Abstract
In the twelve years since the process of RNA interference (RNAi) was first discovered, great progress has been made in understanding its mechanism and exploiting its ability to silence gene expression to study gene function at a genome-wide level. Its extensive use as a screening method has yielded many published lists of genes that play novel roles in higher eukaryotes. However, the usefulness of this information is potentially limited by the occurrence of unintended off-target effects. Here we review the potential causes of off-target effects, and the impact of this phenomenon in interpreting the results of high-throughput RNAi screens. In addition to targeting the intended gene product, artificial short interfering RNAs (siRNAs) can produce off-target effects by down-regulating the expression of multiple messenger RNAs through microRNA-like targeting of the 3′ untranslated region. We examine why this phenomenon can produce high hit rates in siRNA screens, and why independent validation of screening results is critical for the approach to yield new biological insights.
Keywords: RNA interference: A cellular process that downregulates gene expression at a post-transcriptional step. One strand of a small double strand RNA (siRNA or miRNA) integrates in a protein complex called RISC and targets an mRNA with sequence complementarity for cleavage, translation inhibition and/or mRNA decay.; RISC: The RNA-Induced Silencing Complexes are multi-protein complexes containing an Argonaute protein, a small single strand RNA and additional regulatory proteins.; Short interfering RNA (siRNA): An endogenous or artificial (synthetic) small double strand RNA with perfect pairing of the two strands. One strand (called the guide strand) targets RISC to perfectly complementary mRNAs, resulting in endonucleolytic cleavage of the mRNA by the RISC component Ago2.; MicroRNA (miRNA): A mature endogenous small double strand RNA, often with imperfect pairing of the two strands, originating from processing of a precursor RNA molecule by cellular RNases. miRNAs target RISC complexes to sites that have imperfect sequence complementarity, and that are typically located in the 3′ untranslated region of the mRNA.; RNAi off-target effect: Any effect of an RNAi-based treatment that is a consequence of reducing the expression of an unintended target.; siRNA or miRNA seed sequence: The 5′ region of the guide strand of an siRNA or miRNA sequence, extending from nucleotides 2–7 (hexamer) or 2–8 (heptamer).; siRNA multiplicity validation: Validation of a gene-phenotype association by showing that the phenotype can be produced by multiple independent siRNAs that target different regions of the same mRNA
Introduction
The effects of RNA interference (RNAi) were first observed in 1990 in plants (petunia) as an unintended consequence of transgene overexpression (1, 2). The underlying mechanism remained obscure for eight years until the pioneering work of Fire, Mello and colleagues demonstrated that double stranded RNA was critical for suppressing gene expression in C. elegans (3). The availability of the sequenced C. elegans genome, as well as the simplicity of performing RNAi experiments in this organism, led to the first systematic, chromosome-wide analysis of gene function using this method (4, 5), and subsequently to genome-wide screens (6-10). Many screens have also been performed using Drosophila cell culture systems (11-14), where it is straightforward to induce RNAi by treating cells with long double stranded RNA molecules.
Potential adoption of these methods to mammalian cells was limited by the fact that long double stranded RNAs potently activate the innate immune system (15-19), an ancient antiviral response. However, once it became clear that short double stranded RNAs (or short interfering RNAs; siRNAs, 21 nucleotides) could induce gene silencing without strongly activating this response (20), siRNA-based methods have become widely adopted as a tool for studying gene function. Development of libraries of chemically synthesized siRNAs or plasmid-encoded short hairpin RNAs (shRNAs) has enabled the execution of many genome-wide screens in mammalian cells (for reviews, see (refs. 21-25). Despite the transformative potential of this new technology, it is important to recognize that suppression of gene expression by RNAi is not equivalent to inactivation of a gene by mutation. Unlike genetic approaches that modify the DNA of an organism, RNAi acts at the level of mRNA, decreasing mRNA levels or the ability of the mRNA to be translated. As a result, RNAi-based experiments can suffer from a lack of sensitivity due to incomplete suppression of gene expression, or a lack of specificity due to suppression of unintended genes. Here we review the potential challenges of RNAi-based screening, the need for vigilance in designing and interpreting a screen, and the importance of independent validation of screening results. We also examine some potential reasons for the relatively low rate of hit validation from published RNAi screens.
A convergent machinery for the siRNA and microRNA pathways sets the stage for off-target effects
Because experimentally induced RNAi-based gene inactivation co-opts the endogenous cellular RNAi machinery, it is important to understand the normal functions carried out by the RNAi pathway to appreciate how off-target effects can arise. RNAi-based gene regulation can be broadly classified as siRNA-based or microRNA-based (Figure 1; see (26, 27) for review). The key physiological distinction between these pathways is that siRNAs typically silence the expression of the genes from which they are derived, whereas microRNAs typically silence the expression of heterologous genes. For example, the siRNA pathway is important in host defense to suppress the expression of genes encoded by an invading virus. In this case, the siRNAs are derived from double-stranded RNA intermediates that arise during viral replication, and the target RNA is the sequence from which the siRNAs are derived. In contrast, microRNAs are important for normal regulation of gene expression in cells, and downregulate the expression of genes located at loci distinct from the site of expression. However, once the small RNA is generated, the pathways rely on a common downstream machinery to inhibit gene expression. This overlap in function is a key reason that exogenous siRNAs used as research tools can induce nonspecific microRNA-like effects, reducing the expression of unintended genes.
Figure 1.
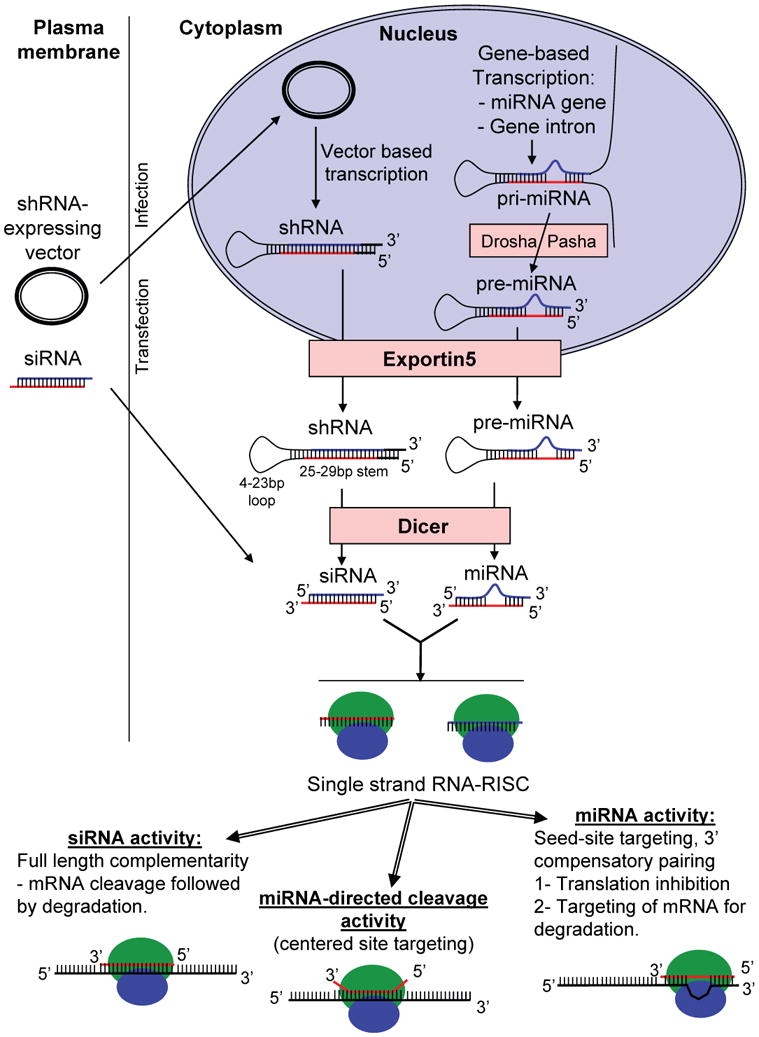
Model of mammalian RNA interference pathways.
RNA molecules involved in RNAi pathways originate from artificial sources including siRNA transfection and shRNA expression from a transfected vector or endogenous sources where miRNA genes encode a pri-miRNA. A protein complex Drosha/Pasha cleaves the pri-miRNA to generate one or more pre-miRNA molecule. shRNAs and pre-miRNAs are transported into the cytoplasm by a mechanism involving Exportin5. The RNase III enzyme Dicer further cleaves shRNAs and pre-miRNAs into mature siRNAs and miRNAs, respectively. siRNA and miRNA single strands incorporate into RISC complexes and serve as templates to target mRNAs for cleavage, translation inhibition and/or mRNA decay. The fate of the targeted mRNA depends on the extent of sequence pairing.
In the siRNA pathway, double stranded RNA that is expressed by an invading virus, or that is expressed by convergent transcription from the genome, is processed by the enzyme Dicer into short 21–22 bp double stranded siRNAs. Next, one strand, called the guide strand, is preferentially incorporated into RISC (RNA-induced silencing complex), and the complementary strand is degraded. However, because strand choice depends on thermodynamic stability of the ends of the double-stranded RNA, in some cases either strand may be incorporated into RISC, leading to a chance of targeting unintended messages. The guide strand then targets RISC to complementary mRNAs, and the Argonaute protein of RISC (Ago2 in mammals) cleaves the target mRNA, rendering it susceptible to degradation by exonucleases. Cleavage by Argonaute requires perfect sequence complementarity at the site of cleavage, 10 bp from the 5′ end of the guide strand. Because an siRNA induces endonucleolytic cleavage of the target mRNA, it can strongly reduce gene expression.
In the microRNA pathway, the genes encoding a microRNA are transcribed by RNA polymerase II and they are capped and polyadenylated like other Pol II transcripts. The resulting transcript, which contains one or more hairpin structures, is processed by the enzyme Drosha in the nucleus, and the resulting product is exported to the cytoplasm, where it is processed by Dicer to yield a mature microRNA. At this point, the siRNA and microRNA pathways converge, as a single strand of RNA derived from the microRNA is incorporated into RISC and guides the selection of silencing targets. However, because microRNAs typically do not have perfect sequence complementarity with their targets, RISC does not generally induce cleavage of the target mRNA. Instead, translational repression is the main outcome, although in many cases the target mRNA is also destabilized by mRNA decay (28). In contrast to the siRNA pathway where complete sequence complementarity is required for target mRNA cleavage, microRNA-induced effects typically require a short region of homology between the microRNA and its target (Figure 2). This region, referred to as the “seed region”, is present at the 5′ end of the guide strand of the microRNA, typically extending from nucleotides 2–8 (29), though recent work indicates that a somewhat longer seed region in the center of the microRNA can lead to mRNA target cleavage in a manner reminiscent to siRNA-directed cleavage (30). As a consequence, targeted genes need only contain a short region of sequence homology to the microRNA, typically seven to eight nucleotides in length. Because of this limited sequence complementarity, a given microRNA has the potential to target a large number of different mRNAs in the cell. The target sites for most endogenous microRNAs are located in the 3′ UTRs of target genes, perhaps because microRNA-RISC complexes are not easily displaced by the translocating ribosome. Repression of gene expression by a microRNA-containing RISC complex is generally not as complete as that induced by a perfectly-matched siRNA-containing complex, perhaps leading to only 50% downregulation (29, 31). Cellular mRNAs may also be targeted by multiple microRNAs, thereby enhancing downregulation of the message. The microRNA pathway is known to regulate many different cellular processes (32, 33). Over 1000 different microRNAs are expressed in human cells (34), and it has been estimated that each microRNA may have an average of 300 different targets, with over half of human protein-coding genes containing conserved microRNA target sequences (35). Because microRNA-based regulation has such a broad reach in cellular biology, its perturbation has the potential to affect almost any type of cellular screen or experiment that might be performed.
Figure 2.
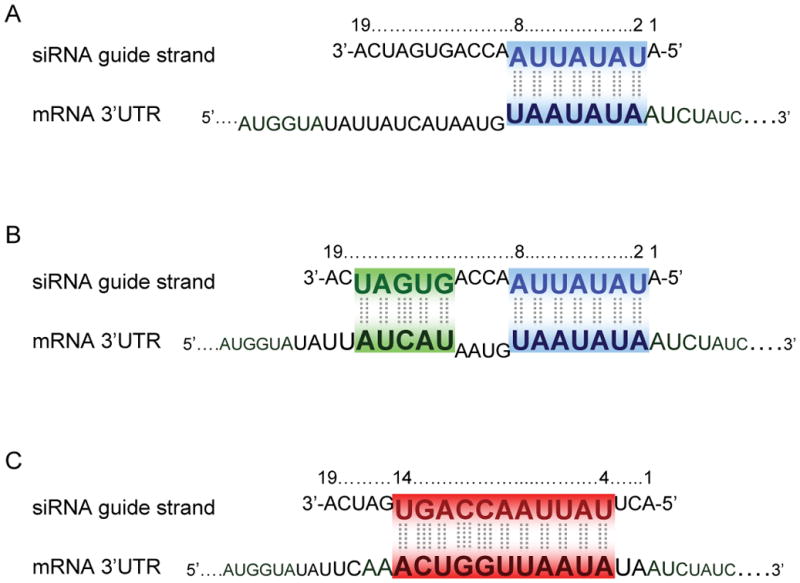
Seed sequence-based interaction of siRNA guide strand with an off-target mRNA.
The 7-nucleotide seed region of the siRNA guide strand (position 2–8) shows full complementarity (blue, A and B) to a target site in the 3′UTR of off-targeted mRNA. Additional siRNA 3′ sequence pairing (green, B) to the target mRNA may contribute to efficient off-targeting by the siRNA. Centered complementarity site (siRNA position 4– 14 or 5–15) to the mRNA target (red, C) can drive a RISC slicer activity-dependent cleavage of the target mRNA.
Sequence-independent off-target effects
There are several mechanisms through which introduction of an exogenous siRNA or shRNA may broadly affect the physiology of a cell, independent of the specific sequence of the reagent used (see (36) for review). First, exogenous expression of short hairpin RNAs (shRNAs) can interfere with endogenous processing of microRNAs. For example, sustained expression of an exogenous shRNA in mice can be fatal, perhaps as a consequence of saturating the pathway that is used to export miRNA precursors from the nucleus (37). Second, introduction of siRNAs into a cell can displace endogenous microRNAs from RISC, thereby altering normal patterns of gene expression. In support of this idea, bioinformatics analysis of published transcriptional profiling experiments revealed that the predicted targets of endogenous microRNAs are expressed at higher levels following transfection of an siRNA designed to target a discrete gene (38). Third, even when using short double stranded RNAs, cells may mount non-specific immune response to transfected synthetic double strand RNA (dsRNA) (39) or viruses used to express shRNAs (40). Fourth, siRNAs have been shown to induce a type of cell-stress response when used at high concentration (41, 42). Therefore, investigators need to be aware that introduction of an exogenous siRNA or shRNA molecule may alter the physiology of a cell in a manner independent of the specific sequence of the siRNA or shRNA.
Sequence-specific off-target effects
In addition to the potential for an exogenously-introduced siRNA or shRNA to globally perturb miRNA-based gene silencing in the cell, each individual siRNA or shRNA may downregulate the expression of a subset of genes in a sequence-dependent manner by acting like a microRNA. In this case, in addition to inducing cleavage of the intended target via perfect base-pairing and Argonaute-induced cleavage, the siRNA may induce microRNA-like effects via interaction with target sequences through its seed region (26, 43-45) (Figure 2). Because this phenomenon requires only a short region of sequence homology, each siRNA has the potential to nonspecifically downregulate the expression of hundreds of different genes in the cell.
Transcriptional profiling has proved to be a useful tool to identify off-target effects at a genome-wide level. However, this approach only detects changes in mRNA level, and therefore may miss off target effects that act only by suppressing translation. In 2003, the first evidence of microRNA-like off-target effect was reported (46). In this study, a series of siRNAs were designed to target two different genes, MAPK14 and IGF1R. Transcriptional profiling revealed that each of the siRNAs produced a distinct pattern of effects on transcription. Though the intended target was downregulated, the expression of many other genes was also affected, in a range of 1.5-fold to 4-fold. This effect was first observed at a high concentration (100 nM) of siRNA, but persisted when the dose was reduced to 4 nM. Kinetic analysis indicated that these changes occurred early and prior to any changes in protein levels of the intended target, suggesting they were not an indirect consequence of knockdown of the intended protein. Analysis of the downregulated genes indicated that as few as eleven contiguous nucleotides of identity to the siRNA were sufficient to induce downregulation, consistent with a microRNA-like effect.
A similar conclusion was reached by a study that analyzed the transcriptional profile of cells treated with 12 different siRNAs targeting three genes (47). This study also used siRNA at 100 nM concentration, and identified 347 genes that were downregulated by only one siRNA but not by other siRNAs targeting the same gene, suggesting they were off-target effects. Similar results were obtained when the concentration of siRNA was reduced to 50 nM. Analysis of the off-targeted genes indicated the frequent presence of one or more perfect sequence matches within the 3′ untranslated region (UTR) matches to the hexamer or heptamer seed region (positions 2– 7 or 2–8) of the guide strand of the siRNA, consistent with a microRNA-like off-target effect. The importance of the seed match region in mediating these effects was confirmed in a later study (48), as alteration of the seed region of the siRNA eliminated the off-target effects but created a new set of off-targeted genes. This study also demonstrated that seed-match mediated off target effects can also arise from expression of shRNAs in cells, and is not restricted to the effect of transfected siRNAs.
Although the evidence for the ability of siRNAs to induce microRNA-like off target effects is strong, it is important to point out that not all studies have reached this conclusion. Whereas one study identified dose-dependent off target effects of an siRNA targeting GFP (49), another transcriptional analysis of cells treated with siRNAs targeting GFP showed no reproducible nonspecific changes in gene expression (50). Other studies have suggested that when a lower concentration of siRNA is used (20 nM), the effects can be quite specific (42). In an analysis of five siRNAs targeting the Rb1 mRNA, transcriptional profiling revealed that the expression of 903/919 genes was altered by three out of five siRNAs; 720 were regulated by all 5 siRNAs (42). Another recent study indicated that selecting for siRNAs with most potent specific knockdown allows using the siRNAs at low nanomolar concentrations which reduced the extent of off-target effects (51). Together these studies suggest that most of the changes in gene expression are a consequence of knockdown of the intended target, but that each siRNA also nonspecifically perturbs the expression of a limited number of additional transcripts.
An important question is whether this level of nonspecific perturbation of gene expression is sufficient to produce changes in cell physiology. This appears to be the case. For example, some siRNAs targeting the gene MEN1 induced changes in expression of p53 and p21 at the mRNA and protein levels in multiple cell lines (52). However, some siRNAs that knocked down MEN1 equally well did not cause changes in p53 or p21 levels, suggesting that alteration of p53 and p21 expression was a result of an off-target effect. This study illustrates why it is essential to correlate a phenotype with the degree of knockdown of the intended target; a lack of correlation strongly suggests that an off-target effect may be involved.
These studies indicate that the ability of an siRNA to produce an off-target effect depends not only on dose of the siRNA but also on the sequence of the specific siRNA used in the study. Based on the ability of an siRNA to induce microRNA-like knockdown through interactions with the seed region of the siRNA, Anderson et al. (53) hypothesized that siRNAs that contain a higher number of seed matches with 3′UTRs in the genome would show a higher frequency of off-target effects. For each siRNA, they calculated a seed complement frequency (SCF), which represents the number of 3′ UTRs in the genome that contain a sequence match to the seed sequence of the siRNA. They first used gene expression profiling to analyze the effects of knockdown of two housekeeping genes by ten different siRNAs each. These siRNAs had a range of SCFs, ranging from high (>3800), to medium (2500–2800) to low (<350). Transcriptional profiling revealed extensive microRNA-like off-target effects for siRNAs with high or medium SCF, but not for siRNAs with low SCF. In addition, the authors demonstrated that siRNAs with low SCF were three-fold less likely to nonspecifically inhibit viability or induce apoptosis via off-target effects. Together these findings indicate the importance of minimizing the SCF when designing siRNAs to study gene function or in generating siRNA libraries for high-throughput screening.
Properties of the mRNA may also impact its susceptibility to off-target effects. Given the similarity in mechanism between seed-match-based off-target effects and microRNA-based targeting, properties of an mRNA that render it sensitive to microRNA targeting may also increase its susceptibility to off-target effects. Grimson et al. performed an extensive analysis of the determinants that contribute to effectiveness of miRNA targeting in addition to seed sequence pairing (54). Target site location in the 3′UTR, their multiplicity and spacing, and additional pairing of the miRNA nucleotides 12-17 were correlated with enhanced miRNA targeting efficiency. The sequence and structural context of mRNA 3′UTR target sites may also play an important role as it was found that higher local AU-richness, potentially weakening secondary structure to increase target site accessibility, was associated with more effective microRNA-based targeting. It is reasonable to expect that such properties would also enhance susceptibility to microRNA-like off-target effects in siRNA library screens.
Although the majority of seed-match-based off-target effects occur by targeting sites in the 3′UTR, recent cross-linking studies suggest the RISC complexes are targeted to coding regions at a surprisingly high frequency. Cross-linking immunoprecipitation (CLIP) of Argonaute and related RNAi proteins has allowed unbiased identification of RISC target sites in a transcriptome-wide manner (55-57). These studies revealed that although most (66-84%) RISC-binding sites occur in exons, only 46-60% of these occurred in 3′ untranslated regions while 38-50% occurred in coding sequences. Only a small fraction (2-4%) mapped to 5′UTR regions. Interestingly, a significant number of RISC binding sites (12-14%) mapped to pre-mRNA intronic regions. The functional significance of RISC targeting to coding sequences or intronic sequences remains unclear, but Hafner et al.(56) determined that targeting to coding regions caused limited but significant destabilization of corresponding mRNAs. These studies indicate that much remains to be elucidated about the function of small RNA molecules, and therefore additional mechanisms of off-target effects, beyond those that target the 3′ UTR, remain possible.
The impact of off-target effects in RNAi screening
Although off-target effects can occur, the degree of gene modulation is typically two-fold or less. An important practical question is whether such off-target effects are sufficient to produce false-positive hits in high-throughput screens, where it might be expected that a greater degree of knockdown of the intended target would be necessary to produce a strong phenotype. However, several investigators have reported that some of the top hits in their screens arise due to microRNA-based off target effects. In a screen to identify novel regulators of the HIF1-α transcription pathway, the top scoring siRNAs did not act through specific knockdown of their intended targets, but rather by targeting the HIF1- α mRNA through microRNA-like seed match effects against sequences in the 3′ UTR (58). In a separate screen for modulators of sensitivity to the Bcl-2 targeting drug ABT-737, the same group determined that many active siRNAs nonspecifically targeted the key anti-apoptotic protein Mcl-1, again through microRNA-like seed match effects (59). Off-target effects through a microRNA-like mechanism were also prominent in an siRNA screen for novel regulators of the TRAIL apoptosis induction pathway (60), suggesting that RNAi screening enriches for siRNAs with relevant off-target effects. Finally, in a screen to identify new components of the spindle checkpoint pathway, we found that the vast majority of active siRNAs acted by targeting the 3′UTR of the known spindle checkpoint gene Mad2 through seed match effects (F. Sigoillot and R. King, unpublished data). Together these results indicate that when evaluating the results from high-throughput screens, investigators should ensure that active siRNAs targeting novel genes do not contain potential seed matches against known components of the pathway.
The role of siRNA multiplicity in siRNA screening
One commonly practiced method to reduce the impact of off-target effects in high-throughput screens is to screen multiple siRNAs or shRNAs per gene, either as a pool or individual reagents, and then to characterize only those genes for which multiple siRNAs yield a phenotype. In this approach, the chance of the phenotype resulting from an off-target effect is believed to be minimized because each siRNA should have a distinct spectrum of off-target effects. Here we examine the effectiveness of this approach in distinguishing true hits from false positive results in siRNA screens.
Table 1 summarizes the results of twenty-nine siRNA or shRNA screens performed in mammalian cells (Table 1 is adapted and expanded from that of (24)). Analysis of Table 1 indicates that the primary hit rate for these screens varies widely, from a low of 0.4% to a high of 25.6%, with a median of 2.3%. For a genome-wide screen involving 20,000 genes, this would yield approximately 460 primary hits. For two of the published screens, there is no validation of the reported hits from the screens. For fourteen of the screens, validation is provided only by showing that at least two or more siRNAs targeting a given gene produce the same phenotype. For the remaining ten screens, additional validation of hits is performed, but typically only a very small fraction of the total hits are validated by independent methods (ranging from 1–5 genes per screen). Therefore, the overall rate of validation relative to the total hit rate is low. The low rate of hit validation may be explained in part by the fact that some validation approaches may be challenging and time-consuming, as described below. Alternatively, it is possible that even genes that score on multiple siRNAs may still represent an off-target effect, thereby reducing the validation rate. In practice, many groups consider a hit to be a true positive if at least two independent siRNAs for a gene produce the phenotype. Here we question the validity of that assumption.
Table 1. Validated genes yield in RNAi high-throughput screens in mammalian cells (adapted from Mohr et al.(24)).
| Cell type | Phenotype | Reagent | SiRNA concentration (nM) |
Number of genes/mRNA targets tested |
Primary screen hits |
Primary Hit Rate (%) |
Number of RNAi molecules scoring | Validated genesa |
References | Year | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | >4 | ||||||||||
| HeLa P4/R5 | Host genes involved in HIV replication | Pooled siRNAs | not provided | 19,709 | 936 | 4.7 | 1 | 9 | 4 | (75) | 2008 | |||
| Jurkat | Host genes involved in HIV replication | Pooled shRNAs | n.a. | 54,509 mRNAs/ESTs | 252 | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | 3 | (86) | 2009 |
| HeLa | Host genes involved in west Nile Virus infection | Pooled siRNAs | 50 | 21,121 | 305 | 1.4 | 180 | 97 | 18 | 2 | (95) | 2008 | ||
| HeLa-derived TZM-b1 | Host genes involved in HIV replication | Pooled siRNAs | 50 | 21,121 | 386 | 1.8 | 191 | 26 | 2 | (84) | 2008 | |||
| HeLa | Spindle assembly checkpoint genes | shRNA | n.a. | 780 | 16 | 2.1 | 6 | 2 | 1b | (89) | 2007 | |||
| HeLa | Regulators of Wnt/β-Catenin pathway | Pooled siRNAs | not provided | 21,125 | 530 | 2.5 | 214 | 16 | 1 | (108) | 2008 | |||
| 293T, HeLa, MCF7 | Regulators of genotoxic stress-induced apoptosis | Pooled shRNAs | n.a. | genome-wide | 30 | n.a. | 29 | 1 | 1c | (92) | 2008 | |||
| Huh7/Rep-Feo | Host genes required for HCV replication | Pooled siRNAs | 50 | 21,094 | 236 | 1.1 | 148 | 35 | 1 | (107) | 2009 | |||
| mouse ES | Regulators of mTERT gene transcription | shRNA | n.a. | 1,360 | 18 | 1.3 | 17 | 1 | 1c | (88) | 2010 | |||
| Huh 7.5.1 | Host genes required for HCV replication | Pooled siRNAs | 50 | 19,470 | 521 | 2.7 | 262 | 5d | (96) | 2009 | ||||
| HeLa | Cell cycle progression and division | esiRNA | 30 | 5,305 | 275 | 5.2 | n.a. | n.a. | n.a. | n.a. | n.a. | 0 | (93, 94) | 2004 |
| H1299 | HIF1 alpha signal transduction | siRNA | 100 | 507 | -- | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | 0e | (58) | 2005 |
| U2OS | Cell cycle progression and division | siRNA | 12.5 | 24,373 and 5,000f | 1,152 | n.a. | 505 | 612 | 0 | (101) | 2006 | |||
| HT29 | Regulators of mitosis | shRNA | n.a. | 1,028 (4903 shRNAs) | 102 | n.a. | 66 | 19 | 2 | 1 | 0 | (100) | 2006 | |
| NCI-H1155 | Modulators of responsiveness to Taxol | Pooled siRNAs | 40 | 21,121 | 87 | 0.4 | 24 | 0 | (109) | 2007 | ||||
| NIH3T3 | Modulators of oxidative stress resistance | Pooled shRNAs | n.a. | genome-wide | 15 | n.a. | 14 | 1 | 0 | (103) | 2007 | |||
| NCI-H196 | Modulators of Bcl-2/Bcl-XL inhibitor ABT-737-induced death | Pooled siRNAs | 25 | 4,000 | 145 | 3.6 | 3 | 0e | (59) | 2007 | ||||
| MCF10A | Genes essential for viability of breast cancer cells | Pooled shRNAs | n.a. | 1000s | 201 | n.a. | 6 | 3 | 0g | (106) | 2008 | |||
| 293T | Host genes involved in HIV replication | Pooled siRNAs | n.a. | 19,628 | 4,555 | 23.2 | 295 | 0 | (85) | 2008 | ||||
| DLD1 | Regulators of Wnt/β-Catenin pathway | Pooled siRNAs | 25 | 20,042 | 740 | 3.7 | 119 | 0 | (99) | 2008 | ||||
| HEK293 | Regulators of cell adhesion | Pooled shRNAs | n.a. | 8500 (43,828 shRNAs) | -- | n.a. | 1 | 0 | (91) | 2008 | ||||
| Jurkat | Resistance to FAS-induced apoptosis | Pooled shRNAs | n.a. | 9500 (45,000 shRNAs) | 11 | n.a. | 5 | 0 | (97) | 2008 | ||||
| MNT-1 | factors involved in melanin biogenesis | Pooled siRNAs | 1.4 | 21,127 | 98 | 0.5 | 25 | 0 | (90) | 2008 | ||||
| RDG3 | Stress granules and Processing bodies assembly | Pooled siRNAs | 40 | 7,317 | -- | n.a. | 171 | 0 | (104) | 2008 | ||||
| BJtsLT | biomarkers for response to an anticancer drug | Pooled shRNAs | n.a. | 8,000 | 100 | 1.3 | 6 | 0 | (102) | 2009 | ||||
| DLD1 | Synthetic lethal interactions with Ras oncogene | Pooled shRNAs | n.a. | 32,293 | 368 | 1.1 | 357 | 11 | 0 | (98) | 2009 | |||
| U2OS | Modulators of circadian clock genes transcription | Pooled siRNAs | 12.5 | 22,468 | 5,751 | 25.6 | 5,383 | 368 | 0 | (110) | 2009 | |||
| HeLa | Modulators of TRAIL-induced apoptosis | siRNA | 2.5pmol/96well | 6,095 | 752 | 12.3 | 721 | 31 | 0 | (60) | 2010 | |||
| GL261 derivative | Regulators of ATF5 transcription | Pooled shRNAs | n.a. | ∼28,000 | 12 | n.a. | 3 | 0 | (105) | 2010 | ||||
Gene validated by rescue expression or with an independent method (not multiplicity of siRNAs)
Bioinformatics database cross-referencing was not considered as validation since it is not clear how information in the databases is itself validated.
TAO1 Kinase siRNA was rescued by expression of a siRNA-resistant form, but other work (80) suggests this may be an off-target effect.
No rescue experiments were performed. assays independent of RNAi were performed (mTERT promoter-luciferase construct were shown to have a functional top hit HIF1alpha consensus site (88); Top hit TAF1 (92) overexpression induced apoptosis).
5 hits were previously reported to interact with HCV core component proteins. No rescue experiments were performed.
Top hits were indentified as seed-match induced off target effects
Two overlapped siRNA libraries were used
The authors report a pooled shRNA methodology and do not focus on gene-phenotype association, thus no validation is attempted
n.a. not available
In Table 2, we consider a hypothetical genome-wide screen (targeting 20,000 genes) using 4 siRNAs per gene. Each siRNA is screened individually, or as a pool and then deconvoluted, to identify which of the four siRNAs is active in the assay. This yields a list of candidate genes, as well as the number of siRNAs that are active for each gene. Published studies have indicated that each siRNA may nonspecifically reduce the expression of 30–100 genes by two-fold or more due to microRNA-like effects (46-49, 61). Based on this information, we assumed that an average siRNA might downregulate 50 genes by two-fold or more through an off-target microRNA-like effect. We then calculated the expected frequency of on-target and off-target genes that would be discovered in a screen, based on the number of genes in the pathway that would yield a phenotype if downregulated by two-fold or more. For the off-target genes, we calculated the frequency with which a gene would be expected to be targeted by one or more of the four different siRNAs.
Table 2.
Estimation of the number of off-target siRNAs that may arise from genome-wide RNAi screens.
In this analysis, we calculate the hit rate and validation rate in an siRNA screen of 20,000 genes using four siRNAs per gene. Genes chosen for validation would include any gene for which at least 2/4 siRNAs produce a phenotype; we assume that at least 2/4 siRNAs that target the true (specific) genes involved in the pathway would yield a phenotype.
Assuming that a given siRNA has on average 50 off-target mRNAs (leading to mRNA knockdown by more than 2 fold) and that these off-targets are random, the probability that an siRNA has a seed match that results in off-targeting for an mRNA out of 20,000 genes is 50/ 20,000 = 1/ 400. Let Y denote the number of genes among the indicated number of genes required for phenotype n ={1, 5, 10, 20, 50} that have a seed match to a given siRNA. If each of the n genes is independent of each other, then P(Y) obeys the binomial distribution B (n, 1/400). Therefore the probability of a given siRNA not to contain a seed match to any of the n genes is given as follows:
P(Y=0) = C10,0 (1/400)0 (1/400)n-0
The probability that a given siRNA has a seed match to at least one of the n genes is
p = 1 - P(Y=0)
The probability P that exactly A = {1, 2, 3, 4} siRNAs out of 4 independent siRNAs tested for a given gene give phenotype as a result of an off-target effect follows the binomial distribution:
P(A) = C4,A pA p4-A
The estimated number of genes with exactly A out of 4 siRNAs giving phenotype as a result of an off-target effect was determined by multiplying the probability P(A) for each n by 20,000, assuming this number of genes was tested in the siRNA screen.
| Number of Genes in a Pathway that Yield Phenotype when Expression Reduced by 50% | 1 | 5 | 10 | 20 | 50 |
|---|---|---|---|---|---|
| A. Number of On-target Genes That Score | 1 | 5 | 10 | 20 | 50 |
| B. Number of Off-target Genes That Score | 200 | 976 | 1906 | 3630 | 7877 |
| B1. Number of genes with exactly 1/4 siRNAs that score | 199 | 958 | 1835 | 3362 | 6465 |
| B2. Number of genes with exactly 2/4 siRNAs that score | 1 | 18 | 70 | 259 | 1293 |
| B3. Number of genes with exactly 3/4 siRNAs that score | 0 | 0 | 1 | 9 | 115 |
| B4. Number of genes with exactly 4/4 siRNAs that score | 0 | 0 | 0 | 0 | 4 |
| Total Number of Genes Identified by Screen (A+B) | 201 | 981 | 1916 | 3650 | 7927 |
| Hit Rate ([A+B]/20,000) | 1% | 4.9% | 9.5% | 18% | 39% |
| Fraction of Genes that are On Target (A/[A+B]) | 0.5% | 0.5% | 0.5% | 0.5% | 0.6% |
| Validation Rate (A/[A+B2+B3+B4]) | 50% | 21.7% | 12.3% | 6.9% | 3.4% |
Inspection of Table 2 reveals several points. First, regardless of the number of genes involved in the pathway, the vast majority of hits are due to off-target effects. Second, the usefulness of siRNA multiplicity for distinguishing between on-target and off-target effects depends strongly on the number of genes that participate in the pathway. For example, if only one gene is involved in a pathway, then it is reasonably likely that any gene for which two of four siRNAs score is due to knockdown of the intended gene rather than to an off-target effect. However, if there are ten or more genes involved in a pathway, the majority of genes that confirm on two out of four siRNAs are instead likely to result from an off-target effect. For example, with ten genes in a pathway, eighty-one genes would score on two or more siRNAs, yet only ten of these genes would represent a specific result of knockdown of the intended gene. The problem becomes worse as the number of genes involved in a pathway increases. Finally, it is also evident that the overall hit rate can become very high, even if there are only 20 genes required for the phenotype. It is likely that many cellular pathways rely on even larger numbers of genes. Based on the likelihood of such off-target effects, it is not surprising that several of the published screens show primary hit rates in excess of twenty percent, with a very low rate of validation of hits.
Approaches for validation
Several reviews (24, 36, 62-64) and editorials (65) have discussed approaches for validating the results of siRNA screens, but as can be seen in Table 1, most hits from siRNA screens are not validated. As we have discussed, caution must be used when employing a criterion of multiple siRNAs to validate a result, especially if the pathway being studied consists of many genes that could contribute to the phenotype. If multiple siRNAs are used to validate a result, it is essential that they be distinct from the siRNAs used in the original screen, because the siRNAs from the screen have been subject to selection by the screening process.
Approaches involving chemical or structural modifications of siRNAs have been developed in an attempt to reduce the likelihood of off-target effects (reviewed in(66)). These modified siRNAs provide an additional tool to validate that knockdown of the intended mRNA is responsible for the observed phenotype. One approach is to modify the passenger strand of the siRNA, to reduce the likelihood that it would be incorporated into RISC, and thereby eliminate any off-target effects due to seed-match effects caused by the passenger strand. Different methods have been developed to neutralize passenger strand incorporation into RISC, including design of asymmetric siRNAs with a shorter 15 nucleotide passenger strand (67) or an internally segmented passenger strand (68). In both cases off-target effects from the passenger strand were reduced while guide strand RNAi efficiency was maintained. Another approach is to chemically modify all nucleotides of the passenger strand with 2′-O-methyl substitution (69). This modification reduces passenger strand incorporation while preserving guide strand incorporation.
Modifications of the guide strand have been developed to reduce the likelihood of seed-match-based off-target effects. One approach is to reduce the thermodynamic stability of the interaction of the seed region with the target mRNA. This can be accomplished by substituting the 8 base pair region at the 5′-end of the guide strand with double strand DNA (70), or by using unlinked nucleic acid (in which the carbon-carbon bond between the 2′ and 3′ of the ribose sugar is broken) at the 7 position of the seed region (71). Using reporter assays, both of these methods have been shown to reduce microRNA-like off-target effects, although the approach has not yet been validated across a range of different siRNAs that target endogenous mRNAs. Alternatively, introduction of 2′-O-methyl substitution at the second position of the seed region has been proposed to alter the manner in which RISC interacts with target mRNAs (72), selectively reducing microRNA-like off-target effects while leaving on-target silencing intact. Jackson et al. (72) found that 2′-O-methyl substitution of the guide strand at positions one and two of the seed region reduced off-target silencing without inducing novel off-targets (72). Out of 10 modified siRNAs tested, all retained strong silencing of the intended target while reducing silencing of 80% of off-target mRNAs by an average of 66%, although the effectiveness varied widely among different mRNAs. Jackson et al. determined (72) that siRNAs with stronger hybridization in the seed region (lower free energy of RNA:RNA binding for bases 2-7) responded less efficiently to 2′-O-methyl modifications than weaker-hybridizing siRNAs, indicating that the modification does not fully abrogate all off-target effects. Because siRNAs with strong seed hybridization may be more effective in reducing expression, they may produce strong phenotypes and may be more likely to be identified in siRNA screens. Therefore, confirmation of a result using the 2′ O-methyl modified version of an siRNA discovered in a screen cannot rule out the possibility that the phenotype is due to an off-target effect. Nevertheless, use of these modified reagents in high-throughput screens may reduce the rate of false-positive hits (73).
Once multiple siRNAs against an intended target have been tested and some shown to induce studied phenotype, a simple approach that can help validate whether an effect is specific is to determine whether the level of knockdown of the intended gene, as measured by protein expression, correlates with phenotype across the series of siRNAs. If siRNAs are identified that cause efficient knockdown of the protein target, but do not induce phenotype, then it is almost certain that an off-target effect contributes to the phenotype.
Rescue experiments represent the gold standard for validation of siRNA screening results. However, as shown in Table 1 most published screens do not include such an approach. Rescue experiments test whether expression of a non-targetable form of the gene is sufficient to rescue the phenotype produced by an siRNA or shRNA. The rescue construct may contain engineered silent mutations that abrogate complementarity to the siRNA, or it may be an evolutionarily diverged form of the gene, such as a mouse gene used to rescue an siRNA-induced phenotype in human cells (74). Alternatively, if siRNAs targeting the 3′ UTR of a gene are used, rescue experiments can be performed with cDNA constructs that lack the 3′ UTR (75). Even when focusing on genes validated using multiple siRNAs, the rate of rescue of phenotype may be lower than expected, consistent with a high frequency of off-target effects even among genes scoring on multiple siRNAs. For example, Zhou et al. performed rescue experiments for nine different genes that were validated by multiple siRNAs in their screen; however, in only four of nine cases could the phenotype be rescued by expression of a non-targetable cDNA (75).
There are several potential pitfalls in designing and executing successful rescue experiments. One issue is that it can be challenging to express the rescue construct at physiological levels. If the phenotype is highly sensitive to gene dose, the experiment can be challenging to interpret. This is a major advantage of using mouse bacterial artificial chromosome (BAC) constructs for rescuing phenotypes in human cells (74, 76), as the protein is more likely to be expressed at physiological levels. Another issue is that it is essential to confirm that knockdown of the endogenous protein is not hampered by expression of the rescue construct. If the rescue construct has even limited complementarity with the siRNA, it can act as a sponge to reduce the effective concentration of the siRNA (77-79). This problem can be controlled for by ensuring that knockdown of the endogenous gene is not reduced when the transgene is expressed, or avoided entirely by a rescue strategy in which cDNAs that lack the 3′UTR are used to rescue the phenotype of siRNAs that target the 3′ UTR of the endogenous gene. Finally, there is a reported case of a false-positive rescue experiment, in which overexpression of the targeted gene rescued the phenotype by sequestration of another regulator of the pathway, reversing an off-target effect (80). Ultimately, the use of methods independent of RNAi machinery should be considered to fully validate the involvement of selected genes in the studied phenotype. For example one can conditionally knockout the genes of interest (81) or test small molecule inhibitors if they are available (82, 83).
Factors influencing the false-negative rate in RNAi-based screens
In designing an RNAi screen, another important issue to consider is how different approaches affect the rate of false-negative results. Although it is difficult to directly compare different screens due to methodological differences or cell types used, four published screens to identify host factors required for HIV replication identified no hits in common across all four screens (75, 84-86). Comparison of each of these screens to each other yielded a higher overlap, from nine to eighteen genes. However, given the fact that each screen identified hundreds (75, 84) or thousands of potential hits (85), the low degree of overlap is surprising. There are several potential explanations for the lack of overlap. First, methodological differences, including the particular cell line that is used, could explain the lack of overlap. Second, as discussed earlier, if the proportion of off-target effects is high, and these effects are dependent on the library or cell line used, a low overlap of genes would be expected. Finally, the screens may simply not be saturating, with a high rate of false-negative results in each screen. This could be explained by differences in the ability of different siRNAs or shRNAs to knockdown the gene of interest. This may depend on characteristics of the siRNA as well as characteristics of the targeted mRNA and protein. Indeed, some proteins with a half-life in days may not be decreased sufficiently when the phenotype is assessed, leading to a false negative result. In theory, it may be possible to achieve more profound knockdown of protein levels using stable expression of shRNAs under conditions of selection, as long as the gene is not essential for cell viability. There may be some genes that are difficult to target with siRNA or shRNA approaches. For example, it has been shown that more abundant mRNAs may tend to dilute out siRNAs, thereby decreasing the efficiency of knockdown (87). In addition, in many cases only a small amount of a target protein may be necessary for adequate function in the cell. Finally, functional redundancy may hamper the identification of genes when evaluating the consequence of knockdown one gene at a time. In this case, a phenotype may be revealed only after simultaneous knockdown of multiple members of a gene family.
Concluding thoughts
RNAi-based approaches have shown tremendous potential for helping us understand gene function in individual experiments. More recently, the advent of genome-wide RNAi-based screens has helped identify new components of various pathways. However, analysis of published screens indicates that the rate of identification of validated genes is less than what might have been expected. Here we have reviewed some potential pitfalls of RNAi-based screening that arise from the potential of off-target effects. Although siRNA and shRNA library design is improving to reduce the magnitude of these effects, it is essential that investigators remain vigilant about the possibility of off-target effects, and design screens and follow-up strategies to validate individual genes to ensure that the screen yields useful insights. Screens against pathways or cellular processes that involve large numbers of different components (cell proliferation, cell death, cell signaling) may be especially prone to a high rate of false positive results. In the future, performing parallel screens with two independent siRNA libraries may help reduce the rate of both false positive and false negative results. Validation of results using independent siRNA reagents as well as rescue experiments is essential to have confidence in the role of a particular gene in the process. Investigators must have an attitude of skepticism toward the role of a novel gene in a process until it has been independently validated. Only under these circumstances will RNAi-based screening meet its tremendous potential as a discovery tool.
Acknowledgments
Work in the King Lab is supported by grants from Sanofi-Aventis, the Lynch Foundation, and NIH grants GM66492 and CA89393 (Dana Farber-Harvard Cancer Center Breast Cancer SPORE).
References
- 1.Napoli C, Lemieux C, Jorgensen R. Introduction of a chimeric Chalcone Synthase gene into Petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 1990;2:279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Krol AR, Mur LA, Beld M, Mol JN, Stuitje AR. Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell. 1990;2:291–299. doi: 10.1105/tpc.2.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 4.Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 5.Gonczy P, Echeverri C, Oegema K, Coulson A, Jones SJ, Copley RR, Duperon J, Oegema J, Brehm M, Cassin E, Hannak E, Kirkham M, Pichler S, Flohrs K, Goessen A, Leidel S, Alleaume AM, Martin C, Ozlu N, Bork P, Hyman AA. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- 6.Hamilton B, Dong Y, Shindo M, Liu W, Odell I, Ruvkun G, Lee SS. A systematic RNAi screen for longevity genes in C. elegans. Genes Dev. 2005;19:1544–1555. doi: 10.1101/gad.1308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim JK, Gabel HW, Kamath RS, Tewari M, Pasquinelli A, Rual JF, Kennedy S, Dybbs M, Bertin N, Kaplan JM, Vidal M, Ruvkun G. Functional genomic analysis of RNA interference in C. elegans. Science. 2005;308:1164–1167. doi: 10.1126/science.1109267. [DOI] [PubMed] [Google Scholar]
- 8.Pothof J, van Haaften G, Thijssen K, Kamath RS, Fraser AG, Ahringer J, Plasterk RH, Tijsterman M. Identification of genes that protect the C. elegans genome against mutations by genome-wide RNAi. Genes Dev. 2003;17:443–448. doi: 10.1101/gad.1060703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simmer F, Moorman C, van der Linden AM, Kuijk E, van den Berghe PV, Kamath RS, Fraser AG, Ahringer J, Plasterk RH. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 2003;1:E12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vastenhouw NL, Fischer SE, Robert VJ, Thijssen KL, Fraser AG, Kamath RS, Ahringer J, Plasterk RH. A genome-wide screen identifies 27 genes involved in transposon silencing in C. elegans. Curr Biol. 2003;13:1311–1316. doi: 10.1016/s0960-9822(03)00539-6. [DOI] [PubMed] [Google Scholar]
- 11.Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, Haas SA, Paro R, Perrimon N. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- 12.DasGupta R, Kaykas A, Moon RT, Perrimon N. Functional genomic analysis of the Wnt-wingless signaling pathway. Science. 2005;308:826–833. doi: 10.1126/science.1109374. [DOI] [PubMed] [Google Scholar]
- 13.Foley E, O'Farrell PH. Functional dissection of an innate immune response by a genome-wide RNAi screen. PLoS Biol. 2004;2:E203. doi: 10.1371/journal.pbio.0020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muller P, Kuttenkeuler D, Gesellchen V, Zeidler MP, Boutros M. Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature. 2005;436:871–875. doi: 10.1038/nature03869. [DOI] [PubMed] [Google Scholar]
- 15.Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 16.Kleinman ME, Yamada K, Takeda A, Chandrasekaran V, Nozaki M, Baffi JZ, Albuquerque RJ, Yamasaki S, Itaya M, Pan Y, Appukuttan B, Gibbs D, Yang Z, Kariko K, Ambati BK, Wilgus TA, DiPietro LA, Sakurai E, Zhang K, Smith JR, Taylor EW, Ambati J. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452:591–597. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marques JT, Williams BR. Activation of the mammalian immune system by siRNAs. Nat Biotechnol. 2005;23:1399–1405. doi: 10.1038/nbt1161. [DOI] [PubMed] [Google Scholar]
- 18.Olejniczak M, Galka P, Krzyzosiak WJ. Sequence-non-specific effects of RNA interference triggers and microRNA regulators. Nucleic Acids Res. 2010;38:1–16. doi: 10.1093/nar/gkp829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sioud M. Innate sensing of self and non-self RNAs by Toll-like receptors. Trends Mol Med. 2006;12:167–176. doi: 10.1016/j.molmed.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 21.Cullen LM, Arndt GM. Genome-wide screening for gene function using RNAi in mammalian cells. Immunol Cell Biol. 2005;83:217–223. doi: 10.1111/j.1440-1711.2005.01332.x. [DOI] [PubMed] [Google Scholar]
- 22.Fuchs F, Boutros M. Cellular phenotyping by RNAi. Brief Funct Genomic Proteomic. 2006;5:52–56. doi: 10.1093/bfgp/ell007. [DOI] [PubMed] [Google Scholar]
- 23.Kittler R, Pelletier L, Buchholz F. Systems biology of mammalian cell division. Cell Cycle. 2008;7:2123–2128. doi: 10.4161/cc.7.14.6322. [DOI] [PubMed] [Google Scholar]
- 24.Mohr S, Bakal C, Perrimon N. Genomic screening with RNAi: results and challenges. Annu Rev Biochem. 2010;79:37–64. doi: 10.1146/annurev-biochem-060408-092949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolters NM, MacKeigan JP. From sequence to function: using RNAi to elucidate mechanisms of human disease. Cell Death Differ. 2008;15:809–819. doi: 10.1038/sj.cdd.4402311. [DOI] [PubMed] [Google Scholar]
- 26.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 28.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 29.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linsen SE, Tops BB, Cuppen E. miRNAs: small changes, widespread effects. Cell Res. 2008;18:1157–1159. doi: 10.1038/cr.2008.311. [DOI] [PubMed] [Google Scholar]
- 32.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 33.Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM, Zhang GZ. Biological functions of microRNAs: a review. J Physiol Biochem. 2010 doi: 10.1007/s13105-010-0050-6. [DOI] [PubMed] [Google Scholar]
- 34.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jackson AL, Linsley PS. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat Rev Drug Discov. 2010;9:57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 37.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 38.Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol. 2009;27:549–555. doi: 10.1038/nbt.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 40.Bridge AJ, Pebernard S, Ducraux A, Nicoulaz AL, Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat Genet. 2003;34:263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- 41.Persengiev SP, Zhu X, Green MR. Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs) RNA. 2004;10:12–18. doi: 10.1261/rna5160904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Semizarov D, Frost L, Sarthy A, Kroeger P, Halbert DN, Fesik SW. Specificity of short interfering RNA determined through gene expression signatures. Proc Natl Acad Sci U S A. 2003;100:6347–6352. doi: 10.1073/pnas.1131959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burchard J, Jackson AL, Malkov V, Needham RH, Tan Y, Bartz SR, Dai H, Sachs AB, Linsley PS. MicroRNA-like off-target transcript regulation by siRNAs is species specifiC. RNA. 2009;15:308–315. doi: 10.1261/rna.1326809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hammond SM. Dicing and slicing: the core machinery of the RNA interference pathway. FEBS Lett. 2005;579:5822–5829. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- 46.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 47.Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J, Marshall WS, Khvorova A. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- 48.Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tschuch C, Schulz A, Pscherer A, Werft W, Benner A, Hotz-Wagenblatt A, Barrionuevo LS, Lichter P, Mertens D. Off-target effects of siRNA specific for GFP. BMC Mol Biol. 2008;9:60. doi: 10.1186/1471-2199-9-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chi JT, Chang HY, Wang NN, Chang DS, Dunphy N, Brown PO. Genomewide view of gene silencing by small interfering RNAs. Proc Natl Acad Sci U S A. 2003;100:6343–6346. doi: 10.1073/pnas.1037853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X, Wang X, Varma RK, Beauchamp L, Magdaleno S, Sendera TJ. Selection of hyperfunctional siRNAs with improved potency and specificity. Nucleic Acids Res. 2009;37:e152. doi: 10.1093/nar/gkp864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scacheri PC, Rozenblatt-Rosen O, Caplen NJ, Wolfsberg TG, Umayam L, Lee JC, Hughes CM, Shanmugam KS, Bhattacharjee A, Meyerson M, Collins FS. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc Natl Acad Sci U S A. 2004;101:1892–1897. doi: 10.1073/pnas.0308698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anderson EM, Birmingham A, Baskerville S, Reynolds A, Maksimova E, Leake D, Fedorov Y, Karpilow J, Khvorova A. Experimental validation of the importance of seed complement frequency to siRNA specificity. RNA. 2008;14:853–861. doi: 10.1261/rna.704708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. Rna. 2008;14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin X, Ruan X, Anderson MG, McDowell JA, Kroeger PE, Fesik SW, Shen Y. siRNA-mediated off-target gene silencing triggered by a 7 nt complementation. Nucleic Acids Res. 2005;33:4527–4535. doi: 10.1093/nar/gki762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin X, Morgan-Lappe S, Huang X, Li L, Zakula DM, Vernetti LA, Fesik SW, Shen Y. ‘Seed’ analysis of off-target siRNAs reveals an essential role of Mcl-1 in resistance to the small-molecule Bcl-2/Bcl-XL inhibitor ABT-737. Oncogene. 2007;26:3972–3979. doi: 10.1038/sj.onc.1210166. [DOI] [PubMed] [Google Scholar]
- 60.Sudbery I, Enright AJ, Fraser AG, Dunham I. Systematic analysis of off-target effects in an RNAi screen reveals microRNAs affecting sensitivity to TRAIL-induced apoptosis. BMC Genomics. 2010;11:175. doi: 10.1186/1471-2164-11-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Myers JW, Chi JT, Gong D, Schaner ME, Brown PO, Ferrell JE. Minimizing off-target effects by using diced siRNAs for RNA interference. J RNAi Gene Silencing. 2006;2:181–194. [PMC free article] [PubMed] [Google Scholar]
- 62.Echeverri CJ, Beachy PA, Baum B, Boutros M, Buchholz F, Chanda SK, Downward J, Ellenberg J, Fraser AG, Hacohen N, Hahn WC, Jackson AL, Kiger A, Linsley PS, Lum L, Ma Y, Mathey-Prevot B, Root DE, Sabatini DM, Taipale J, Perrimon N, Bernards R. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat Methods. 2006;3:777–779. doi: 10.1038/nmeth1006-777. [DOI] [PubMed] [Google Scholar]
- 63.Echeverri CJ, Perrimon N. High-throughput RNAi screening in cultured cells: a user's guide. Nat Rev Genet. 2006;7:373–384. doi: 10.1038/nrg1836. [DOI] [PubMed] [Google Scholar]
- 64.Haney SA. Increasing the robustness and validity of RNAi screens. Pharmacogenomics. 2007;8:1037–1049. doi: 10.2217/14622416.8.8.1037. [DOI] [PubMed] [Google Scholar]
- 65.Editorial. Whither RNAi? Nat Cell Biol. 2003;5:489–490. doi: 10.1038/ncb0603-490. [DOI] [PubMed] [Google Scholar]
- 66.Gaglione M, Messere A. Recent progress in chemically modified siRNAs. Mini Rev Med Chem. 2010;10:578–595. doi: 10.2174/138955710791384036. [DOI] [PubMed] [Google Scholar]
- 67.Sun X, Rogoff HA, Li CJ. Asymmetric RNA duplexes mediate RNA interference in mammalian cells. Nat Biotechnol. 2008;26:1379–1382. doi: 10.1038/nbt.1512. [DOI] [PubMed] [Google Scholar]
- 68.Bramsen JB, Laursen MB, Damgaard CK, Lena SW, Babu BR, Wengel J, Kjems J. Improved silencing properties using small internally segmented interfering RNAs. Nucleic Acids Res. 2007;35:5886–5897. doi: 10.1093/nar/gkm548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kraynack BA, Baker BF. Small interfering RNAs containing full 2′-O-methylribonucleotide-modified sense strands display Argonaute2/eIF2C2-dependent activity. RNA. 2006;12:163–176. doi: 10.1261/rna.2150806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ui-Tei K, Naito Y, Zenno S, Nishi K, Yamato K, Takahashi F, Juni A, Saigo K. Functional dissection of siRNA sequence by systematic DNA substitution: modified siRNA with a DNA seed arm is a powerful tool for mammalian gene silencing with significantly reduced off-target effect. Nucleic Acids Res. 2008;36:2136–2151. doi: 10.1093/nar/gkn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bramsen JB, Pakula MM, Hansen TB, Bus C, Langkjaer N, Odadzic D, Smicius R, Wengel SL, Chattopadhyaya J, Engels JW, Herdewijn P, Wengel J, Kjems J. A screen of chemical modifications identifies position-specific modification by UNA to most potently reduce siRNA off-target effects. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, Marshall W, Khvorova A, Linsley PS. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fedorov Y, Anderson EM, Birmingham A, Reynolds A, Karpilow J, Robinson K, Leake D, Marshall WS, Khvorova A. Off-target effects by siRNA can induce toxic phenotype. RNA. 2006;12:1188–1196. doi: 10.1261/rna.28106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kittler R, Pelletier L, Ma C, Poser I, Fischer S, Hyman AA, Buchholz F. RNA interference rescue by bacterial artificial chromosome transgenesis in mammalian tissue culture cells. Proc Natl Acad Sci U S A. 2005;102:2396–2401. doi: 10.1073/pnas.0409861102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, Espeseth AS. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 76.Sarov M, Stewart AF. The best control for the specificity of RNAi. Trends Biotechnol. 2005;23:446–448. doi: 10.1016/j.tibtech.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 77.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hammond SM. Soaking up small RNAs. Nat Methods. 2007;4:694–695. doi: 10.1038/nmeth0907-694. [DOI] [PubMed] [Google Scholar]
- 79.Loya CM, Lu CS, Van Vactor D, Fulga TA. Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nat Methods. 2009;6:897–903. doi: 10.1038/nmeth.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hubner NC, Wang LH, Kaulich M, Descombes P, Poser I, Nigg EA. Re-examination of siRNA specificity questions role of PICH and Tao1 in the spindle checkpoint and identifies Mad2 as a sensitive target for small RNAs. Chromosoma. 2009;119:149–165. doi: 10.1007/s00412-009-0244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Berdougo E, Terret ME, Jallepalli PV. Functional dissection of mitotic regulators through gene targeting in human somatic cells. Methods Mol Biol. 2009;545:21–37. doi: 10.1007/978-1-60327-993-2_2. [DOI] [PubMed] [Google Scholar]
- 82.Eggert US, Field CM, Mitchison TJ. Small molecules in an RNAi world. Mol Biosyst. 2006;2:93–96. doi: 10.1039/b515335b. [DOI] [PubMed] [Google Scholar]
- 83.Eggert US, Kiger AA, Richter C, Perlman ZE, Perrimon N, Mitchison TJ, Field CM. Parallel chemical genetic and genome-wide RNAi screens identify cytokinesis inhibitors and targets. PLoS Biol. 2004;2:e379. doi: 10.1371/journal.pbio.0020379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 85.Konig R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, Seidel S, Opaluch AM, Caldwell JS, Weitzman MD, Kuhen KL, Bandyopadhyay S, Ideker T, Orth AP, Miraglia LJ, Bushman FD, Young JA, Chanda SK. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yeung ML, Houzet L, Yedavalli VS, Jeang KT. A genome-wide short hairpin RNA screening of jurkat T-cells for human proteins contributing to productive HIV-1 replication. J Biol Chem. 2009;284:19463–19473. doi: 10.1074/jbc.M109.010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Arvey A, Larsson E, Sander C, Leslie CS, Marks DS. Target mRNA abundance dilutes microRNA and siRNA activity. Mol Syst Biol. 2010;6:363. doi: 10.1038/msb.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Coussens M, Davy P, Brown L, Foster C, Andrews WH, Nagata M, Allsopp R. RNAi screen for telomerase reverse transcriptase transcriptional regulators identifies HIF1alpha as critical for telomerase function in murine embryonic stem cells. Proc Natl Acad Sci U S A. 2010;107:13842–13847. doi: 10.1073/pnas.0913834107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Draviam VM, Stegmeier F, Nalepa G, Sowa ME, Chen J, Liang A, Hannon GJ, Sorger PK, Harper JW, Elledge SJ. A functional genomic screen identifies a role for TAO1 kinase in spindle-checkpoint signalling. Nat Cell Biol. 2007;9:556–564. doi: 10.1038/ncb1569. [DOI] [PubMed] [Google Scholar]
- 90.Ganesan AK, Ho H, Bodemann B, Petersen S, Aruri J, Koshy S, Richardson Z, Le LQ, Krasieva T, Roth MG, Farmer P, White MA. Genome-wide siRNA-based functional genomics of pigmentation identifies novel genes and pathways that impact melanogenesis in human cells. PLoS Genet. 2008;4:e1000298. doi: 10.1371/journal.pgen.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang X, Wang JY, Lu X. Systems analysis of quantitative shRNA-library screens identifies regulators of cell adhesion. BMC Syst Biol. 2008;2:49. doi: 10.1186/1752-0509-2-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kimura J, Nguyen ST, Liu H, Taira N, Miki Y, Yoshida K. A functional genome-wide RNAi screen identifies TAF1 as a regulator for apoptosis in response to genotoxic stress. Nucleic Acids Res. 2008;36:5250–5259. doi: 10.1093/nar/gkn506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kittler R, Buchholz F. Functional genomic analysis of cell division by endoribonuclease-prepared siRNAs. Cell Cycle. 2005;4:564–567. [PubMed] [Google Scholar]
- 94.Kittler R, Putz G, Pelletier L, Poser I, Heninger AK, Drechsel D, Fischer S, Konstantinova I, Habermann B, Grabner H, Yaspo ML, Himmelbauer H, Korn B, Neugebauer K, Pisabarro MT, Buchholz F. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 2004;432:1036–1040. doi: 10.1038/nature03159. [DOI] [PubMed] [Google Scholar]
- 95.Krishnan MN, Ng A, Sukumaran B, Gilfoy FD, Uchil PD, Sultana H, Brass AL, Adametz R, Tsui M, Qian F, Montgomery RR, Lev S, Mason PW, Koski RA, Elledge SJ, Xavier RJ, Agaisse H, Fikrig E. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455:242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li Q, Brass AL, Ng A, Hu Z, Xavier RJ, Liang TJ, Elledge SJ. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc Natl Acad Sci U S A. 2009;106:16410–16415. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, Hinkle G, Boehm JS, Beroukhim R, Weir BA, Mermel C, Barbie DA, Awad T, Zhou X, Nguyen T, Piqani B, Li C, Golub TR, Meyerson M, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, Wong KK, Elledge SJ. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Major MB, Roberts BS, Berndt JD, Marine S, Anastas J, Chung N, Ferrer M, Yi X, Stoick-Cooper CL, von Haller PD, Kategaya L, Chien A, Angers S, MacCoss M, Cleary MA, Arthur WT, Moon RT. New regulators of Wnt/beta-catenin signaling revealed by integrative molecular screening. Sci Signal. 2008;1:ra12. doi: 10.1126/scisignal.2000037. [DOI] [PubMed] [Google Scholar]
- 100.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, Carpenter AE, Foo SY, Stewart SA, Stockwell BR, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 101.Mukherji M, Bell R, Supekova L, Wang Y, Orth AP, Batalov S, Miraglia L, Huesken D, Lange J, Martin C, Sahasrabudhe S, Reinhardt M, Natt F, Hall J, Mickanin C, Labow M, Chanda SK, Cho CY, Schultz PG. Genome-wide functional analysis of human cell-cycle regulators. Proc Natl Acad Sci U S A. 2006;103:14819–14824. doi: 10.1073/pnas.0604320103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mullenders J, Fabius AW, Madiredjo M, Bernards R, Beijersbergen RL. A large scale shRNA barcode screen identifies the circadian clock component ARNTL as putative regulator of the p53 tumor suppressor pathway. PLoS One. 2009;4:e4798. doi: 10.1371/journal.pone.0004798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nagaoka-Yasuda R, Matsuo N, Perkins B, Limbaeck-Stokin K, Mayford M. An RNAi-based genetic screen for oxidative stress resistance reveals retinol saturase as a mediator of stress resistance. Free Radic Biol Med. 2007;43:781–788. doi: 10.1016/j.freeradbiomed.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 104.Ohn T, Kedersha N, Hickman T, Tisdale S, Anderson P. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol. 2008;10:1224–1231. doi: 10.1038/ncb1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sheng Z, Li L, Zhu LJ, Smith TW, Demers A, Ross AH, Moser RP, Green MR. A genome-wide RNA interference screen reveals an essential CREB3L2-ATF5-MCL1 survival pathway in malignant glioma with therapeutic implications. Nat Med. 2010;16:671–677. doi: 10.1038/nm.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Silva JM, Marran K, Parker JS, Silva J, Golding M, Schlabach MR, Elledge SJ, Hannon GJ, Chang K. Profiling essential genes in human mammary cells by multiplex RNAi screening. Science. 2008;319:617–620. doi: 10.1126/science.1149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, Chung RT. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe. 2009;5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tang W, Dodge M, Gundapaneni D, Michnoff C, Roth M, Lum L. A genome-wide RNAi screen for Wnt/beta-catenin pathway components identifies unexpected roles for TCF transcription factors in cancer. Proc Natl Acad Sci U S A. 2008;105:9697–9702. doi: 10.1073/pnas.0804709105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Whitehurst AW, Bodemann BO, Cardenas J, Ferguson D, Girard L, Peyton M, Minna JD, Michnoff C, Hao W, Roth MG, Xie XJ, White MA. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature. 2007;446:815–819. doi: 10.1038/nature05697. [DOI] [PubMed] [Google Scholar]
- 110.Zhang EE, Liu AC, Hirota T, Miraglia LJ, Welch G, Pongsawakul PY, Liu X, Atwood A, Huss JW, 3rd, Janes J, Su AI, Hogenesch JB, Kay SA. A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell. 2009;139:199–210. doi: 10.1016/j.cell.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.van Dongen S, Abreu-Goodger C, Enright AJ. Detecting microRNA binding and siRNA off-target effects from expression data. Nat Methods. 2008;5:1023–1025. doi: 10.1038/nmeth.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]


