Abstract
Spontaneous plasmid transformation of Escherichia coli is independent of the DNA uptake machinery for single-stranded DNA (ssDNA) entry. The one-hit kinetic pattern of plasmid transformation indicates that double-stranded DNA (dsDNA) enters E. coli cells on agar plates. However, DNA uptake and transformation regulation remain unclear in this new type of plasmid transformation. In this study, we developed our previous plasmid transformation system and induced competence at early stationary phase. Despite of inoculum size, the development of competence was determined by optical cell density. DNase I interruption experiment showed that DNA was taken up exponentially within the initial 2 minutes and most transforming DNA entered E. coli cells within 10 minutes on LB-agar plates. A half-order kinetics between recipient cells and transformants was identified when cell density was high on plates. To determine whether the stationary phase master regulator RpoS plays roles in plasmid transformation, we investigated the effects of inactivating and over-expressing its encoding gene rpoS on plasmid transformation. The inactivation of rpoS systematically reduced transformation frequency, while over-expressing rpoS increased plasmid transformation. Normally, RpoS recognizes promoters by its lysine 173 (K173). We found that the K173E mutation caused RpoS unable to promote plasmid transformation, further confirming a role of RpoS in regulating plasmid transformation. In classical transformation, DNA was transferred across membranes by DNA uptake proteins and integrated by DNA processing proteins. At stationary growth phase, RpoS regulates some genes encoding membrane/periplasmic proteins and DNA processing proteins. We quantified transcription of 22 of them and found that transcription of only 4 genes (osmC, yqjC, ygiW and ugpC) encoding membrane/periplasmic proteins showed significant differential expression when wildtype RpoS and RpoSK173E mutant were expressed. Further investigation showed that inactivation of any one of these genes did not significantly reduce transformation, suggesting that RpoS may regulate plasmid transformation through other/multiple target genes.
Introduction
Gene transfer through plasmid conjugation or transformation is one of the major cause of antibiotic resistance in bacteria [1]. The emergency of many life killing superbugs, such as NDM-1 bacteria and enterohaemorrhagic Escherichia coli (EHEC), is often a consequence of the transfer of antibiotic resistance genes mediated by plasmids [2], [3]. Although plasmid conjugation was found in E. coli long ago, this species has traditionally been considered not to be naturally transformable because it is transformable only after special treatments (i.e. electric shock or Ca2+ stimulation and heat shock) [4]. While a complete set of competence gene homologs for the assembly of a conserved DNA uptake machinery were found in the genome of E. coli [5]. Moreover, transcription of some competence genes is inducible by a competence regulator homolog Sxy in E. coli [6]. Our work, together with the work from several other groups, showed that E. coli is able to acquire naked plasmid DNA on agar plates at 37°C without the addition of Ca2+ or heat shock [7], [8], [9]. Our further investigation revealed that plasmid transformation on plates is promoted by agar/agarose, a stimulation that is unrelated to divalent cations like Ca2+ , Mg2+ and Mn2+ [10]. Interestingly, none of the DNA uptake gene homologs were found to be involved in mediating spontaneous plasmid transformation of E. coli [10]. The dose-response curve of transformation frequency as a function of DNA concentration showed that E. coli cells acquired plasmid DNA with a single hit kinetics, suggesting that plasmid DNA enters E. coli cells through a different route which allows double-stranded DNA (dsDNA) entry [10]. Entry of dsDNA in E. coli is different from that in other naturally transformable bacteria, which often use the DNA uptake machinery for single-stranded DNA (ssDNA) uptake [11] and DNA binding proteins for processing and integrating the incoming ssDNA [12]. For example, in plasmid transformation of Streptococcus pneumoniae, two strands of ssDNA from two plasmid molecules are taken up into the cytoplasm to re-establish a new plasmid with the assistance from recombinase RecA in the cytoplasm [13].
Our previous work and others showed that E. coli develops competence for spontaneous plasmid DNA uptake at stationary phase [8], [9]. RpoS is an alternative sigma factor which is induced at stationary phase or under conditions of starvation or stress (e.g. temperature, osmolarity or pH) [14], [15]. Whole-genome microarray data reveal that more than 480 genes are potentially regulated by RpoS under different stress conditions [16], [17], [18], [19]. At 37°C, RpoS is degraded by the protease in the exponential growth phase but protected from protease degradation at stationary phase [14], [15]. When E. coli was incubated at a temperature lower than 30°C, RpoS begins accumulating at the exponential phase because its translation is highly promoted by a small RNA DsrA [20]. It remains unknown whether RpoS, the stationary phase master regulator, plays any roles in plasmid transformation of E. coli.
In this study, we further documented natural competence for plasmid transformation in a developed plasmid transformation system. With this transformation system, we first studied the development of competence and examined the effect of inoculum sizes on plasmid transformation. Next, we investigated DNA uptake kinetics and the effect of cell density on plasmid transformation on LB-agar plates. The potential involvement of RpoS in spontaneous plasmid transformation was examined by gene inactivation and complementation, in addition to the effect of a single amino acid in RpoS, lysine 173, which is thought to be important to RpoS selectivity. Finally, to search for new candidates which may be involved in spontaneous plasmid transformation of E. coli, we compared transcription patterns of a series of RpoS-regulated genes in rpoS+ and rpoS− strains through Real-Time PCR (RT-PCR) and examined their potential roles in plasmid transformation.
Results
1. The development of competence for plasmid transformation
Spontaneous plasmid transformation on agar plates has been documented at 37°C [8], [9], [10]. In our previous study, we established a novel transformation system to show that E. coli is naturally transformable by treating cells with static culture in a beaker [8], [10]. To further explore spontaneous plasmid transformation in E. coli, we developed the plasmid transformation system at 30°C, a common temperature in our environment, and static culture was omitted. This transformation system did not require heat shock or the addition of Ca2+. In this developed transformation system, we investigated competence development in wild-type E. coli K-12 strains MC4100 (kindly donated by Dr. Regine Hengge-Aronis) and BW25113 [21] and their derivatives.
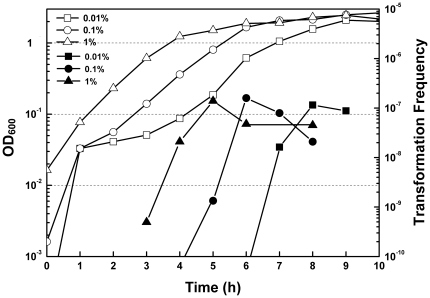
To know competence development during cell growth, we examined transformation patterns as a function of culture time with different inoculum sizes. To prepare recipient cells for plasmid transformation, overnight grown culture in LB broth was inoculated to 100 ml of 1.5× LB (containing yeast extract 7.5 g/L, tryptone 15 g/L and NaCl 7.5 g/L) with a ratio of 1∶100, 1∶1, 000 and 1∶ 10, 000 followed by incubation at 30°C with a low speed (150 rpm). At intervals, 500 µl of the culture was recovered by centrifugation and 450 µl of the supernatant was discarded. Cell pellets were resuspended in the remaining 50 µl supernatant with the addition of plasmid DNA. Transformation was performed by plating the above mixture onto selective plates containing 5% agar. We observed that few exponentially growing E. coli cells were transformed despite of the inoculum sizes (Figure 1). This is in contrast to artificial transformation which requires exponentially growing recipient cells. Instead, competence began to develop at the transition of exponential phase to stationary phase, reached the maximum at the entry of stationary phase and then moderately decreased (Figure 1). Competence development with three different inoculum sizes consistently showed that the highest transformation frequency (1∼2×10−7) occurred when the optical density at 600 nanometer (OD600) reached ∼1.5, no matter how long the cells were cultured (Figure 1). In the following experiments, we investigated plasmid transformation when cells grew to an OD600 of ∼1.5 unless otherwise indicated.
Figure 1. Effect of inoculum size on plasmid transformation.
Overnight grown (13 hr) E. coli MC4100 was inoculated into three triangle flasks each containing 100 ml of fresh LB at ratios of 1∶100 (triangle), 1∶1, 000 (circle) and 1∶100, 000 (rectangle). During incubation, OD600 of the cultures was measured periodically (open symbols). Transformation was performed at intervals as described in Materials and Methods and transformation frequencies were shown (solid symbols). Representative data from three independent experiments were shown.
2. The kinetics of DNA uptake during plasmid transformation on plates
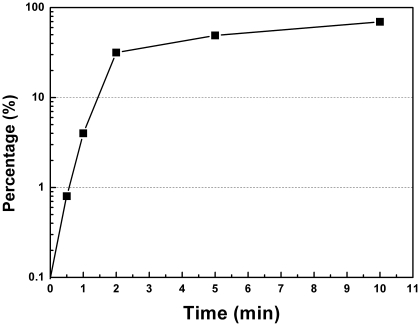
Our previous study and the others showed that plasmid transformation of E. coli occurred on agar plates at 37°C [7], [8], [9]. To test whether DNA uptake occurred prior to plating, excessive DNase I was added after co-incubation of cells from the culture of OD600 of 1.5 and pDsRED in the liquid culture. For both MC4100 and BW25113, no transformants were detected (Table 1). To know when DNA entered competent cells on plates, immediately after plating the mixture of E. coli culture and plasmid DNA, excessive DNase I was spread on the LB-agar plates. No transformants were detected in repeated experiments (n>3). This result indicates that plasmid transformation should occur on LB-agar plates after plating. To explore the kinetics of DNA uptake on agar plates, following the plating of the mixture of competent cells and plasmid DNA, we interrupted DNA uptake by spreading excessive DNase I on LB-agar plates at intervals. We found that DNA was taken up exponentially after spreading competent cells and plasmid DNA on the surface of LB-agar plates. At the start time point (0 minute), no transformants were detected (Figure 2), implying that DNA had not entered cells at that time. Within the first 2 minutes, more than one third of the competent cells acquired plasmid DNA which was not sensitive to DNase I degradation (Figure 2). Five minutes later, the addition of DNase I reduced transformation frequency less than a half (Figure 2). Within the initial 10 minutes on agar plates, 71.9% of the transforming plasmid DNA entered the DNase I insensitive state (Figure 2). The rapid uptake of DNA implies that the route for dsDNA entry might be assembled in the liquid culture before plating but quickly activated on agar plates. Alternatively, stresses introduced by exposure of planktonic cells on plates may contribute to DNA entry. We investigated potential effects of physical stress by plating and oxidative/anti-oxidative stress on plasmid transformation and failed to detect obvious change of transformation frequency by these stresses (see Discussion S1).
Table 1. Effect of DNase I in liquid culture before plating.
| 10 min | 20 min | |||
| +DNase I | −DNase I | +DNase I | −DNase I | |
| MC4100 Tf* | 0 | 1.0×10−6±1.4×10−7 | 0 | 5.6×10−7±9.3×10−8 |
| BW25113 Tf | 0 | 1.7×10−6±5.2×10−8 | 0 | 3.5×10−6±2.0×10−7 |
Tf: Transformation frequency.
Figure 2. Kinetics of DNA entry on LB-agar plates.
E. coli MC4100 cell pellets from the culture at OD600 of 1.5 were resuspended in the supernatant and mixed with plasmid pDsRED at a concentration of 67 µg/ml. The liquid mixture was plated on selective LB-agar plates (50 µl per plate). At intervals, excessive DNase I was spread on one of these plates. The sample without DNase I treatment was set as the control. Percentages of remaining transformants (which indicates DNA uptake kinetics) were calculated by dividing the number of transformants with DNase I treatment by the number of transformants without DNase I treatment. Representative data from three independent experiments were shown.
3. Effect of high cell density on plasmid transformation
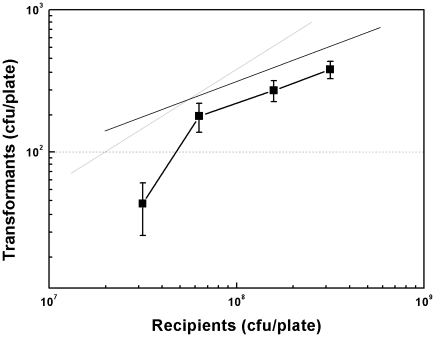
In our previous study, we examined the effect of cell density by serially diluting static culture and observed a non linear relationship between cell density and transformation frequency at low cell density (1.41×105∼3.53×105 CFU/cm2) on medium size plates (diameter of 8.5 cm) [10]. In this study, we investigated the effect of a higher cell density by concentration (with a method described in the Materials and Methods section) and plated competent E. coli cells on smaller agar plates (diameter of 6 cm). When the cell density was concentrated 2-fold, from 3.16×107 CFU/plate (1.12×106 CFU/cm2) to 6.31×107 CFU/plate (2.23×106 CFU/cm2), transformation frequency was increased 4-fold (Figure 3). Surprisingly, when cell density was further increased from 6.31×107 CFU/plate (2.23×106 CFU/cm2) to 3.16×108 CFU/plate (1.12×107 CFU/cm2), a half-order kinetics between recipient cells and transformants was observed (Figure 3).
Figure 3. Effect of cell density on plasmid transformation.
E. coli MC4100 cell pellets from 100 µl, 200 µl, 500 µl and 1 ml of the culture at OD600 of 1.5 were transformed as described in Materials and Methods. The experiment was performed in duplicate. Each point denotes an average of 4 samples. Error bars denote standard deviation. Gray and black lines indicate slopes of 1 and 0.5, respectively.
4. RpoS regulates plasmid transformation
Two independent groups reported spontaneous plasmid transformation at stationary phase [8], [9]. In this study, we again observed that competence was induced at the transition of exponential phase to stationary phase at 30°C (Figure 1). RpoS, an alternative sigma factor [14], [15], activates the transcription of many stress induced genes, including genes encoding membrane and periplasmic proteins and DNA processing genes [16], [17], [18], [19]. In the classical transformation model, membrane proteins and DNA processing proteins are required for DNA uptake and integration [11]. As plasmid transformation of E. coli does not require components of the machinery for classical DNA uptake [9], [10], we questioned whether a new set of DNA uptake and processing genes are involved in mediating plasmid dsDNA transfer and if the stationary phase master regulator RpoS regulates transcription of some components of this set of genes. We first investigated the latter possibility. To check whether RpoS was involved, we compared transformation frequencies between MC4100 and its rpoS mutant derivative FS20 (Table 2). The growth curve patterns of the two strains were almost identical (Figure 4A). Neither did we observe that the inactivation of rpoS had obvious effect on viability during cell growth (data not shown). However, transformation frequency of MC4100 was more than 10-fold higher than FS20 after 4 hours of incubation (OD600, ∼1.0) (Figure 4A). After 5, 6 and 8 hours of incubation, transformation frequencies of MC4100 were systematically more than 3-fold of that of FS20 (Figure 4A). The effect of rpoS on plasmid transformation of E. coli was more evident by over-expressing this gene in an rpoS mutant FS20. At all examined time points (during 3 to 8 hours of incubation), compared with FS20 carrying an empty vector, transformation frequencies were systematically increased ∼10 fold by over-expressing rpoS with pSURpoS (Figure 4B), a plasmid which has ∼10 copies per cell. Additionally, over-expressing rpoS on the plasmid pSURpoS also increased transformation frequency 2.33 to 5.73 folds during the growth of the wild type strain MC4100 (Figure 4C).These data show that rpoS is involved in promotion of plasmid transformation of E. coli.
Table 2. Strains and plasmids used in this study.
| E. coli strain or plasmid | Relevant genotypea and/or description | Source or reference |
| E. coli strains | ||
| ZK126 | W3110 ΔlacU169 tna-2 | [34] |
| MC4100 | F- λ- araD139 Δ(argF-lac)U169 rpsL150 relA deoC1 ptsF25 rbsR flbB5301 | Regine Hengge-Aronis |
| FS20 | MC4100::ΔrpoS::kan; Kanr | Regine Hengge-Aronis |
| FS20-pSU | FS20 containing pSU, Cmr | This study |
| FS20-pSURpoS | FS20 containing pSURpoS, Cmr | This study |
| FS20-pSURpoSK173E | FS20 containing pSURpoSK173E, Cmr | This study |
| MC4100-pSU | FS20 containing pSU, Cmr | This study |
| MC4100-pSURpoS | FS20 containing pSURpoS, Cmr | This study |
| MC4100-pSURpoSK173E | FS20 containing pSURpoSK173E, Cmr | This study |
| BW25113 | F- λ- Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) rph-1 hsdR514 Δ(rhaD-rhaB)568 | [21] |
| JW1477 | BW25113::ΔosmC::kan | [21] |
| JW2992 | BW25113::ΔygiW::kan | [21] |
| JW5516 | BW25113::ΔyqjC::kan | [21] |
| JW3415 | BW25113::ΔugpC::kan | [21] |
| JW5437 | BW25113::ΔrpoS::kan | [21] |
| Plasmids | ||
| pCHAP3100 | p15A replicon; pSU18 carrying ppdD; Cmr | [35] |
| pDsRED | pUC replicon; red fluorescence protein-expressing plasmid; Ampr | [8] |
| pUCRpoS | pUC18 derivative; carrying an rpoS gene; Ampr | This study |
| pSURpoS | pCHAP3100 derivative; carrying an rpoS gene; Cmr | This study |
| pSU | pSURpoS with rpoS deletion; Cmr | This study |
| pSURpoSK173E | pSURpoS with K173E mutation; Cmr | This study |
Cmr, chloramphenicol resistance; Ampr, ampicillin resistance; Kanr, kanamycin resistance.
Figure 4. Plasmid transformation regulated by RpoS.
During 3∼8 hours of incubation at 30°C, at intervals, transformation frequencies (solid symbols) and optimal densities of the cultures (open symbols) of MC4100 (rpoS+) and FS20 (rpoS−) (A); FS20-pSU, FS20-pSURpoS, FS20-pSURpoSK173E (B); and MC4100-pSU, MC4100-pSURpoS, MC4100-pSURpoSK173E (C) were measured. Transformation was performed as described in Materials and Methods. Each point denotes an average of 4 samples. Error bars denote standard deviation. * P value ≤ 0.05; ** P value ≤0.01.
It has been documented that RpoS recognizes promoters through K173 and the mutation of K173 to glutamic acid 173 (K173E) makes RpoS unable to initiate the transcription of targeted genes [22]. To further confirm a role of RpoS on plasmid transformation, we examined the effect of K173E mutation on plasmid transformation. We found that transformation frequencies of FS20-pSURpoSK173E were ∼10-fold lower than that of FS20-pSURpoS and almost identical to that of FS20 carrying an empty vector at all examined time points during cell growth (Figure 4B). Interestingly, over-expressing RpoSK173E reduced the transformation frequency of MC4100 to a level similar to that of FS20 (Figure 4C), showing a dominant effect of RpoSK173E on plasmid transformation. By contrast, the dominant effect of RpoSK173E on plasmid transformation of FS20 was not obvious under such conditions (Figure 4B). Together, we conclude that RpoS is involved in regulating plasmid transformation of E. coli.
5. RpoS-regulated genes and plasmid transformation
Microarray data have shown that expression of a number of genes were activated by RpoS at stationary phase [16], [17], [18], [19]. To accomplish transformation, exogenous plasmid DNA needs to penetrate two membranes and be protected for plasmid establishment in cytoplasm [11], [23]. To search for RpoS regulated genes which play roles in plasmid transformation of E. coli, we quantified the expression of 22 of RpoS-regulated genes which are induced at stationary phase with RT-PCR. Among the 22 genes, products of 5 genes are DNA processing proteins and the other 17 genes encode membrane/periplasmic proteins. Except for potF and osmB, transcription of all the examined genes was lower in FS20-pSU than that in FS20-pSURpoS (Figure 5). Interestingly, transcription of most of these genes was not significantly reduced by K173E mutation (Figure 5). As compared to FS20-pSURpoS, transcripts of all the 5 genes for DNA processing were not significantly reduced (<2-fold or P value>0.05) in FS20-pSURpoSK173E (Figure 5). Whereas among 17 genes encoding membrane/periplasmic associated proteins, the transcription of ugpC was more than 10-fold decreased by the mutation of K173E (Figure 5). K173E mutation also reduced the transcription of stress-inducible membrane protein encoding gene osmC and outer membrane encoding gene yqjC by 2.5 and 2.34 folds respectively (Figure 5). Besides, the transcription of ygiW, which is predicted to encode a periplasmic protein, was reduced 3.83-fold by K173E mutation (Figure 5). The transcript quantification data of RpoS-regulated gene suggest that RpoS may regulate plasmid transformation by controlling membrane/periplasmic proteins which could mediate dsDNA transfer across membranes.
Figure 5. Gene expression patterns in FS20 derivatives.
The expression of putative RpoS regulated genes was quantified in strains FS20-pSU (open column), FS20-pSURpoS (solid column) and FS20-pSURpoSK173E (shaded column) with Real-Time PCR with 16sRNA as a reference. Primers and names of the targeted genes were listed in a table below. Each column denotes an average of 2 samples. Error bars denote standard deviation.
To further screen out transformation related genes, we measured transformation frequencies in E. coli strains defective in RpoS-regulated genes whose transcription was reduced by both rpoS null mutation and K173E mutation (P value ≤0.05) by more than 2 folds. Four genes (osmC, yqjC, ygiW and ugpC) met the above criteria. We investigated their potential involvement in plasmid transformation of E. coli K-12 BW25113 and its mutant derivatives from Keio collection [21]. Construction of these mutants, JW1477 (osmC), JW2992 (ygiW), JW5516 (yqjC) and JW3415 (ugpC), was confirmed by PCR (see Figure S1 and Table S2). Growth curve patterns of the four mutants were similar to the wild type and rpoS mutant (Figure 6). Transformation of all the four mutants did not show significant difference with respect to the wild type while the rpoS mutant showed a noticeable transformation defect (Figure 6). The data show that single gene inactivation of osmC, yqjC, ygiW or ugpC did not affect transformation efficiency.
Figure 6. RpoS regulated genes and plasmid transformation.
BW25113 and its mutant derivatives were transformed as described in Materials and Methods. Cell growths were measured by monitoring OD600 for 8 hours. Each point denotes an average of 2 samples. Error bars denote standard deviation.
Discussion
In this study, we investigated several basic questions in spontaneous plasmid transformation of E. coli on agar plates, which involves competence development and regulation and dsDNA transfer across two cell membranes. First, we observe that competence for plasmid transformation was induced at the beginning of stationary phase in nutrient rich medium under standard culture conditions, and that despite of the inoculum size, the induction of competence was a response to optical cell density rather than the incubation time (Figure 1). Second, the DNA uptake kinetics on agar plates shows that competent cells acquire exogenous plasmid DNA in a short time (Figure 2), suggesting the presence of a presetting DNA entry route in competent cells in liquid culture. Third, the half-order kinetics between recipient cells and transformants is detected when cell density is high on plates (Figure 3). Fourth, effects of rpoS inactivation and over-expression on plasmid transformation show that RpoS is involved in transformation regulation (Figure 4). Additionally, based on the transcription quantification data of RpoS-regulated genes (Figure 5), we found that transcription of 4 genes enconding membrane/periplasmic proteins was reduced by RpoS K173 mutation but inactivation of any one of them did not significantly affect plasmid transformation (Figure 6). Parameters analyzed in this and our previous studies on the plasmid DNA transfer of E. coli were summarized in Table S1.
1. The effect of inoculum size on plasmid transformation
With different inoculum sizes, the induction of competence for plasmid transformation of E. coli was dependent on optical cell density rather than the incubation time (Figure 1). The case in E. coli is different from that in Streptococcus pneumoniae, whose competence development is determined by the incubation time rather than cell density when the inoculum size varies [24]. The different responses to inoculum sizes in the two species are not surprising. First, S. pneumoniae acquires exogenous DNA through a set of proteins that are not functional in plasmid transformation of E. coli and therefore the regulation targets should be different in the two species [10]. Second, competence induction conditions vary largely among species even for the same set of DNA uptake machinery [24], [25], [26], [27]. Different species have evolved their own way to control the development of competence. For E. coli, it seems that this bacterium has evolved its own way to control competence for a different plasmid transformation mode by cell density.
2. DNA uptake on plates and transformation stimulator(s) in agar
To know the kinetics of DNA uptake on agar plates, we interrupted DNA uptake by plating excessive DNase I. The DNase I interruption experiment showed that the first plasmid DNA molecule should be taken up within 30 seconds and that more than one third of plasmid DNA was taken up and protected from DNase I degradation within 2 minutes after plating (Figure 2). The rapid DNA uptake on plates led us think whether a sudden oxidative stress induced DNA uptake or the physical stress during spreading pushed DNA into cells. To test the first possibility, we examined the effect of oxidative reagent H2O2 and anti-oxidant reagents Na2SO3 and NaHSO3 on plasmid transformation. However, we did not detect obvious effects of these oxidant and anti-oxidant reagents on transformation (see Figure S2, S3). To test whether the quick DNA uptake is an artifactual phenomenon triggered by spreading, we compared spreading of the cells on agar surface with a spreader and beads, and did not observe an obvious effect of spreading manners on transformation (see Discussion S1). If DNA uptake is mediated by a specific protein device which is synthesized after plating, such a short time would not be enough for de novo synthesis of a complete set of proteins for DNA uptake apparatus. More likely, the putative DNA uptake apparatus or its components might have been expressed in the liquid culture before activation by certain stimulators (e.g. divalent cations) or molecules available in a short time inside of cells (e.g. small RNA) on agar plates. Our previous work showed that plasmid transformation of E. coli is stimulated by agar/agarose on plates [10]. Although divalent cations chelator EGTA inhibits transformation stimulation by agar/agarose, Ca2+, Fe2+, Mg2+, Mn2+ or Zn2+ in agar was not identified as the transformation stimulator [10]. It remains unknown whether the transformation stimulator triggers DNA uptake within the initial several minutes or plasmid establishment in later stages.
3. Cell density and plasmid transformation
Low cell density: In both this work and our previous study, we observed that the number of transformants increased more rapid than the number of recipient cells at relative low cell density on plates (Fig. 3) [10]. A recent report indicates that cell-to-cell transformation in E. coli may be regulated by a pheromone [28]. In S. pneumoniae, it has been well established that a quorum sensing peptide regulates natural transformation through classical DNA uptake machinery [29], [30]. We tentatively propose that the increase of cell density may help accumulation of a factor that regulates plasmid transformation of E. coli on plates through an unknown mechanism.
High cell density: When cell density was relative high, the increase of the number of transformant was not quicker than the number of recipient cells (Figure 3). It is likely that when the accumulation of the putative competence factor reaches a certain concentration, its effect on plasmid transformation reaches the maximum. Based on the half-order kinetics as a function of cell density (Figure 3), we envision the possibility that cell-to-cell interaction could play a role in the formation of transformants on agar plates. The case would not be unprecedented. For example, a channel called ‘nanotube’, which connects two cells together, has been found to mediate E. coli cell-to-cell plasmid transfer on the solid surface [31], [32].
4. Regulation of plasmid transformation by RpoS
Involvement of RpoS in plasmid transformation: When E. coli cells enter the stationary growth phase, RpoS is highly expressed and many stationary phase genes are switched on [14], [15]. Our previous study showed that plasmid transformation rate is increased remarkably by statically incubating stationary phase E. coli in an open system [8]. Such treatment may help accumulate more RpoS. Below 30°C, the expression of RpoS is promoted by small RNA DsrA which starts accumulating at exponential phase. In the present work, we detected competence at the transition of exponential to stationary phase at 30°C. These phenomena suggest that the accumulation of RpoS may associate with the induction of competence for plasmid transformation. In this study, we confirmed a role of RpoS in spontaneous plasmid transformation of E. coli. Transformation frequency is reduced by inactivating rpoS and over-expressing rpoS remarkably increased transformation (Figure 4). Moreover, the K173E mutation, which alters RpoS selectivity, caused RpoS unable to promote plasmid transformation (Figure 4B), confirming both the importance of K173 to RpoS activity which had been reported [22] and a role of RpoS in plasmid transformation. K173E mutation also reduced transformation frequency in the wild type cell. It is possible that RpoSK173E is able to compete with the active RpoS or other sigma factors (e.g. rpoD) for RNA polymerase or promoters which are important to the transcription of some genes involved in plasmid transformation. The suppression effect of K173E mutaion on plasmid transformation of the rpoS mutant FS20 was not obvious when cell density was high on agar plates (Figure 4B). However, we repeatedly observed a strong suppression effect of K173E mutation on plasmid transformation of the rpoS mutant with non-concentrated cell cultures on LB-agar plates (n>5, data not shown). It seems that when RpoS is absent, high cell density on LB-agar plates could counteract the suppression effect of K173E mutation on plasmid transformation.
Targets for RpoS regulation? In Vibrio cholerae, RpoS regulates natural competence for transformation through a quorum sensing regulator HapR which controls the transcription of a conserved DNA uptake gene [26]. Because our previous study showed that DNA uptake gene orthologs ppdD, hofQ, gspD and ycaI, together with a highly conserved transforming ssDNA processing gene dprA/smf, were not required for spontaneous plasmid transformation of E. coli [10], mechanisms of transformation regulation by RpoS and cell density should be different in E. coli and V. cholerae. Considering that dsDNA enters E. coli cells during spontaneous plasmid transformation [10], we envision that RpoS may regulate the transcription of the gene(s) which belong(s) to a new set of DNA uptake genes encoding membrane/periplasmic proteins for the passage of dsDNA and/or DNA processing genes that are required for plasmid establishment in E. coli. By analyzing documented microarray data, we learned that RpoS regulated the expression of at least 35 genes encoding membrane and periplasmic proteins in stationary-phase E. coli cells [16], [17], [18], [19]. Transcription of 22 of these genes was quantified in this work. Interestingly, transcription of most examined RpoS-regulated genes (17 out of 22) was not significantly affected by K173E mutation and of almost all these genes was higher in a strain expressing RpoSK173E than in an rpoS mutant (Figure 5). It is possible that different promoter elements of a specific gene and their differential sensitivity to complex factors in vivo may complicate gene transcriptional regulation [15]. Transcription of all the five genes for DNA processing was not significantly changed by K173E mutation (Figure 5), indicating that RpoS should not regulate plasmid transformation by affecting DNA processing or plasmid establishment. By contrast, transcription of ugpC (encoding a glycerol ATP-binding transporter) was more than 10-fold reduced by K173E mutation (Figure 5). The other three genes, whose transcription was also significantly affected by K173E mutation (Figure 5), include ygiW and yqjC which potentially encode an outer membrane protein and a periplasmic protein respectively and an osmotically inducible gene osmC which encodes a resistance protein. It seems that RpoS-regulated genes encoding membrane/periplasmic proteins are more prone to be sensitive to K173E mutation. Unfortunately, our further function examination showed that single gene inactivation of ugpC, ygiW, yqjC or osmC did not significantly reduce plasmid transformation frequency (Figure 6). Possibly, RpoS regulates plasmid transformation by targeting on multiple genes which could contribute synergistically to this process. The decrease of plasmid transformation by K173E mutation may reveal that the elements in the promoters of these genes could be quite sensitive to K173 position of RpoS. The high sensitivity of these elements might help screen out genes involved in plasmid transformation of E. coli. By comparing global transcription profiles of competent E. coli cells expressing RpoS and RpoSK173E, together with their counterparts not expressing RpoS as a control, the scope of candidate genes for this new type of plasmid DNA transfer could be defined in further investigations.
5. RpoS, a potential target for controlling gene transfer?
Gene transfer often brings about new intractable pathogens, which are highly virulent and/or resistant to antibiotics. In this study, we have shown that RpoS is able to regulate gene transfer mediated by plasmid in E. coli. RpoS can also regulate gene transfer through chromosomal DNA transformation in V. cholerae [26]. Providing that RpoS is able to regulate different types of gene transfer, it is conceivable to design strategies to control gene transfer by targeting on it. For example, compounds could be designed to block K173 of RpoS or K173 mutation could be applied in the construction of bacterial vaccine strains to prevent virulent/resistant gene transfer. Besides, a group of small RNAs which regulate RpoS translation [15], could be good candidates for direct evaluation of their potential pharmaceutical effects on preventing bacterial gene transfer.
Materials and Methods
1. Bacterial strains, primers and transformation of E. coli on plates
All of the strains and plasmids used in the present study are listed in Table 2. Primers used for cloning and Real-Time PCR (RT-PCR) are listed in Table 3. Plasmid transformation was carried out by using a developed procedure that was previously described and beads were used to spread cells on plates unless otherwise specified [8], [10]. All experiments were performed at 30°C. To prepare a pre-culture, 60 µl of the culture stock from −80°C was inoculated into 3 ml of LB broth in a glass tube and grown overnight (13∼16 hours) to a cell density of OD600 of ∼1.5. To prepare competent cells, unless specified otherwise, 1 ml of the pre-culture was inoculated into 100 ml of 1.5× LB broth (prepared by resolving 7.5 g yeast extract, 15 g tryptone and 7.5 g NaCl in 1 liter H2O) in a triangle glass flask. The culture was horizontally shaken at a speed of 150 rotations per minute. Cell growth was measured by recording the optical cell density at OD600. At intervals, 500 µl of the culture was precipitated and 450 µl of the supernatant was discarded. The cell pellet was resuspended in the remaining 50 µl of the supernatant and 1.5∼3.5 µg of plasmid pDsRED (final concentration, 30∼70 µg ml−1) was added. The mixture was immediately (<1 min) spread with glass beads onto LB plates (diameter, 6 cm) which contained 5% agar and 200 µg/ml of ampicillin, which had been air-dried at 37°C for 24∼48 hours. Transformation frequency was calculated by dividing the number of transformants by viable counts.
Table 3. Primers used in this study.
| Name | Primer Forward | Primer Reverse | Reference |
| rpoS | cTCTAGAtggtccgggaacaacaagaagtta * | cGGGATCccttaattacctgtgtgcgtat | [36], [37] |
| rpoS K173E | cgattcacatcgtaGaggagctgaacgtt & | aacgttcagctcctCtacgatgtgaatcg | [38] |
| dps | tgaaaagttacccgctggac | ggtcaggatatctgcggtgt | This study |
| dinB | ggctgtatccggaacttgaa | ggtggtttgctgaaaatcgt | This study |
| aidB | tgttgcgcgttctcaataag | tgctgctgtaaacgacgaac | This study |
| cbpA | ggcgatctgtatgcggtact | tttcccccaatctttacgtg | This study |
| potF | aattaatggccgggagtacc | agttccgggtcgagattctt | This study |
| bolA | cggctctgaaagccatttta | cagcgcatgaacggtagtag | This study |
| osmE | cctgtggtgaaagacgtcaa | tcacgttgacccaggatgta | This study |
| osmY | aacacaaaatttgcccgttc | cacgttgtcggctttattga | This study |
| osmC | gggaacagtatccaccgaga | aaagcgccattgagaaacat | This study |
| osmB | cggctgttctggcaattact | agtactgcaccgcctaatgc | This study |
| ybgA | cgctgatgcgtatcaaacac | cctgaatggcgcaagttaat | This study |
| hdeB | cactggtgaacgcacaatct | tttcttcatgcagcatccac | This study |
| yqjC | gtgccggtagttatgccact | gggctttattcagaccgtca | This study |
| slp | gggtgaacaacctggcttta | atgcaccatagccgtaatcc | This study |
| ygiW | tcgttgaacgcatctctgac | cgacttcaccctgaatctcaa | This study |
| yphA | ccaatatatggcctcgttgg | gtgaaaaagccaagcacgat | This study |
| yhiD | ttcgctgattggtttgttga | tcagccactcaatgctgttc | This study |
| yebS | gttactgcggatggtgacct | cagccggtcccatagtaaaa | This study |
| yeaY | gtggatttccgtggacaact | aacccgttacttgcatcacc | This study |
| ydcV | ggcagcacagcgtagtgata | ttttgccgaaaaagtctcgt | This study |
| hdeD | gtttatcgtcgggttgctgt | ttatgactgcggttgctgaa | This study |
| ugpC | ataacgaaggcacgcatttc | cgaatgccgagagtcatttt | This study |
| osmC (CHK) | cctgcttatcctcgtgctgt | taacaacgcatcaggcattc | This study |
| ygiW (CHK) | acgcgctctgtacagcacta | caacatcgccacaggtattg | This study |
| yqjC (CHK) | cagtggctgacccggtta | ccagcgtatcggaaagggat | This study |
| ugpC (CHK) | tgttaacgcttatccctccg | gattgacgccagggtgtttt | This study |
| rpoS (CHK) | ggtccgggaacaacaagaagtta | cggattcttaattacctggtgcgtat | This study |
capital letters in bold indicate the restriction enzyme recognizing sequence;
capital letter in bold with underline indicates the position of nucleoside for mutation.
To investigate the effect of inoculum size on plasmid transformation, 0.01, 0.1 and 1 ml of the overnight grown culture were inoculated to three glass flasks. To investigate the effect of cell density on plasmid transformation, cell pellets from 100 µl, 200 µl, 500 µl and 1000 µl of the culture were resuspended in 100 µl of the supernatant and mixed with 6.6 µg of pDsRED, followed by spreading 50 µl of the mixture onto a selective plate. To know whether E. coli was transformed prior to plating, DNase I (∼2 mg/ml, final concentration) was added to the mixture of the culture and pDsRed which had been co-incubated for 10 minutes, then STOP solution (Promega Corp.) was added after 10 or 20 minutes of DNase I treatment. The control was treated in the same manner except that DNase I was not added. To measure the kinetics of DNA uptake on plates, 0 min, 0.5 min, 1 min, 2 min, 5 min and 10 min after plating the mixture of competent cells, 20 µl of 25 mg/ml DNase I was spread on the plate.
2. Recombinant plasmids and strains construction
A DNA fragment containing the rpoS gene together with its 205 bp upstream and 127 bp downstream nucleotides, was amplified from the E. coli ZK126 genome DNA. Primers for cloning the DNA fragment are listed in Table 3. The purified PCR product was cut by XbaI and BamHI and then ligated to the cloning vector pUC18 to form pUCRpoS. The PCR-derived part in pUCRpoS was sequenced. To construct a plasmid expressing rpoS and compatible with the transformation donor plasmid pDsRED, the fragment with the correct rpoS sequence was cut from pUCRpoS and ligated to pCHAP3100 with blunt ends (digested by EcoRI and HindIII to delete the ppdD gene and then treated with mungbean nuclease) to construct pSURpoS. The empty vector pSU was constructed by deleting the rpoS gene in pSURpoS. The two plasmids, pSURpoS and pSU, were transformed to FS20 and MC4100 to create strains FS20-pSU, FS20-pSURpoS, MC4100-pSU and MC4100-pSURpoS.
3. Site-directed mutagenesis of K173 of RpoS
With plasmid pSURpoS DNA as the template, the whole-length plasmid DNA was amplified by two complementary primers K173E-1 and K173E-2 for mutagenesis (Table 3). The PCR product was digested by DpnI to eliminate template pSURpoS DNA and transformed to E. coli ZK126. The PCR-derived parts in the resulting RpoS mutant plasmids were sequenced to confirm the desired mutation. The plasmid with an rpoS derivative gene encoding a protein with K173E mutation was named pSURpoSK173E, which was transformed into FS20 and MC4100 to create the strain FS20-pSURpoSK173E and MC4100-pSURpoSK173E.
4. RNA isolation and real-time PCR
When cells grew to the optical density of OD600 of 1.5 in 100 ml of 1.5× LB broth, RNA was isolated from FS20-pSU, FS20-pSURpoS and FS20-pSURpoSK173E with a protocol provided by the RNA isolation kit (Tiangen Company) and reverse transcribed to cDNA. RT PCR was performed with the CFX-96 sequence detection system (Bio-Rad, USA). SYBR Green Real-Time PCR Mix (Bio-Rad) was used with a primer concentrations of 50 nM. Primer sequences were listed in Table 3. Reaction conditions were 95°C for 3 minutes, followed by 40 cycles of 95°C for 10 seconds and 60°C for 30 seconds. Duplicate PCRs were run for each cDNA sample. 16SrRNA was selected as the reference against which the starting quantity of RNA was normalized. The threshold cycles were calculated with the Bio-Rad CFX-96 manager software. The relative expression of genes were calculated by the formula 2(ΔCt Taget−ΔCt Reference) [33].
5. Examination of mutants from Keio collection
Control PCR experiments confirmed the loss of wild-type gene fragments in the mutant strains from Keio collection [21] and their replacement by a fragment, the size of which was fully consistent with that predicted from simple insertion of antibiotic-resistance gene cassette (see Figure S1 and Table S2).
Supporting Information
Examination of the structure of E. coli mutants tested in Figure 6 (see accompanying Table S2). Analysis of PCR fragments for confirmation of the structure of E. coli mutant strains (Primers used were listed in Table 2). From left to right, each pair of lanes compares wildtype and mutant structures, as follows. Lanes: 1, 4 and 13, DD Marker 2, ugpC mutant; 3, wide type 5, rpoS mutant; 6, wide type 7, yqjC mutant; 8, wide type 9, ygiW mutant; 10, wide type 11, osmY mutant; 12, wide type.
(TIF)
Effect of H2O2 on plasmid transformation. H2O2 was added either at OD600 = 0.1 (A) or OD600 = 1.5 (B). Plasmid transformation was measured either 4, 5 and 6 hours (A) or 10 minutes (B) after the addition of H2O2. Transformation frequency was calculated by dividing the number of transformants per ml by the number of viable counts per ml. Each sample was performed in duplicate. Error bars denote standard deviation.
(TIF)
Effect of anti-oxidant agents on plasmid transformation. Anti-oxidant agents Na2SO3 or NaHSO3 were added to the liquid culture prior to plating (A) or the solid agar plates for plasmid transformation (B). Transformation frequency was calculated by dividing the number of transformants per ml by the number of viable counts per ml. Each sample was performed in duplicate. Error bars denote standard deviation.
(TIF)
Summary of examined factors in plasmid transformation of E. coli .
(DOC)
Examination of the structure of E. coli mutant strains used in this work.
(DOC)
Plasmid transformation and stresses.
(DOC)
Acknowledgments
We thank Dr. Regine Hengge-Aronis, Dr. Steven Finkel, Dr. Anthony Pugsley and Dr. William E. Bentley for kindly providing bacterial strains and plasmids; Dr. Claude Gutierrez, and Dr. Pochi R Subbarayan for useful suggestions; Wei Liu for technical assistance; and Ms. Merritt Tuttle for critically reading the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work is supported by the Natural Science Foudation of China (31100071), the Natural Science Foundation of Zhejiang province (Y3110237), the Doctor's Start-up Grant from Zhejiang Academy of Agricultural Sciences, the Foresight Program from Zhejiang Academy of Agricultural Sciences, and the Zhejiang Open Foundation of the Most Important Subjects. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Nikaido H. Multidrug resistance in bacteria. Annu Rev Biochem. 2009;78:119–146. doi: 10.1146/annurev.biochem.78.082907.145923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venturini C, Beatson SA, Djordjevic SP, Walker MJ. Multiple antibiotic resistance gene recruitment onto the enterohemorrhagic Escherichia coli virulence plasmid. FASEB J. 2010;24:1160–1166. doi: 10.1096/fj.09-144972. [DOI] [PubMed] [Google Scholar]
- 3.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, et al. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Solomon JM, Grossman AD. Who's competent and when: regulation of natural genetic competence in bacteria. Trends Genet. 1996;12:150–155. doi: 10.1016/0168-9525(96)10014-7. [DOI] [PubMed] [Google Scholar]
- 5.Claverys JP, Martin B. Bacterial “competence” genes: signatures of active transformation, or only remnants? Trends Microbiol. 2003;11:161–165. doi: 10.1016/s0966-842x(03)00064-7. [DOI] [PubMed] [Google Scholar]
- 6.Sinha S, Cameron AD, Redfield RJ. Sxy induces a CRP-S regulon in Escherichia coli. J Bacteriol. 2009;191:5180–5195. doi: 10.1128/JB.00476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maeda S, Sawamura A, Matsuda A. Transformation of colonial Escherichia coli on solid media. FEMS Microbiol Lett. 2004;236:61–64. doi: 10.1016/j.femsle.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 8.Sun D, Zhang Y, Mei Y, Jiang H, Xie Z, et al. Escherichia coli is naturally transformable in a novel transformation system. FEMS Microbiol Lett. 2006;265:249–255. doi: 10.1111/j.1574-6968.2006.00503.x. [DOI] [PubMed] [Google Scholar]
- 9.Tsen SD, Fang SS, Chen MJ, Chien JY, Lee CC, et al. Natural plasmid transformation in Escherichia coli. J Biomed Sci. 2002;9:246–252. doi: 10.1007/BF02256071. [DOI] [PubMed] [Google Scholar]
- 10.Sun D, Zhang X, Wang L, Prudhomme M, Xie Z, et al. Transforming DNA uptake gene orthologs do not mediate spontaneous plasmid transformation in Escherichia coli. J Bacteriol. 2009;191:713–719. doi: 10.1128/JB.01130-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen I, Dubnau D. DNA uptake during bacterial transformation. Nat Rev Microbiol. 2004;2:241–249. doi: 10.1038/nrmicro844. [DOI] [PubMed] [Google Scholar]
- 12.Mortier-Barriere I, Velten M, Dupaigne P, Mirouze N, Pietrement O, et al. A key presynaptic role in transformation for a widespread bacterial protein: DprA conveys incoming ssDNA to RecA. Cell. 2007;130:824–836. doi: 10.1016/j.cell.2007.07.038. [DOI] [PubMed] [Google Scholar]
- 13.Saunders CW, Guild WR. Monomer plasmid DNA transforms Streptococcus pneumoniae. Mol Gen Genet. 1981;181:57–62. doi: 10.1007/BF00339005. [DOI] [PubMed] [Google Scholar]
- 14.Hengge R. Proteolysis of sigmaS (RpoS) and the general stress response in Escherichia coli. Res Microbiol. 2009;160:667–676. doi: 10.1016/j.resmic.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 15.Battesti A, Majdalani N, Gottesman S. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol. 2011;65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lacour S, Landini P. SigmaS-dependent gene expression at the onset of stationary phase in Escherichia coli: function of sigmaS-dependent genes and identification of their promoter sequences. J Bacteriol. 2004;186:7186–7195. doi: 10.1128/JB.186.21.7186-7195.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patten CL, Kirchhof MG, Schertzberg MR, Morton RA, Schellhorn HE. Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol Genet Genomics. 2004;272:580–591. doi: 10.1007/s00438-004-1089-2. [DOI] [PubMed] [Google Scholar]
- 18.Vijayakumar SR, Kirchhof MG, Patten CL, Schellhorn HE. RpoS-regulated genes of Escherichia coli identified by random lacZ fusion mutagenesis. J Bacteriol. 2004;186:8499–8507. doi: 10.1128/JB.186.24.8499-8507.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J Bacteriol. 2005;187:1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sledjeski DD, Gupta A, Gottesman S. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 1996;15:3993–4000. [PMC free article] [PubMed] [Google Scholar]
- 21.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006 0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Becker G, Hengge-Aronis R. What makes an Escherichia coli promoter sigmaS dependent? Role of the −13/−14 nucleotide promoter positions and region 2.5 of sigmaS. Mol Microbiol. 2001;39:1153–1165. doi: 10.1111/j.1365-2958.2001.02313.x. [DOI] [PubMed] [Google Scholar]
- 23.Smith HO, Danner DB, Deich RA. Genetic transformation. Annu Rev Biochem. 1981;50:41–68. doi: 10.1146/annurev.bi.50.070181.000353. [DOI] [PubMed] [Google Scholar]
- 24.Claverys JP, Prudhomme M, Martin B. Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu Rev Microbiol. 2006;60:451–475. doi: 10.1146/annurev.micro.60.080805.142139. [DOI] [PubMed] [Google Scholar]
- 25.Dubnau D. Genetic competence in Bacillus subtilis. Microbiol Rev. 1991;55:395–424. doi: 10.1128/mr.55.3.395-424.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meibom KL, Blokesch M, Dolganov NA, Wu CY, Schoolnik GK. Chitin induces natural competence in Vibrio cholerae. Science. 2005;310:1824–1827. doi: 10.1126/science.1120096. [DOI] [PubMed] [Google Scholar]
- 27.Redfield RJ, Cameron AD, Qian Q, Hinds J, Ali TR, et al. A novel CRP-dependent regulon controls expression of competence genes in Haemophilus influenzae. J Mol Biol. 2005;347:735–747. doi: 10.1016/j.jmb.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 28.Etchuuya R, Ito M, Kitano S, Shigi F, Sobue R, et al. Cell-to-cell transformation in Escherichia coli: a novel type of natural transformation involving cell-derived DNA and a putative promoting pheromone. PLoS One. 2011;6:e16355. doi: 10.1371/journal.pone.0016355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Havarstein LS, Coomaraswamy G, Morrison DA. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1995;92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pestova EV, Havarstein LS, Morrison DA. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol Microbiol. 1996;21:853–862. doi: 10.1046/j.1365-2958.1996.501417.x. [DOI] [PubMed] [Google Scholar]
- 31.Dubey GP, Ben-Yehuda S. Intercellular nanotubes mediate bacterial communication. Cell. 2011;144:590–600. doi: 10.1016/j.cell.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Li M, Yan Q, Chen X, Geng J, et al. Across genus plasmid transformation between Bacillus subtilis and Escherichia coli and the effect of Escherichia coli on the transforming ability of free plasmid DNA. Curr Microbiol. 2007;54:450–456. doi: 10.1007/s00284-006-0617-1. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Li F, Sun D, Liu J, Liu N, et al. Molecular analysis shows differential expression of R-spondin1 in zebrafish (Danio rerio) gonads. Mol Biol Rep. 2011;38:275–282. doi: 10.1007/s11033-010-0105-3. [DOI] [PubMed] [Google Scholar]
- 34.Finkel SE, Kolter R. DNA as a nutrient: novel role for bacterial competence gene homologs. J Bacteriol. 2001;183:6288–6293. doi: 10.1128/JB.183.21.6288-6293.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sauvonnet N, Gounon P, Pugsley AP. PpdD type IV pilin of Escherichia coli K-12 can be assembled into pili in Pseudomonas aeruginosa. J Bacteriol. 2000;182:848–854. doi: 10.1128/jb.182.3.848-854.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mulvey MR, Loewen PC. Nucleotide sequence of katF of Escherichia coli suggests KatF protein is a novel sigma transcription factor. Nucleic Acids Res. 1989;17:9979–9991. doi: 10.1093/nar/17.23.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subbarayan PR, Sarkar M. A comparative study of variation in codon 33 of the rpoS gene in Escherichia coli K12 stocks: implications for the synthesis of sigmaS. Mol Genet Genomics. 2004;270:533–538. doi: 10.1007/s00438-003-0944-x. [DOI] [PubMed] [Google Scholar]
- 38.Becker G, Klauck E, Hengge-Aronis R. Regulation of RpoS proteolysis in Escherichia coli: the response regulator RssB is a recognition factor that interacts with the turnover element in RpoS. Proc Natl Acad Sci USA. 1999;96:6439–6444. doi: 10.1073/pnas.96.11.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Examination of the structure of E. coli mutants tested in Figure 6 (see accompanying Table S2). Analysis of PCR fragments for confirmation of the structure of E. coli mutant strains (Primers used were listed in Table 2). From left to right, each pair of lanes compares wildtype and mutant structures, as follows. Lanes: 1, 4 and 13, DD Marker 2, ugpC mutant; 3, wide type 5, rpoS mutant; 6, wide type 7, yqjC mutant; 8, wide type 9, ygiW mutant; 10, wide type 11, osmY mutant; 12, wide type.
(TIF)
Effect of H2O2 on plasmid transformation. H2O2 was added either at OD600 = 0.1 (A) or OD600 = 1.5 (B). Plasmid transformation was measured either 4, 5 and 6 hours (A) or 10 minutes (B) after the addition of H2O2. Transformation frequency was calculated by dividing the number of transformants per ml by the number of viable counts per ml. Each sample was performed in duplicate. Error bars denote standard deviation.
(TIF)
Effect of anti-oxidant agents on plasmid transformation. Anti-oxidant agents Na2SO3 or NaHSO3 were added to the liquid culture prior to plating (A) or the solid agar plates for plasmid transformation (B). Transformation frequency was calculated by dividing the number of transformants per ml by the number of viable counts per ml. Each sample was performed in duplicate. Error bars denote standard deviation.
(TIF)
Summary of examined factors in plasmid transformation of E. coli .
(DOC)
Examination of the structure of E. coli mutant strains used in this work.
(DOC)
Plasmid transformation and stresses.
(DOC)