Abstract
‘Regulator of G-protein Signaling’ (RGS) proteins constitute a class of intracellular signaling regulators that accelerate GTP hydrolysis by heterotrimeric Gα subunits. In recent years, RGS proteins have emerged as potential drug targets for small molecule modulation. Described in this unit are high-throughput screening procedures for identifying modulators of RGS protein-mediated GTPase acceleration (‘GAP activity’), for assessment of RGS domain/Gα interactions (most avid in vitro when Gα is bound by aluminum tetrafluoride), and for validation of candidate GAP-modulatory molecules with the single turnover GTP hydrolysis assay.
Keywords: Regulator of G-protein Signaling (RGS) proteins, heterotrimeric G-protein α subunits, Förster resonance energy transfer (FRET), single-turnover GTP hydrolysis, fluorescence polarization (FP)
Introduction
Regulator of G-protein Signaling’ (RGS) proteins constitute a class of intracellular signaling regulators that accelerate GTP hydrolysis by heterotrimeric Gα subunits. In recent years, RGS proteins have emerged as potential drug targets for small molecule modulation. While ligands for G-protein coupled receptors (GPCRs) comprise a large fraction of currently available small molecule therapeutics, very few modulators of downstream signaling are available. As key regulators of G-protein signaling amplitudes and kinetics, RGS proteins also present a useful class of drug targets. RGS protein inhibitors might be used to enhance cellular responses to upstream GPCR agonists, for example (Kimple et al., 2011). A number of RGS protein inhibitory compounds such as CCG-4986 have been described that react with surface cysteines on RGS4 (Roman et al., 2007; Kimple et al., 2007). Described in this unit are high-throughput screening procedures for identifying modulators of RGS protein-mediated GTPase acceleration (‘GAP activity’). Distinct from previous approaches that measure disruption of the Gα/RGS protein binding interface, the Transcreener® GDP assay examines effects of small molecules on GTP hydrolysis acceleration. As a result, the high throughput screen can identify both inhibitors and activators of RGS protein GAP activity, as well as allosteric modulators. Also described in this unit are a medium-throughput procedure for assessment of RGS domain/Gα interactions (most avid in vitro when Gα is bound by aluminum tetrafluoride or AMF) (Popov et al., 1997), and the more laborious single turnover hydrolysis assay for validation of candidate GAP-modulatory molecules (Berman et al., 1996).
Basic Protocol 1: Identifying candidate RGS protein modulators with the Transcreener GDP assay and a rate-modified Gα subunit
The Transcreener® GDP FP Assay kit measures production of GDP by Gαi1-mediated GTP hydrolysis. RGS4 accelerates hydrolysis, which is the slowest kinetic step in the nucleotide cycle of a Gαi1 double point mutant, Gαi1(R178M/A326S) (Zielinski et al., 2009). The fluorescent nucleotide tracer is displaced from its GDP antibody-binding site as GTP hydrolysis progresses and produces GDP product. In the absence of inhibitors, Gαi1-mediated hydrolysis will decrease polarization, an effect accelerated by RGS4. Compounds that alter the rate of GDP production (i.e., the extent of polarization decrease) may be modulators of RGS4 action. Described is a procedure for screening a small compound library for modulators of RGS4, including plate maps with control wells.
Materials
Solutions and reagents
Compound Library (e.g. Sigma LOPAC or ChemBridge DIVERSet™)
Transcreener® GDP FP Assay kit (Bellbrook Labs Cat no. 3009-1K)
- Assay buffer, final concentration in 20 µL reaction: 10 mM Tris (pH 7.5),
- 1 mM EDTA, 10 mM MgCl2, 10 µM GTP, 10 µg/mL GDP antibody, and 2 nM fluorescent nucleotide tracer; For the protocol below, this will be prepared as a 1.1X solution (18 µL added to a final 20 µL volume)
Recombinant Gαi1(R178M/A326S) subunit (purified as in Zielinski et al., 2009; see Support Protocol 1)
Recombinant RGS4 protein (purified as in Zielinski et al., 2009; see Support Protocol 1)
Black, 384-well assay plates with a non-binding surface (Corning, cat. No. 3676)
Equipment
Fluorescence polarization-capable plate reader (e.g. Tecan Safire2, BMG PHERAstar Plus)
Procedure
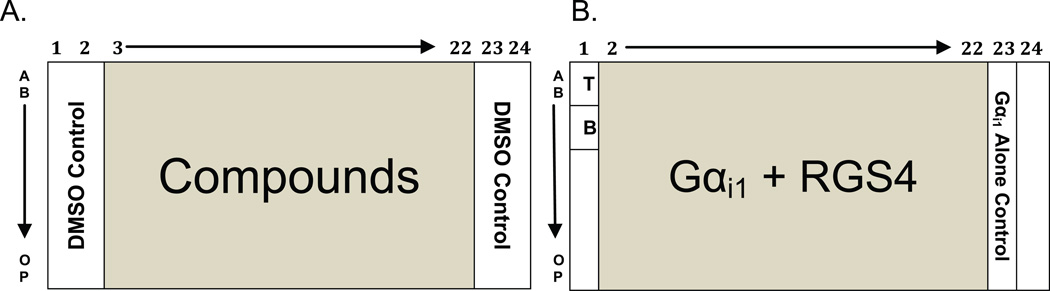
Prepare test agents at 10X the desired assay concentration. In this example, compounds are diluted to 100 µM in 20% DMSO for a final concentration in a 20 µL reaction of 10 µM compound and 2% DMSO. Add 2 µL of 10X compound to wells of a 384 well plate, rows A-P/columns 3-22 (Figure 1A). Add 2 µL of a 20% DMSO control to remainder of the wells.
- Prepare the assay buffer on ice. Prepare Tracer Control (T) by making assay buffer without GDP antibody. Prepare background Buffer Control (B) by making assay buffer without nucleotide tracer. Add 18 µL Tracer Control (T) to rows A-C/column 1. Add 18 µL of Buffer Control (B) to rows D-F/column 1 (Figure 1B).
- The remainder of column 1 and all of column 24 are available for other controls or for standard curves to examine the rate of GDP production with varying concentrations of RGS4 or a known modulatory compound.
Prepare a (Gαi1 alone) control by adding 25 nM Gαi1(R178M/A326S) to chilled assay buffer. Always add Gαi1 (R178M/A326S) immediately before plating, since it initiates nucleotide hydrolysis. Add 18 µL of Gαi1 alone control to rows A-P/column 23 (Figure 1B).
Prepare the (Gαi1 + RGS4) mix by adding 150 nM RGS4 and 25 nM Gαi1(R178M/A326S) to chilled assay buffer. Always add Gαi1 (R178M/A326S) immediately before plating.
Add 18 µL of (Gαi1 + RGS4) mix, at 30-second intervals, to wells in rows A-P/columns 2-22, to initiate the reactions. Column 2 contains the (Gαi1 + RGS4), compound-free control (Figure 1B). Immediately mix the reaction with a plate mixer and incubate at 30°C for ~75 minutes, or a time when the fluorescence polarization has decreased by approximately 100 mP, to allow steady-state accumulation of GDP.
- At 75 minutes (+/− 5 minutes), measure fluorescence polarization using a 635 nm excitation wavelength and 670 nm emission. Assign Tracer Control wells as reference to 20 mP (millipolarization units), and Buffer Control wells as blanks that may be subtracted from sample and reference well values.
- Consistent timing between reaction initiation and fluorescence polarization measurement must be maintained for each plate, because GTP hydrolysis will continue at steady state throughout the experiment.
Figure 1. Example of compound screen setup for Gαi1 and RGS4.
A) Add 2 µL of test compounds to rows A-P/columns 3−22 and add 2 µL DMSO control to rows A-P/columns 1, 2, 23, and 24. B) Add 18 µL of Tracer Control (T) and Buffer Control (B) to rows A-C/column 1 and rows D-F/column 1 respectively. Add 18 uL of Gαi1 alone control to all rows in column 23. Add 18 µL of Gαi1 + RGS4 mix to rows A-P/columns 2−22.
Data analysis
-
7.
The user must predetermine the threshold fluorescence polarization measurement for defining a hit. In general, any compound that falls outside the average mP of (Gαi1 + RGS4) +/− 3 standard deviations (µ +/− 3σ) could be an agent of interest. However, it may be more practical to use a 50% decrease in polarization as the threshold for defining hits. Compounds yielding a polarization less than µ - 3σ are potential enhancers of RGS4 GAP activity, while those yielding polarization greater than µ + 3σ are potential inhibitors of RGS4 GAP activity.
-
8.Assess assay and compound screen robustness by determining the Z’ and Z-factors, respectively (Zhang et al., 1999). Determine the Z’ using the average mP and standard deviation values of the Gαi1 alone control (Column 3) and the (Gαi1 + RGS4) control (Column 23) (Figure 1).
where µ indicates an average value and σ is one standard deviation. Determine the Z-factor by using the average mP and standard deviation values of the Gαi1 alone control (Column 3) and values of the compound wells (rows A-P/columns 2-22).
Basic Protocol 2: Measuring disruption of the RGS domain/Gα interaction by Förster resonance energy transfer (FRET)
Recognition of the Gα GTP hydrolysis transition state is critical for RGS protein function (Berman et al., 1996). Accordingly, RGS domains have high affinity for Gα subunits bound to GDP, magnesium, and aluminum tetrafluoride (AMF), a complex that mimics the GTP hydrolysis transition state. This protocol describes a method for measuring Gα/RGS interactions with FRET (Figure 2), and its potential disruption by small molecule modulators. The approach is a “mix and measure” assay in 96-well format. In contrast with the more laborious, kinetic single turnover hydrolysis assay (see Protocol 3), the Gα/RGS domain interaction FRET assay allows suitable throughput of candidate RGS modulators identified in the Transcreener® GDP assay.
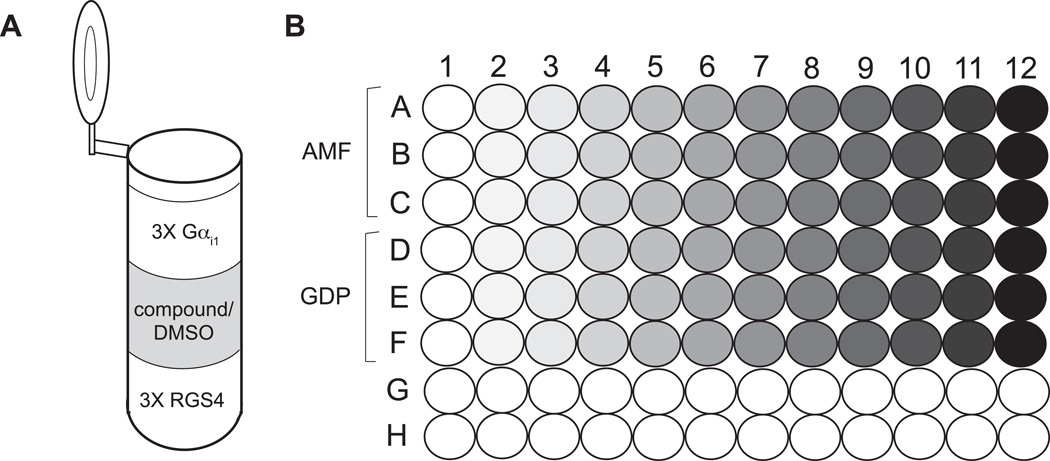
Figure 2. Example RGS protein/Gα FRET experimental setup.
A) Each reaction mixture consists of 150 µL 3X RGS protein solution, 150 µL 3X Gαi1, and compound or DMSO to the desired test concentration. The total reaction volume is 450 µL. B) The reaction mixtures, in both AMF and GDP buffers, are distributed in triplicate across a 96-well plate. The shading gradient represents varying concentrations of test compound.
Materials
Solutions and reagents
GDP buffer: 10 mM HEPES pH 7.5, 150 mM NaCl, 0.0005% NP40 alternative (Calbiochem), 100 µM GDP, 50 µM EDTA
AMF (aluminum, magnesium, and fluoride) buffer: 10 mM HEPES pH 7.5, 150 mM NaCl, 0.0005% NP40 alternative (Calbiochem), 100 µM GDP, 50 µM EDTA, 10 mM MgCl2, 10 mM NaF, 30 µM AlCl3
recombinant YFP-RGS4 fusion protein (purified as in Willard et al., 2004, see Support Protocol 1)
recombinant Gαi1-CFP fusion protein (purified as in Willard et al., 2004, see Support Protocol 1)
candidate modulatory compounds in solution (assay is robust to ~5% (v/v) DMSO)
black polystyrene 96-well plates (e.g. Costar (Corning))
Equipment
fluorescence dual emission plate reader (e.g. POLARstar Omega (BMG Labtech), EnVision (PerkinElmer))
Procedure
Prepare separate 3X RGS protein solutions in both GDP and AMF buffers by diluting YFP-RGS4 fusion protein to 2.7 µM concentration in microcentrifuge tubes. Mix well by flicking the tubes.
Prepare separate 3X Gαi1 solutions in both GDP and AMF buffers by diluting Gαi1-CFP fusion protein to 1.5 µM in microcentrifuge tubes. Mix well by flicking the tubes.
Apportion 150 µL of 3X RGS protein solution in GDP and AMF buffers (from step 1) separately into 12 microcentrifuge tubes each. Typically, parallel GDP and AMF concentration-response curves span a concentration range of 4–5 orders of magnitude for each test agent.
Add candidate compound solutions (or DMSO/vehicle control) to achieve the desired test concentrations in a final volume of 450 µL. Bring each ‘RGS protein and compound’ mixture to 300 µL by diluting in either GDP or AMF buffer. Mix well by flicking the tubes. Allow the ‘RGS and compound mixture’ to incubate at room temperature for 1 hour to optimize their interaction.
Add to each reaction mixture 150 µL of 3X Gαi1 solution in either GDP or AMF buffers (from step 2). Mix well by flicking the tubes and incubate 30 minutes at room temperature. See Figure 3A for a schematic of the reaction mixture components.
Distribute three 125 µL replicates of each reaction across a 96-well polystyrene plate, taking care to avoid air bubbles. See Figure 3B for a schematic of the 96-well plate setup.
- Measure fluorescence at two wavelengths using a compatible plate reader with the following parameters:
Excitation wavelength: 433 nm CFP peak emission wavelength: 474 nm YFP peak emission wavelength: 525 nm Monochrometer-based fluorescence plate readers are optimal but, if filters are used, wavelength cutoffs for excitation and emissions must not overlap. Other plate reader parameters, such as focal height and detector gains, should be adjusted to optimize the FRET ratio difference between GDP and AMF buffer compound-free controls (see Critical Parameters).
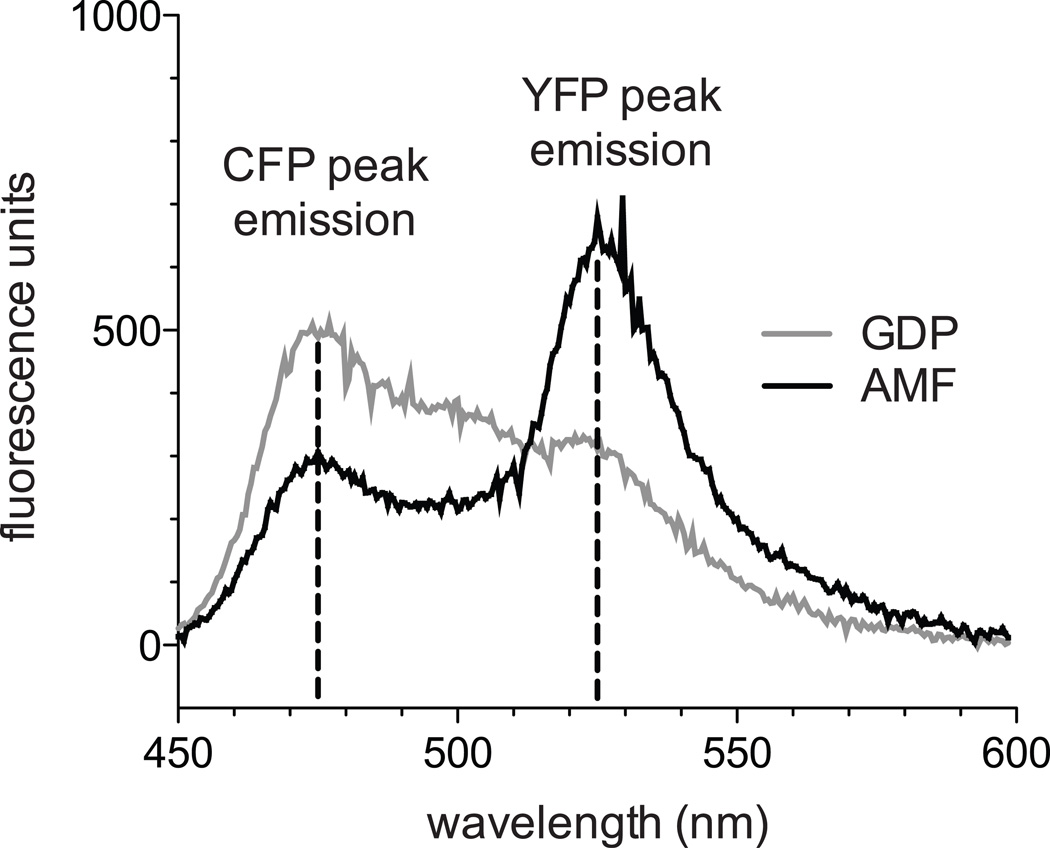
Figure 3. Gαi1-CFP and YFP-RGS4 emission spectra in GDP and AMF buffers.
500 nM Gαi1-CFP and 900 nM YFP-RGS4 were excited at 433 nm and emission spectra measured with a cuvette-based PerkinElmer LS-55 spectrometer. In the presence of AMF, RGS4 and Gαi1 bind with high affinity, leading to FRET between their fluorescent tags and an increased FRET ratio ((525 nm emission)/(474 nm emission)) compared to GDP buffer.
Data analysis
-
9.
Examine the raw fluorescence output for trends that may indicate suboptimal plate reader parameters or compound fluorescence. Fluorescence intensity measurements at 474 and 525 nm should be within the dynamic range of the detectors. Triplicate wells should exhibit highly consistent outputs, and in the absence of compound fluorescence, measurements in the GDP buffer samples should display little variation across the plate.
-
10.
Calculate the FRET ratio ((525 nm emission)/(474 nm emission)) for each well. Plot the averaged FRET ratio for each triplicate reaction against the logarithm of compound concentration.
-
11.
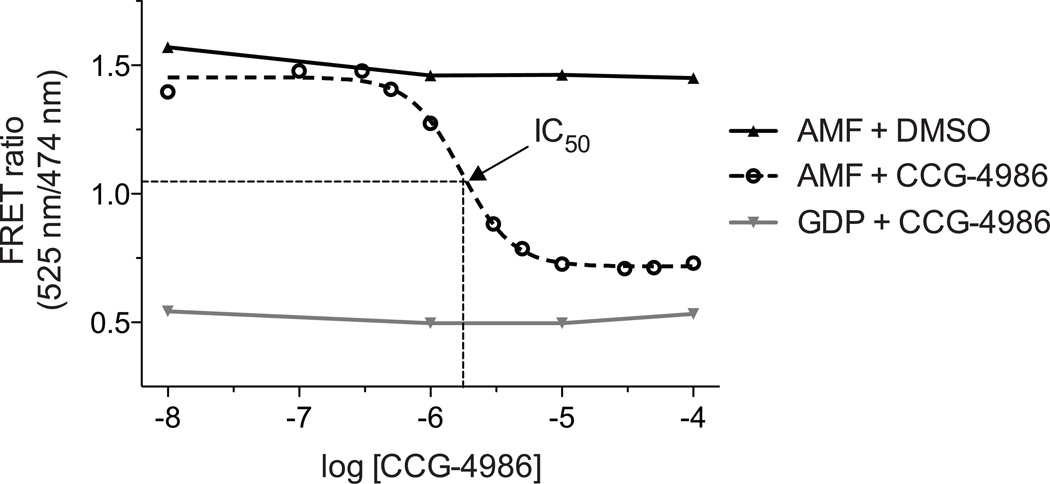
Fit the resulting concentration-response plot (obtained in AMF buffer) with an appropriate semilogarithmic concentration-inhibition curve. For inhibitory compounds, the IC50 value is the compound concentration resulting in a half-maximal FRET signal. In Figure 4, an example concentration-inhibition curve was generated using the cysteine-reactive compound CCG-4986 (Roman et al., 2007; Kimple et al., 2007).
Figure 4. Inhibition of Gαi1/RGS4 interaction by the cysteine-reactive compound CCG-4986.
A previously identified compound, CCG-4986, (Roman et al., 2007) covalently modifies surface cysteines on RGS4, preventing association with Gαi1 (Kimple et al., 2007). The FRET method allows determination of an IC50 for RGS protein inhibition.
Basic Protocol 3: Measuring modulation of GAP activity by single turnover GTP hydrolysis
Single turnover hydrolysis assays for Gα subunits measure phosphate release following a single round of GTP hydrolysis, a process that is accelerated in the presence of an RGS domain. The assay is based on radioactivity, allowing precise measurements and avoiding potential compound fluorescence effects that may occur in the Transcreener® GDP and FRET approaches described above. Like the Transcreener® GDP assay, both inhibition and activation of RGS protein function as well as allosteric effects can be detected. However, the single turnover hydrolysis assay is labor intensive and low-throughput, making it impractical for screening large numbers of compounds. For optimal determination of test compound activity, the following set of experiments is necessary:
Gαi1 alone (indicative of no GAP activity)
Gαi1 with RGS4 and no compound (indicative of full RGS4 GAP activity)
Gαi1 with compound and RGS4 (assessing compound activity on RGS4, the Gα subunit, or both)
Gαi1 with compound and no RGS4 (assessing compound activity directly on the Gα subunit and its intrinsic rate of GTP hydrolysis)
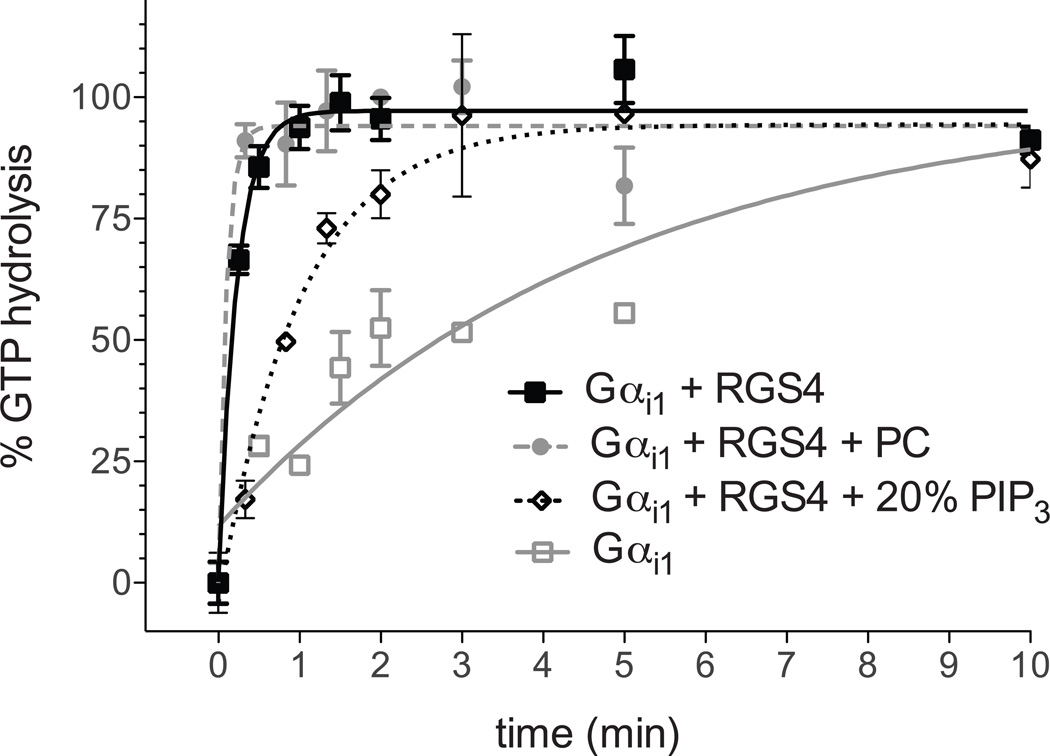
An example of allosteric inhibition of RGS4 GAP activity, by the phospholipid PIP3, is shown in Figure 5. Gαi1 alone possesses a relatively slow rate of GTP hydrolysis that is greatly accelerated in the presence of RGS4. PIP3-containing lipid vesicles specifically inhibit RGS4 GAP activity through an allosteric site within the RGS domain (Ishii and Kurachi, 2004), reducing the rate of GTP hydrolysis toward that of Gαi1 alone as would be expected for an RGS protein inhibitory compound, while phosphatidylcholine-containing vesicles (equivalent to a compound-free control) have no effect.
Figure 5. RGS4 is allosterically modulated by phospholipids in single turnover hydrolysis assays.
Phosphatidyl inositol triphosphate (PIP3) modulates RGS4 activity though an allosteric “B” site, described previously (Ishii and Kurachi, 2004; Tu and Wilkie, 2004). In single turnover assays, inclusion of phospholipid vesicles containing PIP3, but not phosphatidyl choline (PC) only, inhibits RGS4 GAP activity. Similar reversal of RGS4-mediated GTP hydrolysis acceleration would be expected for an inhibitory compound.
Materials
Solutions and reagents
Buffer C: 50 mM Tris pH 7.5, 0.05% C12E10, 1 mM DTT, 10 mM EDTA, 100 mM NaCl, 5 µg/mL bovine serum albumin (BSA)
Buffer D: 40 mM MgCl2, 400 µM GTPγS
recombinant Gαi1 protein (purified as in Kimple et al., 2001; see Support Protocol 1)
recombinant RGS4 protein (purified as in Kimple et al., 2007; see Support Protocol 1)
candidate modulatory compounds (assay is robust to at least 10% DMSO; Blazer et al., 2010)
GTP[γ-32P] (PerkinElmer product no. BLU004Z250UC, specific activity of 6000 Ci/mmol)
5 mL polypropylene test tubes
charcoal slurry: 5% (w/v) activated charcoal (e.g. Sigma cat. no. C4386), 50 mM phosphoric acid, pH 3.0, stored stably long term at 4° C.
scintillation vials and fluid (e.g. Fisher Scintisafe gel)
Equipment
safety equipment and designated space for radioactive materials
water bath (30° C)
refrigerated centrifuge adapted for 5 mL test tubes, suitable for radioactive materials (e.g. Hermle Z400K)
bench-top vortexer
scintillation counter
Procedure
Dilute Gαi1 protein to 133 nM in 1.35 mL of Buffer C.
Dilute RGS4 protein to 200 nM and candidate compound to 4X the desired concentration in 450 µL of Buffer D.
Load 1.35 ml of 133 nM Gαi1 (from step 1) with radionucleotide by adding 1 µL GTP[γ-32P] in the absence of magnesium. Vortex briefly and incubate at 30° C for 10 minutes. Then place on ice for 10 minutes. The remainder of the reaction, and the reaction components, should be on ice.
Apportion 900 µL of ice-cold charcoal slurry into each of 16 test tubes (5 mL test tube capacity), corresponding to 8 reaction time points in duplicate.
Take a zero time point prior to initiation of hydrolysis by transferring 75 µL of the radionucleotide-loaded Gαi1 solution to each of the first two charcoal slurry tubes. Immediately vortex the two tubes.
Initiate GTP hydrolysis by adding 450 µL of the ice-cold Buffer D solution containing RGS4, test compound, and magnesium to the radionucleotide-loaded Gαi1 solution.
- At each of the 7 remaining time points, transfer duplicate 100 µL portions from the reaction to ice-cold charcoal slurry tubes and immediately vortex. A typical time course would be 0 s, 20 s, 40 s, 1 min, 2 min, 3 min, 5 min, and 10 min.
- For accurate kinetic derivations, the initial time points should be closely clustered to capture the rapid initial rate of hydrolysis. At least two observation time points should be included after hydrolysis is essentially complete.
After completion of the time course, centrifuge the charcoal tubes at 3,000 × g and 4° C for 5 minutes to separate charcoal-adsorbed radiolabeled guanine nucleotide from the soluble, free phosphate (Pi) generated by GTP hydrolysis. During centrifugation, distribute 7 mL of scintillation fluid into each of 16 vials.
Place 600 µL of the resulting supernatant into individual vials, each containing 7 mL of scintillation fluid.
Quantify free 32Pi by scintillation counting for 5 minutes.
Data analysis
-
11.
Subtract the average counts per minute (CPM) at time zero from each subsequent time point. Assign the average of the final data point as 100% hydrolysis (saturation), and calculate percent hydrolysis for the remaining data points by division. Plot the average of each duplicate time point against the corresponding time.
-
12.
Fit the resulting kinetic plot with a single-phase exponential association curve. A kinetic constant, kcat, can be derived if enough time points have been taken in the initial stage of the reaction (i.e., before the reaction is ~50% complete), allowing comparison among compound-treated and compound-free reactions containing RGS4 as the accelerator of Gαi1 intrinsic GTPase activity.
COMMENTARY
Background Information
Diverse mammalian cell types respond to extracellular environmental cues through seven transmembrane receptors coupled to heterotrimeric, guanine-nucleotide binding protein (G proteins) composed of Gα, Gβ, and Gγ subunits (Oldham and Hamm, 2008). The Gα subunit cycles between an active, GTP-bound and a resting, GDP-bound conformation. The GDP-bound Gα subunit associates with the obligate Gβγ heterodimer, and the activated receptor catalyzes guanine nucleotide exchange by Gα. When Gα subsequently binds GTP, a conformational change results in separation from Gβγ, allowing interaction with downstream components of second messenger signaling pathways. Signal termination is effected by hydrolysis of GTP through the intrinsic GTPase activity of Gα subunits.
Regulator of G-protein signaling (RGS) proteins accelerate the rate of GTP hydrolysis by Gα proteins, thus acting as negative regulators of G-protein signaling (Kimple et al., 2011). The human genome encodes more than twenty RGS proteins, possessing a highly conserved α-helical RGS domain (or “RGS box”) in combination with numerous other functional domains. The RGS protein family displays a broad diversity of expression profiles and specificities for particular Gα subunits and signaling pathways (Kimple et al., 2011). Furthermore, the GAP activity of a number of RGS proteins can be modulated at allosteric sites (Ishii and Kurachi, 2004; Tu and Wilkie, 2004).
Since many G-protein coupled receptor agonists and antagonists continue to be used as pharmacological agents, and evidence for roles of RGS proteins in multiple disease states is emerging (Kimple et al., 2011), there is a growing interest in identifying small molecule RGS protein modulators that could provide additional pharmacologic control on GPCR signal transduction. In contrast to the standard protein-protein interaction-based approaches to RGS inhibitor screening, the enzymatic approach described herein allows for detection of allosteric modulators and activators of RGS protein GAP activity in addition to direct inhibitors.
Critical parameters
Basic Protocol 1: identifying candidate RGS protein modulators with the Transcreener GDP assay and a rate-modified Gα subunit
In addition to identifying inhibitors or enhancers of RGS4 GAP activity, this type of screen will detect agents with non-specific effects on Gαi1 GTPase activity. The Gα GTP hydrolysis modulators can be identified by counter-screening simultaneously with Gαi1 alone (no RGS4) or by testing hits with secondary screening approaches, such as single turnover hydrolysis assays (see Protocol 3).
The amount of GDP antibody, fluorescent nucleotide tracer, recombinant Gαi1(R178M/A326S) and recombinant RGS4 were optimized for the conditions in this particular screen and may require adjustment based on the specific activities of the enzyme preparations or if different buffers, 384-well plates, or reaction volumes are used. The GDP antibody and fluorescent nucleotide tracer optimization protocols can be found in the Transcreener® GDP FP Assay kit manual. The amounts of Gαi1 and RGS4 to be used are determined by identifying quantities that give an assay signal window of greater than 100 mP (mPGαi1 alone – mPGαi1 + RGS4) at ~75 minutes and also that are within the linear range of GDP product formation (< 10% GTP conversion). The quantity of Gαi1 protein should be minimized because the intrinsic GTPase activity of the Gαi1 protein affects background GDP production.
Although these assays are typically conducted in a kinetic mode, it has been found that 15 mM sodium orthovanadate efficiently quenches the Gαi1(R178M/A326S) + RGS4 reaction. The particular stop solution included with the Transcreener® GDP FP Assay kit (which does not contain sodium orthovanadate), while effective for other GTPases, does not halt this Gα + RGS protein reaction.
Basic Protocol 2: Measuring disruption of the RGS domain/Gα interaction by Förster resonance energy transfer (FRET)
Plate reader parameters, such as focal height and detector gains, should be adjusted to optimize the FRET ratio difference between GDP and AMF buffer compound-free controls. Optimization of these parameters is automated on many plate readers. Alternatively, each parameter can be adjusted incrementally by hand and compound-free GDP and AMF controls measured after each change. Choose the parameter settings that maximize the difference in FRET ratio between GDP and AMF, while giving reproducible fluorescence readings within the detector’s dynamic range.
Basic Protocol 3: Measuring modulation of GAP activity by single turnover GTP hydrolysis
Since single turnover hydrolysis assays are labor-intensive, it is often practical to test only a single concentration of each compound at a time. To observe maximal effects, the concentration of the putative GAP-modulatory compound should be higher than the IC50 (or EC50) value of the compound as first determined in other, higher-throughput assays.
To maximize chances for detection of both inhibitors and allosteric activators of RGS4, single turnover assays must be conducted with an RGS protein concentration lower than the one that saturates the available Gαi1 ‘substrate’, but high enough to provide a significant acceleration of GTP hydrolysis relative to Gαi1 alone. The 100 nM Gαi1 and 50 nM RGS4 concentrations described in this protocol are effective in our experience. However, multiple RGS4 concentrations can be tested to identify this saturation point.
Troubleshooting
Basic Protocol 1: identifying candidate RGS protein modulators with the Transcreener GDP assay and a rate-modified Gα subunit
If the assay signal window is less than 100 mP, or the Z- and Z’-factors are less than required for screening, it may be useful to measure a time course of GDP production (i.e. polarization decrease). Test a matrix of RGS protein and Gαi1(R178M/A326S) concentrations, and measure fluorescence polarization at 5 minute intervals following initiation of the reactions. Choose the protein concentrations and measurement time point that maximize the signal window while remaining in the linear phase of GDP production.
Basic Protocol 2: Measuring disruption of the RGS domain/Gα interaction by Förster resonance energy transfer (FRET)
If a concentration-response curve exhibits a Hill slope significantly different from one, compound fluorescence and/or absorbance is likely. Fluorescent compounds should be assessed with single turnover hydrolysis or other non-fluorescence-based approaches.
Basic Protocol 3: Measuring modulation of GAP activity by single turnover GTP hydrolysis
If saturation hydrolysis is reached by the first time point, an accurate kinetic constant cannot be obtained. To address this problem, the first time point should be measured as quickly as possible after initiation of the reaction. If necessary, the rate of the reaction can be reduced by lowering the RGS protein concentration.
Anticipated Results
Basic Protocol 1: identifying candidate RGS protein modulators with the Transcreener GDP assay and a rate-modified Gα subunit
A robust assay should yield a Z’-factor > 0.7 and a Z-factor > 0.5. The Z-factor results are influenced by the type of compound library examined. In the pilot-screen from Zielinski et al., 2009, the hit rate was 0.7% for compounds from the library that exhibited an inhibitory effect on GDP production that was specific to RGS4-catalyzed GTPase activity. A separate screen of a diverse set of 50,000 compounds (the DIVERSet™ library from ChemBridge) gave a similar result (unpublished data). Further characterization of compounds of hits from this screen can be performed with this assay described above and other protocols detailed in this unit.
Basic Protocol 2: Measuring disruption of the RGS domain/Gα interaction by Förster resonance energy transfer (FRET)
The RGS domains bind with highest affinity to Gα subunits in the transition state-mimetic, AMF form. The FRET ratio in the AMF-containing reactions should decrease with increasing concentrations of a compound that inhibits the RGS domain/Gα interaction, ultimately approaching the value of the FRET ratio recorded in the negative control GDP buffer (i.e., minimal RGS protein/Gα interaction).
Because increasing amounts of any compound should not affect the FRET ratio in GDP buffer (i.e., given the lack of affinity of the RGS domain for inactive, GDP-bound Gα subunits), the concentration-response plot in GDP buffer should be flat. If any trends are observed in the GDP FRET ratio, this likely indicates assay interference by compound absorbance and/or fluorescence.
Basic Protocol 3: Measuring modulation of GAP activity by single turnover GTP hydrolysis
Small molecule allosteric activators of RGS4 will increase the observed kcat beyond the compound-free control as long as the input RGS4 is not saturating. Small molecule inhibitors of RGS4 GAP activity will decrease the observed kcat, approaching that of Gαi1 alone, which represents intrinsic Gα GTPase activity. Compounds that decrease the kcat below that of Gαi1 alone likely exert effects either specifically on the Gα subunit, or non-specifically on both Gαi1 and RGS4. These possibilities can be further investigated with additional single turnover experiments that exclude the RGS protein.
Time Considerations
Basic Protocol 3: Measuring modulation of GAP activity by single turnover GTP hydrolysis
Single turnover hydrolysis assays are laborious and time-consuming. We typically can evaluate only one or two test compounds in a single day. Due to this limitation, one must limit the number of test compounds by utilizing the Transcreener GDP assay and counterscreen, the Gα/RGS FRET assay, and/or other approaches.
Literature Cited
- Berman DM, Wilkie TM, Gilman AG. GAIP and RGS4 are GTPase-activating proteins for the Gi subfamily of G protein alpha subunits. Cell. 1996;86(3):445–452. doi: 10.1016/s0092-8674(00)80117-8. [DOI] [PubMed] [Google Scholar]
- Blazer LL, Roman DL, Chung A, Larsen MJ, Greedy BM, Husbands SM, Neubig RR. Reversible, allosteric small-molecule inhibitors of regulator of G protein signaling proteins. Mol. Pharmacol. 2010;78(3):524–533. doi: 10.1124/mol.110.065128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii M, Kurachi Y. Assays of RGS protein modulation by phosphatidylinositides and calmodulin. Methods Enzymol. 2004;389:105–118. doi: 10.1016/S0076-6879(04)89007-4. [DOI] [PubMed] [Google Scholar]
- Kimple AJ, Willard FS, Giguère PM, Johnston CA, Mocanu V, Siderovski DP. The RGS protein inhibitor CCG-4986 is a covalent modifier of the RGS4 Gα-interaction face. Biochim. Biophys. Acta. 2007;1774(9):1213–1220. doi: 10.1016/j.bbapap.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimple RJ, De Vries L, Tronchere H, Behe CI, Morris RA, Gist Farquhar M, Siderovski DP. RGS12 and RGS14 GoLoco motifs are G alpha(i) interaction sites with guanine nucleotide dissociation inhibitor activity. J. Biol. Chem. 2001;276(31):29275–29281. doi: 10.1074/jbc.M103208200. [DOI] [PubMed] [Google Scholar]
- Oldham WM, Hamm HE. Heterotrimeric G protein activation by G-protein-coupled receptors. Nat. Rev. Mol. Cell. Biol. 2008;9(1):60–71. doi: 10.1038/nrm2299. [DOI] [PubMed] [Google Scholar]
- Popov S, Yu K, Kozasa T, Wilkie TM. The regulators of G protein signaling (RGS) domains of RGS4, RGS10, and GAIP retain GTPase activating protein activity in vitro. Proc. Natl. Acad. Sci. USA. 1997;94(14):7216–7220. doi: 10.1073/pnas.94.14.7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman DL, Talbot JN, Roof RA, Sunahara RK, Traynor JR, Neubig RR. Identification of small-molecule inhibitors of RGS4 using a high-throughput flow cytometry protein interaction assay. Mol. Pharacol. 2007;71(1):169–175. doi: 10.1124/mol.106.028670. [DOI] [PubMed] [Google Scholar]
- Tu Y, Wilkie TM. Allosteric regulation of GAP activity by phospholipids in regulators of G-protein signaling. Methods Enzymol. 2004;389:89–105. doi: 10.1016/S0076-6879(04)89006-2. [DOI] [PubMed] [Google Scholar]
- Willard FS, Kimple RJ, Kimple AJ, Johnston CA, Siderovski DP. Fluorescence-based assays for RGS box function. Methods in Enzymology. 2004;389:56–71. doi: 10.1016/S0076-6879(04)89004-9. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- Zielinski T, Kimple AJ, Hutsell SQ, Koeff MD, Siderovski DP, Lowery RG. Two Galpha(i1) rate-modifying mutations act in concert to allow receptor-independent, steady-state measurements of RGS protein activity. J Biomol Screen. 2009;14:1195–1206. doi: 10.1177/1087057109347473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Key Reference
- Kimple AJ, Bosch DE, Giguère PM, Siderovski DP. Regulators of G-Protein Signaling and Their Gα Substrates: Promises and Challenges in Their Use as Drug Discovery Targets. Pharmacol Rev. 2011;63(3):728–749. doi: 10.1124/pr.110.003038. A recent review of the RGS proteins focused on the rationale for their consideration as valuable drug discovery targets and current efforts to identify chemical probes that modify RGS protein GAP activity.