located in the promoters of miRNAs on the genome.
Supp 5: The miRNA-gene expression correlation value histograms for individual cancer types.
Supp 6: The number of samples with M and R signature in glioblastoma and ovarian cancer.
Supp 7: The first metagene is the list of R genes that occur in both ovarian cancer and GBM at P-value <0.01, sorted in terms of the joint ovarian/GBM average correlation with hsa-miR-142.
Supp 8: The R signature is verified using the first metagene in 11 cancers (glioblastoma, ovarian, breast, colon, kidney clear/papillary cell, lung squamous cell/adenocarcinoma, lower grade glioma, uterine, rectum). In all cancers we ranked the genes based on their correlation to the first metagene.
Supp 9: The second metagene. Derived by averaging the mutual information of each gene to the first metagene (weighted by the number of samples in each cancer) over 11 cancer types and ranking the genes.
Supp 10: The M signature is verified using the second metagene in glioblastoma, ovarian, breast, colon, uterine, rectum, kidney and lung cancer. In all cancers we ranked the methylation sites based on their correlation to the second metagene.
Supp 11: The significance of the microRNAs is verified using the second metagene in glioblastoma and ovarian cancer. In GBM and ovarian we ranked the microRNAs based on their correlation to the second metagene.
Supp 12: Functional annotation clustering of the R genes using the DAVID tool.
Supp 13: Annotations enriched in the M methylation sites using the Broad Institute’s Gene Set Enrichment Analysis Tool (GSEA).