Abstract
p53 is a tumor suppressor gene that is frequently mutated in multiple cancer tissues. Activated p53 protein regulates its downstream genes and subsequently inhibits malignant transformation by inducing cell cycle arrest, apoptosis, DNA repair, and senescence. However, genes involved in the p53-mediated senescence pathway are not yet fully elucidated. Through the screening of two genome-wide expression profile data sets, one for cells in which exogenous p53 was introduced and the other for senescent fibroblasts, we have identified chloride channel accessory 2 (CLCA2) as a p53-inducible senescence-associated gene. CLCA2 was remarkably induced by replicative senescence as well as oxidative stress in a p53-dependent manner. We also found that ectopically expressed CLCA2 induced cellular senescence, and the down-regulation of CLCA2 by small interfering RNA caused inhibition of oxidative stress-induced senescence. Interestingly, the reduced expression of CLCA2 was frequently observed in various kinds of cancers including prostate cancer, whereas its expression was not affected in precancerous prostatic intraepithelial neoplasia. Thus, our findings suggest a crucial role of p53/CLCA2-mediated senescence induction as a barrier for malignant transformation.
Introduction
In nearly half of all human cancers, the p53 gene is mutated or deleted [1]. The p53 protein is induced and activated in the nucleus by a variety of genotoxic stress such as DNA damage, hypoxia, and oxidative stress [2]. Activated p53 functions as a transcriptional factor, inhibits cell growth, and suppresses tumor formation by regulating many genes involved in cell cycle, apoptosis, and cellular senescence [3,4]. Although nearly a hundred p53-inducible genes have been identified so far [5,6], our recent microarray analysis revealed that there are still more than 50 p53-inducible genes that have not been well characterized yet [7]. To provide the detailed critical biologic functions of p53, we have previously identified a number of p53 target genes, such as p53R2, p53AIP1, p53RDL1, and PADI4 [4,8–13].
Senescence is mainly regulated through two major pathways involving p16Ink4a/Rb and p14ARF/p53 [14,15]. Expression of p16Ink4a markedly increases with aging in mouse and human tissues [16,17]. In addition, p16Ink4a expression was shown to increase in senescent fibroblasts [18] as well as in response to oxidative stress or DNA damage [19,20]. p16INK4A functions as a CDK4/CDK6 inhibitor and subsequently leads to a G1 arrest through the regulation of the pRBE2F pathway [21,22].
Conversely, p14ARF controls the level of p53 by inhibiting the p53-specific ubiquitin ligase MDM2 [21,22]. The role of p53 in cellular senescence was clearly demonstrated by the fact that p53-deficient fibroblasts showed resistance to senescence [23]. In addition, reactivation of p53 in murine liver carcinoma cells induced tumor regression through the induction of cellular senescence [24], and mice expressing active mutant p53 displayed an early onset of aging phenotypes [25]. Hence, it is obvious that activated p53 prevents malignant transformation of damaged cells by inducing senescence [26]. Because cellular senescence was observed in precancerous tissues [27,28], senescence is likely to function as a barrier for malignant transformation in the carcinogenesis process [29].
Among a number of transcriptional targets of p53, p21WAF1 was shown to play a principle role in both p53-dependent and independent senescence pathways [30]. p21WAF1 inhibits cell cycle progression through interaction with the cyclin-CDK complex. However, because p21WAF1-null fibroblast presents senescent phenotype [31], it is considered that another p53 target gene(s) may play a pivotal role in p53-dependent senescence induction. In this study, to identify a novel p53 target gene(s) that is essential for p53-dependent senescence induction, we examined the data sets of two genome-wide expression profile analyses. One data set was obtained using cells in which wild-type p53 was exogenously introduced, and the other was obtained using normal human dermal fibroblast (NHDF) cells at several time points from low to high passage (senescent state). Through this screening and subsequent biologic analysis, here we demonstrate a possible role of CLCA2 as a p53-inducible senescence mediator. We also examined the expression of CLCA2 in various cancer tissues. CLCA2 was reported as a p53 target gene that regulates p53-induced apoptotic pathway [32]. In addition, CLCA2 was shown to be downregulated or mutated in breast cancer tissues [33,34]. Recently, Rovillain et al. [35] reported that CLCA2 was strongly induced during senescence. Our findings would further support the crucial role of CLCA2 in the regulation of the senescence pathway as well as human carcinogenesis.
Materials and Methods
Complementary DNA Microarray
Complementary DNA (cDNA) microarray analysis was carried out as previously described [36]. For the senescence microarray, the total RNA of each sample was isolated and amplified using RNeasy spin column kits (Qiagen, Valencia, CA) and T7-Transcription kit (Epicentre Technologies, Madison, MI). For the p53 microarray, poly(A)+ RNAs were isolated from U373MG cells using a standard protocol. Each RNA sample was labeled and hybridized to a micro-array consisting of 36,864 cDNA fragments (http://www.ncbi.nlm.nih.gov/geo/index.cgi, Accession No. GSE14953).
Plasmid Construction
The entire coding sequence of CLCA2 cDNA was amplified by polymerase chain reaction (PCR) using KOD-Plus DNA polymerase (Toyobo, Osaka, Japan) and inserted into pCAGGS vector, which carries a gene conferring neomycin resistance. A Flag epitope tag was placed at the C-terminus of the CLCA2 expression vector. Constructs were confirmed by sequencing.
Cell Culture and Transfections
Each cell line was purchased from American Type Culture Collection, Lonza Biologics, Inc, or Japanese Collection of Research Bioresources. p53-deficient mice were obtained from RIKEN (http://www.cdb.riken.go.jp/arg/, Accession No. CDB0001K) [37]. We prepared mouse embryonic fibroblasts (MEFs) from embryonic day 13.5 embryos by standard protocols and maintained them in Dulbecco modified Eagle medium containing 10% fetal bovine serum. Replication-deficient recombinant adenovirus encoding p53 (Ad-p53) or LacZ (Ad-LacZ) was generated and purified, as previously described [8]. U373MG or H1299 cells were infected with viral solutions at an indicated multiplicity of infection (MOI) and incubated at 37°C until the time of harvest. Cells were transfected with plasmids using FuGENE6 (Roche, Basel, Switzerland). Small interfering RNA (siRNA) oligonucleotides, commercially synthesized by Sigma Genosis (The Woodlands, TX), were transfected with LipofectAMINE 2000 reagent (Life Technologies, Grand Island, NY) for 4 hours. Sequences of oligonucleotides are shown in Table W3. For treatment with genotoxic stress, cells were continuously incubated with hydrogen peroxide (H2O2) or adriamycin for 2 hours or irradiated by γ-ray or ultraviolet as indicated in the respective figure legends. 5-Aza-2′-deoxycytidine (5-Aza) and trichostatin A (TSA) were purchased from Sigma.
Quantitative Real-time PCR and Northern Blot Analysis
Isolation of total RNA from cultured cells was performed using RNeasy spin column kits (Qiagen) according to the manufacturer's instructions. Prostate and colorectal cancer tissues were prepared as previously described [38], and the total RNA of each sample was isolated and amplified. cDNAs were synthesized with the SuperScript Preamplification System (Invitrogen). Quantitative real-time PCR was conducted using the SYBR Green I Master or Probe Master on a Light-Cycler 480 (Roche) according to the manufacturer's recommendations. β2-Microglobulin or β-actin was used for normalization of expression levels. For the Northern blot analysis, a 2-µg aliquot of each poly(A)+ RNA was separated on a 1% agarose gel and transferred to nylon membrane by standard procedures. Hybridization with a random-primed 32P-labeled CLCA2 cDNA probe was carried out according to the manufacturer's instructions. The primer and probe sequences are indicated in Table W3.
Gene Reporter Assay
DNA fragments, including potential p53-binding sites of mouse Clca5, were amplified and subcloned into pGL3-promoter vector (Promega, Madison, WI). To make a series of mutant vectors, a point mutation “T” was inserted into the site of the 4th, the 7th, the 14th, and the 17th nucleotides of the consensus p53-BS using inverse PCR methods. Reporter assay was performed using the Dual Luciferase Assay System (Promega) as described previously [8].
Antibodies
Rabbits were immunized with the recombinant proteins corresponding to the extracellular domain (amino acids 253–899) of CLCA2. Anti-Flag monoclonal (clone M2) and polyclonal (F7425) antibodies, as well as anti-β-actin monoclonal antibody (clone AC15) were purchased from Sigma. Mouse monoclonal anti-p53 (clone DO-1) and anti-p21WAF1 (clone EA10) antibodies were purchased from Calbiochem (San Diego, CA). Anti-HA monoclonal antibody (F-7) was purchased from Santa Cruz Biotechnology, Santa Cruz, CA.
Western Blot Analysis
To prepare whole-cell lysates, cells were collected and lysed in chilled RIPA buffer (50 mM Tris-HCl at pH 8.0, 150 mM sodium chloride, 0.1% SDS, 0.5% DOC, 1% NP-40, and 1 mM phenyl methylsulfonyl fluoride) for 30 minutes on ice and centrifuged at 16,000g for 15 minutes. To concentrate the media, they were mixed with an equal volume of acetone and kept at -80°C for 1 hour. The mixture was then centrifuged at 16,000g for 15 minutes, and the resulting protein pellets were dissolved in SDS sample buffer. Samples were subjected to SDS-PAGE and immunoblot analysis by a standard procedure. Silver staining was carried out using SilverQuest Staining Kit (Invitrogen) according to the manufacturer's instructions.
Immunocytochemistry
Adherent cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) and permeabilized with 0.2% Triton X-100 in PBS for 5 minutes at room temperature. The cells were then covered with blocking solution (3% bovine serum albumin/PBS containing 0.2% Triton X-100) for 30 minutes at room temperature and incubated with rabbit anti-Flag antibody and mouse anti-HA antibody in blocking solution for 60 minutes at room temperature. Primary antibodies were stained with Alexa Fluor 488 antirabbit immunoglobulin G and Alexa Fluor 594 antimouse immunoglobulin G for 1 hour at room temperature and stained with 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI). For cell surface staining, cells were labeled under nonpermeabilized conditions using the buffer without Triton X-100.
Cell Proliferation and Cell Death Assay
Colony formation assays were carried out in six-well culture plates. Cells transfected with pCAGGS/CLCA2 or control plasmid were cultured in the presence of geneticin (Invitrogen) for 2 weeks. As a control plasmid, the entire coding sequence of CLCA2 cDNA was inserted into the vector in the opposite direction. Colonies were stained with crystal violet (Sigma) and scored using Image J software. Soft agar assay was carried out as previously described [8]. For the cell death assay, U373MG cells were infected with either Ad-p53 or Ad-LacZ at 6 hours after transfection of siRNA oligonucleotide. Apoptotic cells were quantified by FACS analysis as previously described [3]. Cell growth was determined by MTT assay using Cell Counting Kit-8 (Dojindo, Kumamoto, Japan) or by BrdU assay using Cell Proliferation ELISA BrdU (Roche).
Senescence-Associated β-Gal Staining
Senescence-associated (SA)-β-gal activity was detected as previously described [39].
Results
CLCA2 Expression Is Increased during Replicative Senescence
To investigate the genes involved in the process of cellular senescence, we compared the mRNA expression profiles of early-and late-passage (senescent) fibroblasts. Newly purchased NHDF cells were serially cultured under the conditions recommended by the distributors. Culture medium was changed every 3 days. When the cells reached 80% to 90% confluence, they were detached and plated at a density of 5 x 105 cell per 10-cm dish (approximately 30% confluence). Cell proliferation ratio of senescent fibroblast cells declined, and the cells became positive for SA-β-gal staining, which is a known senescence marker (Figure W1A). After passaging NHDF cells more than 30 times, they reached the senescence stage and almost stopped to grow. We conducted a cDNA microarray analysis using mRNAs from NHDF cells at the 3rd, 7th, 13th, 19th, and 26th passage levels. Through this analysis, we identified 85 genes that were increased by more than 10-fold in senescent fibroblast after the 26th passage (Table W1).
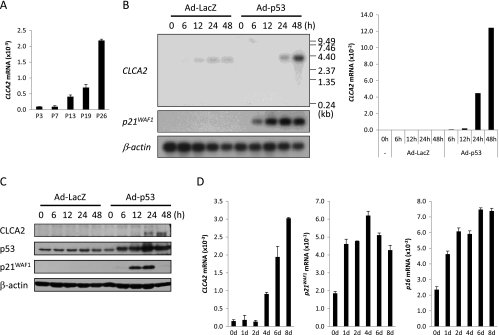
To identify novel p53 target genes, we had previously performed another mRNA expression profile analysis using U373MG p53-mutant glioblastoma cells in which wild-type p53 (Ad-p53) or LacZ (Ad-LacZ) were infected using an adenovirus system [7]. The comparison of these two data sets revealed six possible candidates that were increased by more than 10-fold in both data sets (Table W1). After performing semiquantitative reverse transcription (RT)-PCR for these six genes, we selected CLCA2 for further analysis because CLCA2 was remarkably increased during the cellular senescence process as well as by the ectopic induction of wild-type p53 (Figures 1A and W1, B and C). We also confirmed the p53-mediated CLCA2 induction by quantitative RT-PCR, Northern, and Western blot analyses (Figures 1, B and C, and W2A). The specificity of this antibody was verified by siRNA knockdown (Figure W2, A and B).
Figure 1.
CLCA2 expression is increased during replicative senescence. (A) Quantitative real-time PCR analysis of CLCA2 mRNA in NHDF cells at indicated passage levels. β2-Microglobulin was used for the normalization of expression levels. (B) Northern blot and quantitative real-time PCR analysis of CLCA2 transcript in U373MG cells at indicated times after infection with Ad-p53 or Ad-LacZ at 8 MOIs. β-Actin was used for the normalization of expression levels. p21WAF1 served as a positive control. (C) Western blot analysis of CLCA2 protein in U373MG cells at indicated times after infection with Ad-p53 or Ad-LacZ at 40 MOIs. β-Actin was used for the normalization of expression levels. p53 and p21WAF1 served as a positive control. (D) Quantitative real-time PCR analysis of CLCA2, p21WAF1, and p16Ink4a mRNA in IMR-90 cells at the indicated days after treatment with 400 µM of H2O2. β2-Microglobulin was used for the normalization of expression levels.
To investigate whether CLCA2 is involved in stress-induced pathway, we analyzed the expression of CLCA2 during oxidative stress-induced cellular senescence. We treated IMR-90 normal human diploid fibroblast cells with hydrogen peroxide (H2O2) and examined the expression of CLCA2 by quantitative real-time PCR analysis (Figure 1D). Interestingly, p21WAF1 and p16INK4A expression was increased as early as 1 or 2 days after H2O2 treatment, but the highest level of CLCA2 expression was observed at 8 days in H2O2-treated senescent fibroblasts. These data suggest that CLCA2 is likely to play some role in the later stages of DNA damage response.
Clca5 (Mouse Homologue of Human CLCA2) Is a Target of p53
Because Clca5 (mouse homologue of human CLCA2) was previously shown to be induced by genotoxic stress and suppress cell growth [40,41], we investigated whether Clca5 is associated with the p53-induced senescence induction. When MEFs derived from p53+/+ or p53-/- embryos were treated with oxidative stress, we found decreased staining of SA-β-gal in p53-/- MEF (Figure 2A). We also found that Clca5 expression was induced by oxidative stress in p53+/+ MEF but not in p53-/- MEF (Figure 2B). We then surveyed the genomic sequence of the Clca5 gene and found three putative p53-binding sequences (p53BS 1–3) in the promoter or intron 1 region (Figure 2C). We subcloned a DNA fragment of 1301 base pairs, which included the three putative p53-binding sequences into pGL3 promoter vector (pGL3/p53BR; Promega). We found that cotransfection of pGL3/p53BR with wild-type p53 expression plasmid enhanced the luciferase activity by more than four-fold, whereas the base substitutions within BS1 completely diminished the luciferase activity (Figure 2D). Interestingly, DNA sequence of p53BS1 was highly conserved between mouse Clca5 and human ClCA2 (Figure W3, A–C). These findings indicated that Clca5/CLCA2 could be involved in the p53-induced senescence pathway.
Figure 2.
Clca5 (mouse homologue of human CLCA2) is a target of p53. (A, B) p53+/+ and p53-/- MEFs were treated with 100 µM (A) or 150 µM (B) of H2O2. (A) At 6 days after treatment, the cells were subjected to SA-β-gal staining. (B) Quantitative real-time PCR analysis of Clca5 mRNA at indicated days after treatment. β-Actin was used for the normalization of expression levels. (C) Genomic structure of the mouse Clca5 gene. Gray boxes indicate the locations and relative sizes of the two exons. Arrows indicate the locations of potential p53-binding sites (p53BS1-3) in a p53-binding region (p53BR). Identical nucleotides to the consensus sequence are written in capital letters. The underlined nucleotides were substituted for thymine to examine the specificity of each p53-binding site. (D) Results of luciferase assay of p53BR with or without substitutions at either of the p53BS fragments are shown. Luciferase activity is indicated relative to the activity of mock vector with SDs (n = 2).
CLCA2 Contributes to Oxidative Stress-Induced Senescence
To clarify the role of CLCA2 in genotoxic stress-induced senescence pathway, we analyzed the expression of CLCA2 in H2O2-treated IMR-90 cells that were transfected with siRNA against p53 (sip53) or CLCA2 (siCLCA2-1 or siCLCA2-2). The result of quantitative real-time PCR analysis showed the drastic suppression of CLCA2 expression by p53 knockdown to the same extent as siCLCA2 treatment (Figure 3A). Moreover, inhibition of CLCA2 or p53 expression significantly suppressed H2O2-induced cellular senescence (Figure 3B). We also examined whether ectopic expression of CLCA2 could induce cellular senescence. MCF7 cells were stained with SA-β-gal 3 days after introduction of CLCA2-expression plasmid. As a result, we found an increase in positive staining for SA-β-gal in cells expressing CLCA2 compared with Mock-transfected cells (Figure 3, C and D). Taken together, our findings clearly indicated that p53-CLCA2 pathway contributes to genotoxic stress-induced senescence.
Figure 3.
CLCA2 is a key mediator of oxidative stress-induced senescence. (A, B) IMR-90 cells were transfected with siRNA oligonucleotides designed to suppress the expression of CLCA2 or p53 at 7 hours before treatment with 400 µM of H2O2. siEGFP was used as a control. At 8 days after treatment, the cells were subjected to quantitative real-time PCR analysis of CLCA2 mRNA (A) and SA-β-gal staining (B). β2-Microglobulin was used for the normalization of expression levels. The proportion of cells positive for SA-β-gal staining is indicated (B, upper panels). The representative images of cells are shown (B, lower panels). Asterisks indicate P value obtained by Student's t test: *P < .05 and **P < .01. (C, D) MCF7 cells were transfected with CLCA2 expression plasmid. (C) After 3 days, the cells were subjected to SA-β-gal staining. (D) The proportion of cells positive for SA-β-gal staining is indicated. Asterisk indicates P value obtained by Student's t test: **P < .01.
Down-regulation of CLCA2 in Prostate and Colorectal Cancer
Because CLCA2 was shown to be a potential breast tumor suppressor [34], we examined the expression of CLCA2 in various cancers using gene expression database that consists of more than 1000 cancer tissues [36,38,42–44]. The expression of CLCA2 was remarkably decreased in breast, bladder, esophageal, lung, and prostate cancer tissues (Table 1). We performed quantitative real-time PCR analyses and found that CLCA2 mRNA was frequently decreased in prostate and colorectal cancers (<20% of control in 9 of 10 prostate cancer tissues and in 10 of 17 colorectal cancer tissues; Figures 4A and W4A). Cellular senescence was shown to be observed in precancerous tissues and is likely to function as a barrier for malignant transformation [45]. Therefore, we investigated the expression of CLCA2 in premalignant tissues of the prostate (prostatic intraepithelial neoplasia [PIN]). As a result, the expression of CLCA2 in PIN is comparable with that in their corresponding normal prostate tissues but significantly higher than that in cancer tissues (Figure 4B).
Table 1.
Decreased Expression of CLCA2 in Cancer Tissues Examined by cDNA Microarray Analyses.
| Cancer Tissues | Suppression of CLCA2 (<1/3), n (%) | Total Samples (n) | References |
| Lung cancer (SCLC) | 7 (100) | 7 | Taniwaki et al. [36] |
| Prostate cancer | 21 (81) | 26 | Ashida et al. [38] |
| Esophageal cancer | 10 (53) | 19 | Yamabuki et al. [42] |
| Breast cancer | 30 (46) | 66 | Nishidate et al. [44] |
| Bladder cancer | 9 (28) | 32 | Takata et al. [43] |
Figure 4.
Down-regulation of CLCA2 in prostate cancer. (A) Relative CLCA2 expression in prostate cancer tissues compared with normal tissues was examined by quantitative real-time PCR analysis. β-Actin was used for the normalization of expression levels. (B) CLCA2 expressions in prostate cancer tissues, PIN, and normal tissues were examined by quantitative real-time PCR analysis. β-Actin was used for the normalization of expression levels. (C) CLCA2 expressions in prostate cancer cell lines were examined by quantitative real-time PCR analysis. β2-Microglobulin was used for the normalization of expression levels. (D) Three prostate cancer cell lines were treated with 5 µM of 5-Aza and/or 0.5 µM of TSA. CLCA2 expressions were examined by quantitative real-time PCR analysis. β2-Microglobulin was used for the normalization of expression levels. Asterisks indicate P value obtained by Student's t test: *P < .05, **P < .01, and N.S. (not statistically significant).
Because expression of CLCA2 is regulated by p53, inactivation of p53 is likely to cause the decrease of CLCA2 expression in cancer cells. Therefore, we examined the CLCA2 expression and the p53 gene mutation in five prostate cancer cell lines as well as one normal prostate epithelial cell line. As a result, the expression of CLCA2 was down-regulated in cancer cell lines compared with that of normal prostate epithelial cell line (P < .01), but we found no significant difference in CLCA2 expression between wild-type and mutant p53 cell lines (Figure 4C; P = .86). These data suggested that there may be another mechanism that suppresses CLCA2 expression in cancer cells other than p53 mutation. Because epigenetic change is one of the major causes of gene silencing in cancer tissues, we treated three prostate cancer cell lines with TSA (inhibitor of histone deacetylation) and/or 5-Aza (inhibitor of DNA methylation). We found that, after treatment, the CLCA2 expression was partially restored in all three cell lines (Figure 4D), suggesting the possible role of epigenetic alteration in CLCA2 inactivation. Similarly, 5-Aza treatment increased CLCA2 expression in two colorectal cancer cell lines among seven examined (Figure W4B). Nevertheless, we found that HCT116 p53-/- cells exhibited lower CLCA2 expression than HCT116 p53+/+ cells (Figure W4C). In addition, cells carrying wild-type p53 exhibited relatively higher CLCA2 expression than cells with p53 mutation, although the difference was not statistically significant (P =.47; Figure W4D and Table W2). Because p53 was frequently inactivated by various mechanisms in cancer tissues, both epigenetic alteration and p53 inactivation would play crucial roles in the deregulation of CLCA2 expression in prostate and colorectal cancer tissues.
Discussion
Here we show that CLCA2 expression is remarkably increased during cellular senescence. Ectopic expression of CLCA2 induced senescence in cells, and knockdown of CLCA2 as well as p53 inhibited genotoxic stress-induced cellular senescence. These findings revealed that CLCA2 is a mediator of p53-induced senescence.
We have previously demonstrated the distinct mechanisms by which p53 determined cell fate through the induction of different sets of its target genes. At an early time point after repairable DNA damage, p53 induces p21WAF1 and p53R2, which cooperatively modulate cell survival through the induction of cell cycle arrest and subsequent repair of damaged DNA [4]. However, when DNA damage is too severe to be repaired, specifically activated p53 induces genes such as p53AIP1, a crucial mediator of apoptotic pathway, to eliminate the damaged cells [8]. Because senescence is also an irreversible process like apoptosis, our finding suggested that p53 regulates cell fate by inducing senescence associate gene(s) when cells are severely damaged. Actually, CLCA2 expression was peaked at a later time point during replicative senescence compared with p16Ink4a and p21WAF1, suggesting the activation of CLCA2 in the aging tissues. Because most prostate cancer affects elderly men, decreased expression of CLCA2 due to epigenetic alteration or p53 inactivation would play an important role in prostate carcinogenesis.
Walia et al. reported that CLCA2 regulates intracellular pH. Under physiological conditions, the ion channels maintain the body pH within a range of 7.35 to 7.45. However, the pH of tumor cells was found to be neutral or alkaline [46]. Higher pH was associated with the apoptosis resistance and the invasive property of tumor cells, and inhibition of the Na(+)/H(+) exchanger isoform 1 (NHE1) induced apoptosis in cancer cells [47]. Conversely, aging caused the change of the composition and function of cell membranes and subsequently decreased the pH [48]. Degenerative neural diseases such as Parkinson disease and Alzheimer disease indicate a decreased pH in cells and tissues [49]. Thus, intracellular pH is strongly associated with oncogenic and senescence phenotype. Taken together, CLCA2 could function as a barrier for malignant transformation by inducing cellular senescence and tumor regression through the regulation of intracellular pH.
N-terminal CLCA2 was shown to be cleaved and secreted into the culture medium [50]. Although several studies indicated the growth suppressive effect of membrane-localized CLCA2, the role of soluble CLCA2 was not well characterized. The physiological significance of processing at its extracellular domain varies among substrates proteins. Tumor necrosis factor α is cleaved by TACE and exhibits strong systemic effects [51]. In contrast, soluble MICA blocks natural killer cell activity [52], whereas membrane-bound MICA stimulates immune response mediated by natural killer cell [53]. E-cadherin is cleaved by metalloproteinase and a residual membrane-tethered product is degraded by intracellular proteolytic pathway [54]. Because senescent human fibroblasts were shown to stimulate proliferation of epithelial cells in vitro and in vivo [55], soluble CLCA2 secreted by the senescence-activated fibroblasts might exhibit growth-promoting activity in contrast to the membrane-localized CLCA2. We would like to analyze the role of membrane-localized and soluble CLCA2 on the regulation of cellular senescence pathway in the future study. Many organ-specific serum biomarkers for aging such as N-telopeptide of type I collagen and MT1-MMP for bone resorption [56], CD4/CD8 ratio for immune system, albumin, and growth hormone for nutrition [57] are clinically valuable [58]. Telomere length is a good marker for biologic aging [59], but serum biomarkers of human aging have not yet been clinically validated. Because CLCA2 was increased in response to genotoxic stress as well as replicative senescence, serum CLCA2 would be a good biomarker for aging. Although we have not yet been able to establish an ELISA test to detect soluble CLCA2 owing to the high background of antibody, we would like to investigate the role of soluble CLCA2 as a senescence biomarker in the future.
Although further functional analyses, especially for the role of the cleaved N-terminal fragment, are essential to fully elucidate the role of CLCA2 in senescence and carcinogenesis, we believe that our findings may lead to the development of a novel cancer therapy by reactivating CLCA2 signaling.
Supplementary Material
Acknowledgments
The authors thank T. Katagiri for helpful discussions and K. Makino for technical assistance.
Footnotes
This work was supported in part by grant 18687012 from the Japan Society for the Promotion of Science and Ministry of Education, Culture, Sports, Science, and Technology of Japan (to K.M.). The authors declare no conflict of interest.
This article refers to supplementary materials, which are designated by Tables W1 to W3 and Figures W1 to W4 and are available online at www.neoplasia.com.
References
- 1.Soussi T, Asselain B, Hamroun D, Kato S, Ishioka C, Claustres M, Beroud C. Meta-analysis of the p53 mutation database for mutant p53 biological activity reveals a methodologic bias in mutation detection. Clin Cancer Res. 2006;12:62–69. doi: 10.1158/1078-0432.CCR-05-0413. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 3.Matsuda K, Yoshida K, Taya Y, Nakamura K, Nakamura Y, Arakawa H. p53AIP1 regulates the mitochondrial apoptotic pathway. Cancer Res. 2002;62:2883–2889. [PubMed] [Google Scholar]
- 4.Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukuda S, Matsui K, Takei Y, Nakamura Y. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 5.el-Deiry WS. Regulation of p53 downstream genes. Semin Cancer Biol. 1998;8:345–357. doi: 10.1006/scbi.1998.0097. [DOI] [PubMed] [Google Scholar]
- 6.Nakamura Y. Isolation of p53-target genes and their functional analysis. Cancer Sci. 2004;95:7–11. doi: 10.1111/j.1349-7006.2004.tb03163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanikawa C, Furukawa Y, Yoshida N, Arakawa H, Nakamura Y, Matsuda K. XEDAR as a putative colorectal tumor suppressor that mediates p53-regulated anoikis pathway. Oncogene. 2009;28:3081–3092. doi: 10.1038/onc.2009.154. [DOI] [PubMed] [Google Scholar]
- 8.Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, Nishimori H, Tamai K, Tokino T, Nakamura Y, et al. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell. 2000;102:849–862. doi: 10.1016/s0092-8674(00)00073-8. [DOI] [PubMed] [Google Scholar]
- 9.Tanikawa C, Matsuda K, Fukuda S, Nakamura Y, Arakawa H. p53RDL1 regulates p53-dependent apoptosis. Nat Cell Biol. 2003;5:216–223. doi: 10.1038/ncb943. [DOI] [PubMed] [Google Scholar]
- 10.Tanikawa C, Ueda K, Nakagawa H, Yoshida N, Nakamura Y, Matsuda K. Regulation of protein citrullination through p53/PADI4 network in DNA damage response. Cancer Res. 2009;69:8761–8769. doi: 10.1158/0008-5472.CAN-09-2280. [DOI] [PubMed] [Google Scholar]
- 11.Tanikawa C, Ri C, Kumar V, Nakamura Y, Matsuda K. Crosstalk of EDA-A2/XEDAR in the p53 signaling pathway. Mol Cancer Res. 2010;8:855–863. doi: 10.1158/1541-7786.MCR-09-0484. [DOI] [PubMed] [Google Scholar]
- 12.Kidokoro T, Tanikawa C, Furukawa Y, Katagiri T, Nakamura Y, Matsuda K. CDC20, a potential cancer therapeutic target, is negatively regulated by p53. Oncogene. 2008;27:1562–1571. doi: 10.1038/sj.onc.1210799. [DOI] [PubMed] [Google Scholar]
- 13.Tanikawa C, Espinosa M, Suzuki A, Masuda K, Yamamoto K, Tsuchiya E, Ueda K, Daigo Y, Nakamura Y, Matsuda K. Regulation of histone modification and chromatin structure by the p53-PADI4 pathway. Nat Commun. 2012;3:676. doi: 10.1038/ncomms1676. [DOI] [PubMed] [Google Scholar]
- 14.Schmitt CA, Fridman JS, Yang M, Lee S, Baranov E, Hoffman RM, Lowe SW. A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell. 2002;109:335–346. doi: 10.1016/s0092-8674(02)00734-1. [DOI] [PubMed] [Google Scholar]
- 15.Agger K, Cloos PA, Rudkjaer L, Williams K, Andersen G, Christensen J, Helin K. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene-and stress-induced senescence. Genes Dev. 2009;23:1171–1176. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zindy F, Quelle DE, Roussel MF, Sherr CJ. Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene. 1997;15:203–211. doi: 10.1038/sj.onc.1201178. [DOI] [PubMed] [Google Scholar]
- 17.Ressler S, Bartkova J, Niederegger H, Bartek J, Scharffetter-Kochanek K, Jansen-Durr P, Wlaschek M. p16INK4A is a robust in vivo biomarker of cellular aging in human skin. Aging Cell. 2006;5:379–389. doi: 10.1111/j.1474-9726.2006.00231.x. [DOI] [PubMed] [Google Scholar]
- 18.Wu J, Xue L, Weng M, Sun Y, Zhang Z, Wang W, Tong T. Sp1 is essential for p16 expression in human diploid fibroblasts during senescence. PLoS One. 2007;2:e164. doi: 10.1371/journal.pone.0000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quereda V, Martinalbo J, Dubus P, Carnero A, Malumbres M. Genetic cooperation between p21Cip1 and INK4 inhibitors in cellular senescence and tumor suppression. Oncogene. 2007;26:7665–7674. doi: 10.1038/sj.onc.1210578. [DOI] [PubMed] [Google Scholar]
- 20.Ksiazek K, Piwocka K, Brzezinska A, Sikora E, Zabel M, Breborowicz A, Jorres A, Witowski J. Early loss of proliferative potential of human peritoneal mesothelial cells in culture: the role of p16INK4a-mediated premature senescence. JAppl Physiol. 2006;100:988–995. doi: 10.1152/japplphysiol.01086.2005. [DOI] [PubMed] [Google Scholar]
- 21.Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- 22.Romagosa C, Simonetti S, Lopez-Vicente L, Mazo A, Lleonart ME, Castellvi J, Ramon y Cajal S. p16Ink4a overexpression in cancer: a tumor suppressor gene associated with senescence and high-grade tumors. Oncogene. 2011;30:2087–2097. doi: 10.1038/onc.2010.614. [DOI] [PubMed] [Google Scholar]
- 23.Harvey M, Sands AT, Weiss RS, Hegi ME, Wiseman RW, Pantazis P, Giovanella BC, Tainsky MA, Bradley A, Donehower LA. In vitro growth characteristics of embryo fibroblasts isolated from p53-deficient mice. Oncogene. 1993;8:2457–2467. [PubMed] [Google Scholar]
- 24.Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H, Lu X, Soron G, Cooper B, Brayton C, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 26.Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 28.Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, Benguria A, Zaballos A, Flores JM, Barbacid M, et al. Tumour biology: senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 29.Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 30.Herbig U, Jobling WA, Chen BP, Chen DJ, Sedivy JM. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a) Mol Cell. 2004;14:501–513. doi: 10.1016/s1097-2765(04)00256-4. [DOI] [PubMed] [Google Scholar]
- 31.Wang YA, Elson A, Leder P. Loss of p21 increases sensitivity to ionizing radiation and delays the onset of lymphoma in atm-deficient mice. Proc Natl Acad Sci USA. 1997;94:14590–14595. doi: 10.1073/pnas.94.26.14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walia V, Ding M, Kumar S, Nie D, Premkumar LS, Elble RC. hCLCA2 is a p53-inducible inhibitor of breast cancer cell proliferation. Cancer Res. 2009;69:6624–6632. doi: 10.1158/0008-5472.CAN-08-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gruber AD, Pauli BU. Tumorigenicity of human breast cancer is associated with loss of the Ca2+-activated chloride channel CLCA2. Cancer Res. 1999;59:5488–5491. [PubMed] [Google Scholar]
- 34.Li X, Cowell JK, Sossey-Alaoui K. CLCA2 tumour suppressor gene in 1p31 is epigenetically regulated in breast cancer. Oncogene. 2004;23:1474–1480. doi: 10.1038/sj.onc.1207249. [DOI] [PubMed] [Google Scholar]
- 35.Rovillain E, Mansfield L, Caetano C, Alvarez-Fernandez M, Caballero OL, Medema RH, Hummerich H, Jat PS. Activation of nuclear factor-κ B signalling promotes cellular senescence. Oncogene. 2011;30:2356–2366. doi: 10.1038/onc.2010.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taniwaki M, Daigo Y, Ishikawa N, Takano A, Tsunoda T, Yasui W, Inai K, Kohno N, Nakamura Y. Gene expression profiles of small-cell lung cancers: molecular signatures of lung cancer. Int J Oncol. 2006;29:567–575. [PubMed] [Google Scholar]
- 37.Tsukada T, Tomooka Y, Takai S, Ueda Y, Nishikawa S, Yagi T, Tokunaga T, Takeda N, Suda Y, Abe S, et al. Enhanced proliferative potential in culture of cells from p53-deficient mice. Oncogene. 1993;8:3313–3322. [PubMed] [Google Scholar]
- 38.Ashida S, Nakagawa H, Katagiri T, Furihata M, Iiizumi M, Anazawa Y, Tsunoda T, Takata R, Kasahara K, Miki T, et al. Molecular features of the transition from prostatic intraepithelial neoplasia (PIN) to prostate cancer: genome-wide gene-expression profiles of prostate cancers and PINs. Cancer Res. 2004;64:5963–5972. doi: 10.1158/0008-5472.CAN-04-0020. [DOI] [PubMed] [Google Scholar]
- 39.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beckley JR, Pauli BU, Elble RC. Re-expression of detachment-inducible chloride channel mCLCA5 suppresses growth of metastatic breast cancer cells. J Biol Chem. 2004;279:41634–41641. doi: 10.1074/jbc.M408334200. [DOI] [PubMed] [Google Scholar]
- 41.Elble RC, Pauli BU. Tumor suppression by a proapoptotic calcium-activated chloride channel in mammary epithelium. J Biol Chem. 2001;276:40510–40517. doi: 10.1074/jbc.M104821200. [DOI] [PubMed] [Google Scholar]
- 42.Yamabuki T, Daigo Y, Kato T, Hayama S, Tsunoda T, Miyamoto M, Ito T, Fujita M, Hosokawa M, Kondo S, et al. Genome-wide gene expression profile analysis of esophageal squamous cell carcinomas. Int J Oncol. 2006;28:1375–1384. [PubMed] [Google Scholar]
- 43.Takata R, Katagiri T, Kanehira M, Tsunoda T, Shuin T, Miki T, Namiki M, Kohri K, Matsushita Y, Fujioka T, et al. Predicting response to methotrexate, vinblastine, doxorubicin, and cisplatin neoadjuvant chemotherapy for bladder cancers through genome-wide gene expression profiling. Clin Cancer Res. 2005;11:2625–2636. doi: 10.1158/1078-0432.CCR-04-1988. [DOI] [PubMed] [Google Scholar]
- 44.Nishidate T, Katagiri T, Lin ML, Mano Y, Miki Y, Kasumi F, Yoshimoto M, Tsunoda T, Hirata K, Nakamura Y. Genome-wide gene-expression profiles of breast-cancer cells purified with laser microbeam microdissection: identification of genes associated with progression and metastasis. Int J Oncol. 2004;25:797–819. [PubMed] [Google Scholar]
- 45.Majumder PK, Grisanzio C, O'Connell F, Barry M, Brito JM, Xu Q, Guney I, Berger R, Herman P, Bikoff R, et al. A prostatic intraepithelial neoplasia-dependent p27 Kip1 checkpoint induces senescence and inhibits cell proliferation and cancer progression. Cancer Cell. 2008;14:146–155. doi: 10.1016/j.ccr.2008.06.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reshkin SJ, Bellizzi A, Albarani V, Guerra L, Tommasino M, Paradiso A, Casavola V. Phosphoinositide 3-kinase is involved in the tumor-specific activation of human breast cancer cell Na(+)/H(+) exchange, motility, and invasion induced by serum deprivation. JBiolChem. 2000;275:5361–5369. doi: 10.1074/jbc.275.8.5361. [DOI] [PubMed] [Google Scholar]
- 47.Rich IN, Worthington-White D, Garden OA, Musk P. Apoptosis of leukemic cells accompanies reduction in intracellular pH after targeted inhibition of the Na(+)/H(+) exchanger. Blood. 2000;95:1427–1434. [PubMed] [Google Scholar]
- 48.Marinho CF, Costa-Maia J, Pinto-de-Barros J, Oliveira CR. Correlation between human platelet cytoplasmic membrane outer leaflet fluidity, Na+/H+ exchanger activity and aging. Eur Arch Psychiatry Clin Neurosci. 1997;247:275–277. doi: 10.1007/BF02900306. [DOI] [PubMed] [Google Scholar]
- 49.Xiong Z-G, Pignataro G, Li M, Chang S-Y, Simon RP. Acid-sensing ion channels (ASICs) as pharmacological targets for neurodegenerative diseases. Curr Opin Pharmacol. 2008;8:25–32. doi: 10.1016/j.coph.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elble RC, Walia V, Cheng HC, Connon CJ, Mundhenk L, Gruber AD, Pauli BU. The putative chloride channel hCLCA2 has a single C-terminal transmembrane segment. JBiol Chem. 2006;281:29448–29454. doi: 10.1074/jbc.M605919200. [DOI] [PubMed] [Google Scholar]
- 51.Kriegler M, Perez C, DeFay K, Albert I, Lu SD. A novel form of TNF/cachectin is a cell surface cytotoxic transmembrane protein: ramifications for the complex physiology of TNF. Cell. 1988;53:45–53. doi: 10.1016/0092-8674(88)90486-2. [DOI] [PubMed] [Google Scholar]
- 52.Bauer S, Groh V, Wu J, Steinle A, Phillips JH, Lanier LL, Spies T. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285:727–729. doi: 10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- 53.Groh V, Wu J, Yee C, Spies T. Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature. 2002;419:734–738. doi: 10.1038/nature01112. [DOI] [PubMed] [Google Scholar]
- 54.Ito K, Okamoto I, Araki N, Kawano Y, Nakao M, Fujiyama S, Tomita K, Mimori T, Saya H. Calcium influx triggers the sequential proteolysis of extracellular and cytoplasmic domains of E-cadherin, leading to loss of β-catenin from cell-cell contacts. Oncogene. 1999;18:7080–7090. doi: 10.1038/sj.onc.1203191. [DOI] [PubMed] [Google Scholar]
- 55.Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc Natl Acad Sci USA. 2001;98:12072–12077. doi: 10.1073/pnas.211053698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ren XH, Peng XD, Wu XP, Liao EY, Sun ZQ. Association between serum soluble membrane type matrix metalloproteinase-1 (MT1-MMP) levels and bone mineral density, and biochemical markers in postmenopausal women. Clin Chim Acta. 2008;390:44–48. doi: 10.1016/j.cca.2007.12.022. [DOI] [PubMed] [Google Scholar]
- 57.Yang SC, Chiang CK, Hsu SP, Hung KY. Relationship between interdialytic weight gain and nutritional markers in younger and older hemodialysis patients. J Ren Nutr. 2008;18:210–222. doi: 10.1053/j.jrn.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 58.Thompson HJ, Voss JG. Health-and disease-related biomarkers in aging research. Res Gerontol Nurs. 2009;2:137–148. doi: 10.3928/19404921-20090401-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bekaert S, De Meyer T, Van Oostveldt P. Telomere attrition as ageing biomarker. Anticancer Res. 2005;25:3011–3021. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.