Abstract
Elevated levels of p130Cas/BCAR1 (Crk-associated substrate/breast cancer antiestrogen resistance 1) are found in aggressive breast tumors and are associated with tamoxifen resistance of mammary cancers. p130Cas promotes the integration of protein complexes involved in multiple signaling pathways frequently deregulated in breast cancer. To elucidate mechanisms leading to p130Cas up-regulation in mammary carcinomas and during acquired tamoxifen resistance, the regulation of p130Cas/BCAR1 was studied. Because multiple putative binding motifs for the inducible transcription factor EGR1 were identified in the 5′ region of BCAR1, the p130Cas/BCAR1 regulation by EGR1 and its coregulator NAB2 was investigated. Overexpression or short interfering RNA (siRNA)-mediated down-regulation of EGR1 or NAB2, and chromatin immunoprecipitations indicated that EGR1 and NAB2 act in concert to positively regulate p130Cas/BCAR1 expression in breast cancer cells. p130Cas depletion using siRNA showed that, in tamoxifen-sensitive MCF-7 cells, p130Cas regulates EGR1 and NAB2 expression, whereas in the derivative tamoxifen-resistant TAM-R cells, only NAB2 levels were influenced. BCAR1 messenger RNA and p130Cas protein were upregulated by phorbol esters following the kinetics of late response genes in MCF-7 but not in TAM-R cells. Thus, in MCF-7 cells, we identified a positive feedback loop where p130Cas positively regulates EGR1 and NAB2, which in turn induce p130Cas expression. Importantly, compared with MCF-7, enhanced NAB2 expression and increased EGR1 binding to the BCAR1 5′ region observed in TAM-R may lead to the constitutively increased p130Cas/BCAR1 levels in TAM-R cells. The uncovered differences in this EGR1/NAB2/p130Cas network in MCF-7 versus TAM-R cells may also contribute to p130Cas up-regulation during acquired tamoxifen resistance.
Introduction
p130 Crk-associated substrate (p130Cas) is an adapter protein that was first isolated from v-Src- and v-Crk-transformed rat fibroblasts [1]. Its gene was independently identified as breast cancer antiestrogen resistance 1 (BCAR1) [2,3]. Here, we refer to the gene and messenger RNA (mRNA) as BCAR1 and to the protein as p130Cas. p130Cas is the founding member of the Cas family, which includes HEF1 (NEDD9), Efs/Sin, and HEF1-Efs-p130Cas-Like (HEPL) [4–6]. Proteins of this family function as scaffolds integrating large multiprotein complexes in response to stimuli, such as growth factor and hormone release, and integrin engagement [5,7–9]. Evidence shows that signaling pathways activated by several of these stimuli are frequently enhanced in breast cancer, for example, by overexpression of receptor tyrosine kinases (RTKs) of the ErbB family (ErbB1 [EGF receptor] and ErbB2 [Her2/neu]) [10,11], integrin receptors (α3β1 and αvβ3 integrins) [8,12], and estrogen [13].
In breast cancer patients, elevated p130Cas levels are associated with an increased rate of relapse and aggressiveness of the disease [14]. p130Cas expression is higher in pleural effusions of breast cancer patients compared with primary tumors [15]. Also, in feline and canine models of breast cancer, p130Cas levels positively correlate with advanced breast carcinomas [16]. Increased p130Cas protein expression has been implicated in mediating resistance to Adriamycin and tamoxifen [17,18]. Recent in vitro studies have shown elevated p130Cas levels in tamoxifen-resistant breast cancer cells (TAM-R) that were derived from tamoxifen-sensitive MCF-7 cells [19]. Resistance to antiestrogens such as tamoxifen and fulvestrant has also been associated with enhanced growth factor signaling involving the upregulation of RTKs of the ErbB family and augmented Akt pathway activation [19–21]. Disruption of the p130Cas signaling node leads to attenuation of the extracellular signal-regulated kinase (ERK) and phosphatidylinositol 3-kinase (PI3K)/Akt survival pathways, reduced migration, and resensitization of TAM-R cells to tamoxifen [19].
Members of the early growth response (EGR) transcription factor (TF) family have been implicated in breast cancer progression and antiestrogen resistance [22,23]. The zinc finger factors EGR1 (NGFI-A), EGR2 (Krox-20), and EGR3 are immediate early response genes that are important for the regulation of differentiation, proliferation, and cell death in response to environmental stimuli [24,25]. Extracellular signals such as cytokines, growth factors, and hormones that lead to the activation/phosphorylation of p130Cas also rapidly and transiently induce the expression of the EGR family members [26,27]. EGR target genes comprise, for example, vascular endothelial growth factor (VEGF) [28], PDGF [29], as well as cyclin-dependent kinase 1 inhibitorp21Waf1/Cip1 [22]. The activity of the EGR TFs is modulated in part through the gene product of its own target, the delayed early response gene NGFI-A binding protein 2 (NAB2) [30–32].
Given the number of studies that link p130Cas levels to breast cancer progression and the fact that tamoxifen is still a commonly used drug in hormonal adjuvant therapy, understanding the p130Cas gene regulation in mammary carcinomas is of great importance. However, the regulatory mechanisms that upregulate p130Cas expression in mammary carcinomas and during acquired tamoxifen resistance in human breast tumors are elusive. Here, we demonstrate the presence of two BCAR1 mRNAs, using alternative first exons, in normal human mammary gland tissue and in breast tumors. The results suggest that, in breast cancer cells, p130Cas expression/signaling upregulates the TF EGR1 and its coregulator NAB2, which, in turn, are involved in the induction of p130Cas expression. This regulatory network was altered in tamoxifen-resistant breast cancer cells, which exhibited higher NAB2 levels. Therefore, our studies identified a transcriptional circuit that controls p130Cas expression in breast cancer cells and a p130Cas-controlled regulatory network that may contribute to acquired tamoxifen resistance.
Materials and Methods
Cell Lines and Culture Conditions
Cell lines were purchased from the American Type Culture Collection (ATCC, Manassas, VA) or obtained as indicated. The following cell lines were used: BT-20 (Sam Lee, Massachusetts General Hospital; human breast carcinoma, estrogen receptor [ER] negative); ZR-75-1, MCF-7, and the MCF-7-derivative TAM-R (Robert Nicholson, Cardiff, United Kingdom; human breast carcinoma, ER positive). MCF-7 cells were grown in RPMI-1640 supplemented with 10% fetal bovine serum (FBS), 1 mM sodium pyruvate, 2 mM l-glutamine, 100 U/ml penicillin, and 100 U/ml streptomycin. TAM-R cells were cultured in phenol red-free RPMI-1640 and 10% charcoal-stripped FBS, supplemented with 4 mM glutamine and antibiotics/sodium pyruvate as previously mentioned. BT-20 and ZR-75-1 cells were cultured in Dulbecco modified Eagle medium, 10% FBS, and supplemented as previously mentioned. Cells were maintained at 37°C and 5% CO2 and tested for Mycoplasma contamination.
For phorbol myristate acetate (PMA) treatment, 2 x 105 cells were seeded in six-well plates and, after 24 hours of plating, serum starved (0.5% serum) for 16 hours before stimulation by the addition of PMA (final concentration 10 ng/ml; Sigma, St Louis, MO) for indicated times.
Generation of Expression Constructs
Human wild-type (wt) and dominant negative (dn) NAB2 complementary DNAs (cDNA) [32,33] were subcloned into pCXbsr digested with BamHI. Human EGR1 cDNA was obtained by reverse transcription (RT) of total RNA prepared from MCF-7 cells stimulated for 2 hours with PMA using the EGR1-specific reverse primer EGR1-NotI, including a NotI site (Table 1, for primer sequences). The obtained EGR1 cDNA was amplified by polymerase chain reaction (PCR) using Pfu polymerase with the EGR1-EcoRI forward primer and the EGR1-NotI reverse primer and subcloned into the EcoRI/NotI-digested pCXbsr vector. Constructs were validated by sequencing. QIAGEN Plasmid Maxi Kit (Qiagen, Valencia, CA) was used for DNA preparation.
Table 1.
Primer Sequences.
| Name | Forward (5′-3′) | Name | Reverse (5′-3′) |
| EGR1-EcoRI | ATAGAATTCATGGCCGCGGCCAAGGCCG | EGR1-NotI | ATAGCGGCCGCTTAGCAAATTTCAATTGTCCTGG |
| BCAR1 total | TGGAGCAGGACACGCAGGG | BCAR1-rev | GGTGGGCACCACCACCTTTG |
| BCAR1-1 | CGGACACCATGAACCACCTGAA | ||
| BCAR1-1′ | CCAAGATGTCCGTGCCTAAC | ||
| BCAR1-1q | GACACCATGAACCACCTGAA | BCAR1q-rev | TCATACATGCCCACCAAGAT |
| BCAR1-1′q | CGGCCAAGATGTCCGTGC | ||
| BCAR1 totalq-f | GTCCACCTACAGGGGAAGGA | BCAR1 totalq-rev | CGTGACACCTCCTGTTCCAG |
| GAPDHq-f | TGCACCACCAACTGCTTAGC | GAPDHq-r | GGCATGGACTGTGGTCATGAG |
| PPIAq-f | TTGGGCCAATAAGAAGTTGTGAA | PPIAq-r | AACCGACTGGCTAAAATCCAC |
| RPLP0q-f | GCTGCTGCCCGTGCTGGTG | RPLP0q-r | TGGTGCCCCTGGAGATTTTAGTGG |
| RN18S1q-f | CGGCTACCACATCCAAGGAA | RN18S1q-r | TTTTCGTCACTACCTCCCCG |
| BCAR1-1 -226 | CTCGGGCAGGGGCCTGGAG | BCAR1-1 -58 | AGCCCGATCCCAGCATGC |
| BCAR1_1′ -360 | TCCTCTTCCCTGGGGGTCTC | BCAR1-1′ -162 | TCTGGGTCGCCCAACCAGTC |
| control-f | ATGGTTGCCACTGGGGATCT | control-r | TGCCAAAGCCTAGGGGAAGA |
In Silico Analysis
Analyses of BCAR1 and the exon 1 mRNA and protein variants were conducted using database resources and software from AceView [34], the EMBL-European Bioinformatics Institute, Genomatix [35], and the National Center for Biotechnology Information. In silico analysis of putative promoter regions was performed using the Genomatix suite of programs.
Retroviral Transduction
Retroviral transduction was carried out as described [19,36]. Briefly, transient expression of EGR1, wt or dn NAB2, and/or empty vector (ev) control in MCF-7, TAM-R, and BT-20 cells was achieved by transduction with equal volumes of supernatant containing amphotropic CXbsr virus particles that carry either the cDNAs for EGR1, wt or dn NAB2, or ev. Equal amounts of ev control supernatant were added to the single-gene transductions to control for DNA amounts/virus titer. To enrich for cells expressing the introduced cDNAs, cells were selected with 10 µg/ml Blasticidin (Invitrogen, Grand Island, NY) for 3 days before harvesting.
RNA Interference
Transfection of p130Cas short-interfering RNAs (siRNAs) was performed as previously described [19]. Depletion of human EGR1 was achieved by transfection of pooled siRNA (50 nM) in 12-well plates after 38 hours using Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendations. ON-TARGETplus SMART pool siRNA targeting EGR1 (catalog no. L-006526-00-0005) and negative control (catalog no. D-001810-01-05) were purchased from Dharmacon (Chicago, IL). Sequences of siRNA-specific for EGR1 were as follows: 5′-GAUGAACGCAAGAGGCAUA-3′; 5′-CGACAGCAGUCCCAUUUAC-3′; 5′-GGACAUGACAGCAACCUUU-3′; 5′-GACCUGAAGGCCCUCAAUA-3′. Down-regulation of human NAB2 was achieved by transfection of pooled siRNAs (50 nM) in 12-well plates after 72 hours using Lipofectamine 2000. siRNA sequences targeting NAB2 (catalog no. sc-36014; Santa Cruz Biotechnology, Santa Cruz, CA) were as follows: 5′-CCAACCUCCUUUCCUACUAtt-3′; 5′-CAACCUCCUUUCCUACUAUtt-3′; 5′-CCUACUUGUCCUCCUUGAAtt-3′.
Protein Analysis
Whole cell extracts (WCEs) were prepared and analyzed by Western blot analysis (WB) as described [37]. Briefly, cells were incubated in HNTG lysis buffer (50 mM HEPES [pH 7.4], 150 mM NaCl, 1% Triton X-100, 10% glycerol, 10 mM EDTA) supplemented with protease inhibitor cocktail (Roche, Nutley, NJ) and phosphatase inhibitors (1 mM sodium vanadate and 1 mM sodium fluoride) for 30 minutes on ice followed by centrifugation at 16,000g and 4°C for 30 minutes. Equal amounts of protein were analyzed by SDS-PAGE. Antibodies (Abs) used were mAbs against β-actin (Sigma), NAB2 [38], and p130Cas (clone 21; BD Transduction Laboratories, Sparks, MD). Polyclonal rabbit Abs against EGR1 (588; sc-110) were purchased from Santa Cruz Biotechnology.
Reverse Transcription-PCR
Cells were washed twice with cold PBS and RNA was extracted using TRIzol reagent (Invitrogen). Total RNA (2 µg) was reverse transcribed using SuperScript II Reverse Transcriptase (Invitrogen) and random hexamer primers (Invitrogen) according to the manufacturer's recommendations. cDNAs reverse transcribed using oligo-dT primer from human breast tissue mRNA were purchased from Biochain Institute (Hayward, CA). PCR was performed using 0.5 µl of undiluted cDNA in a 20-µl PCR with the use of Taq DNA polymerase and reaction buffer from Biochain Institute. To check for linear amplification, each PCR was performed at different cycle numbers. Exon overlapping PCR primers were applied to exclude amplification of genomic DNA and cDNA generated from unspliced precursor mRNA. Primer sequences are found in Table 1. Reverse primer BCAR1-rev was used for BCAR1 total, BCAR1-1, and BCAR1-1′ amplifications. Forward primers and amplicon sizes were as follows: BCAR1 total, 551 bp; BCAR1-1, 657 bp; BCAR1-1′, 653 bp. Reaction conditions (annealing temperature and cycle number) were as follows: BCAR1 total, 62°C and 25; BCAR1-1, 64°C and 27; and BCAR1-1′, 64°C and 27. The BCAR1 total reaction amplifies parts of exon 2 and exon 3, which are present in both exon 1 variants. To control for the integrity and uniformity of the sample preparations, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was amplified as described previously [33]. Products were separated by electrophoresis through 1.2% agarose gels and validated by sequencing.
Quantification of Gel Bands
Densitometric analysis of gel bands was performed using Quantity One software (Bio-Rad). Equal adjustments of contrast and brightness of gel pictures and scanned films were applied to all parts of the image using Adobe Photoshop CS2 version 9.0. Vertical lines in Figures 1D, 3, 4A and B, and 8A indicate that results from different WB are shown.
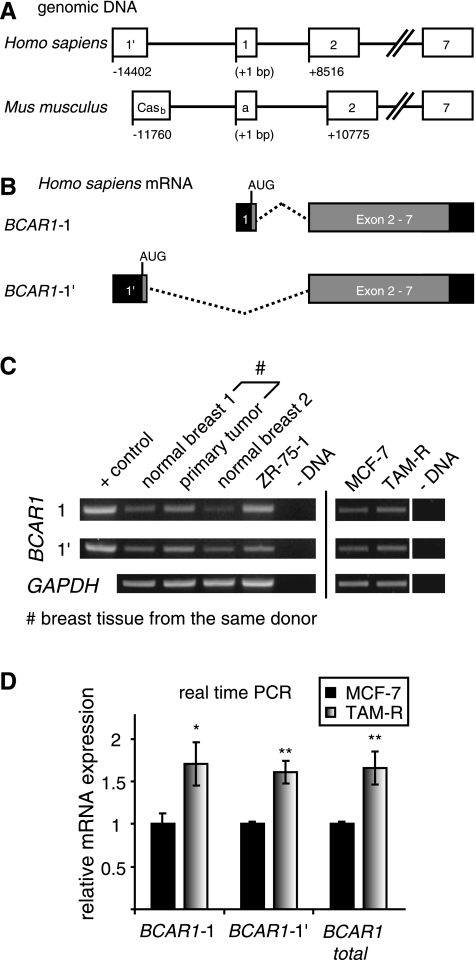
Figure 1.
BCAR1 uses alternative first exons in normal and cancerous breast tissues. (A) Comparison of the human and mouse BCAR1 genes. The human BCAR1-1 and BCAR1-1′ reflect the murine p130Cas isoforms Casa and Casb, respectively. Boxes represent exons. // indicates that sequences of exons 3 through 6 were omitted. Numbering indicates the TSS of variant 1′ relative to exon 1 TSS (+1). (B) Schematic of the BCAR1 Exon 1 mRNA variants. Relative position of the translation start site (AUG) for each variant is indicated. Black boxes indicate untranslated regions; gray boxes, translated regions; dashed lines, spliced-out intron region. (C) Levels of BCAR1-1, BCAR1-1′, and GAPDH (control) mRNA in human breast tissues and ZR-75-1, MCF-7 (tamoxifen-sensitive), and TAM-R (tamoxifen-resistant) mammary carcinoma cell lines were determined by RT-PCR as described previously. +control, positive control for PCR; - DNA, negative control for PCR. Results are from one representative experiment of two performed. (D) Expression of BCAR1 mRNAs is increased in tamoxifen-resistant TAM-R cells. Tamoxifen-sensitive MCF-7 and its derivative tamoxifen-resistant TAM-R cells were cultured similarly for 2 weeks and seeded at 3 x 105 cells in six-well plates. After 3 days at a confluence of 70% to 90%, cells were harvested and mRNA was prepared and reverse transcribed as described previously. Levels of BCAR1-1, BCAR1-1′, total BCAR1, and the internal controls GAPDH, PPIA, and RPLP0 mRNA in MCF-7 cells and TAM-R were determined by real-time PCR as described previously. Shown are the results after normalization to GAPDH. Similar results were obtained after normalization to PPIA and RPLP0. Average levels relative to the expression in MCF-7 cells (black bar, set to 1) and SD from triplicate were determined as described previously. P values were calculated using t test. *P < .05. **P < .01. Results are from one representative experiment of two performed.
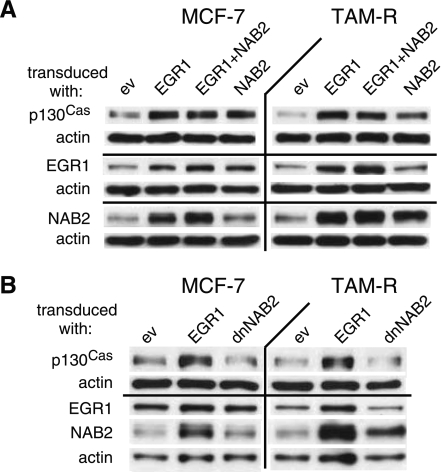
Figure 3.
EGR1 and NAB2 are involved in the regulation of p130Cas expression. (A and B) MCF-7 and TAM-R cells were transduced with the indicated cDNAs for 2 days and selected for 3 days with blasticidin as described previously. WCEs (20 µg) were separated by SDS-PAGE, subjected to WB, and probed with the indicated antibodies. Data are presented from one experiment of at least three performed with similar results.
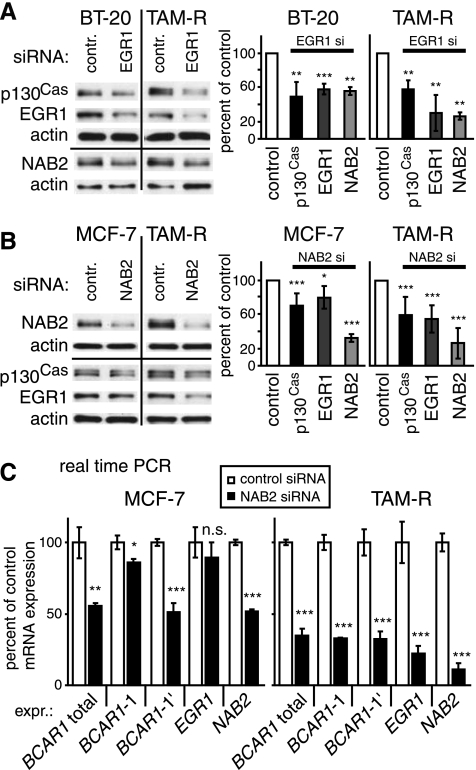
Figure 4.
Endogenous EGR1 and NAB2 are important regulators of p130Cas expression. (A) BT-20 and TAM-R cells were transfected with siRNAs for EGR1 or control siRNA. WCEs (20 µg) were analyzed for p130Cas, EGR1, NAB2, and actin (loading control) expression by WB 38 hours after treatment (left panel). Right panel: Densitometric analysis of protein expression. (B) MCF-7 and TAM-R cells were transfected with siRNAs for NAB2 or control siRNA. WCEs (20 µg) were analyzed for p130Cas, EGR1, NAB2, and actin (loading control) expression by WB 72 hours after treatment. Right panel: Densitometric analysis of protein expression. (A and B) Average expression levels relative to negative control (black bar, set to 100%) and SD from three representative experiments performed were determined as described previously. (C) MCF-7 and TAM-R cells were transfected with siRNAs for NAB2 or control siRNA. After 72 hours, mRNA levels of BCAR1-1, BCAR1-1′, total BCAR1, NAB2, EGR1, and the internal controls RPLP0, PPIA, and RN18S1 in MCF-7 and TAM-R cells were determined by real-time PCR as described previously. Shown are the results after normalization to RPLP0. Similar results were obtained after normalization to PPIA and RN18S1. Average levels relative to the expression in MCF-7 cells (black bar, set to 100%) and SD from triplicate were determined as described previously. Results are from one representative experiment of two performed. (A–C) P values were calculated using t test. *P < .05. **P < .01. ***P < .001. n.s. indicates not significant.
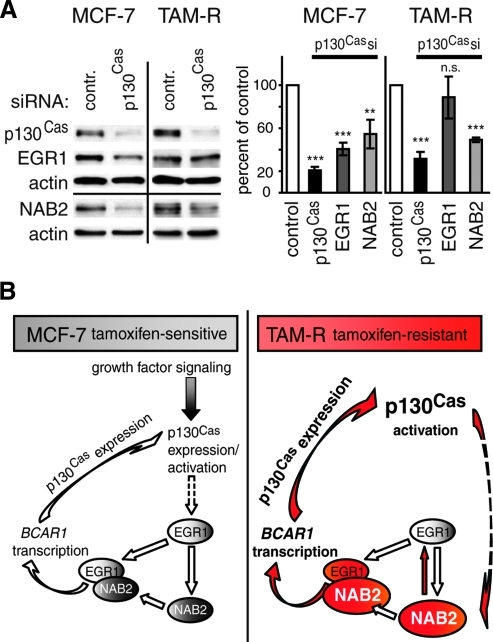
Figure 8.
p130Cas is involved in the regulation of EGR1 and NAB2 expression. (A) MCF-7 and TAM-R cells were transfected with BCAR1-specific siRNAs or control siRNA. Left panel: WCEs (20 µg) were analyzed for p130Cas, EGR1, NAB2, and actin (control) expression by WB 48 hours after treatment. Results are presented from one representative experiment of three performed. Right panel: Densitometric analysis of p130Cas, EGR1, and NAB2 protein expression in siRNA-treated cells. Average expression levels relative to negative control (black bar, set to 100%) and SD from three experiments performed were determined. P values were calculated using t test. **P < .01. ***P < .001. n.s. indicates not significant. (B) Model of the positive feedback loop regulating p130Cas expression in response to environmental stimuli in tamoxifen-sensitive MCF-7 and tamoxifen-resistant TAM-R breast cancer cells. Solid arrows depict direct interactions or up-regulation/activation; dashed arrows indicate additional undefined steps in between leading to an up-regulation of expression. Right panel: Red color indicates enhanced activity or expression (as shown by larger size symbols) of the depicted molecule or process in TAM-R versus MCF-7 cells.
Real-time PCR
Real-time PCR was performed in 20-µl reactions in triplicate using ABI PRISM 7300 Sequence Detection System Version 1.4.0 (Applied Biosystems, Carlsbad, CA) and 2x FastStart Universal SYBR Green PCR Master (ROX; Roche Applied Science, Indianapolis, IN) as recommended by the manufacturer using 2 ng of cDNA, reverse transcribed as described previously. Comparative Ct method was applied for relative quantification of each gene expression between samples [39] and normalized to GAPDH, peptidylprolyl isomerase A (PPIA), ribosomal protein, large, P0 (RPLP0), and RNA 18S ribosomal 1 (RN18S1) levels, as indicated. PCR specificity and amplicon size were validated by electrophoresis through a 2% agarose gel and subsequent sequencing. Efficiency of each PCR was more than 98% as determined by performing the PCRs for each primer pair with serial DNA dilutions. Primer sequences are presented in Table 1. Reverse primer BCAR1q-rev was used for BCAR1-1 and BCAR1-1′ amplifications. Forward primers and amplicon sizes were as follows: BCAR1-1q, 209 bp; BCAR1-1′q, 211 bp. BCAR1 total levels were detected by amplification of a region from exon 5 to exon 7, which is present in both exon 1 variants, using the primers BCAR1 totalq-f and BCAR1 totalq-rev. Primers used in GAPDH, PPIA, RPLP0, and RN18S1 PCRs are shown in Table 1. Primers for EGR1 and NAB2 were described previously [33].
Chromatin Immunoprecipitation Real-time PCR
Briefly, 2.5 x 106 (MCF-7) or 3 x 106 (TAM-R) cells were seeded in 10-cm dishes and after 1 day of plating serum starved (0.5% serum) for 16 hours before harvesting. Subsequently, chromatin immunoprecipitation (ChIP) was performed using EZ-ChIP kit (Millipore, Billerica, MA) following the manufacturer's protocol using 4 µg of antibody. Abs used were as follows: rabbit anti-EGR1 (588, sc-110; Santa Cruz Biotechnology) and normal rabbit IgG control (sc-2027; Santa Cruz Biotechnology). ChIP real-time PCRs were performed as described previously using 3 µl of purified DNA. For amplification of the BCAR1-1 and BCAR1-1′ 5′ regions, 4 µl of 5 M betaine (Sigma) was added to the PCRs. Primers (for sequences, see Table 1) and amplicon sizes were as follows: BCAR1-1 -226 and BCAR1-1 -58, 169 bp; BCAR1-1′ -360 and BCAR1-1′ -162, 199 bp. As control for chromatin bound nonspecifically, a human negative control primer set (control-f and control-r; amplicon size, 174 bp; Active Motif, Carlsbad, CA) was used. Specificity of the PCRs was validated by electrophoresis through 1.6% agarose gels and subsequent sequencing. The enrichment of EGR1 to the genomic BCAR1 5′ or negative control regions was calculated as followed. The percentage of the input DNA (1%of the nuclear extract used) that was recovered was calculated for each sample, and the EGR1 values were related to the percentage computed for the rabbit IgG control. Subsequently, the resulting fold of IgG values obtained by the EGR1-ChIP for the BCAR1 5′ regions were divided by the fold of IgG retrieved by the EGR1-ChIP for the genomic negative control region to account for possible unspecific precipitation of genomic DNA by the EGR1 antibody in different samples/cell lines.
Results
BCAR1—Alternative First Exon Usage in Normal and Cancerous Breast Tissues
Recent studies have shown elevated p130Cas protein levels in tamoxifen-resistant breast cancer cells [19] and, more importantly, in antiestrogen-resistant breast cancers in patients [40]. Since to date the regulation of p130Cas/BCAR1 expression remains enigmatic, a comprehensive analysis of the BCAR1 gene was performed to decipher mechanisms that may contribute to the observed up-regulation of p130Cas expression.
Database searches and gene structure analysis revealed that the human BCAR1 gene uses an alternative first exon (BCAR1-1′, NM_001170718) located 14,402 bp upstream of the first described exon 1 (BCAR1-1, RefSeq NM_014567.3) (Figure 1, A and B). Comparative genomics indicated that BCAR1-1 and BCAR1-1′ reflect the p130Cas isoforms Casa and Casb described in mice, respectively [41]. Both first exons encode for only three distinct amino acids (aa) (NHL [BCAR1-1] and SVP [BCAR1-1′]) and the resulting isoforms are conserved in mammals. The genomic distance between the two first exons points to the use of alternative promoters.
To study the regulation of BCAR1 in mammary carcinomas, we examined the expression of BCAR1 exon 1 variant mRNA in two normal human breast tissues, a human primary tumor, and in the human breast cancer cell line ZR-75-1 (Figure 1C). Furthermore, to elucidate differences in acquired tamoxifen resistance, the tamoxifen-sensitive MCF-7 breast cancer cells and its resistant derivative, TAM-R, expressing higher p130Cas protein levels [19], were used as a model system and therefore also included. In all samples, both BCAR1 variants were detected. Compared with normal breast tissue 1, the levels of BCAR1-1 as well as of BCAR1-1′ were increased in the primary tumor sample, which was derived from the same donor. Higher levels of BCAR1-1′ over BCAR1-1 or vice versa were found in normal breast tissue 2 and ZR-75-1 cells, respectively, suggesting a distinct regulation of each BCAR1/p130Cas variant. In MCF-7 and TAM-R cells, a similar expression of both variants was detected. However, a slight increase in both BCAR1 variants was observed in TAM-R versus MCF-7 cells.
To further investigate whether the enhanced protein levels in TAM-R cells correlate with higher mRNA expression, BCAR1-1, BCAR1-1′, and total BCAR1 (determined by amplification of a region spanning exons 5 to 7) mRNA expression levels were analyzed by real-time PCR in MCF-7 and TAM-R cells (Figure 1D). Significant increases in BCAR1-1 (1.71-fold, P < .05), BCAR1-1′ (1.61-fold, P < .01), and total BCAR1 (1.66-fold, P < .01) were observed in TAM-R compared to MCF-7 cells.
Taken together, these results show that BCAR1-1 and BCAR1-1′ are coexpressed in human breast tissues and breast cancer cell lines, and both variants seem to be differentially regulated in some circumstances. Furthermore, tamoxifen-resistant cells have elevated mRNA levels compared to tamoxifen sensitive.
In Silico Analysis of the Flanking Regions of the BCAR1 Exon 1 Variants Reveals Multiple Clusters of Binding Sites for Transcription Factors of the EGR Family
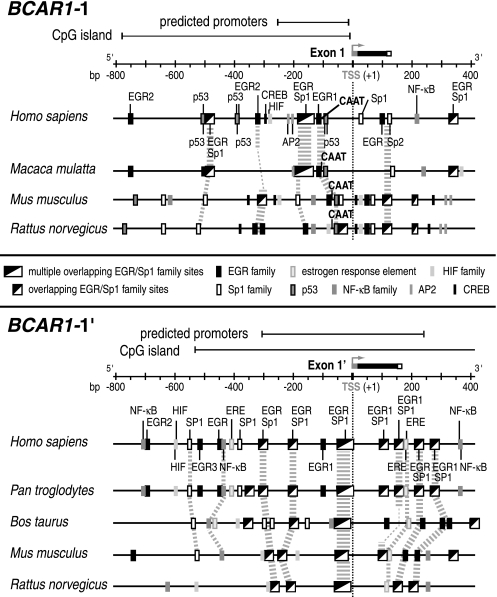
To gain insight into the mechanisms regulating the p130Cas variant expression, a comprehensive phylogenetic analysis of the 5′ and 3′ flanking regions of the alternative first exons of BCAR1 was performed (Figure 2).
Figure 2.
Schematic representation of putative regulatory elements in the flanking regions of the alternative first exons of BCAR1 in mammalian species, as indicated. Comparative genomics were performed using database resources from AceView and the Genomatix suite of programs. Different species were used for the analysis of the BCAR1-1 and BCAR1-1′ regions owing to a restricted availability and gaps in the genomic sequence. Predicted promoter regions, CpG islands, and TF binding sites are indicated for BCAR1-1 and BCAR1-1′. Conserved cis regulatory elements are emphasized by gray dashed lines/blocks. Numbering is relative to TSS (+1, gray boxes); exon 1: black boxes, untranslated regions; small white boxes, translated regions.
Putative promoters colocated with CpG islands were identified in the regions flanking human exons 1 and 1′. A CAAT box was only found at bp -92 of exon 1 and no TATA box or initiator element is present in both regions. The lack of these core promoter elements points to the involvement of specific TFs in the regulation of BCAR1 expression. Owing to the elevated p130Cas levels in breast cancer putative binding sites for TFs including members of the EGR, NF-κB, and hypoxia-inducible factor (HIF) family, p53, AP2, estrogen response elements (ERE), and cAMP-responsive elements binding proteins (CREB), involved in tumor development and progression, were explored. The region bp -800 to +400 relative to the transcription start site (TSS, +1) of exon 1, contains 14 putative binding sites for the EGR family, most of which overlap with nine SP1 motifs, seven p53, two AP2 sites, and a single HIF and CREB motif. Of note is a conserved cluster of overlapping binding sites (bp -196 to -125) including six EGR and five SP1 motifs. Several EGR/Sp1 motifs within bp -500 to +150 and two p53 binding sites near the CAAT box are conserved among the different species (Figure 2).
In the 5′ region of exon 1′, an evolutionary conserved cluster of four EGR and three overlapping SP1 sites at bp -29 to +2, one EGR1/SP1 site, and two ERE at bp +150 to +180 were predicted. In addition, nine EGR, five SP1, four NF-κB, two HIF sites, and one ERE were identified (bp -800 to +400).
These results point to a complex regulation of the expression of the BCAR1-1 and BCAR1-1′ variants involving members of the EGR family and other TFs, such as NF-κB, and p53.
EGR1 and Its Coregulator NAB2 Are Involved in the Regulation of p130Cas Expression in Human Breast Cancer Cells
To elucidate a potential role of EGR1 and the influence of its coregulator NAB2 on p130Cas expression, MCF-7 and TAM-R cells were transduced with EGR1, NAB2 (wt or dn), and ev control (Figure 3). NAB2 is a target gene of EGR1 and, in turn, can act as corepressor [31,32] and coactivator [30] of EGR1-mediated transcription. dnNAB2 contains an aa exchange that prevents the binding to EGR1. dnNAB2 is still able to form complexes with endogenous NAB2, thereby greatly abolishing the binding of also endogenous NAB2 to EGR1 [42]. Protein levels were analyzed by WB after 5 days, which included 3 days of selection with blasticidin to enrich for cells expressing the introduced cDNAs. Cells transduced with EGR1 had profoundly higher amounts of p130Cas and NAB2. The influence of NAB2 on EGR1-mediated p130Cas expression was examined by cotransduction of EGR1 and NAB2. p130Cas was not altered by coexpression of EGR1 and NAB2 compared to EGR1 single transduction, suggesting that NAB2 does not inhibit EGR1-mediated p130Cas expression. Cells transduced with NAB2 alone showed also elevated p130Cas levels. Importantly, ectopic expression of dnNAB2 did not result in higher p130Cas expression in MCF-7 and TAM-R cells (Figure 3B). These experiments suggest that EGR1 and NAB2 complex formation is required to increase p130Cas levels in breast cancer cell lines.
EGR1 and NAB2 Depletion Attenuates p130Cas/BCAR1 Expression at the mRNA and Protein Levels
Subsequently, the importance of endogenous EGR1 and NAB2 in the regulation of p130Cas expression was further investigated using siRNAs. MCF-7, TAM-R, or BT-20 breast cancer cells were transfected with siRNAs directed against EGR1, NAB2, or a negative control siRNA, and the effects on p130Cas, EGR1, and NAB2 were assessed by WB (Figure 4, A and B). Results in Figure 4A are presented for TAM-R and BT-20 cells because no adequate depletion of EGR1 levels was achieved in MCF-7 cells. Transfection of EGR1 siRNA resulted not only in a reduction of EGR1 levels by 70% and 41% in TAM-R and BT-20 cells, respectively, but also of p130Cas expression by 41% (P < .01) and 50% (P < .01). NAB2 levels were also significantly (P < .01) reduced by 73% (TAM-R) and 44% (BT-20).
Depletion of NAB2 by siRNA targeting NAB2 by 63% (MCF-7) and 74% (TAM-R) also led to a reduction of p130Cas levels (P < .001) by 30% and 40% in MCF-7 and TAM-R cells, respectively (Figure 4B), underscoring the importance of NAB2 in regulating p130Cas expression. Furthermore, EGR1 expression was reduced in TAM-R cells by 47% (P < .001), whereas in MCF-7 cells, a reduction of only 20% (P < .05) was observed.
To investigate whether endogenous EGR1 and NAB2 are involved also in the transcriptional regulation of p130Cas expression, the BCAR1 mRNA levels were analyzed after depletion of NAB2 by siRNA (Figure 4C). After NAB2 siRNA transfection, the mRNA levels of BCAR1-1, BCAR1-1′, total BCAR1, NAB2, and EGR1 were determined in MCF-7 and TAM-R cells by real-time PCR. Transfection of NAB2 siRNA resulted in a reduction of NAB2 levels by 49% and 88% in MCF-7 and TAM-R cells, respectively, correlating with reductions in total BCAR1 expression by 56% (P < .01) and 65% (P < .001). Corresponding to the minor effects of NAB2 depletion on EGR1 protein levels in MCF-7 (Figure 4B), EGR1 mRNA expression was only significantly reduced in TAM-R cells (by 78%, P < .001). Interestingly, BCAR1-1′ levels were substantially depleted (P < .001) by 49% (MCF-7) and 68% (TAM-R) in both cell lines, whereas BCAR1-1 expression was reduced to a much greater extent in TAM-R by 67% (P < .001) than in MCF-7 cells by 14% (P < .05).
Taken together, these results indicate that the TF EGR1 and its coregulator NAB2 are positive regulators of p130Cas/BCAR1 expression in human breast cancer cells. Importantly, NAB2 seems to be involved in the regulation of BCAR1-1 and BCAR1-1′ expression in the tamoxifen-resistant TAM-R cells, whereas NAB2 influences mostly BCAR1-1′ levels in the tamoxifen-sensitive MCF-7 cells.
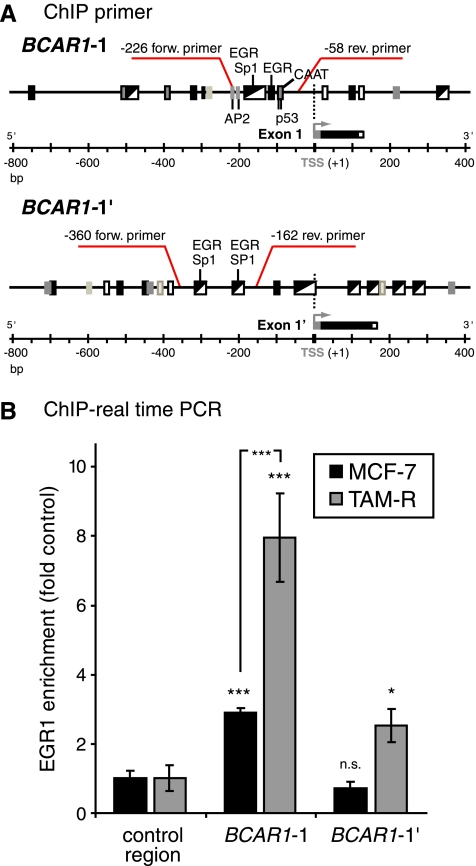
Increased Binding of EGR1 to the BCAR1 5′ Regions in Tamoxifen-Resistant TAM-R Compared to Tamoxifen-Sensitive MCF-7 Cells
Because RNA interference experiments suggested that EGR1 regulates BCAR1 expression, EGR1 was tested for its ability to directly bind to the potential BCAR1 promoter regions. ChIP assays were performed on isolated chromatin from MCF-7 and TAM-R cells and evaluated by real-time PCR for the genomic DNA 5′ regions of BCAR1-1 and BCAR1-1′ that contain multiple putative EGR1 binding sites (Figure 5A). Binding to these specific regions was investigated because they are located in the predicted promoter regions, and similar clusters of EGR/Sp1 sites in a comparable distance from the TSS were identified as crucial in multiple other promoters [28,29,32]. To account for nonspecific binding of EGR1, the recovery of DNA by IgG control serum and enrichment to a genomic negative control region were included in the calculation as described in the Materials and Methods. In MCF-7 cells, EGR1 binding was found located within the 5′ region from bp -226 to -58 of BCAR1-1 (2.9-fold enrichment, P < .001) containing a cluster of multiple overlapping EGR/Sp1 and a single EGR1 site, whereas no binding was detectable to the 5′ region of BCAR1-1′ from bp -360 to -162 (Figure 5B). This region similarly contains two clusters of overlapping EGR/Sp1 motifs. In contrast, in TAM-R cells, binding of EGR1 to BCAR1-1 (7.96-fold enrichment, P < .001) and BCAR1-1′ (2.53-fold enrichment, P < .05) was observed. The binding to BCAR1-1 in TAM-R cells was significantly higher (P < .001) than in MCF-7 cells.
Figure 5.
Distinct binding of EGR1 to the BCAR1-1 and BCAR1-1′ 5′ regions in MCF-7 and TAM-R cells. ChIP real-time PCR was used to investigate EGR1 binding to the BCAR1 5′ regions as described in the Materials and Methods. (A) Schematic of the flanking regions of the alternative first exons of BCAR1. Positions of primers used for ChIP analyses and putative TF binding sites within the amplified regions are indicated. (B) Immunoprecipitated BCAR1-1, BCAR1-1′, and negative control regions were amplified by real-time PCR as described previously. Shown is the average relative fold EGR1 enrichment at the BCAR1 5′ regions in relation to the EGR1 binding at the negative control DNA region (control region, set to 1) and SD calculated from triplicate as explained in detail in the Materials and Methods. Control region: PCR to control for chromatin bound nonspecifically. Results are from one representative experiment of at least three performed. P values of negative control region versus BCAR1 5′ regions, or as indicated, were calculated using t test. *P < .05. ***P < .001. n.s. indicates not significant.
These results suggest differential regulation of the p130Cas variant expression in MCF-7 versus TAM-R cells by EGR1.
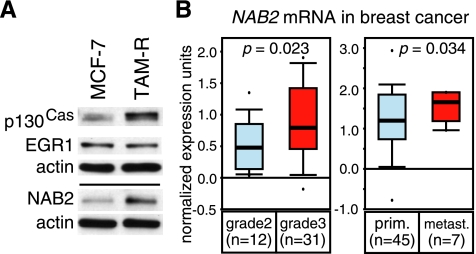
Aggressive and Late-Stage Breast Cancers Display Elevated NAB2 Levels
As more significant effects by depletion of the EGR1 coregulator NAB2 were observed in TAM-R versus MCF-7 cells, NAB2 expression in both cell lines was compared. MCF-7 and TAM-R cells were cultured similarly for 2 weeks to eliminate differences in the basal expression of EGR1 and NAB2 induced by varied culture conditions. WB of basal p130Cas, EGR1, and NAB2 protein levels (Figure 6A) showed similar EGR1 levels in both cell lines but profoundly elevated NAB2 levels in tamoxifen-resistant TAM-R cells. Commonly, breast cancers with acquired tamoxifen resistance represent more advanced tumors that are difficult to treat. Therefore, we investigated whether increased NAB2 levels correlate with mammary carcinoma grade and/or malignancy by analyzing microarray gene expression data sets available at www.oncomine.org. For microarray studies to be included, we required a minimum of five samples for each group and a similar trend in expression differences if multiple reporters were used in these studies. The NAB2 mRNA levels significantly increased from grade 2 to grade 3 specimens of human invasive ductal breast cancers in the Ginestier study (P = .023, reporter number 216017_s_at) [43] (Figure 6B, left panel). A similar increase was observed in the study of Zhao et al. [44] (P = .033, reporter IMAGE:1926749; not shown), whereas no significant changes from grade 2 to grade 3 were detected in the Radvanyi study (reporter number U48361) [45] (not shown). Thus, the analyses were extended to compare NAB2 levels in primary sites and metastases of breast cancer patients. The levels of NAB2 mRNA were significantly higher in metastasis versus primary site in the Radvanyi study (P = .023, reporter number U48361) [45] (Figure 6B, right panel). Similar results were found in two other microarray studies (not shown) that fulfilled our requirements, supporting these findings. These data suggest that advanced human breast cancers express higher levels of NAB2.
Figure 6.
NAB2 expression is elevated in aggressive and late-stage breast cancers. (A) MCF-7 and TAM-R cells were cultured similarly for 2 weeks and seeded at 3 x 105 cells in six-well plates. After 3 days at a confluence of 70% to 90%, cells were harvested and WCEs (20 µg) were subjected to WB to compare the basal p130Cas, EGR1, and NAB2 protein levels. Actin was used as a loading control. Results are presented from one representative experiment of three performed. (B) Box plots of data from breast carcinoma microarray data sets using the Oncomine Cancer Profiling Database (www.oncomine.org) plotted on a log scale. The t test was performed directly through the Oncomine 4.4 software. n indicates the number of samples in each group. Left panel: a box plot of data from the Ginestier study (reporter number 216017_s_at) [43] including grade 2 and grade 3 human invasive ductal breast cancer samples. The difference in NAB2 mRNA expression between the two groups was significant (P = .023). Right panel: data from the Radvanyi study (reporter number U48361) [45] including samples of human breast cancer obtained from the primary site (prim.) or metastasis (metast.). The difference in NAB2 expression between the two groups was significant (P = .034).
p130Cas/BCAR1 Expression Is Induced by Phorbol Esters Following the Kinetics of a Late Response Gene in Human Breast Cancer Cells
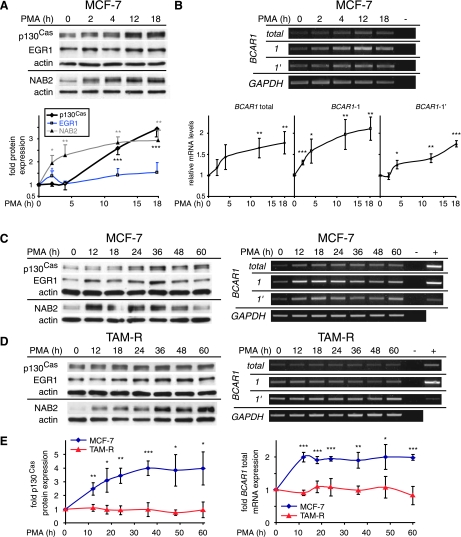
The immediate early gene EGR1 and its coregulator, the delayed early response gene NAB2, are highly inducible by PMA [24,31,38], which is an activator of protein kinase C, thereby mimicking growth factor signaling. Accordingly, depletion of growth factors leads to downregulation of EGR1 and NAB2, which can vary in time and strength in different cell types. To elucidate whether BCAR1/p130Cas expression levels can be influenced by environmental signals, an analysis of the expression kinetics of p130Cas in response to PMA was performed in MCF-7 and TAM-R cells. Breast cancer cells were serum starved for 16 hours followed by PMA treatment for indicated times and the p130Cas, EGR1, and NAB2 protein expression and total BCAR1, BCAR1-1, and BCAR1-1′ mRNA levels were monitored by WB and RT-PCR, respectively (Figure 7). Initial treatment of MCF-7 cells for up to 18 hours (Figure 7, A and B) showed a weak induction of total BCAR1 (1.18-fold, not significant) and BCAR1-1 (1.3-fold, P < .001) mRNA levels at 2 hours continuously increasing until 18 hours (1.77-fold [P < .01] and 2.11-fold [P < .01], respectively). A delayed up-regulation of BCAR1-1′ was observed at 4 hours (1.25-fold, P < .05) further increasing until 18 hours (1.74-fold, P < .001). Accordingly, major p130Cas protein induction occurred at 12 hours, which was stable for up to 18 hours. Coinciding with the initial up-regulation of total BCAR1 and BCAR1-1, a transient induction of EGR1 at 2 hours and increasing levels of NAB2 protein were detected. EGR1 returned to basal levels at 4 hours and was reinduced at 12 and 18 hours, whereas NAB2 expression constantly increased until 18 hours. Because a strong up-regulation of p130Cas levels was still present, at 18 hours, the time of the PMA treatment was extended to 60 hours to elucidate the late expression kinetics. This study included also the tamoxifen-resistant TAM-R (Figure 7, C–E) and ER-negative BT-20 breast cancer (not shown) cells. In MCF-7 cells, the total BCAR1 mRNA induction remained similar for up to 60 hours, whereas a continuous increase in protein levels was observed, reaching its maximum at 36 hours (4-fold), which was stable for up to 60 hours (Figure 7C). Whereas the induction of BCAR1-1′ mRNA was relatively stable for 60 hours, BCAR1-1 levels slightly declined from 24 to 60 hours. The late p130Cas protein and mRNA expression patterns also correlated with the protein levels of EGR1 and NAB2 in MCF-7 cells. Similar results were obtained in BT-20 cells (not shown).
Figure 7.
p130Cas mRNA and protein expression are inducible in response to phorbol esters. (A-D) MCF-7 and TAM-R cells, as indicated, were serum starved for 16 hours followed by stimulation with PMA for indicated times up to 60 hours or left untreated (0 h). (A and B, upper panel, and C and D) Results are presented from one experiment of three performed with similar results. (A) Upper panel: WCEs (20 µg) were analyzed for p130Cas, EGR1, and NAB2 protein expression by WB. Lower panel: Densitometric analysis of protein expression. (B) Upper panel: expression of total BCAR1 mRNA and of BCAR1-1, BCAR1-1′, and GAPDH (control) mRNAs was determined by RT-PCR as described previously. - DNA indicates the negative control for PCR. Lower panel: Densitometric analysis of total BCAR1 mRNA, BCAR1-1, and BCAR1-1′ expression. (C and D) Left panels: WCEs (20 µg) were analyzed for p130Cas, EGR1, and NAB2 protein expression by WB. Right panels: Expression of total BCAR1 mRNA and of BCAR1-1, BCAR1-1′, and GAPDH (control) mRNAs was determined by RT-PCR as described previously. + control indicates positive control for PCR; - DNA, negative control for PCR. (E) Densitometric analysis of p130Cas protein (left panel) and total BCAR1 mRNA (right panel) expression in MCF-7 and TAM-R cells. (A and B, lower panel, and E) Average expression levels in relation to untreated cells (set to 1) and SD from three experiments performed were determined. Statistically significant changes compared with untreated cells are indicated: *P < .05, **P < .01, and ***P < .001. P values were calculated using t test.
In TAM-R cells, no major changes of total p130Cas protein and BCAR1 mRNA levels were observed (Figure 7D). Increased EGR1 expression was observed from 24 to 60 hours, whereas NAB2 levels were induced earlier further increasing until reaching a plateau at ∼36 hours. These results indicate that p130Cas protein and mRNA expression is inducible in response to PMA treatment in tamoxifen-sensitive MCF-7 cells, following the kinetics of a late response gene, but not in tamoxifen-resistant TAM-R cells (as compared in Figure 7E).
Differential p130Cas Regulation of EGR1 and NAB2 Expression in Tamoxifen-Sensitive MCF-7 and Tamoxifen-Resistant TAM-R Cells
In response to numerous stimuli, p130Cas mediates the activation of multiple signaling pathways including the Ras/mitogen-activated protein kinase and PI3K/Akt cascades [5,19]. Because EGR1 expression is regulated through mitogen-activated protein kinase and PI3K-dependent pathways [46], the influence of p130Cas on endogenous EGR1 levels was investigated. MCF-7 and TAM-R cells were transiently transfected with siRNAs specific for BCAR1 or a control siRNA (Figure 8A). In MCF-7 cells depletion of p130Cas levels by 79% led to a reduction of EGR1 and NAB2 expression of 60% (P < .001) and 45% (P < .01), respectively. Interestingly, a comparable downregulation of p130Cas in TAM-R cells did not significantly reduce EGR1 levels but decreased NAB2 expression by 50% (P < .001). This difference was also indirectly observed in MCF-7 and TAM-R cells as similar EGR1 levels were found in both cell lines but profoundly elevated NAB2 levels in TAM-R cells (Figure 6A). This suggests that the higher p130Cas expression in TAM-R cells correlates with increased NAB2 levels.
In summary, these findings suggest that in MCF-7 cells p130Cas signaling induces the expression of EGR1 and NAB2, thereby establishing a positive feedback loop to activate its own expression. Importantly, this mechanism seems to be partially altered in the tamoxifen-resistant derivative TAM-R (as compared in Figure 8B).
Discussion
Here, we studied the regulation of p130Cas/BCAR1 expression in mammary carcinomas. We show for the first time that p130Cas mRNA and protein are upregulated in response to PMA treatment, mimicking growth factor signaling. Our results indicate that, in breast cancer cells, p130Cas mediates the induction of the TF EGR1 and its coregulator NAB2. Moreover, EGR1 and NAB2 are, in turn, involved in the up-regulation of p130Cas/BCAR1, pointing to the existence of a positive feedback loop in mammary cancers. Importantly, we discovered significant differences in the p130Cas/EGR1/NAB2 network among tamoxifen-sensitive and tamoxifen-resistant breast cancer cells. We demonstrated that NAB2 expression is elevated in tamoxifen-resistant breast cancer cells and in advanced and more aggressive human breast cancers, suggesting that NAB2 contributes to the enhanced levels of p130Cas found in these cancers.
Previously, we showed that p130Cas protein levels are elevated in vitro in tamoxifen-resistant breast cancer cells (TAM-R) [19]. Our experiments now revealed increased BCAR1 mRNA expression in the same tamoxifen-resistant TAM-R model system compared with the parental tamoxifen-sensitive MCF-7 cells [19,20]. These data suggest that increased expression of BCAR1 may contribute to the previously observed elevated p130Cas protein levels in tamoxifen-resistant cells and possibly in breast cancer. Corresponding to the alternative first exon variants Casa and Casb of murine p130Cas that did not show altered activity in binding to focal adhesion kinase [41], we identified the human mRNA orthologs, referred to as BCAR1-1 and BCAR1-1′, respectively. The large genomic distance between the first exon of each variant supports the potential usage of alternative promoters.
The in silico analysis of the 5′ regions of both BCAR1 variants revealed several prominent clusters of overlapping EGR/SP1 sites. Similar cis regulatory elements present in the promoters of, for example, the vascular endothelial growth factor (VEGF) [28], PDGF-A [29], and NAB2 [32], have been shown to be essential for driving EGR1-induced gene expression. With knockdown approaches and overexpression studies, we showed that EGR1 and its coregulator NAB2 control BCAR1/p130Cas expression in breast cancer cells. The EGR1 target NAB2 can act as both a corepressor [31,32] and a coactivator [30] of EGR1-mediated transcription. Our studies suggest that NAB2 coactivates the EGR1-mediated increase in p130Cas levels and that complex formation of EGR1 and NAB2 may be required to positively regulate p130Cas expression. More potent NAB2 regulatory effects on BCAR1/p130Cas and EGR1 levels in TAM-R versus MCF-7 cells were revealed by NAB2 knockdown experiments. In MCF-7 cells, NAB2 depletion primarily affected BCAR1-1′ levels, whereas in TAM-R cells, BCAR1-1, BCAR1-1′ mRNAs, and EGR1 mRNA and protein were significantly reduced. This suggests that NAB2 is important for BCAR1-1′ expression in both cell lines and, in addition, in TAM-R cells for maintaining EGR1 and BCAR1-1 levels. Importantly, higher basal NAB2 expression was found in TAM-R versus MCF-7 cells, and consistently, in ChIP assays performed with these cell lines, significantly increased binding of EGR1 to the BCAR1 5′ regions in TAM-R cells was observed. Therefore, our studies suggest that high constitutive NAB2 levels may result in sustained EGR1/NAB2 complex formation important for the recruitment of EGR1 to the BCAR1 regulatory 5′ regions and subsequently enhanced BCAR1/p130Cas levels in tamoxifen-resistant TAM-R cells. Correspondingly, elevated total BCAR1 mRNA levels and a similar increase in the expression of both exon 1 variants were found in TAM-R cells in comparison to MCF-7 cells (Figure 1, C and D). This suggests that, in this tamoxifen-resistant model, both BCAR1 variants are regulated by likewise mechanisms. However, the different expression patterns of the two mRNA variants in additional breast tissues and a breast cancer cell line (Figure 1C) suggest that additional mechanisms may distinctly regulate the expression of the two p130Cas isoforms by, for example, tissue-specific TFs or in response to different signaling pathways altered during tumor development/progression. Such mechanisms have been observed for the aromatase (Cyp19) [47] and ERα genes (ESR1) [48]. Because increased NAB2 expression was also found in breast cancer specimens derived from higher-grade tumors and metastases [43–45], this mechanism may contribute not only to the elevated BCAR1/p130Cas levels observed in tamoxifen-resistant but also in aggressive/advanced mammary carcinomas [14,15,40].
Binding sites for members of additional TF families, such as NF-κB, p53, and HIF, that are inducible or activated by multiple signaling pathways, were predicted in the 5′ regions of the alternative first BCAR1 exons by in silico analysis. At this point, their importance in BCAR1 regulation cannot be ruled out, and these sites further point to the inducibility of p130Cas/BCAR1 expression. However, p130Cas/BCAR1 levels have not yet been linked to cell cycle progression or to be upregulated by extracellular stimuli such as growth factors as it is known for the family member HEF1 [7]. Previous studies primarily focused on the early posttranslational modifications of the p130Cas protein in response to different stimuli. Our comprehensive investigation of the expression kinetics in response to phorbol esters, mimicking growth factor signaling, indicated for the first time that p130Cas protein and BCAR1 mRNA are inducible following the kinetics of a late response gene in MCF-7 cells. However, BCAR1/p130Cas levels were not majorly influenced in TAM-R cells, which may in part be explained by the high basal BCAR1/p130Cas expression in these cells. In addition, this implies that the elevated BCAR1/p130Cas levels in TAM-R cells might be maintained by intrinsic mechanisms. These altered activities in MCF-7 and TAM-R suggest changes in the regulation of BCAR1 expression during acquired tamoxifen resistance.
In summary, based on the results presented here, we propose the following models for the regulation of BCAR1 expression in tamoxifen-sensitive and -resistant cells (Figure 8B). In the tamoxifen-sensitive MCF-7 cells, upon extracellular stimuli, p130Cas is activated/phosphorylated, leading to the activation of downstream signaling pathways [7] that induce the expression of the immediate early gene EGR1 [46]. Subsequently, EGR1 activates the transcription of the delayed early response gene NAB2 [33]. In turn, the EGR1/NAB2 complex mediates the late induction of BCAR1, thereby establishing a positive feedback loop. In contrast, in the tamoxifen-resistant TAM-R cells, p130Cas levels are not majorly influenced by phorbol esters, mimicking growth factor signaling. Therefore, we suggest that the constitutively high expression and phosphorylation levels of p130Cas [19] lead to enhanced downstream signaling and possibly to a constant induction of NAB2. Because p130Cas signaling does not influence EGR1 levels in TAM-R cells, other TFs of, for example, the EGR family [32] may induce NAB2 expression. High NAB2 expression is important for maintaining basal EGR1 levels in these cells and, subsequently, results in enhanced EGR1/NAB2 complex formation and thus the activation of BCAR1 expression. This reciprocal circuit may in part explain why p130Cas and NAB2 expression is constitutively upregulated in TAM-R cells.
The results of our studies in identifying EGR1 and NAB2 as regulators of p130Cas/BCAR1 may have an impact not only on breast cancer but also on other malignancies. Recently, p130Cas/BCAR1 was identified as a key player in prostate carcinomas, and increased levels were associated with aggressiveness and progression of this disease [49,50]. Because EGR1 overexpression has been described in multiple studies to be crucial for driving prostate cancer progression [51–53], it may also contribute to the up-regulation of p130Cas in this type of cancer. It would be interesting to see whether the p130Cas/EGR1/NAB2 circuit or alterations of this network may play a role in promoting these carcinomas as well.
Further deciphering of the precise mechanism of the p130Cas/EGR1/NAB2 feedback loop may help to understand the molecular biology not only of breast cancers but also of other malignancies and to identify new targets for the development of novel treatment strategies.
Acknowledgments
The authors thank Robert I. Nicholson for providing TAM-R cells as well as Matthew D. Layne, Barbara D. Smith, and Judith P. Johnson for helpful comments and critical reading of the article. The authors also thank Nora D. Mineva and Mathilde Romagnoli for expert assistance with the Oncomine database and Shefali Soni for her technical expertise and helpful advice.
Abbreviations
- BCAR1
breast cancer antiestrogen resistance 1
- ChIP
chromatin immunoprecipitation
- p130Cas
Crk-associated substrate
- dn
dominant negative
- EGR
early growth response
- ev
empty vector
- ER
estrogen receptor
- ERE
estrogen response element
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HIF
hypoxia-inducible factor
- NAB2
NGFI-A binding protein 2
- PPIA
peptidylprolyl isomerase A
- PI3K
phosphatidylinositol 3-kinase
- RT-PCR
reverse transcription-polymerase chain reaction
- RPLP0
ribosomal protein, large, P0
- RN18S1
RNA 18S ribosomal 1
- siRNA
short interfering RNA
- TF
transcription factor
- TSS
transcription start site
- WB
Western blot analysis
- WCE
whole cell extract
- wt
wild-type
Footnotes
This work was supported by Public Health Service grants CA106468 and CA143108 from the National Cancer Institute and the Susan G. Komen for the Cure Breast Cancer Foundation grant KG101208.
References
- 1.Sakai R, Iwamatsu A, Hirano N, Ogawa S, Tanaka T, Mano H, Yazaki Y, Hirai H. A novel signaling molecule, p130, forms stable complexes in vivo with v-Crk and v-Src in a tyrosine phosphorylation-dependent manner. EMBO J. 1994;13:3748–3756. doi: 10.1002/j.1460-2075.1994.tb06684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brinkman A, van der Flier S, Kok EM, Dorssers LC. BCAR1, a human homologue of the adapter protein p130Cas, and antiestrogen resistance in breast cancer cells. J Natl Cancer Inst. 2000;92:112–120. doi: 10.1093/jnci/92.2.112. [DOI] [PubMed] [Google Scholar]
- 3.Dorssers LC, van Agthoven T, Dekker A, van Agthoven TL, Kok EM. Induction of antiestrogen resistance in human breast cancer cells by random insertional mutagenesis using defective retroviruses: identification of bcar-1, a common integration site. Mol Endocrinol. 1993;7:870–878. doi: 10.1210/mend.7.7.8413311. [DOI] [PubMed] [Google Scholar]
- 4.Singh MK, Dadke D, Nicolas E, Serebriiskii IG, Apostolou S, Canutescu A, Egleston BL, Golemis EA. A novel Cas family member, HEPL, regulates FAK and cell spreading. Mol Biol Cell. 2008;19:1627–1636. doi: 10.1091/mbc.E07-09-0953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouton AH, Riggins RB, Bruce-Staskal PJ. Functions of the adapter protein Cas: signal convergence and the determination of cellular responses. Oncogene. 2001;20:6448–6458. doi: 10.1038/sj.onc.1204785. [DOI] [PubMed] [Google Scholar]
- 6.Singh M, Cowell L, Seo S, O'Neill G, Golemis E. Molecular basis for HEF1/NEDD9/Cas-L action as a multifunctional co-ordinator of invasion, apoptosis and cell cycle. Cell Biochem Biophys. 2007;48:54–72. doi: 10.1007/s12013-007-0036-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tikhmyanova N, Little JL, Golemis EA. CAS proteins in normal and pathological cell growth control. Cell Mol Life Sci. 2010;67:1025–1048. doi: 10.1007/s00018-009-0213-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabodi S, Pilar Camacho-Leal M, Di Stefano P, Defilippi P. Integrin signalling adaptors: not only figurants in the cancer story. Nat Rev Cancer. 2010;10:858–870. doi: 10.1038/nrc2967. [DOI] [PubMed] [Google Scholar]
- 9.Kumbrink J, Kirsch KH. Targeting Cas family proteins as a novel treatment for breast cancer. In: Gunduz E, Gunduz M, editors. Breast Cancer — Current and Alternative Therapeutic Modalities. 2011. pp. 37–62. InTech. [Google Scholar]
- 10.Marcotte R, Muller WJ. Signal transduction in transgenic mouse models of human breast cancer—implications for human breast cancer. J Mammary Gland Biol Neoplasia. 2008;13:323–335. doi: 10.1007/s10911-008-9087-3. [DOI] [PubMed] [Google Scholar]
- 11.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009;360:790–800. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 12.Vellon L, Menendez JA, Lupu R. αVβ3 integrin regulates heregulin (HRG)-induced cell proliferation and survival in breast cancer. Oncogene. 2005;24:3759–3773. doi: 10.1038/sj.onc.1208452. [DOI] [PubMed] [Google Scholar]
- 13.Henderson BE, Feigelson HS. Hormonal carcinogenesis. Carcinogenesis. 2000;21:427–433. doi: 10.1093/carcin/21.3.427. [DOI] [PubMed] [Google Scholar]
- 14.van der Flier S, Brinkman A, Look MP, Kok EM, Meijer-van Gelder ME, Klijn JG, Dorssers LC, Foekens JA. Bcar1/p130Cas protein and primary breast cancer: prognosis and response to tamoxifen treatment. J Natl Cancer Inst. 2000;92:120–127. doi: 10.1093/jnci/92.2.120. [DOI] [PubMed] [Google Scholar]
- 15.Konstantinovsky S, Smith Y, Zilber S, Tuft SH, Becker AM, Nesland JM, Reich R, Davidson B. Breast carcinoma cells in primary tumors and effusions have different gene array profiles. J Oncol. 2010;2010:969084. doi: 10.1155/2010/969084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scibelli A, d'Angelo D, Pelagalli A, Tafuri S, Avallone L, Della MR, Staiano N. Expression levels of the focal adhesion-associated proteins paxillin and p130Cas in canine and feline mammary tumors. Vet Res. 2003;34:193–202. doi: 10.1051/vetres:2002066. [DOI] [PubMed] [Google Scholar]
- 17.Dorssers LC, Van der Flier S, Brinkman A, van Agthoven T, Veldscholte J, Berns EM, Klijn JG, Beex LV, Foekens JA. Tamoxifen resistance in breast cancer: elucidating mechanisms. Drugs. 2001;61:1721–1733. doi: 10.2165/00003495-200161120-00004. [DOI] [PubMed] [Google Scholar]
- 18.Ta HQ, Thomas KS, Schrecengost RS, Bouton AH. A novel association between p130Cas and resistance to the chemotherapeutic drug adriamycin in human breast cancer cells. Cancer Res. 2008;68:8796–8804. doi: 10.1158/0008-5472.CAN-08-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soni S, Lin BT, August A, Nicholson RI, Kirsch KH. Expression of a phosphorylated p130(Cas) substrate domain attenuates the phosphatidylinositol 3-kinase/Akt survival pathway in tamoxifen resistant breast cancer cells. J Cell Biochem. 2009;107:364–375. doi: 10.1002/jcb.22136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knowlden JM, Hutcheson IR, Jones HE, Madden T, Gee JM, Harper ME, Barrow D, Wakeling AE, Nicholson RI. Elevated levels of epidermal growth factor receptor/c-erbB2 heterodimers mediate an autocrine growth regulatory pathway in tamoxifen-resistant MCF-7 cells. Endocrinology. 2003;144:1032–1044. doi: 10.1210/en.2002-220620. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Su H, Rahimi M, Tochihara R, Tang C. EGFRvIII-induced estrogen-independence, tamoxifen-resistance phenotype correlates with PgR expression and modulation of apoptotic molecules in breast cancer. Int J Cancer. 2009;125:2021–2028. doi: 10.1002/ijc.24540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim CG, Choi BH, Son SW, Yi SJ, Shin SY, Lee YH. Tamoxifen-induced activation of p21Waf1/Cip1 gene transcription is mediated by early growth response-1 protein through the JNK and p38 MAP kinase/Elk-1 cascades in MDA-MB-361 breast carcinoma cells. Cell Signal. 2007;19:1290–1300. doi: 10.1016/j.cellsig.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki T, Inoue A, Miki Y, Moriya T, Akahira J, Ishida T, Hirakawa H, Yamaguchi Y, Hayashi S, Sasano H. Early growth responsive gene 3 in human breast carcinoma: a regulator of estrogen-meditated invasion and a potent prognostic factor. Endocr Relat Cancer. 2007;14:279–292. doi: 10.1677/ERC-06-0005. [DOI] [PubMed] [Google Scholar]
- 24.Thiel G, Cibelli G. Regulation of life and death by the zinc finger transcription factor Egr-1. J Cell Physiol. 2002;193:287–292. doi: 10.1002/jcp.10178. [DOI] [PubMed] [Google Scholar]
- 25.Carter JH, Lefebvre JM, Wiest DL, Tourtellotte WG. Redundant role for early growth response transcriptional regulators in thymocyte differentiation and survival. J Immunol. 2007;178:6796–6805. doi: 10.4049/jimmunol.178.11.6796. [DOI] [PubMed] [Google Scholar]
- 26.Beckmann AM, Wilce PA. Egr transcription factors in the nervous system. Neurochem Int. 1997;31:477–510. doi: 10.1016/s0197-0186(96)00136-2. [DOI] [PubMed] [Google Scholar]
- 27.Honkaniemi J, Zhang JS, Longo FM, Sharp FR. Stress induces zinc finger immediate early genes in the rat adrenal gland. Brain Res. 2000;877:203–208. doi: 10.1016/s0006-8993(00)02673-1. [DOI] [PubMed] [Google Scholar]
- 28.Liu D, Evans I, Britton G, Zachary I. The zinc-finger transcription factor, early growth response 3, mediates VEGF-induced angiogenesis. Oncogene. 2008;27:2989–2998. doi: 10.1038/sj.onc.1210959. [DOI] [PubMed] [Google Scholar]
- 29.Khachigian LM, Williams AJ, Collins T. Interplay of Sp1 and Egr-1 in the proximal platelet-derived growth-factor-A-chain promoter in cultured vascular endothelial-cells. J Biol Chem. 1995;270:27679–27686. doi: 10.1074/jbc.270.46.27679. [DOI] [PubMed] [Google Scholar]
- 30.Sevetson BR, Svaren J, Milbrandt J. A novel activation function for NAB proteins in EGR-dependent transcription of the luteinizing hormone beta gene. J Biol Chem. 2000;275:9749–9757. doi: 10.1074/jbc.275.13.9749. [DOI] [PubMed] [Google Scholar]
- 31.Svaren J, Sevetson BR, Apel ED, Zimonjic DB, Popescu NC, Milbrandt J. NAB2, a corepressor of NGFI-A (Egr-1) and Krox20, is induced by proliferative and differentiative stimuli. Mol Cell Biol. 1996;16:3545–3553. doi: 10.1128/mcb.16.7.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumbrink J, Kirsch KH, Johnson JP. EGR1, EGR2, and EGR3 activate the expression of their coregulator NAB2 establishing a negative feedback loop in cells of neuroectodermal and epithelial origin. J Cell Biochem. 2010;111:207–217. doi: 10.1002/jcb.22690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumbrink J, Gerlinger M, Johnson JP. Egr-1 induces the expression of its corepressor nab2 by activation of the nab2 promoter thereby establishing a negative feedback loop. J Biol Chem. 2005;280:42785–42793. doi: 10.1074/jbc.M511079200. [DOI] [PubMed] [Google Scholar]
- 34.Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7(suppl 1):S12–S14. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 36.Min C, Kirsch KH, Zhao Y, Jeay S, Palamakumbura AH, Trackman PC, Sonenshein GE. The tumor suppressor activity of the lysyl oxidase propeptide reverses the invasive phenotype of Her-2/neu-driven breast cancer. Cancer Res. 2007;67:1105–1112. doi: 10.1158/0008-5472.CAN-06-3867. [DOI] [PubMed] [Google Scholar]
- 37.Kirsch KH, Georgescu MM, Hanafusa H. Direct binding of p130(Cas) to the guanine nucleotide exchange factor C3G. J Biol Chem. 1998;273:25673–25679. doi: 10.1074/jbc.273.40.25673. [DOI] [PubMed] [Google Scholar]
- 38.Kirsch KH, Korradi Y, Johnson JP. Mader: a novel nuclear protein over expressed in human melanomas. Oncogene. 1996;12:963–971. [PubMed] [Google Scholar]
- 39.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 40.van der Flier S, Chan CM, Brinkman A, Smid M, Johnston SR, Dorssers LC, Dowsett M. BCAR1/p130Cas expression in untreated and acquired tamoxifen-resistant human breast carcinomas. Int J Cancer. 2000;89:465–468. doi: 10.1002/1097-0215(20000920)89:5<465::aid-ijc11>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 41.Polte TR, Hanks SK. Interaction between focal adhesion kinase and Crk-associated tyrosine kinase substrate p130Cas. Proc Natl Acad Sci USA. 1995;92:10678–10682. doi: 10.1073/pnas.92.23.10678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Svaren J, Sevetson BR, Golda T, Stanton JJ, Swirnoff AH, Milbrandt J. Novel mutants of NAB corepressors enhance activation by Egr transactivators. EMBO J. 1998;17:6010–6019. doi: 10.1093/emboj/17.20.6010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ginestier C, Cervera N, Finetti P, Esteyries S, Esterni B, Adelaide J, Xerri L, Viens P, Jacquemier J, Charafe-Jauffret E, et al. Prognosis and gene expression profiling of 20q13-amplified breast cancers. Clin Cancer Res. 2006;12:4533–4544. doi: 10.1158/1078-0432.CCR-05-2339. [DOI] [PubMed] [Google Scholar]
- 44.Zhao HJ, Langerod A, Ji Y, Nowels KW, Nesland JM, Tibshirani R, Bukholm IK, Karesen R, Botstein D, Borresen-Dale AL, et al. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol Biol Cell. 2004;15:2523–2536. doi: 10.1091/mbc.E03-11-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Radvanyi L, Singh-Sandhu D, Gallichan S, Lovitt C, Pedyczak A, Mallo G, Gish K, Kwok K, Hanna W, Zubovits J, et al. The gene associated with trichorhinophalangeal syndrome in humans is overexpressed in breast cancer. Proc Natl Acad Sci USA. 2005;102:11005–11010. doi: 10.1073/pnas.0500904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumahara E, Ebihara T, Saffen D. Nerve growth factor induces zif268 gene expression via MAPK-dependent and -independent pathways in PC12D cells. J Biochem. 1999;125:541–553. doi: 10.1093/oxfordjournals.jbchem.a022319. [DOI] [PubMed] [Google Scholar]
- 47.Wang H, Li R, Hu Y. The alternative noncoding exons 1 of aromatase (Cyp19) gene modulate gene expression in a posttranscriptional manner. Endocrinology. 2009;150:3301–3307. doi: 10.1210/en.2008-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kos M, Reid G, Denger S, Gannon F. Minireview: genomic organization of the human ERα gene promoter region. Mol Endocrinol. 2001;15:2057–2063. doi: 10.1210/mend.15.12.0731. [DOI] [PubMed] [Google Scholar]
- 49.Fromont G, Vallancien G, Validire P, Levillain P, Cussenot O. BCAR1 expression in prostate cancer: association with 16q23 LOH status, tumor progression and EGFR/KAI1 staining. Prostate. 2007;67:268–273. doi: 10.1002/pros.20516. [DOI] [PubMed] [Google Scholar]
- 50.Fromont G, Cussenot O. The integrin signalling adaptor p130Cas is also a key player in prostate cancer. Nat Rev Cancer. 2011;11:227. doi: 10.1038/nrc2967-c1. [DOI] [PubMed] [Google Scholar]
- 51.Abdulkadir SA, Carbone JM, Naughton CK, Humphrey PA, Catalona WJ, Milbrandt J. Frequent and early loss of the EGR1 corepressor NAB2 in human prostate carcinoma. Hum Pathol. 2001;32:935–939. doi: 10.1053/hupa.2001.27102. [DOI] [PubMed] [Google Scholar]
- 52.Abdulkadir SA, Qu Z, Garabedian E, Song SK, Peters TJ, Svaren J, Carbone JM, Naughton CK, Catalona WJ, Ackerman JJ, et al. Impaired prostate tumorigenesis in Egr1-deficient mice. Nat Med. 2001;7:101–107. doi: 10.1038/83231. [DOI] [PubMed] [Google Scholar]
- 53.Ma J, Ren Z, Ma Y, Xu L, Zhao Y, Zheng C, Fang Y, Xue T, Sun B, Xiao W. Targeted knockdown of EGR-1 inhibits IL-8 production and IL-8-mediated invasion of prostate cancer cells through suppressing EGR-1/NF-κB synergy. J Biol Chem. 2009;284:34600–34606. doi: 10.1074/jbc.M109.016246. [DOI] [PMC free article] [PubMed] [Google Scholar]