Abstract
Toll-like receptors (TLRs) signaling pathways are the first lines in defense against Salmonella enteritidis (S. enteritidis) infection but the molecular mechanism underlying susceptibility to S. enteritidis infection in chicken remains unclear. SPF chickens injected with S. enteritidis were partitioned into two groups, one consisted of those from Salmonella-susceptible chickens (died within 5 d after injection, n = 6), the other consisted of six Salmonella-resistant chickens that survived for 15 d after injection. The present study shows that the bacterial load in susceptible chickens was significantly higher than that in resistant chickens and TLR4, TLR2-1 and TLR21 expression was strongly diminished in the leukocytes of susceptible chickens compared with those of resistant chickens. The induction of expression of pro-inflammatory cytokine genes, IL-6 and IFN-β, was greatly enhanced in the resistant but not in susceptible chickens. Contrasting with the reduced expression of TLR genes, those of the zinc finger protein 493 (ZNF493) gene and Toll-interacting protein (TOLLIP) gene were enhanced in the susceptible chickens. Finally, the expression of TLR4 in peripheral blood mononuclear cells (PBMCs) infected in vitro with S. enteritidis increased significantly as a result of treatment with 5-Aza-2-deoxycytidine (5-Aza-dc) while either 5-Aza-dc or trichostatin A was effective in up-regulating the expression of TLR21 and TLR2-1. DNA methylation, in the predicted promoter region of TLR4 and TLR21 genes, and an exonic CpG island of the TLR2-1 gene was significantly higher in the susceptible chickens than in resistant chickens. Taken together, the results demonstrate that ZNF493-related epigenetic modification in leukocytes probably accounts for increased susceptibility to S. enteritidis in chickens by diminishing the expression and response of TLR4, TLR21 and TLR2-1.
Introduction
Toll-like receptors (TLRs) signaling pathways are the first lines in defense against Salmonella infection. The TLRs are broadly distributed on a variety of leukocytes [1], where they function as the primary sensors to initiate innate immune responses by responding to pathogen-associated molecular patterns (PAMPs) from bacteria, viruses, fungi or parasites [2], [3], [4]. The transcription factor NF-κB [5] is subsequently activated to induce the expression of immune and pro-inflammatory genes such as tumor necrosis factor alpha (TNF-α), interleukins 6, 1 beta, 8 and 12 (IL-6, IL-1β, IL-8, IL-12), and interferon (IFN), etc. [6], [7], [8].
Four TLRs (TLR4, TLR2, TLR9 and TLR5) are responsible for recognition of antigens from S. enteritidis in humans and mice. The dominant TLR involved in the host response to Salmonella infection is TLR4 [9]. Mutations in the TLR4 gene increase the risk of Gram-negative infections in humans and mice [10], [11], [12] and mice deficient in both TLR4 and TLR2, or TLR4 and TLR9, were highly susceptible to Salmonella typhimurium [13]. Several specific avian TLR genes have been described. Avian TLR2A (TLR2-1) and TLR2B (TLR2-2) seem to have arisen from a duplication of TLR2 found in other vertebrates [14] and avian TLR21 is a functional homolog of mammalian TLR9 [15].
Signaling pathways mediated by TLRs are tightly regulated to balance the activation and inhibition of inflammatory responses [16]. Multiple layers, involving many diverse factors, participate in negative regulation of TLR signaling. For example, suppressor of cytokine signaling 1 (SOCS1), phosphatidylinositol 3 kinase (PI3K), toll interacting protein (TOLLIP), and zinc finger protein A20 (A20) are intracellular negative regulators suppressing the signaling of TLR2, TLR4 and TLR9 in multiple pathways [17]. Transcriptional regulation of TLRs can also influence the inflammatory responses. In the clinical course of cystic fibrosis (CF), increased expression of TLR2 caused chronic inflammation [18]. Diminished expression and function of TLR1, TLR2 and TLR4 accounts for T cell hyporesponsiveness in human filarial infection [19].
Little is known about the underlying mechanisms of transcriptional regulation of TLRs beyond ZNF160-dependent epigenetic regulation decreasing the expression of TLR4 in intestinal epithelial cells [20], [21], [22]. While the ZNF160 gene has not been identified in chicken, a Blastn search identified an avian homolog (ZNF493). Whether or not the same mechanism plays a role in modulating the immune response of the host to S. enteritidis infection remains unclear. Hypermethylation of promoter CpG dinucleotides has been associated with decreased expression of the gene [23], [24]. Some reports have indicated that methylation status of exonic CpG islands correlates with transcriptional activity [25]. In order to analyze the regulatory mechanism of TLRs, the methylation status in the promoter region and exonic CpG islands of TLRs were investigated.
Chickens are carriers of S. enteritidis that colonize the alimentary tract of chickens and, through excrement, can contaminate food products and water [26]. It was considered to be important to delineate part of the molecular mechanisms underlying differences in susceptibility of chickens to infection with S. enteritidis. The present study confirmed that the aberrant expression of TLR4, TLR21, and TLR2-1 in peripheral blood leukocytes was associated with the susceptibility to S. enteritidis infection in chickens. More interestingly, it was demonstrated that the dysregulation of TLR4, TLR21, and TLR2-1 was probably due to ZNF493-related epigenetic modification, including histone acetylation and DNA methylation.
Results
Increased bacterial load in susceptible chickens
The bacterial load in the blood at 0 h (before bacteria challenge), 8 h, 16 h, 24 h and 3 d post infection (TPI) were compared in six chickens that died within 5 d after infection with S. enteritidis (susceptible group) and six chickens that survived until 15 d TPI (resistant group). Results are presented in Table 1. S. enteritidis was not detected in any of the samples until 8 h TPI. From 16 h to 3 d TPI, the number of S. enteritidis in susceptible chickens was significantly higher (P<0.05) than that in resistant chickens. Notably, the bacterial load in susceptible chickens increased more dramatically at 16 h TPI and declined less significantly at 3 d TPI than that in resistant chickens. The results indicate that increased bacterial load is associated with susceptibility to S. enteritidis in chickens.
Table 1. Kinetics of Salmonella Enteritidis loads in inoculated SPF chickens determined by qPCR across all the times.
| 0 h | 8 h | 16 h | 24 h | 3 d | 12 d | |
| S | 0.00±0.00 | 6.54±0.32 | 7.05±0.23 | 6.96±0.06 | 6.87±0.21 | |
| R | 0.00±0.00 | 6.30±0.08 | 6.37±0.59 | 6.49±0.10 | 5.75±0.32 | 6.09±0.17 |
| P value | P<0.05 | P<0.05 | P<0.05 |
Data are presented as the mean bacterial loads and is expressed as log10 of the bacterial genome copy number per ml of blood (± SD) obtained from susceptible (S) and resistant (R) chickens.
Decreased expression of TLR4, TLR21 and TLR2-1 genes in susceptible chickens
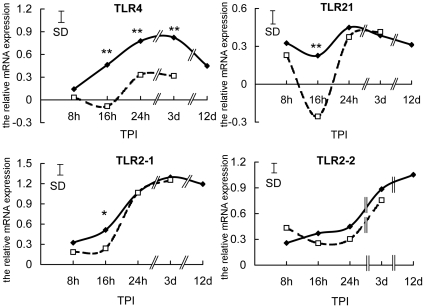
In order to explore the molecular mechanisms of susceptibility to S. enteritidis infection, the expression levels of Toll-like receptors (TLRs) were examined in susceptible chickens. The abundance of TLR4, TLR21, TLR2-1 and TLR2-2, and transcripts and changes at all times post-inoculation were compared by q-RT-PCR in susceptible and resistant chickens. There were no significant differences in the expression of TLRs at 0 h (data not shown) and 8 h TPI between these two groups (Fig. 1), but, at later times, susceptible chickens had depressed expression of TLRs genes compared with resistant chickens. This was most evident at 16 h TPI, when TLR4, TLR21 and TLR2-1 transcripts were all significantly lower in the susceptible group than in the resistant group. Only TLR2-2 mRNA did not differ between the two groups across all sampling times, whereas TLR4 expression in resistant chickens was persistently and significantly higher than in susceptible chickens from 16 h to 3 d (Fig. 1). The results suggest that higher susceptibility to S. enteritidis and increased bacterial load might result from depressed expression of TLR4, TLR21 and TLR2-1 at the early stage of infection.
Figure 1. Decreased expression of TLR4, TLR21 and TLR2-1 genes in susceptible chickens.
The relative expression of TLR4, TLR21, TLR2-1 and TLR2-2 in leukocytes of susceptible (□ ---- □) and resistant (⧫——⧫) chickens at 8 h, 16 h, 24 h, 3 d, and 12 d after infection with S.enteritidis is shown. Relative values, normalized using β-actin mRNA levels and the average expression levels in both groups at 0 h, are shown. The data are means (SD shown by the vertical bars) of 6 birds (*P<0.05; **P<0.01). TPI is time post-infection.
Partially diminished inflammatory response in susceptible chickens
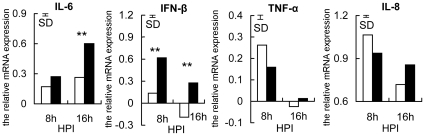
Four pro-inflammatory cytokine genes (IL-6, IFN-β, TNF-α, and IL-8) were used to investigate if the decreased expression of TLR4, TLR21 and TLR2-1 at 16 h in susceptible chickens resulted in a mitigated inflammatory response. Consistent with the expression of TLRs in the resistant and susceptible groups, the induction of IL-6 and IFN-β transcription was greatly enhanced in the resistant, but not in the susceptible chickens at 16 h post-infection (Fig. 2). These results indicate that diminished expression of TLR4, TLR21 and TLR2-1 in the susceptible chickens leads to a decreased inflammatory response. The similar levels of IL-8 in both groups demonstrated that only some of the pro-inflammatory cytokines showed down-regulation in susceptible chickens, perhaps because IL-8 was regulated by other TLRs. In contrast, there was no obvious difference in the expression of TNF-α at 16 h, indicating that not all pro-inflammatory cytokine genes are induced at this early stage of infection (Fig. 2). Collectively, the results were consistent with higher susceptibility to S. enteritidis in birds being due to the partially diminished inflammatory response associated with decreased expression of TLR4, TLR21 and TLR2-1.
Figure 2. Partially diminished inflammatory response in susceptible chickens.
The relative expression of IL-6, IFN-β, TNF-α and IL-8 in susceptible (open bars) and resistant (filled bars) chickens at 8 h and 16 h after infection with S.enteritidis is shown. Data are means (n = 6), normalized to β-actin mRNA and the average expression at 0 h (**P<0.01). The vertical bar is the SD from the error mean square of the ANOVA. HPI is hours post-infection.
Enhanced expression of TOLLIP and ZNF493 genes in susceptible chickens
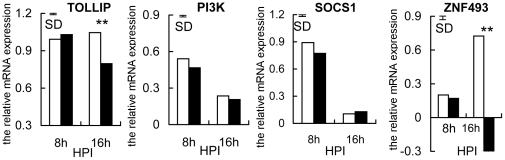
An attempt was then made to identify the molecular mechanisms responsible for decreased expression of TLR4, TLR21 and TLR2-1 in the susceptible chickens. The expression of four negative regulators of TLR2, TLR4 and TLR21 signaling pathways (TOLLIP, PI3K, SOCS1 and ZNF493, a chicken homolog of mammalian ZNF160) was compared between susceptible and resistant chickens at 8 h and 16 h. There were no differences (P>0.05) between susceptible and resistant chickens in expression of TOLLIP, PI3K, SOCS1 and ZNF493 before infection or at 8 h TPI, when expression was increased in all birds. At 16 h, however, expression of TOLLIP and ZNF493 in susceptible chickens was pronounced and exceeded that in the resistant chickens (P<0.01), while the other genes were up-regulated to lesser degrees and there were no differences between the two groups of chickens. Note the substantial increase in ZNF493 transcripts between 8 h and 16 h in the susceptible chickens when those in resistant chickens changed in the opposite direction (Fig. 3).
Figure 3. Enhanced expression of TOLLIP and ZNF493 genes in susceptible chickens.
The relative expression of TOLLIP, PI3K, SOCS1 and ZNF493 genes in susceptible (open bars) and resistant (filled bars) chickens at 8 h and 16 h after infection with S. enteritidis is shown. Data are means (n = 6), normalized to β-actin mRNA and the average expression at 0 h (**P<0.01). The vertical bar is the SD from the error mean square of the ANOVA. HPI is hours post-infection.
DNA methyltransferase inhibitor 5-Aza-dc and/or the histone deacetylase inhibitor TSA increased expression of TLR4, TLR21 and TLR2-1
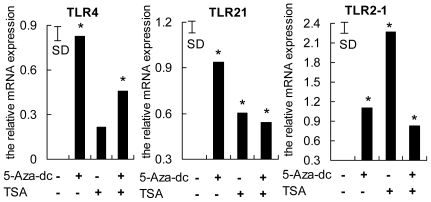
The possibility that TLR4, TLR21 and TLR2-1 gene expression was regulated by epigenetic modification (histone acetylation and/or DNA methylation) in leukocytes infected with S. enteritidis was examined. Isolated PBMCs from SFP chickens were infected with S. enteritidis in the absence and presence of combinations of 5-Aza and TSA in the culture media. As shown in Fig. 4, 5-Aza-dc provoked a significant increase in TLR4 expression. Either 5-Aza-dc or TSA was effective in up-regulating the expression of TLR21 and TLR2-1; the effect of 5-Aza-dc was greater in the case of TLR21 and that of TSA was greater for TLR2-1. No cooperative effects of 5-Aza-dc and TSA on the expression of the genes were observed. These results indicate that histone acetylation and DNA methylation are involved in the repression of TLR4, TLR21 and TLR2-1 expression in PBMCs of chickens during S. enteritidis infection.
Figure 4. DNA methyltransferase inhibitor 5-Aza-dc and/or the histone deacetylase inhibitor TSA increased TLR4, TLR21 and TLR2-1 expression.
Peripheral blood mononuclear cells were inoculated with S. enteritidis without additions (controls) or in the presence of 5-Aza-dc, TSA or TSA plus 5-Aza-dc. Relative abundances of TLR4, TLR21 and TLR2-1 mRNA were analyzed by qPCR and normalized to β-actin mRNA. Data are means (n = 3) and comparisons were made to expression in the controls (-,-). The vertical bar is the SD from the error mean square of the ANOVA, * indicates P<0.05.
Higher methylation in the predicted promoter region of TLR4 and TLR21 gene, and an exonic CpG island of TLR2-1 gene in susceptible chickens
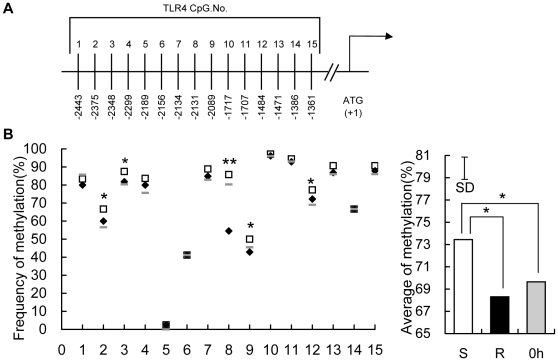
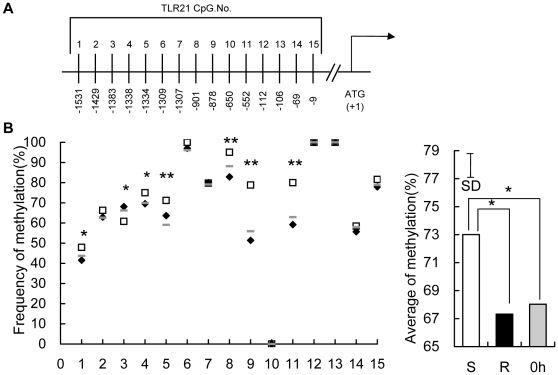
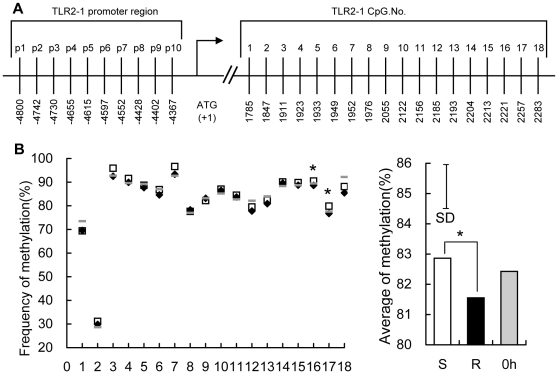
The possibility that diminished expression of TLR4, TLR21 and TLR2-1 at 16 h TPI might be due to differences in methylation was explored at multiple locations within each of these genes, using leukocyte DNA at 0 h and 16 h TPI. For both TLR4 (Fig. 5A) and TLR21 (Fig. 6A), 15 CpG motifs in the predicted promoter regions were assessed. In the case of TLR2-1, 10 CpG motifs within the promoter and 18 CpG motifs in an exonic CpG island (Fig. 7A) were evaluated. There were no differences (P>0.05) in the methylation of TLR4, TLR21 and TLR2-1 genes between the susceptible and resistant chickens at 0 h (data not shown), and the average methylation level of all the 12 chickens before infection is shown as the basic methylation status. Interestingly, the average methylation levels of TLR4 and TLR21 at 16 h rose dramatically from the basic level at 0 h in susceptible chickens whereas it fell slightly (around 1%) in resistant chickens. Thus, higher methylation in the predicted promoter region of the TLR4 and TLR21 genes, was evident in susceptible versus resistant chickens at 16 h (Fig. 5B, Fig. 6B). This trend was also evident in several CpG sites (5 sites for TLR4, 7 for TLR21). No significant differences were observed in the promoter region of the TLR2-1 gene between these two groups at 16 h (data not shown), but an exonic CpG island of the TLR2-1 gene showed higher methylation level in susceptible than resistant chickens, although the difference was not as great as that in TLR4 and TLR21 (Fig. 7B).
Figure 5. Methylation of 15 CpG motifs in the predicted promoter region of the TLR4 gene.
(A) The distribution of the 15 CpG dinucleotides from −2443 to −1361 in the upstream region of the TLR4 gene relative to the translation start site (+1). (B) Genomic DNA from peripheral blood leukocytes of uninfected chickens at 0 h (⁃), susceptible (□) and resistant (⧫) chickens at 16 h TPI was modified with sodium bisulfite, amplified by PCR, cloned, and 12–16 independent clones were sequenced. The frequency of methylated CpGs in each CpG site (data are means of 12 birds for uninfected chicken and 6 birds for susceptible and resistant chickens, respectively) are shown and comparisons were made between susceptible and resistant chickens. The average of % methylation at each CpG site within all 15 CpGs in peripheral blood leukocytes of uncharged chickens (0 h, filled grey bars), susceptible (S, open bars) and resistant (R, filled black bars) chickens at 16 h after infection with S. enteritidis are presented. The vertical bar is the SD from the error mean square of the ANOVA, * indicates P<0.05, ** indicates P<0.01.
Figure 6. Methylation of 15 CpG motifs in the predicted promoter region of TLR21 gene.
(A) The distribution of the 15 CpG dinucleotides from −1531 to −9 in the upstream region of the TLR21 gene relative to the translation start site (+1). (B) Genomic DNA from peripheral blood leukocytes of uninfected chickens at 0 h (⁃), susceptible (□) and resistant (⧫) chickens at 16 h TPI was modified with sodium bisulfite, amplified by PCR, cloned, and 12–16 independent clones were sequenced. The frequency of methylated CpGs in each CpG site (data are means of 12 birds for uninfected chicken and 6 birds for susceptible and resistant chickens, respectively) are shown and comparisons were made between susceptible and resistant chickens. The average of % methylation at each CpG site within all 15 CpGs in peripheral blood leukocytes of uncharged chickens (0 h, filled grey bars), susceptible (S, open bars) and resistant (R, filled black bars) chickens at 16 h after infection with S. enteritidis are presented. The vertical bar is the SD from the error mean square of the ANOVA, * indicates P<0.05, ** indicates P<0.01.
Figure 7. Methylation of 18 CpG motifs in the exon and 10 CpG motifs in the predicted promoter region of TLR2-1 gene.
(A) the distribution of the 18 CpG dinucleotides from 1785 to 2283 in the exon region and 10 CpG dinucleotides from −4800 to −4367 in the predicted promoter region of the TLR2-1 gene relative to the translation start site (+1). (B) Genomic DNA from peripheral blood leukocytes of uninfected chickens at 0 h (⁃), susceptible (□) and resistant (⧫) chickens at 16 h TPI was modified with sodium bisulfite, amplified by PCR, cloned, and 12–16 independent clones were sequenced. The frequency of methylated CpGs in each CpG site (data are means of 12 birds for uninfected chicken and 6 birds for susceptible and resistant chickens, respectively) are shown and comparisons were made between susceptible and resistant chickens. The average of % methylation at each CpG site within all 18 CpGs in peripheral blood leukocytes of uncharged chickens (0 h, filled grey bars), susceptible (S, open bars) and resistant (R, filled black bars) chickens at 16 h after infection with S. enteritidis are presented. The vertical bar is the SD from the error mean square of the ANOVA, * indicates P<0.05.
Collectively, the results presented here show that diminished expression and response of TLR4, TLR21 and TLR2-1 in peripheral blood leukocytes, due to epigenetic modification, likely account for increased susceptibility to S. enteritidis in chickens.
Discussion
Although the physiological importance of transcriptional regulation of TLRs is unclear, several reports indicate that it directly influences the immune response of the host. The expression of TLRs, specifically TLR2 and TLR4, is induced by various PAMPs from bacteria, viruses, fungi or parasites for inflammatory responses in macrophages, epithelia, cecum and spleen [27], [28], [29]. Dysregulated expression of TLRs can impair the immune response of the host, resulting in various diseases. In the clinical course of cystic fibrosis (CF), dysregulated expression of TLR2 caused chronic inflammation [18]. Diminished expression and function of TLR1, TLR2 and TLR4 accounts for T cell hyporesponsiveness in human filarial infection [19]. The fact that various expression patterns of TLRs appear in tissues with different immune responses and function demonstrates the important role of transcriptional regulation of TLRs in the signaling of TLRs. For example, in enterocytes, depressed expression of TLR4 contributes to maintenance of intestinal homeostasis [22]. The downregulation of TLR5 expression was observed in cecum by S. enteritidis infection, which might be beneficial to protect host cells from overstimulation by bacterial flagellin [29]. In addition, genetic line has significant effect on TLR expression, which may partly explain genetic variability in immune response to S. enteritidis [30]. In this study, the reduced expression of the pro-inflammatory cytokines TNF-α and IL-6 in leukocytes of susceptible chickens (Fig. 2), confirmed that reduced expression of TLR4, TLR21 and TLR2-1 constrained the immune response to S. enteritidis, which is consistent with the human studies.
While not previously described for S. enteritidis infection, epigenetic regulation of TLR4 and TLR21 involving ZNF493 in chickens, participates in the negative regulation of TLRs. The avian ZNF493 examined here (and the mammalian homolog ZNF160) are Kruppel-related zinc finger proteins with an N-terminal repressor domain, the Kruppel associated box (KRAB), a potent repressor of transcription [31]. The mechanism involves recruiting KRAB-associated protein 1 (KAP1), triggering de novo DNA methylation [32], and the forming of a multimolecular complex comprising histone deacetylases, which induces transcriptional repression through the formation of heterochromatin [33], [34], [35]. The present study shows a dramatic enhancement of ZNF493 expression in susceptible chickens at 16 h, contrasting with diminished expression of TLR4 and TLR21 (Fig. 3). This finding prompted the experiment using chicken PBMCs infected with S. enteritidis in vitro, which demonstrated that expression of TLR4 and TLR21 was significantly promoted by inhibitors of DNA methyltransferase and histone deacetylase (Fig. 4). In addition, the susceptible, but not the resistant, chickens had increased methylation of TLR4 and TLR21 genes at 16 h compared with their basal levels at 0 h, which is consistent with the increased expression of ZNF493 in the susceptible chickens at 16 h (Fig. 5, Fig. 6). All of these findings indicate that ZNF493-related epigenetic regulation of TLR4 and TLR21 in leukocytes plays a role in the negative regulation of TLRs in chickens. Two possibilities might explain the differences at the transcriptional level of ZNF493 gene in susceptible and resistant chickens: (1) polymorphisms in the regulatory regions, including promoter of the ZNF493 gene; (2) polymorphisms of regulatory genes of the ZNF493 gene. White Leghorn chickens are known to have genetic variability in resistance to S. enteritidis among different strains [36]. The SPF chicken used in the present study is a commercial Babcock® White Leghorn line, which is very likely to have multiple genetic origins and genetic variability in susceptibility to S. enteritidis. The present study, however, shows that no polymorphisms in the promoter region of avian ZNF493 gene were detected in susceptible and resistant chickens (data not shown). It implies that diminished expression of ZNF493 gene might result from the polymorphisms of its regulatory genes or other regulatory regions (introns, 3′-UTR…).
There is little known about the overall transcriptional regulatory mechanism of TLRs. Based on the known reports, it can be inferred that positive transcriptional regulation of TLR by cytokines to augment TLR signaling and negative feedback control from negative regulators to terminate activation of TLRs are the basic mechanisms of transcriptional regulation of TLRs [17], [37], [38]. Moreover, transcriptional regulation of TLRs varies in different tissues, indicating that tissue-specific genes modify the regulatory system [22], [29]. In addition, the pathogen probably can also exploit and modulate the regulatory system, disturbing the normal expression of TLRs [18], [19], [39], [40]. In the present study, the expression of TLRs showed a common trend in obviously rising to the maximal level at around 3 d, followed by a fall by 12 d (Fig. 1). This trend indicates positive regulation by cytokines played a role in the initial upregulation stage and negative feedback control in the later downregulation stage. The epigenetic modification of TLRs in this study seems to be driven by two opposite mechanism, methylation and demethylation, depending on the particular TLR. TLR4 and TLR21, but not TLR2-1 showed an obvious downregulation and increase in methylation level in susceptible chickens at 16 h TPI, which probably resulted from ZNF493-related negative epigenetic modification. For all the three genes, the abundance of mRNA increased significantly compared with that at 0 h and the methylation level in resistant chickens similarly declined slightly from the basic level (Fig. 5, Fig. 6, Fig. 7), indicating that demethylation was widely involved in the regulation of TLRs. This demethylation happened in resistant chickens with higher expression of inflammatory proinflammatory cytokines, indicating that it could be one of the positive regulatory mechanisms of the cytokines.
The role for ZNF493-related epigenetic regulation of TLRs in the response to infection with S. enteritidis, however, seems to be quite different from the basic negative regulatory mechanism of the TLR signaling pathway. Immune signaling pathways mediated by TLRs are tightly regulated to avoid over-activation of inflammatory responses and most negative regulators use a mode of negative feedback to terminate TLRs activation. They are induced by the activation of TLRs, or are constitutively expressed, but could possibly exert their functions only when TLRs are over-activated [41], [42]. Since the diminished expression of TLRs and induction of ZNF493 in the susceptible chickens occurred at the early stage of S. enteritidis invasion when induction of inflammatory response genes was even lower than in the resistant chickens (Fig. 2), and expression of TLR4, TLR21 and TLR2-1 remained at low levels (Fig. 1), it is not reasonable to account for the induction of ZNF493 by negative feedback control from the host and, instead, it might have been provoked by S. enteritidis. S. enteritidis almost certainly benefits from the diminished expression of TLR4, TLR21 and TLR2-1 for its successful invasion and colonization of the susceptible host. Indeed, S. enteritidis secretes virulence factors to temporarily turn off TLR signaling to aid in colonization of host cells by inactivating IRAK, a kinase in the signaling pathway [39], [40].
All of the findings described here, comparing blood bacterial load, transcript abundance and DNA methylation in leukocytes of susceptible and resistant chickens, along with the effects of inhibitors for epigenetic modification on transcript abundance in isolated PBMCs, are consistent with S. enteritidis being able to provoke epigenetic regulation of the transcription of TLR4, TLR21 and TLR2-1 as an important strategy for weakening host defenses.
Materials and Methods
Bacterial Strains and Infections
S. enteritidis (50041) was obtained from the China Institute of Veterinary Drugs Control (IVDC, Beijing, China) and was used for all infections. Bacteria were resuscitated overnight in Luria–Bertani (LB) broth at 37°C in an orbital shaking incubator at 150 rpm. The number of CFU of S. enteritidis was determined by plating serial dilutions.
SPF Chickens and In Vivo Infections
Animal studies were performed according to protocols approved by the Beijing Laboratory Animal Use and Care office (approval number: SYXK 2006-0027). Specific-pathogen-free White Leghorn chickens were purchased from the Beijing Laboratory Animal Research Center (BLARC, Beijing, China). Birds were reared in separate cages in the SPF chicken experimental center of Beijing Academy of Agriculture and Forestry Sciences (Beijing, China) and given ad libitum access to water and a diet specifically designed for SPF chickens (BLARC). Birds were confirmed to be free of Salmonella by culturing faecal samples in buffered peptone water (BPW) overnight with shaking at 150 rpm followed by spreading and culture (37°C, 18–24 h) on brilliant green agar containing 100 mg/ml nalidixic acid [43].
Chickens (n = 20) aged 30 d, were blood sampled (0 h) then injected intramuscularly into the breast with 0.5 ml PBS containing 8.7×108 CFU S. enteritidis (50041) and additional blood samples were taken at 8 h, 16 h, 24 h, 3 d, and 12 d. Blood from the wing vein (0.5 ml) was taken into EDTA-coated syringes and held on ice for ∼1 h before lysing and isolating leukocytes (see below). Nine chickens died within 5 days, 4 died between the 5th and the 8th day after injection and the remaining 7 survived until 15 d. Before detailed analyses were performed, the chickens and their samples were partitioned into two groups, one consisted of those from Salmonella-susceptible chickens (died within 5 d after injection, n = 6), the other consisted of six of the total of seven Salmonella-resistant chickens that survived for 15 d.
Quantitative RT-PCR (qPCR)
Erythrocytes in blood samples were lysed with Red Blood Cell Lysis Buffer (Roche, Shanghai, China) to isolate peripheral blood leukocytes. Total RNA from leukocytes or cultured PBMCs was prepared using Trizol reagent (Invitrogen, USA) and purified with an RNA cleaning kit (Tiangen, Beijing, China) after treatment with RNase-free DNase to eliminate any gDNA contamination. Total RNA was quantified with a NanoDrop 2000 Spectrophotometer (Thermo Scientific, USA) and formaldehyde gel electrophoresis, and adjusted to the 500 ng/µl. First-strand cDNA was synthesized from 2 µg total RNA (Promega, Beijing, China). Specific mRNAs were quantified by qPCR with an ABI 7500 Real-time Detection System (Applied Biosystems, USA) using a SYBR® Premix Ex Taq™ II kit (Takara, Dalian, China); the primers used (Beijing Genome Institute, Beijing, China), based on chicken sequences, were designed by Primer Premier 5.0 and are listed in Table 2. The amplification was performed in a total volume of 20 µl, containing 10 µl of 2× SYBR Green I real-time PCR Master Mix (ABI), 0.4 µl ROX, 2 µl of the 3×diluted cDNA, 1 µl of each primer(10 µmol), and 5.6 µl ddH2O. The concentrations of primers and cDNA were optimized to ensure similar PCR efficiencies (close to 100%) between the target genes and the reference gene (β-actin), if needed. The real-time PCR program started with denaturing at 95°C for 1 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. Dissociation analysis of amplification products was performed after each PCR to confirm that only one PCR product was amplified and detected. Data were analyzed with ABI 7500 SDS software (ABI) with the baseline being set automatically by the software and values of average dCT (normalized using β-actin) was exported into Excel for the calculation of relative mRNA expression. The comparative CT method was used [44], to determine fold-changes in gene expression, calculated as 2−▵▵CT using average expression levels in samples of both groups at 0(h as the calibrator (assigned an expression level of 0). Results were expressed as relative mRNA expression which was log(2−▵▵CT) at each time, from triplicate analyses.
Table 2. qPCR primers used in this study.
| Gene name | Sequence | GenBank No. |
| β-actin | f-5′-GAGAAATTGTGCGTGACATCA-3′ | L08165 |
| r-5′-CCTGAACCTCTCATTGCCA-3′ | ||
| TLR4 | f-5′-AGTCTGAAATTGCTGAGCTCAAAT-3′ | AY064697 |
| r-5′-GCGACGTTAAGCCATGGAAG-3′ | ||
| TLR2-1 | f-5′-TTACCGGTGCTTCATTCACA-3′ | NM_204278 |
| r-5′-CATATCCCATGCTCCTTTCC-3′ | ||
| TLR2-2 | f-5′-TGTACACTCTTGGGCACTGG-3′ | NM_001161650 |
| r-5′-CATGGCACCAGAAACACCTT-3′ | ||
| TLR21 | f-5′-GATGGAGACAGCGGAGAA-3′ | NM_001030558 |
| r-5′-GCGGAAGTACAAAGGTGC-3′ | ||
| TNF-α | f-5′-GAAGCAGCGTTTGGGAGT-3′ | AY765397 |
| r-5′-GTTGTGGGACAGGGTAGG-3′ | ||
| IL-8 | f-5′-AACAAGCCAAACACTCCT-3′ | NM_205498 |
| r-5′-AGGGTGGATGAACTTAGAAT-3′ | ||
| IL-6 | f-5′-GCAGGACGAGATGTGCAA-3′ | NM_204628 |
| r-5′-CCAGGTAGGTCTGAAAGGC-3′ | ||
| IFN-β | f-5′-CCAGCTCCTTCAGAATACG-3′ | NM_001024836 |
| r-5′-TGCGGTCAATCCAGTGTT-3′ | ||
| PI3K | f-5′-AACATCTGGCAAAACCAAGG-3′ | NM_001004410 |
| r-5′-CTGCAATGCTCCCTTTAAGC-3′ | ||
| SOCS1 | f-5′-GCCCATGAGAAGCTGAAGTC-3′ | NM_001137648.1 |
| r-5′-GGGGTGACCAATACCTTCCT-3′ | ||
| TOLLIP | f-5′-AAGGCAGGGTGATGACAAAG-3′ | NM_001006471 |
| r-5′-AGGAGGTGGTATTGCCACAG-3′ | ||
| A20 | f-5′-GACAGGCTGATGCAACTTGA-3′ | XR_026935 |
| r-5′-CAAACCCAGAACCGTTCACT-3′ | ||
| ZNF493 | f-5′-CGGAGCACAACGACTGTAGA-3′ | XM_001236375 |
| r-5′-GAGAAGCACAGGGGTTGAAG-3′ |
Determination of bacterial load in blood
The bacterial load in the blood of chicken was estimated by serovar-specific qPCR assay as described previously [45], [46]. The probe (5′-FAM-TGCAGCGAGCATGTTCTGGAAAGC-TAMRA-3′) and primers set (the forward primer, 5′-TCCCTGAATCTGAGAAAGAAAAACTC-3′; the reverse primer, 5′-TTGATGTGGTTGGTTCGTCACT-3′) were designed from the SdfI gene (Gen-Bank Accession No. AF370707.1). Bacterial DNA isolated from peripheral blood of chickens at 0h, 8h, 16h, 24h, 3d, and 12d was amplified using a real-time PCR core kit (R-PCR version 2.1, Takara, Dalian, China) in a 25µL reaction mixture containing 0.6µL of each primer (10µmol/L), 0.75µL of deoxyribonucleotide triphosphates (10mmol/L), 1.25 U of Ex Taq DNA Polymerase (Ex Taq Hot Start Version, Takara), 5µL of 5× PCR buffer (Mg2+ free), 0.8µL of TaqMan probe (5µmol/L), 0.5µL of Mg2+ (250mmol/L), and 5µL of templates. Each PCR run consisted of a 5(min hot start at 95°C, followed by 40 cycles consisting of 30 s of denaturation at 94°C, 30 s of annealing at 55°C, and a fluorescence read step. S. enteritidis DNA was isolated from bacterial cultures and CFU of S. enteritidis in the bacterial cultures was quantified by serially dilutions in BPW and plating, as the number of genomic copies. To extrapolate the bacterial number in each blood sample, serial dilutions of the genomic DNA were amplified (copy number ranging from 102 to 108).
Isolation and culture of peripheral blood mononuclear cells (PBMCs)
Peripheral blood mononuclear cells (PBMCs) were isolated from a separate group of six 30 d-old SPF chickens using Ficoll-Hypaque, specific gravity 1.077 (Tian Jin Hao Yang Biological Manufacturing Co., Tianjin, China). Briefly, fresh, non-coagulated blood, diluted 1∶1 in Ca++, Mg++-free Hanks' balanced salt solution (Sigma, Shanghai) was overlaid and centrifuged at 1500 rpm for 30 min. to obtain the 1.077 band. The PBMCs were collected and washed twice in RPMI 1640 medium (Invitrogen, USA) and resuspended in fresh RPMI 1640. The cell concentration was adjusted to 1.5×107 PBMCs/ml and 2 ml were cultured in 1640 medium containing 10% (v/v) fetal bovine serum (Biowest; Beijing, China). Cells were cultured at 37°C in a humidified incubator under 5% CO2.
Inhibition of histone deacetylase and DNA methyltransferase in PBMCs
Cells, prepared as above, were inoculated with 1×105 CFU S. enteritis in PBS and treated as follows: 10 µM 5-aza-2-deoxycytidine (5-Aza-dc; Sigma, Shanghai, China) for 3 d; 80 nM trichostatin A (TSA; Shanghai, Sigma) for 24 h, or with TSA plus 5-Aza-dc for an additional 24 h after initial treatment with just 5-Aza-dc for 2 d. Cells were then harvested to obtain total RNA. Transcripts of TLR4, TLR21, and TLR2-1 were quantified by qPCR as described below.
Bisulfite conversion reaction and DNA sequencing
Genomic DNA from peripheral blood leukocytes of susceptible (n = 6) and resistant (n = 6) chickens at 16 h after infection with S. enteritidis was prepared using the phenol/chloroform method. To analyze methylation of CpG motifs, 500 ng of genomic DNA was denatured at 98°C for 10 min, modified by the conversion reagent (bisulfite) at 64°C for 2.5 h, and then purified using an EZ DNA Methylation-Gold Kit™ (Zymo Research, Beijing, China).
The promoter region (including core promoter, proximal promoter and distal promoter) of the TLR4 and TLR21 genes were amplified by PCR from the sulfite-modified genomic DNA using two pairs of primers of TLR4 (TLR4-P1, TLR4-P2) and TLR21 (TLR21-P1, TLR21-P2, TLR21-P3). The promoter region and a predicted CpG island in the exon of TLR2-1 were amplified using PCR primer pairs TLR2-1-P1 and TLR2-1-P2 (Table 3). CpG islands were found (http://www.uscnorris.com/cpgislands2/cpg.aspx) using 50% GC; ObsCpG/ExpCpG, 0.60; length, 300 bp; and gap between adjacent islands, 100 bp. PCR amplifications were performed using the GoTaq® Hot Start Colorless Master Mix (Promega). Following purification of PCR products, they were cloned into the pMD-18T vector for sequencing; 12–16 clones from each sample were analyzed.
Table 3. Primers for methylation detection.
| Primer name | Sequence | Gene ID |
| TLR4-P1 | f-5′-AAAAGTAGATTGATTTTTAATGTGGA-3′ | 417241 |
| r-5′-TGTTTTTTTTTGTAGA GTTTAGG-3′ | ||
| TLR4-P2 | f-5′-AGAGATTTTTGATGATTTTATTAGA-3′ | 417241 |
| r-5′-GTAATTGTAAAGTTATTTTTGGG-3′ | ||
| TLR21-P1 | f-5′-GTTGTTAGTAGATATTTTTTGGTAGG-3′ | 415623 |
| r-5′-AATATCTAATTCCCTTCATCAATAA-3′ | ||
| TLR21-P2 | f-5′-TTATTTAGTGGGTAGTGGGGTT-3′ | 415623 |
| r-5′-AACAAAACTAAAAAAAACCAATAA-3′ | ||
| TLR21-P3 | f-5′-TAGAGTATTAGGGAGGTGGTATAG-3′ | 415623 |
| r-5′ACTCAATAACACCATCCCAATA-3′ | ||
| TLR2-1-P1 | f-5′-AAATTTTGTTTTTAGATTTGTGATT-3′ | 374141 |
| r-5′-CTACAACCCTCTCATCCTACCA-3′ | ||
| TLR2-1-P2 | f-5′-AGGTTGGGAGTGTGTAGTTGTTAT-3′ | 374141 |
| r-5′-AGAAGTAGTTTTTTGGTGAGGT-3′ |
Statistical analysis
When needed for normality and homogeneity of variance, data were log-transformed. Analyses were by two-way GLM ANOVA (in vivo study) or one-way (in vitro study) ANOVA using SAS (version 8.0). The models were:
| (1) |
| (2) |
| (3) |
| (4) |
where: y = relative mRNA expression (log-transformed); l = bacterial load; m = the frequency of methylated CpG; μ = population mean; G = the effect of 2 different groups (resistant and susceptible chickens); T = the effect of time (8 h, 16 h, 24 h, 3 d or 12 d after infection); Tr = the effect of 4 treatment combinations (5-Aza-dc, TSA: −,−; +,−; −,+; +,+); Gt = the effect of three different groups (uninfected, resistant and susceptible chickens); P = the effect of CpG positions (15 CpG positions for TLR4 and 18 for TLR2-1) and e was the random error. Multiple comparisons of means were performed using Duncan's multiple range tests and the SD derived from the Error Mean Squares. Significance was set at p<0.05 (highly significant if p<0.01).
Acknowledgments
The authors thank W. Bruce Currie, Emeritus Professor, Cornell University, for suggestions on the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by National Natural Science Foundation of China (No. 30871776) and the National Key Technology R&D Program (2011BAD28B03) and the China Agriculture Research System (CARS-42). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kabelitz D. Expression and function of Toll-like receptors in T lymphocytes. Curr Opin Immunol. 2007;19:39–45. doi: 10.1016/j.coi.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov R, Janeway CA Innate immunity: the virtues of a nonclonal system of recognition. Cell. 1997;91:295–298. doi: 10.1016/s0092-8674(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 3.Janeway CA, Medzhitov R. Innate immunity: Lipoproteins take their Toll on the host. Current Biology. 1999;9:R879–R882. doi: 10.1016/s0960-9822(00)80073-1. [DOI] [PubMed] [Google Scholar]
- 4.Hoffmann JA, Kafatos FC, Janeway CA, Ezekowitz RAB. Phylogenetic perspectives in innate immunity. Science. 1999;284:1313–1318. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]
- 5.Akira S, Takeda K. Toll-like receptor signalling. Nature Reviews Immunology. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 6.Ulevitch RJ, Tobias PS. Receptor-Dependent Mechanisms of Cell Stimulation by Bacterial-Endotoxin. Annual Review of Immunology. 1995;13:437–457. doi: 10.1146/annurev.iy.13.040195.002253. [DOI] [PubMed] [Google Scholar]
- 7.Flynn JAL, Lazarevic V, Nolt D. Long-term control of Mycobacterium tuberculosis infection is mediated by dynamic immune responses. Journal of Immunology. 2005;175:1107–1117. doi: 10.4049/jimmunol.175.2.1107. [DOI] [PubMed] [Google Scholar]
- 8.Monack DM, Mueller A, Falkow S. Persistent bacterial infections: The interface of the pathogen and the host immune system. Nature Reviews Microbiology. 2004;2:747–765. doi: 10.1038/nrmicro955. [DOI] [PubMed] [Google Scholar]
- 9.Weiss DS, Raupach B, Takeda K, Akira S, Zychlinsky A. Toll-like receptors are temporally involved in host defense. Journal of Immunology. 2004;172:4463–4469. doi: 10.4049/jimmunol.172.7.4463. [DOI] [PubMed] [Google Scholar]
- 10.Poltorak A, He XL, Smirnova I, Liu MY, Van Huffel C, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 11.Lorenz E, Mira JP, Frees KL, Schwartz DA. Relevance of mutations in the TLR4 receptor in patients with gram-negative septic shock. Arch Intern Med. 2002;162:1028–1032. doi: 10.1001/archinte.162.9.1028. [DOI] [PubMed] [Google Scholar]
- 12.Hue NT, Lanh MN, Phuong LT, Vinh H, Chinh NT, et al. Toll-like receptor 4 (TLR4) and typhoid fever in Vietnam. Proc. ASM Conference ‘Salmonella: From Pathogenesis to Therapeutics’ Am Soc Microbiol. 2006;B38:56. [Google Scholar]
- 13.Arpaia N, Godec J, Lau L, Sivick KE, McLaughlin LM, et al. TLR Signaling Is Required for Salmonella typhimurium Virulence. Cell. 2011;144:675–688. doi: 10.1016/j.cell.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Werling D, Jann OC, Offord V, Glass EJ, Coffey TJ. Variation matters: TLR structure and species-specific pathogen recognition. Trends Immunol. 2009;30:124–130. doi: 10.1016/j.it.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Brownlie R, Zhu J, Allan B, Mutwiri GK, Babiuk LA, et al. Chicken TLR21 acts as a functional homologue to mammalian TLR9 in the recognition of CpG oligodeoxynucleotides. Mol Immunol. 2009;46:3163–3170. doi: 10.1016/j.molimm.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 16.Cook DN, Pisetsky DS, Schwartz DA. Toll-like receptors in the pathogenesis of human disease. Nat Immunol. 2004;5:975–979. doi: 10.1038/ni1116. [DOI] [PubMed] [Google Scholar]
- 17.Liew FY, Xu D, Brint EK, O'Neill LA. Negative regulation of toll-like receptor-mediated immune responses. Nat Rev Immunol. 2005;5:446–458. doi: 10.1038/nri1630. [DOI] [PubMed] [Google Scholar]
- 18.Shuto T, Furuta T, Oba M, Xu H, Li JD, et al. Promoter hypomethylation of Toll-like receptor-2 gene is associated with increased proinflammatory response toward bacterial peptidoglycan in cystic fibrosis bronchial epithelial cells. FASEB J. 2006;20:782–784. doi: 10.1096/fj.05-4934fje. [DOI] [PubMed] [Google Scholar]
- 19.Babu S, Blauvelt CP, Kumaraswami V, Nutman TB. Cutting edge: diminished T cell TLR expression and function modulates the immune response in human filarial infection. J Immunol. 2006;176:3885–3889. doi: 10.4049/jimmunol.176.7.3885. [DOI] [PubMed] [Google Scholar]
- 20.Melmed G, Thomas LS, Lee N, Tesfay SY, Lukasek K, et al. Human intestinal epithelial cells are broadly unresponsive to Toll-like receptor 2-dependent bacterial ligands: implications for host-microbial interactions in the gut. J Immunol. 2003;170:1406–1415. doi: 10.4049/jimmunol.170.3.1406. [DOI] [PubMed] [Google Scholar]
- 21.Abreu MT, Vora P, Faure E, Thomas LS, Arnold ET, et al. Decreased expression of Toll-like receptor-4 and MD-2 correlates with intestinal epithelial cell protection against dysregulated proinflammatory gene expression in response to bacterial lipopolysaccharide. J Immunol. 2001;167:1609–1616. doi: 10.4049/jimmunol.167.3.1609. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi K, Sugi Y, Hosono A, Kaminogawa S. Epigenetic regulation of TLR4 gene expression in intestinal epithelial cells for the maintenance of intestinal homeostasis. J Immunol. 2009;183:6522–6529. doi: 10.4049/jimmunol.0901271. [DOI] [PubMed] [Google Scholar]
- 23.Herman JG, Civin CI, Issa JP, Collector MI, Sharkis SJ, et al. Distinct patterns of inactivation of p15INK4B and p16INK4A characterize the major types of hematological malignancies. Cancer Res. 1997;57:837–841. [PubMed] [Google Scholar]
- 24.Gonzalez-Zulueta M, Bender CM, Yang AS, Nguyen T, Beart RW, et al. Methylation of the 5′ CpG island of the p16/CDKN2 tumor suppressor gene in normal and transformed human tissues correlates with gene silencing. Cancer Res. 1995;55:4531–4535. [PubMed] [Google Scholar]
- 25.Hisano M, Ohta H, Nishimune Y, Nozaki M. Methylation of CpG dinucleotides in the open reading frame of a testicular germ cell-specific intronless gene, Tact1/Actl7b, represses its expression in somatic cells. Nucleic Acids Res. 2003;31:4797–4804. doi: 10.1093/nar/gkg670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wigley P, Berchieri A, Jr, Page KL, Smith AL, Barrow PA. Salmonella enterica serovar Pullorum persists in splenic macrophages and in the reproductive tract during persistent, disease-free carriage in chickens. Infect Immun. 2001;69:7873–7879. doi: 10.1128/IAI.69.12.7873-7879.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hausmann M, Kiessling S, Mestermann S, Webb G, Spottl T, et al. Toll-like receptors 2 and 4 are up-regulated during intestinal inflammation. Gastroenterology. 2002;122:1987–2000. doi: 10.1053/gast.2002.33662. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Wang Y, Yamakuchi M, Isowaki S, Nagata E, et al. Upregulation of toll-like receptor 2 gene expression in macrophage response to peptidoglycan and high concentration of lipopolysaccharide is involved in NF-kappa b activation. Infect Immun. 2001;69:2788–2796. doi: 10.1128/IAI.69.5.2788-2796.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abasht B, Kaiser MG, Lamont SJ. Toll-like receptor gene expression in cecum and spleen of advanced intercross line chicks infected with Salmonella enterica serovar Enteritidis. Vet Immunol Immunopathol. 2008;123:314–323. doi: 10.1016/j.vetimm.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 30.Abasht B, Kaiser MG, van der Poel J, Lamont SJ. Genetic lines differ in Toll-like receptor gene expression in spleens of chicks inoculated with Salmonella enterica serovar Enteritidis. Poult Sci. 2009;88:744–749. doi: 10.3382/ps.2008-00419. [DOI] [PubMed] [Google Scholar]
- 31.Halford S, Mattei MG, Daw S, Scambler PJ. A novel C2H2 zinc-finger protein gene (ZNF160) maps to human chromosome 19q13.3-q13.4. Genomics. 1995;25:322–323. doi: 10.1016/0888-7543(95)80149-g. [DOI] [PubMed] [Google Scholar]
- 32.Wiznerowicz M, Jakobsson J, Szulc J, Liao S, Quazzola A, et al. The Kruppel-associated box repressor domain can trigger de novo promoter methylation during mouse early embryogenesis. J Biol Chem. 2007;282:34535–34541. doi: 10.1074/jbc.M705898200. [DOI] [PubMed] [Google Scholar]
- 33.Jakobsson J, Cordero MI, Bisaz R, Groner AC, Busskamp V, et al. KAP1-mediated epigenetic repression in the forebrain modulates behavioral vulnerability to stress. Neuron. 2008;60:818–831. doi: 10.1016/j.neuron.2008.09.036. [DOI] [PubMed] [Google Scholar]
- 34.Sripathy SP, Stevens J, Schultz DC. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol Cell Biol. 2006;26:8623–8638. doi: 10.1128/MCB.00487-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ayyanathan K, Lechner MS, Bell P, Maul GG, Schultz DC, et al. Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes Dev. 2003;17:1855–1869. doi: 10.1101/gad.1102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindell KA, Saeed AM, McCabe GP. Evaluation of resistance of four strains of commercial laying hens to experimental infection with Salmonella enteritidis phage type eight. Poult Sci. 1994;73:757–762. doi: 10.3382/ps.0730757. [DOI] [PubMed] [Google Scholar]
- 37.Yang H, Wei J, Zhang H, Song W, Wei W, et al. Upregulation of Toll-like Receptor (TLR) expression and release of cytokines from mast cells by IL-12. Cell Physiol Biochem. 2010;26:337–346. doi: 10.1159/000320557. [DOI] [PubMed] [Google Scholar]
- 38.Yang H, Wei J, Zhang H, Lin L, Zhang W, et al. Upregulation of Toll-like receptor (TLR) expression and release of cytokines from P815 mast cells by GM-CSF. BMC Cell Biol. 2009;10:37. doi: 10.1186/1471-2121-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collier-Hyams LS, Zeng H, Sun J, Tomlinson AD, Bao ZQ, et al. Cutting edge: Salmonella AvrA effector inhibits the key proinflammatory, anti-apoptotic NF-kappa B pathway. J Immunol. 2002;169:2846–2850. doi: 10.4049/jimmunol.169.6.2846. [DOI] [PubMed] [Google Scholar]
- 40.Haraga A, Ohlson MB, Miller SI. Salmonellae interplay with host cells. Nat Rev Microbiol. 2008;6:53–66. doi: 10.1038/nrmicro1788. [DOI] [PubMed] [Google Scholar]
- 41.Kinjyo I, Hanada T, Inagaki-Ohara K, Mori H, Aki D, et al. SOCS1/JAB is a negative regulator of LPS-induced macrophage activation. Immunity. 2002;17:583–591. doi: 10.1016/s1074-7613(02)00446-6. [DOI] [PubMed] [Google Scholar]
- 42.Nakagawa R, Naka T, Tsutsui H, Fujimoto M, Kimura A, et al. SOCS-1 participates in negative regulation of LPS responses. Immunity. 2002;17:677–687. doi: 10.1016/s1074-7613(02)00449-1. [DOI] [PubMed] [Google Scholar]
- 43.Li P, Xia P, Wen J, Zheng M, Chen J, et al. Up-regulation of the MyD88-dependent pathway of TLR signaling in spleen and caecum of young chickens infected with Salmonella serovar Pullorum. Vet Microbiol. 2010;143:346–351. doi: 10.1016/j.vetmic.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 44.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 45.Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology. 2002;148:257–266. doi: 10.1099/00221287-148-1-257. [DOI] [PubMed] [Google Scholar]
- 46.Deng SX, Cheng AC, Wang MS, Cao P. Serovar-specific real-time quantitative detection of Salmonella enteritidis in the gastrointestinal tract of ducks after oral challenge. Avian Dis. 2008;52:88–93. doi: 10.1637/8102-090107-Reg. [DOI] [PubMed] [Google Scholar]