Abstract
A hallmark of the eukaryotic cell is the actin cytoskeleton, involved in a wide array of processes ranging from shape determination and phagocytosis to intracellular transport and cytokinesis. Recently, we reported the discovery of an actin-based cytoskeleton also in Archaea. The archaeal actin ortholog, Crenactin, was shown to belong to a conserved operon, Arcade (actin-related cytoskeleton in Archaea involved in shape determination), encoding an additional set of cytoskeleton-associated proteins. Here, we elaborate on the implications of these findings for the evolutionary relation between archaea and eukaryotes, with particular focus on the possibility that eukaryotic actin and actin-related proteins have evolved from an ancestral archaeal actin gene. Archaeal actin could thus have played an important role in cellular processes essential for the origin and early evolution of the eukaryotic lineage. Further exploration of uncharacterized archaeal lineages is necessary to find additional missing pieces in the evolutionary trajectory that ultimately gave rise to present-day organisms.
Keywords: actin, arcade, archaea, ARPs, crenactin, cell shape, cytoskeleton, eukaryogenesis, MreB, phagocytosis
Discovery of an actin-based cytoskeleton in Archaea
In eukaryotes, actin filaments constitute key components of the cytoskeleton, involved in pivotal processes such as determination and maintenance of cell shape and cellular junctions, cytokinesis, and vesicle-mediated transport such as endocytosis and phagocytosis.1 Recently, bona fide archaeal actin orthologs were identified in several crenarchaeal genomes, as well as in ‘Candidatus Korarchaeum cryptofilum’2 (Fig. 1). The archaeal actin ortholog, denoted Crenactin, was shown to polymerize into a cytoskeletal structure in the hyperthermophilic crenarchaeon Pyrobaculum calidifontis, and therefore inferred to be involved in cell shape determination.3 Immunofluorescence microscopy imaging revealed that Crenactin formed helical filaments that traversed the length of the rod-shaped cells.3 In a cell subpopulation, these filaments had been remodelled into band-like structures, presumably in preparation for cell division, in this respect resembling the bacterial cell-shape-determining protein MreB,4 which belongs to the same ATPase superfamily as actin and Crenactin. In addition, the phylogenetic distribution of the Crenactin-encoding gene (cren-1) across archaeal genomes correlated with rod- or filamentous cell morphologies, supporting an involvement in cell shape formation for the archaeal actin orthologs.3
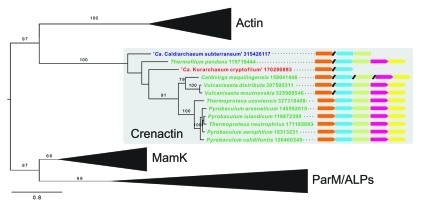
Figure 1.
An archaeal actin ortholog. Schematic overview of actin protein family phylogeny, demonstrating strong support (bootstrap value 97) for common ancestry of archaeal Crenactin and eukaryotic actin proteins. The tree was generated as described previously,3 except that distant members (MreB and Hsp70) of the actin protein family were omitted and novel archaeal sequences were added (Crenactin orthologs from ‘Ca. Caldiarchaeum subterraneum’, Vulcanisaeta distributa, Vulcanisaeta moutnovskia and Thermoproteus uzoniensis). For clarity, the actin, ParM/ALPs and MamK clades are displayed as triangles (for full details of included sequences, see3). The gray shading highlights the Crenactin clade and, for each species, the gene organization of the arcade gene cluster is indicated, with the genes encoding Arcadin-1 (rkd-1), Crenactin (cren-1), Arcadin-2 (rkd-2), Arcadin-3 (rkd-3) and Arcadin-4 (rkd-4) depicted in orange, blue, green, magenta and orange, respectively. The tree was rooted according to the topology obtained previously,3 with bootstrap values shown for branches with a support above 70 (out of 100 replicates). Sequences are denoted by species name and gene identifier, with crenarchaeal, korarchaeal and aigarchaeal Crenactin orthologs indicated in green, red and blue fonts, respectively. Note that the Crenactin ortholog of the aigarchaeon ‘Ca. Caldiarchaeum subterraneum’ represents the deepest branch in the Crenactin tree, although with low support.
The cren-1 gene was found to belong to a conserved gene cluster, which, in agreement with the involvement in cytoskeletal processes, was denoted Arcade (actin-related cytoskeleton in Archaea involved in shape determination; gray panel in Figure 1). In addition to cren-1, the gene cluster comprised several genes (rkd genes) whose products, Arcadins, were shown also to assemble into helical structures, presumably in conjunction with Crenactin filaments.3
The Arcadin-2 gene product, in contrast, displayed a punctuated distribution across P. calidifontis cells, and was found to localize between segregated nucleoids in a cell subpopulation and might, hence, play a cytokinesis-associated role in this organism. The presence of an Arcadin-2 ortholog in the composite genome of ‘Candidatus Caldiarchaeaum subterraneum’, belonging to the recently proposed Aigarchaeota phylum5 is, however, puzzling since this genome also encodes components of both the FtsZ and CdvBC cell division machineries,5 and might instead point toward a role in the genome segregation process, or in coordination of mitosis with cytoskeletal remodeling (above).
Irrespective, a picture emerges from the P. calidifontis study in which a core cytoskeleton is formed by Crenactin and Arcadin-1, with Arcadin-3 and -4 performing auxiliary roles, while the precise function of Arcadin-2 in the Thermoproteales3 needs to be further investigated. Detailed biochemical and molecular characterization of Crenactin and Arcadins is also of potential value for biotechnological exploitation, due to the intrinsic heat-stability of all gene products from the hyperthermophilic P. calidifontis species.
Importantly, the identification of Crenactin-based shape-determining structures in Archaea indicates that an actin-based cytoskeleton was established prior to the divergence of the archaeal and eukaryotic lineages. In the following, we discuss the implications of this observation with respect to the process of eukaryogenesis and the emergence of the eukaryotic cell.
Crenactin and eukaryogenesis
Recent phylogenomic studies have provided support for an evolutionary scenario in which the eukaryotic lineage emerged from within the archaeal domain.6,7 In particular, evidence has been presented for a model in which eukaryotes are suggested to have originated from a lineage that also gave rise to the Crenarchaeota phylum.8 In light of this, the discovery of an actin-based cytoskeleton in crenarchaea, as well as in other deep archaeal lineages, adds momentum to discussions concerning the origin of the eukaryotic cell and early stages of eukaryogenesis. The role of actin as a key-player in invagination-based processes such as endocytosis and phagocytosis is well documented, and a requirement for a phagocytotic machinery to engulf a putative α-proteobacterial ancestor of the mitochondrion has been put forward as a prerequisite in several models to explain the origin of the eukaryotic cell (see9 for overview).
In certain evolutionary models, it is argued that the proto-eukaryote was relatively complex at the cellular level in order to execute engulfment.9,10 The phagocytotic machinery present in extant eukaryotes is, indeed, highly complex and comprises a wide variety of associated proteins,2 in addition to actin. We envision that Crenactin could have provided primitive phagocytotic capabilities to an ancestral archaeal lineage that contributed to the emergence of the eukaryotic cell, by facilitating the fusion with the proto-mitochondrion3 in a far less complex process than that performed by the fully developed phagocytotic machineries present in extant eukaryotes. Interestingly, several members of the Thermoproteales, all of which contain the Crenactin-encoding gene, display bent or even branched cells, occasionally with extended cellular protrusions.11,12 Assuming these structures are not methodological artifacts, this suggests a certain degree of cytoskeletal flexibility in these species, and, by inference, in ancestral archaeal lineages. It is, thus, conceivable that, in an archaeal lineage from which the eukaryotic lineage would have emerged, such actin-mediated cytoskeletal flexibility may have allowed for primitive phagocytosis that occasionally sustained cellular fusions.
In addition, a recent study of the deeply-branching microbial eukaryote Giardia intestinalis revealed that the actin cytoskeleton was capable of facilitating receptor-mediated endocytosis in absence of canonical actin-binding proteins.13 Whether the relatively simple cytoskeleton in Giardia represents a relic of an ancient past, or whether it is the result of reductive genome evolution remains to be elucidated. Yet, it does indicate that actin is capable of sustaining membrane invagination in absence of the array of proteins usually involved in this process.
Actin proliferation and sub-functionalization during early stages of eukaryogenesis
Apart from a potential role in cellular fusion events that may have given rise to the eukaryotic cell, actin has functionally diversified during evolution and is, in addition to the well-characterized cytoskeletal roles, implicated in other key processes that may have promoted eukaryogenesis. For example, recent studies have provided evidence for nuclear localization of an actin sub-fraction where, in conjunction with other proteins, it is involved in the assembly of the nuclear envelope and in processes influencing transcriptional activity.14,15
Further support for the importance of actin in the evolution of the eukaryotic cell may be inferred from the excessive proliferation of actin homologs across all branches of the eukaryotic tree. Most eukaryotes contain at least eight actin paralogs, denoted actin-related proteins (ARPs). ARPs share overall structure and low-level sequence homology with conventional actin, and phylogenetic analyses have revealed that ARPs can be divided into several distinct groups, which, inferred from their distribution, have formed via ancient gene duplication events, most of which predate the establishment of the major eukaryotic lineages.16 As for actin, the functions of ARPs are not merely restricted to a role in cytoskeleton modulation. Whereas certain ARPs, such as ARP2 and ARP3, play crucial roles in actin polymerization, biochemical and genetic evidence is mounting for other ARPs to function largely, or exclusively, in the nucleus.14 For example, several ARPs have been identified as members of chromatin remodeling complexes that locally modulate chromatin structure and activity by re-locating or removing nucleosomes.17 Overall, a picture emerges in which actin, and ancient actin paralogs, have played important roles throughout the emergence and diversification of the eukaryotic lineage, and it is possible that a progenitor of Crenactin played a key role in founding the broad family of actins and ARPs present in extant eukaryotes.
Additional eukaryotic signatures in archaeal phyla
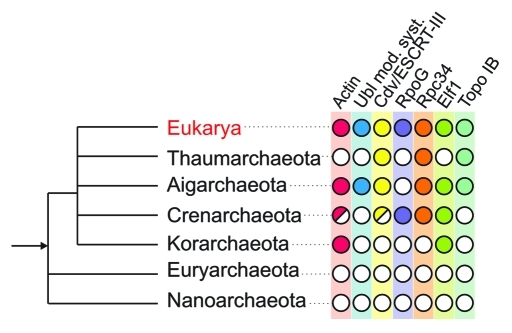
The identification of a Crenactin-based cytoskeleton in Archaea necessitates a re-evaluation of the hypothesis of the actin cytoskeleton as specific to the eukaryotic cell, and the presence of actin orthologs in members of the Crenarchaeota and other proposed archaeal phyla lends support to scenarios that associate a progenitor of this phylum with the origin of the eukaryotic cell. Interestingly, recent studies have revealed a series of additional presumed eukaryotic signature proteins (ESPs) in different established, or inferred, archaeal phyla (Fig. 2). For example, orthologs of the eukaryotic ESCRT-III membrane remodeling system have been found to be part of the Cdv cell division machinery in crenarchaeal species18-20 and, in addition, several eukaryotic transcription machinery components (Rpc34,21 RpoG22 and Elf123) have been identified in crenarchaeal, korarchaeal and thaumarchaeal genomes (Fig. 2). More recently, an analysis of the ‘Candidatus Caldiarchaeaum subterraneum’ composite genome revealed the presence of a presumably fully functional ubiquitin-like protein modifier system.5 Clearly, further exploration of uncharacterized deep archaeal lineages may well reveal additional eukaryotic-like features, and thus provide new insights into events that may have been fundamental for the origin and early evolution of the eukaryotic cell.
Figure 2.
Eukaryotic signature genes in archaeal phyla. Schematic overview of established, as well as inferred, archaeal phyla, with Crenarchaeota, Thaumarchaeota, Korarchaeota and the recently proposed Aigarchaeota forming a monophyletic group putatively including eukaryotes (red font), with the bacterial root depicted as an arrow. Comparative analysis of archaeal genomes24 has revealed the presence of eukaryotic signature proteins within these phyla, as indicated by the shading pattern: red, actin; blue, ubiquitin-type protein modifier system; yellow, Cdv (ESCRT-III-like) cell division machinery; purple, eukaryotic RNA polymerase-subunit RPB8 (RpoG); orange, eukaryotic RNA polymerase III subunit RPC34; green, eukaryotic transcription elongation factor Elf1; turquoise, eukaryotic-type topoisomerase 1B.25 Open (white) circles indicate absence of a given ortholog. Note that actin and Cdv proteins are present in a subset of crenarchaeal genomes, displaying an anti-correlated phylogenetic distribution, as indicated by the partly shaded circles.
Acknowledgments
This work was supported by grants 621–2010–5551 and 621–2009–4813 from the Swedish Research Council to RB and TJGE, respectively, a Strong Research Environment grant (Uppsala Microbiomics Center) from the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning to RB, and by an ERG grant (European Union) to TJGE.
Footnotes
Previously published online: www.landesbioscience.com/journals/cib/article/16974
References
- 1.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–12. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yutin N, Wolf MY, Wolf YI, Koonin EV. The origins of phagocytosis and eukaryogenesis. Biol Direct. 2009;4:9. doi: 10.1186/1745-6150-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ettema TJ, Lindås AC, Bernander R. An actin-based cytoskeleton in archaea. Mol Microbiol. 2011;80:1052–61. doi: 10.1111/j.1365-2958.2011.07635.x. [DOI] [PubMed] [Google Scholar]
- 4.Vats P, Rothfield L. Duplication and segregation of the actin (MreB) cytoskeleton during the prokaryotic cell cycle. Proc Natl Acad Sci USA. 2007;104:17795–800. doi: 10.1073/pnas.0708739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nunoura T, Takaki Y, Kakuta J, Nishi S, Sugahara J, Kazama H, et al. Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res. 2011;39:3204–23. doi: 10.1093/nar/gkq1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox CJ, Foster PG, Hirt RP, Harris SR, Embley TM. The archaebacterial origin of eukaryotes. Proc Natl Acad Sci USA. 2008;105:20356–61. doi: 10.1073/pnas.0810647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foster PG, Cox CJ, Embley TM. The primary divisions of life: a phylogenomic approach employing composition-heterogeneous methods. Philos Trans R Soc Lond B Biol Sci. 2009;364:2197–207. doi: 10.1098/rstb.2009.0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lake JA, Henderson E, Oakes M, Clark MW. Eocytes: a new ribosome structure indicates a kingdom with a close relationship to eukaryotes. Proc Natl Acad Sci USA. 1984;81:3786–90. doi: 10.1073/pnas.81.12.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poole AM, Penny D. Evaluating hypotheses for the origin of eukaryotes. Bioessays. 2007;29:74–84. doi: 10.1002/bies.20516. [DOI] [PubMed] [Google Scholar]
- 10.Cavalier-Smith T. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int J Syst Evol Microbiol. 2002;52:297–354. doi: 10.1099/00207713-52-2-297. [DOI] [PubMed] [Google Scholar]
- 11.Amo T, Paje ML, Inagaki A, Ezaki S, Atomi H, Imanaka T. Pyrobaculum calidifontis sp. nov., a novel hyperthermophilic archaeon that grows in atmospheric air. Archaea. 2002;1:113–21. doi: 10.1155/2002/616075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Itoh T, Suzuki K, Sanchez PC, Nakase T. Caldivirga maquilingensis gen. nov., sp. nov., a new genus of rod-shaped crenarchaeote isolated from a hot spring in the Philippines. Int J Syst Bacteriol. 1999;49:1157–63. doi: 10.1099/00207713-49-3-1157. [DOI] [PubMed] [Google Scholar]
- 13.Paredez AR, Assaf ZJ, Sept D, Timofejeva L, Dawson SC, Wang CJ, et al. An actin cytoskeleton with evolutionarily conserved functions in the absence of canonical actin-binding proteins. Proc Natl Acad Sci USA. 2011;108:6151–6. doi: 10.1073/pnas.1018593108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bettinger BT, Gilbert DM, Amberg DC. Actin up in the nucleus. Nat Rev Mol Cell Biol. 2004;5:410–5. doi: 10.1038/nrm1370. [DOI] [PubMed] [Google Scholar]
- 15.Blessing CA, Ugrinova GT, Goodson HV. Actin and ARPs: action in the nucleus. Trends Cell Biol. 2004;14:435–42. doi: 10.1016/j.tcb.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 16.Goodson HV, Hawse WF. Molecular evolution of the actin family. J Cell Sci. 2002;115:2619–22. doi: 10.1242/jcs.115.13.2619. [DOI] [PubMed] [Google Scholar]
- 17.Dion V, Shimada K, Gasser SM. Actin-related proteins in the nucleus: life beyond chromatin remodelers. Curr Opin Cell Biol. 2010;22:383–91. doi: 10.1016/j.ceb.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 18.Ettema TJ, Bernander R. Cell division and the ESCRT complex: A surprise from the archaea. Commun Integr Biol. 2009;2:86–8. doi: 10.4161/cib.7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindås AC, Karlsson EA, Lindgren MT, Ettema TJ, Bernander R. A unique cell division machinery in the Archaea. Proc Natl Acad Sci USA. 2008;105:18942–6. doi: 10.1073/pnas.0809467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT system in cell division in archaea. Science. 2008;322:1710–3. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blombach F, Makarova KS, Marrero J, Siebers B, Koonin EV, van der Oost J. Identification of an ortholog of the eukaryotic RNA polymerase III subunit RPC34 in Crenarchaeota and Thaumarchaeota suggests specialization of RNA polymerases for coding and non-coding RNAs in Archaea. Biol Direct. 2009;4:39. doi: 10.1186/1745-6150-4-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koonin EV, Makarova KS, Elkins JG. Orthologs of the small RPB8 subunit of the eukaryotic RNA polymerases are conserved in hyperthermophilic Crenarchaeota and “Korarchaeota”. Biol Direct. 2007;2:38. doi: 10.1186/1745-6150-2-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daniels JP, Kelly S, Wickstead B, Gull K. Identification of a crenarchaeal orthologue of Elf1: implications for chromatin and transcription in Archaea. Biol Direct. 2009;4:24. doi: 10.1186/1745-6150-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ettema TJ, de Vos WM, van der Oost J. Discovering novel biology by in silico archaeology. Nat Rev Microbiol. 2005;3:859–69. doi: 10.1038/nrmicro1268. [DOI] [PubMed] [Google Scholar]
- 25.Brochier-Armanet C, Gribaldo S, Forterre P. A DNA topoisomerase IB in Thaumarchaeota testifies for the presence of this enzyme in the last common ancestor of Archaea and Eucarya. Biol Direct. 2008;3:54. doi: 10.1186/1745-6150-3-54. [DOI] [PMC free article] [PubMed] [Google Scholar]