Abstract
RseA sequesters RpoE (σE) to the inner membrane of Escherichia coli when envelope stress is low. Elevated envelope stress triggers RseA cleavage by the sequential action of two membrane proteases, DegS and RseP, releasing σE to activate an envelope stress reducing pathway. Revertants of a ΔdegP ΔbamB strain, which fails to grow at 37°C due to high envelope stress, harbored mutations in the rseA and rpoE genes. Null and missense rseA mutations constitutively hyper-activated the σE regulon and significantly reduced the major outer membrane protein (OMP) levels. In contrast, a novel rpoE allele, rpoE3, resulting from the partial duplication of the rpoE gene, increased σE levels greater than that seen in the rseA mutant background but did not reduce OMP levels. A σE-dependent RybB::LacZ construct showed only a weak activation of the σE pathway by rpoE3. Despite this, rpoE3 fully reversed the growth and envelope vesiculation phenotypes of ΔdegP. Interestingly, rpoE3 also brought down the modestly activated Cpx envelope stress pathway in the ΔdegP strain to the wild type level, showing the complementary nature of the σE and Cpx pathways. Through employing a labile mutant periplasmic protein, AcrAL222Q, it was determined that the rpoE3 mutation overcomes the ΔdegP phenotypes, in part, by activating a σE-dependent proteolytic pathway. Our data suggest that a reduction in the OMP levels is not intrinsic to the σE-mediated mechanism of lowering envelope stress. They also suggest that under extreme envelope stress, a tight homeostasis loop between RseA and σE may partly be responsible for cell death, and this loop can be broken by mutations that either lower RseA activity or increase σE levels.
Introduction
DegP is the major periplasmic protease in Escherichia coli [1]–[2]. Its proteolytic activity towards outer membrane proteins (OMPs) was initially observed against a mutant LamB protein with a temperature sensitive folding defect [3]. Subsequent studies showed an absolute or conditional requirement for DegP in cells either expressing folding-defective OMPs [4]–[5] or lacking a factor involved in OMP assembly [6]–[9].
Spiess et al. [10] first demonstrated that DegP possesses both protease and chaperone activities. A series of structural and biochemical studies on DegP provided important mechanistic clues [11]–[14]. These studies showed that the binding of unfolded substrate proteins to DegP triggers its reversible oligomerization into a cage-like structure and membrane association greatly influences DegP's assembly and activity. A full rescue of the temperature sensitive growth phenotype of ΔdegP requires DegP's protease activity [10]. The expression of degP is under the control of the σE and Cpx regulons [15]–[18], which together constitute the two major envelope stress response pathways [19]–[21]. Both pathways are required to elevate degP expression to overcome the potentially lethal envelope stress caused by aberrant OMP assembly [9].
Compensatory mutations have been isolated that overcome the temperature sensitive growth phenotype of cells lacking DegP. The first of this was isolated by Baird and Georgopoulos [22] by selecting for revertants that grew at 42°C. One of the revertants displayed a cold sensitive growth phenotype and was found to affect a gene named sohA (suppressor of htrA) [22]. The mechanism by which sohA reversed the growth phenotype of ΔdegP could not be determined. Prior to this study, Kiino and Silhavy [23] identified suppressor mutations of a LamB::LacZ hybrid protein in a locus they termed prlF and hypothesized that it was involved in some aspect of protein localization. The prlF gene, which turned out to be the site of the sohA mutation, together with yhaV, was recently determined to constitute a toxin-antitoxin module [24]. However, despite the new finding, the mechanism by which the sohA/prlF alleles overcome the growth defects of ΔdegP remains unknown.
The second ΔdegP suppressor mutation was obtained somewhat serendipitously and mapped to the cyaA gene [25]. It was concluded that the lower cAMP levels in the mutant cyaA ΔdegP background overcame the temperature sensitive growth phenotype by inducing the σE and Cpx stress regulons. While it remains to be determined exactly how reduced cAMP levels leads to up-regulation of the envelope stress response systems, another alarmone, ppGpp, was shown to up-regulate σE activity in a RseA-independent manner [26]. In a follow-up study, the authors concluded that ppGpp affects σE activity both directly, by influencing σE-dependent transcription, and indirectly, via altering competition for the core polymerase in favor of σE [27]. From these studies, it is apparent that the metabolic state of the bacterial cytoplasm also influences the envelope stress response systems.
In this study, we sought revertants of a ΔdegP strain also lacking bamB, a nonessential component of the BAM complex [28]–[29]. The simultaneous absence of degP and bamB confers a severe growth phenotype even at 30°C [8]. We reasoned that the absence of DegP in conjunction with defective BAM machinery would yield potentially new kinds of suppressors that could give a novel insight into the envelope stress response. Indeed, envZ was identified as one of the genes in which suppressor alterations improved the growth phenotype of the ΔdegP ΔbamB strain [30]. EnvZ and OmpR constitute a two-component signal transduction system [31], whose activity is modulated by a newly identified inner membrane protein, MzrA [30], [32]. MzrA directly interacts with EnvZ and changes its enzymatic activities so as to elevate the steady-state levels of OmpR∼P. Consequently, overexpression of MzrA or certain alterations in EnvZ constitutively activate the EnvZ/OmpR regulon. The activated EnvZ/OmpR regulon directly or indirectly (via inducing the expression of small regulatory RNAs) inhibits the synthesis of several OMPs and reduces envelope stress [30], [33].
Here we report two new sites of suppressor mutations that overcome the growth defects of a ΔdegP ΔbamB strain. Mutations in rseA constitutively activate the σE regulon and reduce envelope stress in part by lowering the major OMP levels. In contrast, an rpoE mutation, which causes a novel duplication/truncation of the rpoE gene, does not reverse the growth defect by lowering the major OMP levels; instead, it appears to activate a proteolytic pathway that partly compensates for the loss of DegP.
Results
Mutations that overcome the temperature sensitivity phenotype of ΔdegP
One of the ways to understand the consequences of a conditional lethal mutation is by isolating and characterizing revertants that have acquired so-called suppressor mutations and overcome the harmful effects of the original mutation. We have previously shown that a mutant simultaneously lacking degP and bamB displays a synthetic and conditional lethal phenotype at or above 37°C and grows poorly even at 30°C [8]. Because of this phenotype, the double mutant provides an ideal background in which to isolate suppressor mutations that reverse the growth defect, presumably by improving the OMP assembly environment and/or reducing envelope stress.
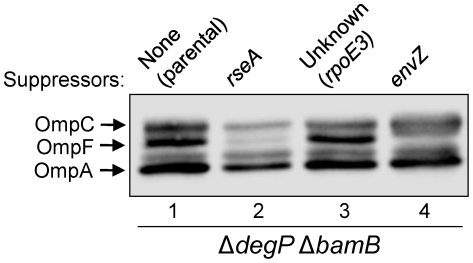
Liquid cultures of the ΔdegP ΔbamB double mutant were grown in LB at 30°C, diluted, spread onto LB agar plates, and incubated at 37°C until colonies appeared (between 24 and 36 hours). Temperature resistant colonies, which arose at a frequency of around 10−7, were then purified at 30°C and suppressor mutants categorized based on growth robustness and OMP levels. Using these criteria, the suppressors were classified into three groups (Fig. 1). One group included suppressor mutations affecting envZ of the EnvZ/OmpR two-component regulon (Fig. 1, lane 4; [30]).
Figure 1. Profiles of major OMPs from the parental strain (ΔdegP ΔbamB) and representative revertants carrying three different types of suppressor mutations.
OMP levels were examined by Western blot analysis from overnight grown cultures at 30°C. Each lane contains protein samples from equal number of cells, based on OD600.
The second group of revertants were distinct from those carrying missense mutations in envZ in that the levels of all the major OMPs (Fig. 1, lane 2), not just that of OmpF as seen in the envZ mutants, were significantly reduced. Since this phenotype is reminiscent of an rseA null mutation, we determined the nucleotide sequence of the rseAB-rpoE region. Three different types of mutations were found in the rseA gene, resulting in (a) a W33R substitution, which was previously shown to directly affect the RseA-σE binding pocket [34], (b) a frame-shift mutation after the sixteenth codon of rseA, and (c) an IS1 insertion element at nucleotide 454 of rseA. In each case, the mutations presumably abrogated RseA's ability to sequester σE at the inner membrane, thereby elevating the σE-mediated envelope stress response. Thus, rseA mutations suppressed the ΔdegP ΔbamB-mediated conditional lethal phenotype, in part, by lowering the load of OMPs in the envelope, and additionally by increasing the synthesis of periplasmic chaperones.
The last group was represented by one suppressor, which was noted to have the unusual ability to suppress lethality without significantly altering steady-state OMP levels (Fig. 1; lane 3). Through P1 transduction-mediated marker replacement, it was determined this third suppressor could fully correct the temperature sensitive growth defect of a ΔdegP bamB + strain. Because of these two properties, we chose to investigate this suppressor further in the ΔdegP bamB + strain and not in the original ΔdegP ΔbamB strain, which grows very poorly and prone to accumulating suppressors.
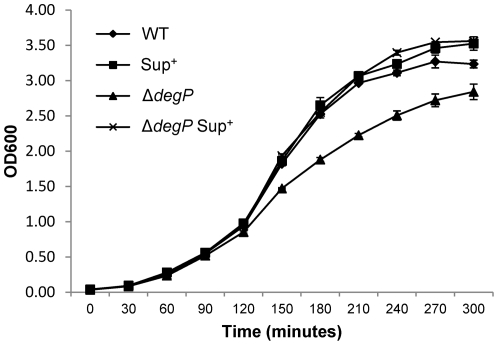
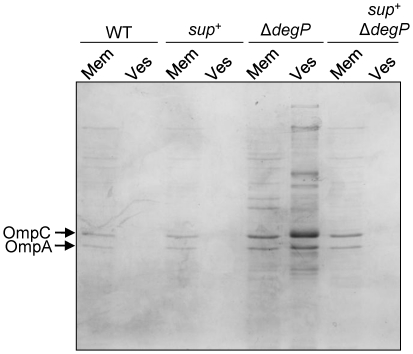
To quantify the extent to which the third suppressor mutation reversed the growth defect in ΔdegP cells, growth was measured by culturing cells in LB for five hours at 39°C, which is the sub-lethal growth temperature for the ΔdegP mutant, with cell density measured every thirty minutes. In contrast to ΔdegP-only cells, ΔdegP cells containing the suppressor mutation grew at a rate indistinguishable from the degP + strain (Fig. 2). degP + cells containing the suppressor mutation grew just like the degP + parental strain (Fig. 2). It is known that ΔdegP cells vesiculate profusely [35]. We asked whether the suppressor mutation can reverse this phenotype of ΔdegP. Membranes and vesicles obtained from cultures grown at 39°C were analyzed by SDS-PAGE and proteins were visualized after Coomassie blue staining (Fig. 3). As expected, ΔdegP cells released a large amount of vesicles containing a variety of proteins, including OmpC and OmpA. However, the presence of the suppressor mutation almost completely reversed this phenotype of ΔdegP. Hardly any proteins were visible from the vesicle fraction of wild-type and suppressor-containing cells (Fig. 3).
Figure 2. Growth curves of bacterial cultures grown at 39°C.
Wild type and ΔdegP cells, with or without the suppressor mutation, were grown overnight at 30°C. Next day, overnight cultures were diluted to 1∶100 in flasks containing fresh, pre-warmed, Luria broth and growth was resumed at 30°C and 39°C. OD600 was measured from bacterial samples withdrawn every 30 minutes. Only 39°C growth curves, obtained from two independent experiments, are shown. All strains grew almost identically at 30°C.
Figure 3. The suppressor mutation reverses the vesiculation phenotype of a ΔdegP strain.
Cultures were grown at 39°C in Luria broth for five hours, after which membranes and vesicles were prepared as described in the Experimental Procedure section. Samples were analyzed by SDS-PAGE and proteins were visualized after Coomassie blue staining. Abbreviations: mem, membrane; ves, vesicle; sup +, the unknown suppressor mutation.
Identification of the suppressor mutation
Before determining the mechanism of ΔdegP suppression, we set out to identify the third suppressor mutation. For this, we introduced by P1 transduction a random Tn10 (Tetr) insertion library into a ΔdegP strain harboring the suppressor mutation and selected for tetracycline resistant (TetR) transductants. Over a thousand TetR transductants were screened for the reversion to temperature sensitivity (i.e., loss of suppressor) by replica plating at 40°C and 30°C. One colony became temperature sensitive, indicating that either the insertion of Tn10 in a gene confers the growth defect or the Tn10 brought in the flanking wild type DNA and replaced the suppressor mutation. To distinguish the two possibilities, we determined the linkage of Tn10 to the temperature sensitivity phenotype by transducing the TetR marker into the ΔdegP strain containing the suppressor mutation. Approximately 50% of the time the resulting TetR transductants became temperature sensitive, indicating that the Tn10 is closely linked to the suppressor mutation and not inserted in a gene that confers a temperature sensitive growth phenotype.
To conclusively determine the chromosomal location of the Tn10, arbitrarily primed polymerase chain reaction (AP-PCR) was utilized. PCR products obtained after the second, high-stringency reaction were analyzed on a 4% w/v agarose gel, excised and sequenced using the appropriate Tn10 specific primers used in the reaction. Using this method, the Tn10 was found to disrupt yfiF located at 58.5′ on the chromosome, at nucleotide 883. A known Kanr insertion in glrK (57.9′ on the E. coli chromosome) was utilized to determine which side of yfiF::Tn10 the suppressor mutation was located. ΔglrK::Kanr was introduced into ΔdegP suppressor-yfiF::Tn10 cells by P1 transduction, and Kanr transductants were screened for Tets and temperature sensitive phenotypes. Using this method, the gene order was determined to be glrK-suppressor-yfiF.
Identification of the gene affected by the suppressor mutation
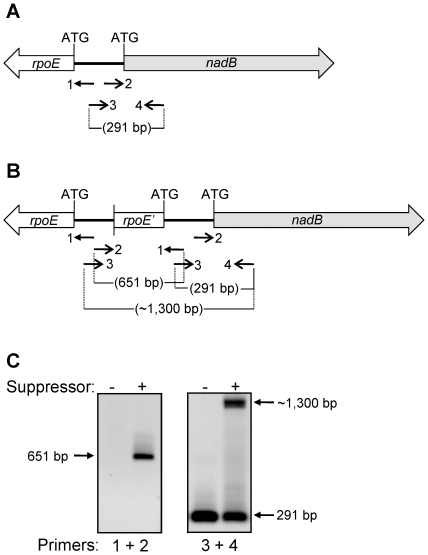
Several sets of primers were designed to amplify 1 to 3 kb long DNA from the 57.93 to 58.53 minute region of the chromosome from wild-type and suppressor-containing strains and the products were sequenced. When amplifying the rpoE-nadB region, we noted the presence of a PCR product from the suppressor strain that was approximately 1 kb larger than that amplified from the wild-type strain (data not shown). Sequence analysis indicated the presence of duplicated DNA in the suppressor strain.
In order to conclusively determine the molecular nature of the suppressor mutation, a primer set was designed which consisted of one primer landing upstream of rpoE and reading into rpoE (primer 1) and one primer landing upstream of nadB and reading into nadB (primer 2) (Fig. 4A). Because rpoE and nadB are divergently transcribed, the only way to obtain PCR product is if there was truly a duplication of rpoE or nadB, such that the two “forward” primers were able to prime the reaction by facing each other in the chromosome. Indeed, when PCR was performed in the wild-type and suppressor strains, only the suppressor strain gave rise to a fragment using the divergent primer set (Fig. 4C). Another set of primers that points towards each other and lands 157 bp upstream of nadB ATG (primer 3) and 124 bp downstream from the nadB ATG (primer 4) produced an expected 291-bp long PCR fragment from wild type strain (Fig. 4C). However, the suppressor containing strain yielded two fragments of 291 bp and about 1.3 kb (Fig. 4C), confirming the presence of duplicated DNA (Fig. 4B).
Figure 4. Identification of the genetic rearrangement in the suppressor strain.
PCR amplifications were carried out to narrow down the site of possible genetic rearrangement in the rpoE-nadB region of the chromosome. (A, B) Approximate drawings showing the rpoE and nadB genes from the wild-type (A) and suppressor (B) strains, as well as the positions of primers (numbered 1 to 4), their orientations, and size of the amplified DNA fragments. (C) Agarose gels showing the results of PCR amplifications from the wild-type and suppressor strains. Numbers 1 to 4 refer to the primers shown in (A) and (B).
The purified 651-bp long PCR product was sequenced using the same primers used for amplification, revealing a novel duplication-truncation of rpoE (Fig. 4B). The mutation consisted of rpoE truncated at nucleotide 396, corresponding to amino acid F122. After F122, the amino acid sequence continued with the non-native MVWYA sequence before reaching a stop codon (UAG). In addition to coding for five non-native amino acids, the region immediately after F122 was identical to the DNA sequence from 21 bases upstream of the nadB translation start to the translation start site of the native, full-length copy of rpoE was reached (Fig. 4B). Thus, the native copy of rpoE gene, present downstream of the 3′-tructaed copy of rpoE, is likely transcribed by the native promoter as well as the truncated rpoE gene promoter. We refer to the suppressor mutation as rpoE3, named after the suppressor isolate numbered 3.
RpoE (σE) levels are elevated in the rpoE3 background
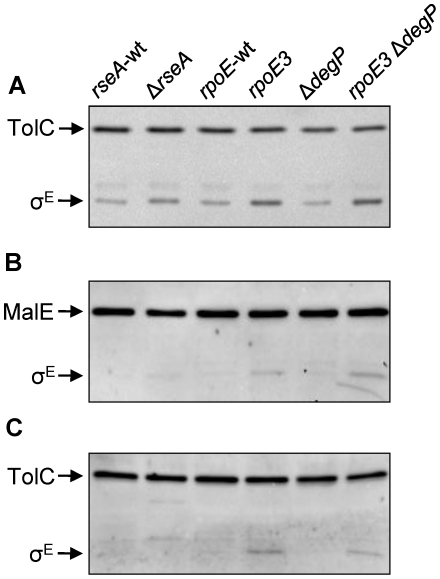
Because of the partial duplication of rpoE in rpoE3, we asked whether the steady-state σE levels were elevated in cells harboring rpoE3. Wild-type and ΔdegP cells containing rpoE3 were grown to late exponential phase, and whole-cell extracts were isolated to analyze σE levels by Western blot using σE-specific antibodies (Fig. 5). In strains containing the rpoE3 allele, σE levels were four-fold (in degP + background) to six-fold (in ΔdegP background) higher than in their rpoE + counterparts (Fig. 5A). This is likely due to elevated rpoE expression resulting from the rpoE3-mediated promoter duplication (in degP + background) and increased envelope stress (in ΔdegP background). Despite these increases in σE levels, the steady-state levels of OMPs were not reduced in the rpoE3 mutant (Fig. 1). In comparison, the σE level in a rseA null strain went up by 2.5 fold (Fig. 5A), which is lower than that seen in the rpoE3 strain, yet OMP levels were drastically reduced in the rseA mutant background (Fig. 1). It is noteworthy that despite the existence of a truncated rpoE open reading frame in addition to the full length open reading frame, no small molecular weight bands reacted with the σE-specific antibodies. The reason for this could be that either the antibodies used did not recognize the truncated σE protein, the truncated protein was not produced, or it was highly unstable.
Figure 5. Examination of the σE (RpoE) levels from different genetic backgrounds and cell fractions.
σE was detected by Western blots from protein samples obtained from whole cells (A), soluble (B, periplasm and cytoplasm) and insoluble (C, membranes) fractions. TolC-specific and σE-specific antibodies were used in (A), while antibodies raised against σE and a MalE-TolC fusion protein were used in (B) and (C). The MalE-TolC fusion antibodies were used to verify the purity of soluble (MalE) and insoluble or membrane (TolC) fractions. Relevant genotypes of the strains are shown at the top.
σE must be released into the cytoplasm in order to become active. One possible explanation for the increase in σE levels without a concomitant reduction in OMPs in the rpoE3 mutant is that, since rseA itself is a member of the σE regulon, the rpoE3 mutation increases the level of membrane-bound σE rather than the soluble σE, thus limiting the σE response. To test this, cells were grown at 37°C to late exponential phase, lysed by French press, and soluble and insoluble fractions were separated by centrifugation at 100 000 g for an hour. σE from the two fractions were analyzed by Western blot using anti-σE antibodies (Fig. 5B and C). Purity of the two fractions was affirmed by probing for TolC (insoluble) and MalE (soluble) proteins. Surprisingly, σE levels increased in both fractions (Fig. 5B and C). Quantification revealed that soluble σE levels increased 1.38 fold (Fig. 5B) and insoluble σE levels decreased 0.78 fold (Fig. 5C) in the rpoE3 ΔdegP strain compared to the rpoE3 strain. Thus, despite the increase of σE in both fractions, there appears to be a small but preferential increase in the soluble active σE form in the rpoE3 ΔdegP strain. This is expected, since additional envelope stress caused by the loss of DegP would trigger a further release of σE from the envelope to combat stress.
rpo3 triggers a modest σE response
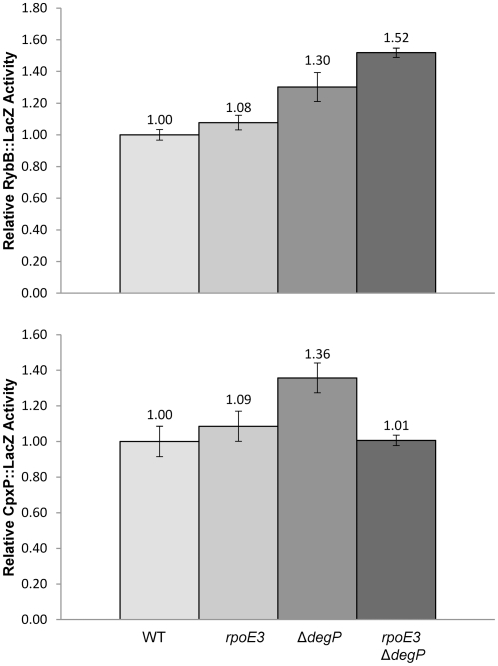
Based on the results obtained above, we hypothesized that the suppressor mutation exerted its effect by slightly activating the σE response without reducing the steady-state OMP levels (Fig. 1). To test this, rpoE3 was introduced into cells containing a RybB::LacZ fusion. RybB, whose expression is controlled by σE, encodes a small regulatory RNA responsible for down-regulating synthesis of several OMPs, including OmpC, OmpF and OmpA [36]–[38].
RybB::LacZ activity was determined from wild type, rpoE3, ΔdegP and rpoE3 ΔdegP cells grown at 39°C to mid- to late-exponential phase (Fig. 6A). In cells harboring the rpoE3 allele, RybB::LacZ activity increased by about 10% relative to wild type, indicating only a slight activation of the σE response at this growth temperature. The ΔdegP cells, which grew slightly slower than the parental strain (Fig. 2), showed a 30% increase in the RybB::LacZ compared to the parental strain. In the rpoE3 ΔdegP double mutant, which does not show a growth defect, an additive effect was observed: RybB::LacZ activity increased by 50% over wild type. This indicates that the two mutations elevated rpoE expression by independent mechanisms: rpoE3 directly increased σE levels, while the ΔdegP mutation indirectly activated σE by causing envelope stress. Only a modest increase in RybB::LacZ expression would help explain why there was no significant reduction in steady-state OMP levels in the rpoE3 ΔdegP background compared to the parental rpoE + degP + strain (Fig. 1). In contrast, the RybB::LacZ activity in an rseA null background went up almost six-fold (Gerken and Misra, unpublished data). This increase correlates well with a significant reduction in the OMP levels (Fig. 1). We also measured RybB::LacZ activity from cells grown at 37°C since there is a possibility that the relatively small difference observed between wild type and mutant strains grown at 39°C could be due to the basal RybB::LacZ activity in wild type strain being already high at 39°C. The results from 37°C growth experiments were similar to those obtained from cells grown at 39°C, with a relative increase of 20% (rpoE3), 20% (ΔdegP), and 70% (rpoE3 ΔdegP) (Fig. S1). The additive effect of rpoE3 and ΔdegP was once again observed, indicating the two mutations activate RybB::LacZ expression by independent mechanisms.
Figure 6. Effects of ΔdegP and rpoE3 mutations on RybB::LacZ (A) and CpxP::LacZ (B) activities.
For each strain, two independent cultures were grown at 39°C to a mid-log phase and used for β-galactosidase assays. LacZ activities are relative to the wild-type strain. The relevant genotypes of the strains are labeled at the bottom.
rpo3 restores envelope homeostasis by lowering ΔdegP-mediated activation of the Cpx pathway
Both Cpx and σE pathways are important in reducing envelope stress caused by aberrant β-barrel OMP assembly [9]. The Cpx-mediated upregulation of degP is critical for this reduction in envelope stress [9]. Consistent with this view, expression of cpxP, a prototypical member of the Cpx regulon, is modestly upregulated in ΔdegP cells grown at 39°C (Fig. 6B; [9]). We asked whether the elevated σE response by rpoE3 will help reduce the activated Cpx response observed in ΔdegP cells. As shown in Fig. 6B, the ΔdegP-mediated elevated CpxP::LacZ activity returned to the wild type level in the rpoE3 ΔdegP double mutant background. These results showed the complementary nature of the two envelope stress response pathways controlled by the σE and Cpx systems.
Potential activation of a DegP-independent proteolysis pathway in the rpoE3 mutant
One of the major functions of the σE response is to promote the proper folding and/or destruction of aberrantly folded OMPs in the envelope. In a ΔdegP background, rpoE3 obviously cannot simply increase degP transcription to compensate for increased stress and abolish the temperature sensitive growth phenotype. We therefore asked whether rpoE3 caused the activation of another proteolytic pathway, thus allowing for normal growth in a ΔdegP background.
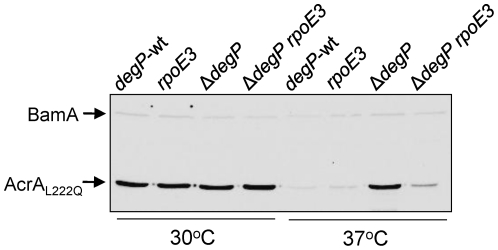
For this, we employed AcrAL222Q, an unstable variant of AcrA that is rapidly degraded in stationary phase grown cultures at 37°C, but not at 30°C, in a DegP-dependent manner [39]. Because of its instability properties, AcrAL222Q makes an ideal substrate to assay envelope proteolysis activity in vivo. The chromosomal allele of acrA expressing AcrAL222Q was introduced into cells harboring rpoE3 and lacking tolC (ΔtolC destabilizes the mutant AcrA protein). In order to test for proteolysis in this background, degP was also removed by a degP::Kanr insertion.
Cells were grown overnight in LB broth at 37°C and 30°C, after which samples were equalized according to final cell densities and boiled in SDS sample buffer. Samples were analyzed by Western blot using antibodies recognizing AcrA. At 30°C, the presence of degP::Kanr or rpoE3 did not significantly affect AcrAL222Q levels (Fig. 7, lanes 1–4). However, at 37°C AcrAL222Q levels increased six folds in a degP::Kanr background compared to the degP + strain (Fig. 7, lanes 5 and 7). In an rpoE3 degP + background, the protein was also degraded due to the presence of intact DegP (Fig. 7, lane 6). Interestingly, the presence of rpoE3 in the degP::Kanr background caused a three-fold reduction in the AcrAL222Q levels compared to that present in the degP::Kanr background alone (Fig. 7, lane 8), reflecting decreased stability of the protein, presumably due to an increase in proteolysis in the envelope. Consistent with the involvement of a σE-mediated, DegP-independent proteolytic pathway, AcrAL222Q levels were also reduced in a ΔrseA ΔdegP background where σE was fully activated (Fig. S2A).
Figure 7. Levels of a labile AcrA variant, AcrAL222Q, in cells with various genetic backgrounds.
Cultures were grown at 30°C and 37°C and AcrAL222Q levels were determined from whole cell extracts by Western blot analysis. BamA, detected by BamA antibodies, was used as a gel loading control.
acrA is not known to be a member of the σE regulon. Indeed, we have previously shown that the wild-type AcrA protein level did not change in a bamA mutant background in which the σE response was strongly induced [40]. Nevertheless, to eliminate the possibility that the expression of the acrA gene is somehow strongly affected by the induction of σE in the rpoE3 background, we created an isogenic set of strains to those mentioned above, except that this set contained the wild type acrA gene. Western analysis data showed that at 37°C in the rpoE3 degP::Kanr background, the level of wild type AcrA decreased only about 10% compared to the rpoE + ΔdegP cells (Fig. S2B). In contrast, AcrAL222Q levels were reduced over 300% in the rpoE3 degP::Kanr background compared to rpoE + degP::Kanr cells (Fig. 7). These data support the notion that a post-synthesis event, most likely an activated proteolytic pathway of the σE regulon, is primarily responsible for the observed destabilization/degradation of AcrAL222Q in the absence of DegP. This pathway, in part, contributes to the mechanism of suppression by rpoE3.
Discussion
One of the hallmarks of the fully activated σE system is the severely reduced major OMP levels resulting from high expression of the σE-controlled small regulatory RNAs, RybB and MicA [36]–[38]. Indeed, rseA mutations that constitutively activate the σE stress response pathway also dramatically lower the major OMP levels (Fig. 1). Even though the rpoE3 mutation significantly elevates σE in the cell—over two and fourfold compared to ΔrseA and rpoE + strains, respectively—no significant changes in the levels of major OMPs, OmpA and OmpC, were observed, indicating that the σE stress response pathway was not fully activated. This was also reflected by a mere 10% and 20% increase in σE-controlled RybB::LacZ activity at 39°C and 37°C, respectively. We suspect that in the rpoE3 background the continued presence of RseA at presumably slightly higher levels prevents σE from becoming fully active. In the rpoE3 ΔdegP background, the σE pathway is more active than in the rpoE3 or rpoE + background, based on the elevated RybB::LacZ activity and a complete reversal of the vesiculation and temperature sensitive growth phenotypes of ΔdegP. Yet, the absence of a significant effect on the major OMP levels suggests the extent of σE activation still remains lower in the rpoE3 background than in a ΔrseA background.
In contrast to ΔdegP cells grown at 39°C, a greater σE and Cpx activation is observed in cells with impaired OMP assembly pathways stemming from the loss of a major periplasmic chaperone, SurA, or the presence of a defective BAM complex [9], [40]. It is conceivable that differences in the biochemical nature of OMP assembly intermediates and their levels account for some of the disparities in envelope stress response in different genetic backgrounds. For example, it is well established that during aberrant OMP assembly, exposure of the C-terminal OMP residues in the periplasm activates the σE pathway by promoting RseA proteolysis [41]. This likely occurs in a background lacking SurA or expressing a defective BAM complex. In contrast, there is no evidence that OMP assembly is significantly defective in ΔdegP cells, since we do not see any decrease in OmpA and OmpC levels from the envelopes. Nevertheless, it is likely that there is some increase in the nascent, unassembled OMPs that have fallen off the proper assembly pathway and are normally captured and/or degraded by DegP. Also, the levels of other unstable/unfolded envelope proteins, including those that normally reside in the periplasm, must increase in ΔdegP cells grown at 39°C. Thus, envelope stress is likely to be created in ΔdegP cells—enough to induce to a robust stress response through activating the σE or Cpx pathways. The predominant response to this stress appears to be the release of outer membrane vesicles, but this is clearly not sufficient when growth temperature reaches 40°C where viability of ΔdegP cells becomes severely compromised. The fact that rpoE3 completely reverses the phenotypes of ΔdegP suggests that it must lower the levels of stress-causing envelope proteins. However, unlike the rseA null mutations, which significantly lower OMP levels, the effect of rpoE3 on OMPs alone in not sufficient to account for its protective phenotype.
Since OMP levels are not reduced by rpoE3, alternative stress responses must account for the reversal of the ΔdegP phenotypes. Our results indicate that up-regulation of a DegP-independent, σE-dependent proteolytic pathway can partly accounts for this reversal. To test this pathway, we used a mutant AcrA protein, AcrAL222Q, which is rapidly degraded in a DegP-dependent manner [39]. In the absence of DegP, the mutant AcrA protein level rose significantly, but in the presence of rpoE3, AcrAL222Q levels went down again. The level of AcrAL222Q was also reduced in the absence of RseA, indicating the involvement of σE. With no reduction in wild type AcrA levels in the rpoE3 background, it appears that a post-synthesis step, most likely involving increased proteolysis, accounts for the reduced AcrAL222Q level. We do not currently know the identity of the protease(s) responsible for AcrAL222Q degradation in the absence of DegP in the ropE3 and ΔrseA strains.
Overexpression of SohB and DegQ from plasmids has been reported to rescue the temperature sensitive growth phenotype of ΔdegP [42]–[43]. However, neither sohB nor degQ is known to be regulated by σE and there is no experimental data showing SohB is actually a protease. yfgC, a gene regulated by σE, is predicted to encode a periplasmic metalloprotease [44]. However, deletion or overexpression of YfgC did not influence AcrAL222Q stability or the ΔdegP phenotypes (data not shown). It should be noted that activation of a proteolytic pathway likely represents only a component of the broader defensive response triggered by σE. Therefore, we believe that the collective action of increased proteolysis and other defensive measures most likely account for the rpoE3-mediated reversal of the ΔdegP phenotypes.
The fact that mutations required to increase σE levels and rescue the ΔdegP phenotypes are either in rseA, a gene encoding for the negative regulator of σE, or the rpoE gene itself suggests that even under high envelope stress conditions the amount of free σE becomes limiting. If growth at high temperatures without DegP indeed leads to the accumulation of misfolded OMPs, then these accumulated OMPs should trigger the DegS/RseP-mediated proteolytic pathway to degrade RseA and elevate free σE levels [45]. The fact that ΔdegP cells begin to die at 39°C suggests that the defensive response system may have maxed out, perhaps because newly released and activated σE allows for a greater synthesis of RseA, which in turn re-captures σE. In other words, under extreme envelope stress, a tight homeostasis loop between RseA and σE may partly be responsible for cell death. This loop can be broken by mutations that either lower RseA activity or increase σE levels. An alternative means to overcoming the growth phenotype of ΔdegP without directly influencing the RseA-σE loop would be through mutations that influence the cAMP-CRP [25] or EnvZ/OmpR [30] pathway. The prlF/sohA mutations likely provide yet another mechanism to overcome the temperature sensitive growth phenotype of ΔdegP.
Materials and Methods
Bacterial strains, media and reagents
Escherichia coli K-12 bacterial strains used in this study were constructed in an MC4100 background [46] and are shown in the Table S1. Luria-Bertani broth (LB) and Luria-Bertani agar (LBA) were prepared as previously described [47]. When required, media were supplemented with 50 µg ml−1 ampicillin, 25 µg ml−1 kanamycin, 12.5 µg ml−1 chloramphenicol, 10 µg ml−1 tetracycline and 0.2% L-(+) arabinose (Sigma). All other chemicals were of analytical grade. Bacterial growth curves were performed by diluting overnight cultures 1∶100 into 25 ml fresh pre-warmed media supplemented with appropriate antibiotics. Cultures were vigorously shaken at 39°C in baffle-bottom flasks in a water incubator with cell density checked every 30 min.
DNA manipulation
Standard bacterial genetic methods were carried out as previously described [47]. Chromosomal deletion of degP, followed by the removal of the antibiotic resistance cassette was carried out using the λ-red mediated gene deletion method as previously described [48]. Primer sequences are available upon request.
The unknown Tn10 marker was identified by the arbitrary-primed PCR (AP-PCR) method [49]–[50]. The first, low-stringency round of PCR was carried out with wild-type cells and cells containing the unknown Tn10 marker (see below). That round of PCR used both ARB1 and ARB6 primers [50], as well as either Tn10 left or Tn10 right primers to amplify fragments on either side of the Tn10. First round products were analyzed on an 0.8% agarose gel, and fragments unique to the Tn10 strain were excised using a QIAquick gel extraction kit (QIAGEN) and subjected to a second, high-stringency round of PCR using the appropriate Tn10 primer and ARB2 [50]. Amplified products were cleaned up using a QIAquick PCR purification kit and sequenced using the appropriate Tn10 primer or ARB2. Sequences were analyzed to obtain non-Tn10 sequence, and unassigned sequences were analyzed using BLASTn to determine the location of the Tn10.
Cell fractionation
Fresh cultures were grown by sub-culturing overnight cultures into fresh media with appropriate antibiotics at a starting OD600 of 0.025. Cell pellets, collected from freshly grown cells, were resuspended in a lysis buffer containing 20 mM Tris-HCl pH 7.5, 2 mM phenylmethylsulfonylfluoride (PMSF), 10 mM MgCl2 and 25 µg ml−1 DNase I. Samples were passed twice through a French pressure cell, followed by low-speed centrifugation of lysates to remove unlysed cells. Crude lysates were the centrifuged at 100,000 g for 1 h at 4°C to pellet membranes. Periplasm was extracted using the gentle osmotic shock method [51]. Prior to French Press lysis, cell pellets were resuspended in periplasm extraction buffer (10 mM Tris-HCl pH 7.5, 500 mM sucrose, 10 mM EDTA and 0.2 mg ml−1 lysozyme) and incubated on ice for 30 min. Samples were centrifuged at 100,000 g for 15 min and periplasm drawn off as soluble fraction. Spheroplast pellets were then washed with cold 10 mM Tris-HCl pH 7.5 and subjected to French press lysis as described above. After high speed spin, cytoplasm was harvested from the soluble fraction. Membranes pellets were routinely washed in 10 mM Tris-HCl pH 7.5 and resuspended according to cell density in the same buffer.
Protein methods
For Western blot analysis from whole cells, aliquots of cultures, normalized to cell density, were pelleted in a microcentrifuge. Pellets were resuspended in sample buffer (62.5 mM Tris-HCl pH 6.8, 10% v/v glycerol, 100 µg ml−1 bromophenol blue, 5% v/v β-mercaptoethanol and 2% w/v sodium dodecyl sulfate [SDS]). Samples were boiled for 5 min and analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE). When required, 4 M urea was added in order to resolve OmpC and OmpF. Membrane, periplasmic and cytoplasmic fractions were prepared in the same buffer as whole-cell samples. After electrophoresis, proteins were transferred to Immobilon-P polyvinylidene difluoride (PVDF) membrane (Millipore) using a Bio-Rad mini-transblot. Membranes were blotted with antibodies raised against OmpF (1∶16 667), LamB (1∶10 000), AcrA (1∶10 000), TolC (1∶10 000), MalE (1∶10 000) RpoE (1∶5 000) or GroEL (1∶25 000). Blots were developed as described previously [40]. Protein bands were quantified using Bio-Rad Quantity One software. When appropriate, SDS-PAGE gels were stained with Coomassie brilliant blue R-250 (Sigma).
Enzymatic assays
β-galactosidase activity was determined by an established method [52], modified for use with microtiter plates. Kinetic analysis of β-galactosidase activity was carried out using a Versamax microtiter plate reader (Molecular Dynamics). Activity was calculated as the rate of ONPG (ortho-Nitophenyl-β-D-galactopyranoside) cleavage (OD420) normalized to cell density (OD562) in each well. All assays were performed in quadruplicate.
Supporting Information
Effects of Δ degP and rpoE3 mutations on RybB::LacZ activity. Two independent cultures were grown at 37°C to a mid-log phase and used for β-galactosidase assays. LacZ activities are relative to the wild-type strain. Relevant genotypes are shown at the bottom.
(TIF)
Levels of AcrAL222Q (A) and wild type acrA (B) in different genetic backgrounds. AcrA levels were determined by Western blot analysis of whole cell lysates prepared from overnight grown cultures at 37°C. GroEL, LamB and AcrA were detected using specific antibodies. GroEL served as a gel loading control. Each lane contains protein samples from equal number of cells, based on OD600. Relevant genotypes are shown at the top.
(TIF)
A list of Escherichia coli K-12 strains used in this study.
(DOC)
Acknowledgments
We are grateful to Dr. Carol A. Gross for RpoE antibodies. We thank Phu Vuong and Leanne Misra for reading the manuscript.
Footnotes
Competing Interests: The author, Rajeev Misra, is a PLoS One Editorial Board member. This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials.
Funding: National Institutes of Health; GM048167. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lipinska B, Fayet O, Baird L, Georgopoulos C. Identification, characterization, and mapping of the Escherichia coli htrA gene, whose product is essential for bacterial growth only at elevated temperatures. J Bacteriol. 1989;171:1574–1584. doi: 10.1128/jb.171.3.1574-1584.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strauch KL, Johnson K, Beckwith J. Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J Bacteriol. 1989;171:2689–2696. doi: 10.1128/jb.171.5.2689-2696.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Misra R, Peterson A, Ferenci T, Silhavy TJ. A genetic approach for analyzing the pathway of LamB assembly into the outer membrane of Escherichia coli. J Biol Chem. 1991;266:13592–13597. [PubMed] [Google Scholar]
- 4.Misra R, CastilloKeller M, Deng M. Overexpression of protease deficient DegPS210A rescues the lethal phenotype of Escherichia coli OmpF assembly mutants in a degP background. J Bacteriol. 2000;182:4882–4888. doi: 10.1128/jb.182.17.4882-4888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CastilloKeller M, Misra R. Protease-deficient DegP suppresses lethal effects of a mutant OmpC protein by its capture. J Bacteriol. 2003;185:148–154. doi: 10.1128/JB.185.1.148-154.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rizzitello AE, Harper JR, Silhavy TJ. Genetic evidence for parallel pathways of chaperone activity in the periplasm of Escherichia coli. J Bacteriol. 2001;183:6794–6800. doi: 10.1128/JB.183.23.6794-6800.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Onufryk C, Marie-Laure C, Fang FC, Gross CA. Characterization of the six lipoproteins in the σE regulon. J Bacteriol. 2005;187:4552–4561. doi: 10.1128/JB.187.13.4552-4561.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charlson ES, Werner JN, Misra R. Differential effects of yfgL mutation on the biogenesis of Escherichia coli outer membrane proteins and lipopolysaccharide. J Bacteriol. 2006;188:7186–7194. doi: 10.1128/JB.00571-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerken H, Leiser OP, Bennion D, Misra R. Involvement and necessity of the Cpx regulon in the event of aberrant β-barrel outer membrane protein assembly. Mol Microbiol. 2010;75:1033–1046. doi: 10.1111/j.1365-2958.2009.07042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spiess C, Bell A, Ehrmann M. A temperature-dependent switch from chaperones to protease in a widely conserved heat shock protein. Cell. 1999;97:339–347. doi: 10.1016/s0092-8674(00)80743-6. [DOI] [PubMed] [Google Scholar]
- 11.Krojer K, Garrido-Franco M, Huber R, Ehrmann M, Clausen T. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature. 2002;416:455–459. doi: 10.1038/416455a. [DOI] [PubMed] [Google Scholar]
- 12.Krojer T, Sawa J, Schäfer E, Saibil HR, Ehrmann M, et al. Structural basis for the regulated protease and chaperone function of DegP. Nature. 2008;453:885–891. doi: 10.1038/nature07004. [DOI] [PubMed] [Google Scholar]
- 13.Jiang J, Zhang X, Chen Y, Wu Y, Zhou ZH, et al. Activation of DegP chaperone-protease via formation of large cage-like oligomers upon binding to substrate proteins. Proc Natl Acad Sci USA. 2008;105:11939–11944. doi: 10.1073/pnas.0805464105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen Q-T, Bai X-C, Chang L-F, Wu Y, Wang H-W, et al. Bowl-shaped oligomeric structures on membranes as DegP's new functional forms in protein quality control. Proc Natl Acad Sci U S A. 2009;106:4858–4863. doi: 10.1073/pnas.0811780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lipinska B, Sharma S, Georgopoulos C. Sequence analysis and regulation of the htrA gene of Escherichia coli: a σ32-independent mechanism of heat-inducible transcription. Nuc Acid Res. 1988;16:10053–10067. doi: 10.1093/nar/16.21.10053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erickson J, Gross CA. Identification of the σE subunit of Escherichia coli RNA polymerase: a second alternate sigma factor involved in high-temperature gene expression. Genes Dev. 1989;3:1462–1471. doi: 10.1101/gad.3.9.1462. [DOI] [PubMed] [Google Scholar]
- 17.Danese PN, Snyder WB, Cosma CL, Davis LJB, Silhavy TJ. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev. 1995;9:387–398. doi: 10.1101/gad.9.4.387. [DOI] [PubMed] [Google Scholar]
- 18.Pogliano J, Lynch AS, Belin D, Lin ECC, Beckwith J. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 1997;11:1169–1182. doi: 10.1101/gad.11.9.1169. [DOI] [PubMed] [Google Scholar]
- 19.Danese PN, Silhavy TJ. The σE and the Cpx signal transduction systems control the synthesis of periplasmic protein-folding enzymes in Escherichia coli. Genes Dev. 1997;11:1183–1193. doi: 10.1101/gad.11.9.1183. [DOI] [PubMed] [Google Scholar]
- 20.Connolly L, Peñas ADL, Alba BM, Gross CA. The response to extracytoplasmic stress in Escherichia coli is controlled by partially overlapping pathways. Genes Dev. 1997;11:2012–2021. doi: 10.1101/gad.11.15.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raivio TL, Silhavy TJ. The σE and Cpx regulatory pathways: overlapping but distinct envelope stress responses. Curr Opin Microbiol. 1999;2:159–165. doi: 10.1016/S1369-5274(99)80028-9. [DOI] [PubMed] [Google Scholar]
- 22.Baird L, Georgopoulos C. Identification, cloning, and characterization of the Escherichia coli sohA gene, a suppressor of the htrA (degP) null phenotype. J Bacteriol. 1990;172:1587–1594. doi: 10.1128/jb.172.3.1587-1594.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiino DR, Silhavy TJ. Mutation prlF1 relieves the lethality associated with export of β-galactosidase hybrid proteins in Escherichia coli. J Bacteriol. 1984;154:878–883. doi: 10.1128/jb.158.3.878-883.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt O, Schuenemann VJ, Hand NJ, Silhavy TJ, Martin J, et al. prlF and yhaV encode a new toxin–antitoxin system in Escherichia coli. J Mol Biol. 2007;372:894–905. doi: 10.1016/j.jmb.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strozen TG, Langen GR, Howard SP. Adenylate cyclase mutations rescue the degP temperature-sensitive phenotype and induce the sigma E and Cpx extracytoplasmic stress regulons in Escherichia coli. J Bacteriol. 2005;187:6309–6316. doi: 10.1128/JB.187.18.6309-6316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Costanzo A, Ades SE. Growth phase-dependent regulation of the extracytoplasmic stress factor, σE, by guanosine 3′,5′-bispyrophosphate (ppGpp). J Bacteriol. 2006;188:4627–4634. doi: 10.1128/JB.01981-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Costanzo A, Nicoloff H, Barchinger SE, Banta AB, Gourse RL, et al. ppGpp and DksA likely regulate the activity of the extracytoplasmic stress factor σE in Escherichia coli by both direct and indirect mechanisms. Mol Microbiol. 2008;67:619–632. doi: 10.1111/j.1365-2958.2007.06072.x. [DOI] [PubMed] [Google Scholar]
- 28.Vuong P, Bennion D, Mantei J, Frost D, Misra R. Analysis of YfgL and YaeT interactions through bioinformatics, mutagenesis, and biochemistry. J Bacteriol. 2008;190:1507–1517. doi: 10.1128/JB.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, et al. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 30.Gerken H, Charlson ES, Cicirelli EM, Kenney LJ, Misra R. MzrA, a novel modulator of the EnvZ/OmpR two-component regulon. Mol Microbiol. 2009;72:1408–1422. doi: 10.1111/j.1365-2958.2009.06728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall MN, Silhavy TJ. The ompB locus and the regulation of the major outer membrane porin proteins of Escherichia coli K-12. J Mol Biol. 1981;146:23–43. doi: 10.1016/0022-2836(81)90364-8. [DOI] [PubMed] [Google Scholar]
- 32.Gerken H, Misra R. Structure-function analysis of MzrA, a modulator of the EnvZ/OmpR regulatory system. J Bacteriol. 2010;192:6271–6278. doi: 10.1128/JB.00855-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guillier M, Gottesman S. Remodeling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol Microbiol. 2006;59:231–247. doi: 10.1111/j.1365-2958.2005.04929.x. [DOI] [PubMed] [Google Scholar]
- 34.Campbell EA, Tupy JL, Gruber TM, Wang S, Sharp MM, et al. Crystal structure of Escherichia coli σE with the cytoplasmic domain of its anti-σ RseA. Cell. 2003;11:1067–1078. doi: 10.1016/s1097-2765(03)00148-5. [DOI] [PubMed] [Google Scholar]
- 35.McBroom AJ, Kuehn MJ. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol Microbiol. 2007;63:545–558. doi: 10.1111/j.1365-2958.2006.05522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guillier M, Gottesman S, Storz G. Modulating the outer membrane with small RNAs. Genes Dev. 2006;20:2338–2348. doi: 10.1101/gad.1457506. [DOI] [PubMed] [Google Scholar]
- 37.Johansen J, Rasmussen AA, Overgaard M, Valentin-Hansen P. Conserved small non-coding RNAs that belong to the σE regulon: role in down-regulation of outer membrane proteins. J Mol Biol. 2006;364:1–8. doi: 10.1016/j.jmb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 38.Papenfort K, Pfeiffer V, Mika F, Lucchini S, Hinton JCD, et al. σE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol Microbiol. 2006;62:1674–1688. doi: 10.1111/j.1365-2958.2006.05524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerken H, Misra R. Genetic evidence for functional interactions between TolC and AcrA proteins of a major antibiotic efflux pump of Escherichia coli. Mol Microbiol. 2004;54:620–631. doi: 10.1111/j.1365-2958.2004.04301.x. [DOI] [PubMed] [Google Scholar]
- 40.Bennion D, Charlson ES, Coon E, Misra R. Dissection of β-barrel outer membrane protein assembly pathways through characterizing BamA POTRA 1 mutant of Escherichia coli. Mol Microbiol. 2010;77:1153–1171. doi: 10.1111/j.1365-2958.2010.07280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walsh NP, Alba BM, Bose B, Gross CA, Sauer RT. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell. 2003;113:61–71. doi: 10.1016/s0092-8674(03)00203-4. [DOI] [PubMed] [Google Scholar]
- 42.Baird L, Lipinska B, Raina S, Georgopoulos C. Identification of the Escherichia coli sohB gene, a multicopy suppressor of the HtrA (DegP) null phenotype. J Bacteriol. 1991;173:5763–5770. doi: 10.1128/jb.173.18.5763-5770.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waller PRH, Sauer RT. Characterization of degQ and degS, Escherichia coli genes encoding homologs of the DegP protease. J Bacteriol. 1996;178:1146–1153. doi: 10.1128/jb.178.4.1146-1153.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. Conserved and variable functions of the σE stress response in related genomes. PLoS Biol. 2006;4:43–59. doi: 10.1371/journal.pbio.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alba BM, Leeds JA, Onufryk C, Lu CZ, Gross CA. DegS and YaeL participate sequentially in the cleavage of RseA to activate the σE-dependent extracytoplasmic stress response. Genes Dev. 2002;16:2156–2168. doi: 10.1101/gad.1008902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Casadaban MJ. Transposition and fusion of the lac genes to select promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;141:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 47.Silhavy TJ, Berman ML, Enquist LW. Experiments with gene fusions. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1984. [Google Scholar]
- 48.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nuc Acid Res. 1990;18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fontaine F, Stewart EJ, Lindner AB, Taddei F. Mutations in two global regulators lower individual mortality in Escherichia coli. Mol Microbiol. 2008;67:2–14. doi: 10.1111/j.1365-2958.2007.05988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arié J-P, Sassoon N, Betton J-M. Chaperone function of FkpA, a heat shock prolyl isomerase, in the periplasm of Escherichia coli. Mol Microbiol. 2001;39:199–210. doi: 10.1046/j.1365-2958.2001.02250.x. [DOI] [PubMed] [Google Scholar]
- 52.Miller JH. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1992. pp. 71–74. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effects of Δ degP and rpoE3 mutations on RybB::LacZ activity. Two independent cultures were grown at 37°C to a mid-log phase and used for β-galactosidase assays. LacZ activities are relative to the wild-type strain. Relevant genotypes are shown at the bottom.
(TIF)
Levels of AcrAL222Q (A) and wild type acrA (B) in different genetic backgrounds. AcrA levels were determined by Western blot analysis of whole cell lysates prepared from overnight grown cultures at 37°C. GroEL, LamB and AcrA were detected using specific antibodies. GroEL served as a gel loading control. Each lane contains protein samples from equal number of cells, based on OD600. Relevant genotypes are shown at the top.
(TIF)
A list of Escherichia coli K-12 strains used in this study.
(DOC)