Abstract
Nucleus-encoded regulatory factors are major contributors to mitochondrial biogenesis and function. Several act within the organelle to regulate mitochondrial transcription and translation while others direct the expression of nuclear genes encoding the respiratory chain and other oxidative functions. Loss-of-function studies for many of these factors reveal a wide spectrum of phenotypes. These range from embryonic lethality and severe respiratory chain deficiency to relatively mild mitochondrial defects seen only under conditions of physiological stress. The PGC-1 family of regulated coactivators (PGC-1α, PGC-1β and PRC) plays an important integrative role through their interactions with transcription factors (NRF-1, NRF-2, ERRα, CREB, YY1 and others) that control respiratory gene expression. In addition, recent evidence suggests that PGC-1 coactivators may balance the cellular response to oxidant stress by promoting a pro-oxidant environment or by orchestrating an inflammatory response to severe metabolic stress. These pathways may serve as essential links between the energy generating functions of mitochondria and the cellular REDOX environment associated with longevity, senescence and disease.
1. Introduction
Mitochondria are the sites of an array of biochemical activities but are best known for their role in biological oxidations. In particular, they house enzyme systems that convert the chemical bond energy derived mainly from the oxidation of carbohydrates and lipids to NADH and FADH2. These powerful reducing agents are utilized by the electron transport chain and oxidative phosphorylation system of the mitochondrial inner membrane to generate an electrochemical proton gradient across the membrane. This is accomplished by a series of electrogenic proton pumps that couple electron flow to the extrusion of protons to the cytosolic side of the membrane. The resulting proton motive force, comprised of both a voltage potential and a pH gradient, is used by the membrane bound ATP synthase to drive the synthesis of ATP [1,2] or by uncoupling proteins to generate heat [3]. Although the mitochondrial electron carriers are highly efficient in delivering electron pairs to molecular oxygen, the terminal acceptor, occasionally molecular oxygen can become partially reduced by a single electron forming superoxide anion, a highly reactive and toxic species. This occurs predominately at complexes I and III presumably under conditions of excess reducing power and abundant oxygen. The resulting superoxide anion can be converted to hydrogen peroxide via superoxide dismutases localized to both the cytosol and the mitochondrial matrix. However, cells contain a number of powerful scavenging systems to deal with these toxic products making it unlikely that they accumulate in large amounts under normal conditions. Hydrogen peroxide is converted to water by catalase, peroxiredoxin or glutathione peroxidase accounting for the protection of cellular constituents from large transient amounts of free radicals [4].
Mitochondria and their chloroplast cousins are unique among eukaryotic organelles in having their own genetic systems. Mammalian mitochondria have a covalently closed circular genome of approximately 16 kilobases that encodes 37 genes. In contrast to the nuclear genome, where long and short interspersed repeats, introns and vast intergenic regions account for more than 95 percent of the total DNA, the mtDNA of mammals and other vertebrates exhibits a highly economical sequence organization. In mammals, mitochondrial genes are lacking introns and are arranged head to tail with little or no intergenic regions. The only protein coding genes direct the synthesis of 13 mRNAs for essential respiratory chain subunits while the remaining 24 genes encode the 22 tRNAs and 2 rRNAs required for translation of these mRNAs within the mitochondrial matrix. Thus, the entire mitochondrial genetic system is retained solely for the purpose of providing 13 essential protein subunits of respiratory complexes I, III, IV and V. These are expressed through the bi-directional synthesis of multigenic transcripts, followed by the processing, polyadenylation and translation of individual mRNAs [5,6]. Surprisingly, the organizational economy found in vertebrate mtDNA is not observed in plants and fungi where the mitochondrial genomes are much larger and contain intergenic regions, introns and multiple promoters and transcriptional units [7,8]. Nevertheless, nearly the same small complement of mitochondrial genes exists over the entire evolutionary spectrum necessitating that nuclear genes control mitochondrial transcription, translation and DNA replication as well as provide the vast majority of gene products required for the biochemical functions and molecular architecture of the organelle.
2. Phenotypic similarities associated with loss of essential nucleus-encoded mitochondrial functions
Because of the semiautonomous nature of mitochondria, interest has focused on a number of nucleus-encoded gene products that act exclusively within the organelle to regulate the mitochondrial genetic system (Fig. 1). In particular, loss-of-function studies provide insights into the phenotypic effects of ablating the expression of nuclear gene products that regulate mitochondrial gene expression. The first of these was Tfam, a high mobility group (HMG)-box protein that stimulates bi-directional transcription through specific promoter recognition [9]. Tfam has been recovered in vertebrate nucleoids in association with other proteins required for mtDNA genomic integrity and expression [10-12]. A germ line Tfam knockout mouse exhibited embryonic lethality at E10.5, a severe oxidative phosphorylation defect and a marked reduction in mtDNA content, demonstrating a requirement for Tfam in mtDNA maintenance in vivo [13]. Interestingly, the hearts of a cardiac/skeletal muscle-specific Tfam knockout, in addition to having respiratory chain defect and diminished mtDNA levels, also had abundant atypical mitochondria with tubular cristae [14]. Skeletal muscle-specific Tfam knockout mice had several features characteristic of mitochondrial myopathy in humans including cytochrome oxidase-deficient ragged red muscle fibers that derive their appearance from the accumulation of abundant abnormal mitochondria [15].
Fig. 1.
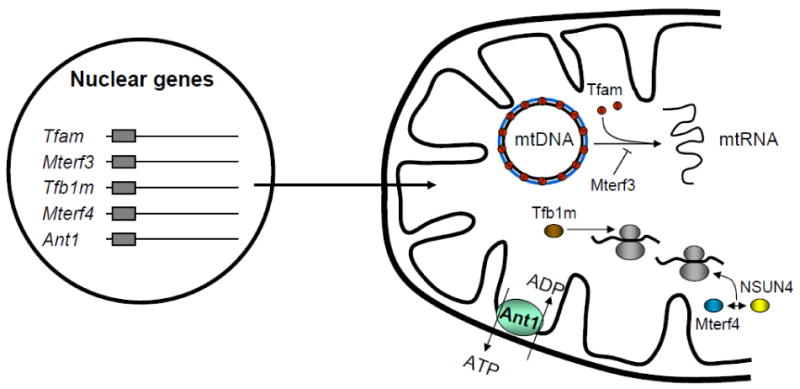
Summary of the sites of action for nuclear gene products that act within the mitochondria to control mitochondrial transcription and translation. Mouse knockouts of the indicated nuclear genes share common phenotypes consisting of embryonic lethality and severe respiratory chain deficiency accompanied by abundant abnormal cardiac mitochondria. Tfam (red spheres) binds the mitochondrial chromosome at multiple sites and functions in both mtDNA maintenance and transcription initiation. Mter3 serves as a negative regulator of this process. Tfb1m (brown ellipse) and Mterf4 (blue ellipse) participate in mitochondrial ribosome assembly. Tfb1m is a dimethytransferase that catalyzes the adenine dimethylation of the small ribosomal RNA required for ribosome assembly and translation. Similarly, a complex containing Mterf4 and the rRNA methyltransferase, NSUN4 (yellow ellipse), participates in the assembly of the large ribosomal subunit. Although not a direct regulator of gene expression, mouse knockouts of the adenine nucleotide translocator, Ant1 (green ellipse), share several phenotypic features with Tfam, Tfb1m, Mterf3 and Mterf4 knockouts. Thus, ablations of different nuclear genes whose products function exclusively within the mitochondria have similar consequences for the biogenesis and function of the organelle.
The basic pattern of embryonic lethality at approximately E8.5 in germ line knockouts and severe respiratory chain defects associated with abundant atypical mitochondria in the heart-specific counterparts has been observed upon the ablation of other nucleus encoded factors that supply diverse functions within the mitochondria (Fig. 1). This is true for Mterf3, encoding a negative regulator of mitochondrial transcription initiation [16], Tfb1m, encoding a dimethyltransferase [17], and, most recently, Mterf4, encoding a regulator of ribosome biogenesis and translation [18]. Loss of these functions is often accompanied by defects in respiratory complexes I, III, IV and V which rely upon mitochondria-encoded subunits. Interestingly, mice with a homozygous germ line knockout of Ant1, encoding an adenine nucleotide translocator (Fig. 1), also had ragged red muscle fibers with abundant abnormal mitochondria and their hearts displayed cardiac hypertrophy with massive mitochondrial proliferation [19]. It is also notable that a germ line mouse knockout of PolgA, encoding a subunit of mitochondrial DNA polymerase, also resulted in mid gestation lethality [20].
These findings illustrate that ablation of nuclear genes, whose products provide diverse functions within the mitochondria to maintain electron transport and oxidative phosphorylation share similar phenotypic features in both germ line and tissue-specific knockout mice. Embryonic lethality at mid-gestation is common to most of the germ line knockouts. Although multiple genes contribute diverse regulatory functions, the muscle-specific knockouts of each resulted in an increased abundance of morphologically and functionally defective cardiac mitochondria. This suggests that there is a similar adaptive response to genetic lesions affecting non-redundant nuclear functions that act exclusively to maintain the biogenesis and function of the mitochondrial respiratory apparatus.
3. Nuclear transcription factors
Characterization of cytochrome c and cytochrome oxidase promoters led to the identification of nuclear respiratory factors, NRF-1 and NRF-2, as activators of nuclear genes providing multiple mitochondrial functions. NRF-1binds a palindromic recognition site in the cytochrome c promoter as a homodimer and functions as a positive regulator of transcription [21-24]. Serine phosphorylation of the protein in proliferating cells enhances both its DNA binding [24] and trans-activation functions [25]. NRF-1 has been implicated in the expression of many nuclear genes required for expression of the mitochondrial respiratory chain [26-28]. In addition to respiratory chain subunits, NRF-1 is linked to the expression of nuclear genes necessary for mitochondrial transcription [29,30], heme biosynthesis [31,32], and protein import and assembly [33-35]. In addition, NRF-1 is among seven transcription factors whose recognition sites are most frequently found in the proximal promoters of ubiquitously expressed genes [36]. Chromatin immunoprecipitations (ChIP) coupled with microarray assay (ChIP-on-chip) identified 691 genes whose promoters are occupied by NRF-1 in living cells [28]. The majority are involved in mitochondrial biogenesis and metabolism. A major subclass of the NRF-1 targets also bound the growth regulatory transcription factor E2F and this subset was enriched in genes required for DNA replication, mitosis and cytokinesis. An NRF-1 siRNA reduced expression of several E2F target genes along with Tfam and cytochrome c confirming the biological significance of these observations.
Physiological studies support a role for NRF-1 in integrating respiratory chain expression with diverse cellular functions. Elevated NRF-1 expression or DNA binding activity has been associated with exercise-related changes in mitochondrial biogenesis [37-39]. NRF-1 DNA binding activity, cytochrome c content and mitochondrial density all increase upon activation of AMP-activated protein kinase by a creatine analog that induces muscle adaptations resembling those observed during exercise [40]. Muscle-specific COX genes depend upon MEF-2 and/or E-box consensus elements for their tissue-specific expression [41]. NRF-1 regulates the expression of MEF-2A in muscle providing a potential mechanism for indirect NRF-1 control of muscle-specific COX subunits and other muscle-specific MEF-2 target genes [42]. Interestingly, the transcriptional expression of NRF-1 is under the control of estradiol in a breast adenocarcinoma cell line mediated by the direct interaction of estrogen receptor with its recognition site in the NRF-1 promoter [43]. The induction of NRF-1 by estrogen was followed by the Tfam-regulated induction of mitochondrial transcripts and subsequent increases in oxygen consumption and mitochondrial DNA levels. The results implicate NRF-1 as a primary estrogen receptor target gene involved in the increased mitochondrial activity in estrogen-responsive cells. In addition, a link between NRF-1 and the lipopolysaccharide induced inflammatory response was demonstrated by the identification of NF-κB-responsive elements in the NRF-1 promoter region [44]. This response is also facilitated by CREB binding to the promoter and the functional interaction between NF-κB and CREB in controlling NRF-1-dependent mitochondrial biogenesis may help mitigate inflammatory damage. CREB is known to combine with NRF-1 in the regulated expression of cytochrome c in response to serum growth factors [25]. Its association with the NRF-1 promoter during the inflammatory response may be suggestive of a general CREB-dependent mechanism for the control of respiratory chain expression. Additionally, NRF-1 acts on the expression of the chemokine receptors CXCR4 and CXCR7 which bind stromal-derived factor-1, a proinflammatory α-chemokine regulating the metastatic behavior of several cancers [45,46]. These results are indicative of a link between respiratory gene expression and pro-inflammatory pathways.
A nuclear protein designated as NRF-2 was purified to homogeneity as a multisubunit transcription factor that bound essential cis-acting elements in the cytochrome oxidase subunit IV (COXIV) promoter [47,48]. These sites had the GGAA core motif that is recognized by the ETS-domain family of transcription factors. NRF-2 has an α subunit that binds DNA and four others (β1, β2, γ1 and γ2,) that complex with α to provide an activation domain and to modulate binding affinity [48,49]. Molecular cloning of the five subunits [49] revealed that NRF-2 is the human homologue of mouse GABP [50]. Both NRF-1 and NRF-2 have been linked directly to the expression of all ten nucleus-encoded cytochrome oxidase subunits [51-53]. Chromatin immunoprecipitations demonstrated that NRF-2 associates in vivo with multiple COX subunit promoters [51]. Moreover, expression of all ten nucleus-encoded COX subunits was abrogated by expression of either a dominant negative NRF-2 allele or a siRNA directed against NRF-2α [52] pointing to an important regulatory function for NRF-2 in the control of cytochrome oxidase expression. NRF-2 sites have been found in many other nuclear genes related to respiratory chain expression (for compilations see [26,27] including genes for Tfam [29] and the TFB isoforms where both NRF-1 and NRF-2α occupy both TFB promoters in living cells as determined by chromatin immunoprecipitation [30]. Similar control by both NRFs extend to three of the four human succinate dehydrogenase (complex II) subunit genes [54-56]. Like NRF-1, NRF-2 participates in the expression of the mitochondrial protein import machinery with functional recognition sites in TOMM70 [57,58] and TOMM20 [34] promoters. NRF-2 has also been linked to the expression of several mitochondrial translation initiation factors [59]. Involvement of both NRFs in the expression of mitochondrial transcription factors and the subunits of protein import receptor complexes is consistent with a mechanism for coordinating the expression of the respiratory chain with the biogenesis of the organelle itself. This is consistent with the induction of several members of the PGC-coactivator-NRF regulatory axis along with MyoD during mitochondrial biogenesis that occurs early in a program of skeletal muscle regeneration [60].
In addition to the NRFs, the orphan nuclear receptors, estrogen-related receptors α, β and γ (ERRs), have emerged as important regulators of many genes involved in oxidative metabolism [61-63]. In particular, ERRα is expressed at high levels in oxidative tissues and promotes β-oxidation through its control of the medium chain acyl-coenzyme A dehydrogenase (MCAD) gene [64-66]. ERRα is an important target in the control of mitochondrial biogenesis by PGC-1α [67,68] and may provide a regulatory link between fatty acid oxidation and the respiratory chain [61]. ERRα engages in a direct regulatory interaction with NRF-2 (GABP) and the same computational analysis places NRF-1 downstream from ERRα in the regulatory cascade [68]. It is notable that genome wide surveys of ERRα binding suggests that ERRα can bind the putative GABPα promoter region and that NRF-1 sites are significantly enriched among genomic sequences showing ERRα occupancy [69]. More recently, the induced expression of PGC-1β during myogenic differentiation was found to activate a number of respiratory genes by direct targeting of both NRF-1 and ERRα [70]. Immunoprecipitation experiments demonstrated that both NRF-1 and ERRα interact directly with PGC-1β. Moreover, the deletion of NRF-1 and ERRα recognition sites within respiratory gene promoters or the siRNA silencing of either transcription factor attenuated the induction of respiratory gene expression or mitochondrial biogenesis by PGC-1β. Finally, functional genomic analysis involving the genome-wide location of transcription factor occupancy has revealed that the ERRs are associated with hundreds of genes controlling all aspects of mitochondrial function [62,63]. On this basis it has been postulated that the ERRs are master regulators of mitochondrial biogenesis and function.
Additional nuclear transcription factors have been linked to the control of mitochondrial respiratory function. The cAMP response element binding protein, CREB, along with NRF-1 is involved in the growth-regulated expression of cytochrome c [25,71,72]. Both factors participate in the induction of cytochrome c in response to serum stimulation of quiescent cells [25]. CREB has been rediscovered as an activator of the cytochrome c promoter and is required for activation of the promoter by ERRα [69]. CREB sites are not generally found in the proximal promoters of nuclear respiratory genes [25] and there is no selective enrichment among genomic fragments containing ERR binding sites [69]. The initiator element binding factor, YY1, is engaged in both the positive and negative control of certain cytochrome oxidase subunit genes [73,74]. YY1 recognition sites along with those for the NRFs and ERRα were enriched among genes encoding OXPHOS subunits and assembly factors [75]. However, YY1 and CREB showed less specificity for respiratory genes than NRF-1 or NRF-2. Nevertheless, YY1 has been identified as a target for the nutrient sensor mTOR and YY1 silencing in myotubules results in diminished mitochondrial gene expression [76]. Finally, c-myc directs the expression of certain NRF-1 target genes [77] and myc null fibroblasts are deficient in mitochondrial content [78]. Interestingly, c-myc positively regulates mitochondrial biogenesis and the respiratory apparatus through PGC-1β, an effect that is repressed by HIF-1 in the absence of the von Hippel-Landau tumor supressor. This pathway may be part of a molecular switch from oxidative phosphorylation to aerobic glycolysis mediated by HIF-1 in certain cancers [79].
4. In vivo phenotypes associated with loss of nuclear transcription factor function
Homozygous knockout mice have been generated for many of the nuclear transcription factors associated with mitochondrial biogenesis and respiratory function. Many of these exhibit peri-implantation lethality. This is the case for NRF-1 [80], GABPα (NRF-2α) [81] and YY1 [82]. These severe phenotypes are not surprising given the fact that the factors act on hundreds of genes affecting a range of cellular functions. In the case of NRF-1, the null blastocysts had depleted mitochondrial DNA levels and diminished mitochondrial membrane potential consistent with a respiratory chain deficiency [80]. However, they failed to progress beyond E6.5 and also failed to differentiate in vitro. This likely reflects the fact that NRF-1 targets include genes encoding metabolic enzymes, components of signaling pathways and gene products necessary for chromosome maintenance and nucleic acid metabolism among others [23]. In keeping with a potential link between mitochondrial biogenesis and cell cycle progression, mouse embryo fibroblasts lacking GABPα (NRF-2α) failed to proliferate because of an inability to replicate DNA [83]. Only a small subset of GABP target genes showed reduced expression and these did not include genes related to mitochondrial function. This was ascribed to the fact that other transcription factors that contribute to the expression of GABP targets were normally expressed and may compensate for the defect. Interestingly, the Drosophila homologue of NRF-2α is required for the expression of most genes encoding mitochondrial proteins and also links mitochondrial abundance to cell growth [84]. Finally, a homozygous knockout of c-myc results in lethality between E9.5 and E10.5 [85]. This is consistent with a role for c-myc in the control of cell cycle progression and metabolic networks [86]. These in vivo results suggest that key factors implicated in the biogenesis of mitochondria also function in the control of the cell cycle.
Mouse knockouts of other nuclear factors required for mitochondrial function have surprisingly mild phenotypes. PPARα null mice have diminished expression of mitochondrial fatty acid oxidation enzymes [87] while ERRα knockouts have reduced lipogenesis and exhibit cold intolerance [88], but both are viable and fertile with no gross phenotypic abnormalities. This might be explained by the fact that, unlike YY1 and the NRFs, both of these factors have multiple isoforms that may compensate for the loss of a single family member. For example, ERRγ and ERRα bind the same recognition sites within essentially the same collection of target genes [62] although the ERRγ knockouts exhibit early postnatal lethality associated with a defect in oxidative phosphorylation in the heart [89]. It is important to note that many target genes for ERR and the other nuclear factors have been identified by functional genomic analysis and there is a general paucity of information regarding the direct effects of a given factor on protein expression, complex assembly or metabolic flux through a specific pathway. This may be especially significant for the respiratory complexes which are thought to be in excess and thus must suffer substantial reductions in expression before the threshold for a respiratory defect is crossed [90]. In addition to cold intolerance [91], the ERRα null mice display a number of cardiac deficiencies when stressed by pressure overload but apparently lack any robust defect in organelle biogenesis that might be expected from the loss of a master regulator [92].
5. The PGC-1 coactivator family
The PGC-1coactivator family is a small family of nuclear transcriptional cofactors consisting of PGC-1α, PGC-1β and PRC. PGC-1α, the founding member of the family, was discovered based on its induction during adaptive thermogenesis in brown fat and its interaction with PPARγ, an important regulator of adipocyte differentiation [93]. PGC-1α lacks histone-modifying activities but it interacts through a potent amino terminal activation domain with coactivators such as SRC-1 and CBP/p300 which have their own intrinsic ability to remodel chromatin [94,95]. The association of PGC-1α with an increasing number of coactivator complexes is consistent with the many hormonal and nutrient signaling pathways associated with PGC-1α. In addition to PPARγ, PGC-1α binds a large complement of transcription factors and nuclear hormone receptors. Among those directly associated with mitochondrial respiratory function are NRF-1, ERRα, YY1, PPARα and MEF2C. A homologue of PGC-1α was designated as PGC-1β or PERC [96,97]. Both PGC-1α and β are enriched in tissues that contain abundant mitochondria such as heart, skeletal muscle and brown fat but PGC-1β is not induced in brown fat in response to cold exposure [96,97]. PGC-1α and β share sequence similarities along their entire lengths consistent with a relatively recent gene duplication [96]. PRC is more divergent from the other two family members with sequence conservations restricted to discreet domains [98]. These shared domains include the amino terminal activation domain and RNA recognition motif shared by all three family members and a central proline rich region and carboxy terminal R/S domain shared only by PGC-1α and PRC. The sequence conservation and spatial arrangement of these domains is consistent with related function.
NRF-1 was an early transcription factor target for PGC-1α in the regulation of mitochondrial biogenesis (Fig. 2). PGC-1α can trans-activate NRF-1 target genes and a dominant negative allele of NRF-1 blocks the effects of PGC-1α on mitochondrial biogenesis [99]. PGC-1β resembles PGC-1α in its functional interaction with NRF-1 and in its ability to direct the expression of nuclear respiratory genes and mitochondrial mass when expressed from viral vectors [100,101]. PGC-1α also targets estrogen-related receptor α (ERRα) in conjunction with GABPα (NRF-2α) in regulating respiratory genes including cytochrome c and β-ATP synthase [67,68]. A role for PGC-1α as a regulator of mitochondrial biogenesis is consistent with gain of function experiments in cultured cells [99] and in transgenic mice [102]. Ectopic PGC-1α expression in cultured cells increases COXIV and cytochrome c protein levels as well as the steady-state level of mtDNA [99]. Cardiac-specific over expression in transgenic mice results in massive increases in mitochondrial content in cardiac myocytes leading to edema and dilated cardiomyopathy [102].
Fig. 2.
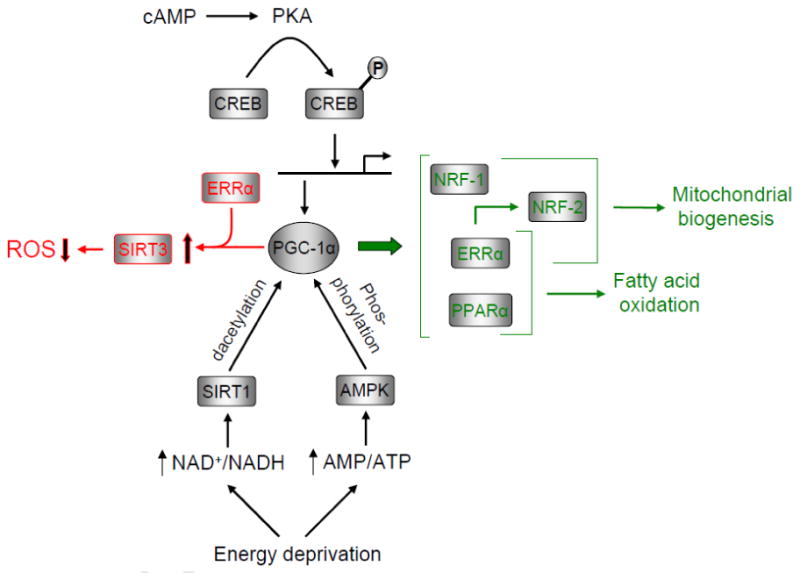
Potential integration of mitochondrial oxidative and antioxidant pathways by PGC-1α. PGC-1α gene expression is induced by cAMP signaling through protein kinase A (PKA) phosphorylation of the transcription factor CREB. This pathway is observed upon β-adrenergic receptor activation during thermogenesis in brown fat. PGC-1α is also activated by posttranscriptional phosphorylation by AMPK or by deacetylation by SirT1 in response to nutrient deprivation. Induction or activation of the coactivator can enhance mitochondrial biogenesis and oxidative function through the coactivation of multiple transcription factors involved in respiratory gene expression (green pathway). PGC-1α activation may also promote an antioxidant environment by combining with ERRα in the induction of SirT3, a mitochondrial sirtuin that can trigger ROS detoxification (red pathway).
Despite its sequence divergence from the other PGCs, PRC is similar to PGC-1α and β in its binding to NRF-1, CREB and ERRα and in its ability to trans-activate nuclear genes required for mitochondrial respiratory function (Fig. 3A) including cytochome c, 5-aminolevulinate synthase, Tfam and the TFB1M and TFB2M isoforms [30,72,98,103]. For each of these promoters, maximal activation by PRC requires functional NRF-1 recognition sites. Despite these similarities, PRC mRNA shows little induction in brown fat under conditions where PGC-1α is markedly elevated [98]. However, PRC levels are higher in proliferating cells compared to those that are growth arrested because of serum withdrawal or contact inhibition [72,98]. PRC mRNA is rapidly induced upon serum stimulation of quiescent fibroblasts in the absence of de novo protein synthesis. This, along with its relatively short mRNA half-life, is characteristic of the class of immediate early genes implicated in the proliferative response to growth factor stimulation (Fig. 3A). These genes encode chemokines, growth factors, proto-oncogenes, serine-threonine kinases, nucleic acid enzymes and transcription factors, among others [104,105]. Recently, PRC has been shown to direct mitochondrial biogenesis through its interaction with ERRα in cultured thyroid cells [106]. PRC was a potent coactivator of ERRα-mediated gene expression and PRC enhanced the effect of ERRα on both mitochondrial mass and cytochrome oxidase activity. In addition, PRC silencing by siRNA had a negative effect on both mitochondrial mass and oxygen consumption in highly oxidative cells. Thus, PRC has the characteristics of a growth regulatory factor that can affect both respiratory chain expression and the biogenesis of mitochondria.
Fig. 3.
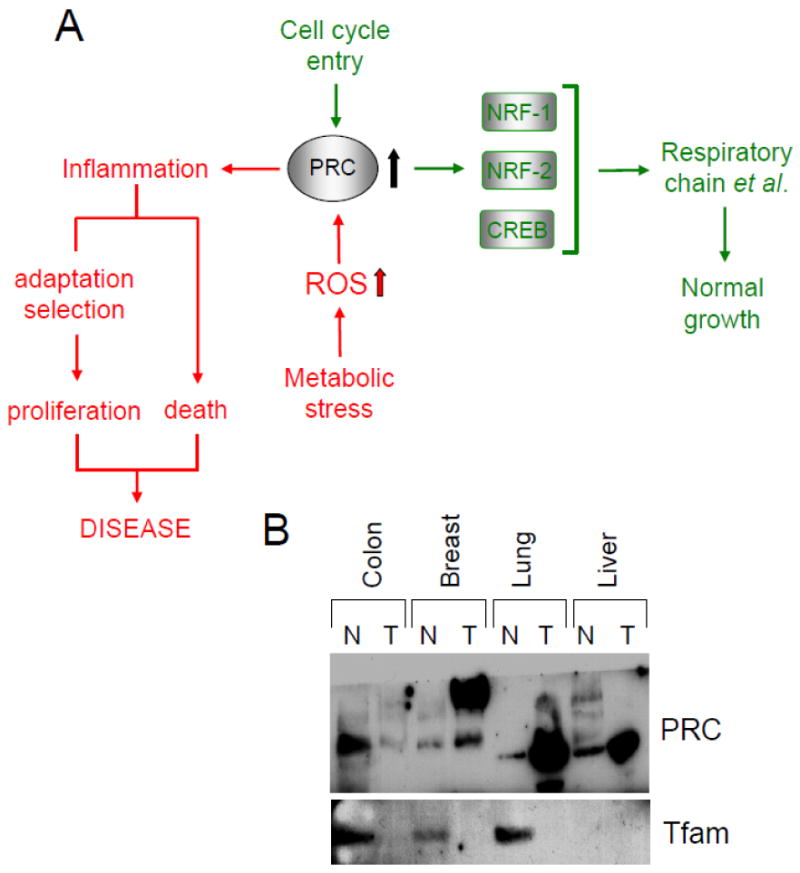
A potential role for PRC in orchestrating a balance between cell cycle entry and the inflammatory response to metabolic stress. (Panel A) Under normal conditions (green pathway), PRC protein levels are transiently upregulated during entry to the cell cycle. This coincides with the normal initiation of cell growth or as a response to serum stimulation of quiescent fibroblasts. In this capacity PRC is thought to activate genes encoding the respiratory chain and other growth-related functions through interactions with its target transcription factors. Alternatively, PRC protein levels can be induced in response to severe metabolic stress (red pathway) brought about by nutrient starvation or chemical uncoupling. This is associated with the activation of an antioxidant-sensitive inflammatory/stress program possibly leading to abnormal proliferation or cell death. Genes from this program have been associated with the tumor microenvironment and with cellular senescence. (Panel B) Evidence for elevated PRC protein levels in some solid tumors. A commercial immunoblot from DNA Technologies containing protein samples from several different tumors and the adjacent normal tissue was probed with affinity-purified rabbit anti-PRC antibody or rabbit anti-Tfam. The results show a robust PRC signal in breast, lung and liver tumors compared to modest expression in the surrounding normal tissue. Surprisingly, the PRC signal from breast tumor migrated at a higher molecular mass. The Tfam control was expressed at much reduced levels in several tumor samples compared to the surrounding tissue possibly reflecting the decreased mitochondrial mass associated with the glycolytic nature of many tumors.
6. Mitochondrial phenotypes associated with loss of PGC-1 family coactivator function
Over expression of PGC-1α or PGC-1β in cultured cells and in mouse tissues results in substantial increases in mitochondrial content [95,99,102,107]. In contrast, mice with germ line targeted disruptions of either PGC-1α [108,109] or PGC-1β [110,111] were viable and fertile with essentially normal energy expenditure, mitochondrial abundance and morphology. However, under physiological stress the PGC-1 knockouts exhibit more severe phenotypes. The PGC-1α null mouse was unable to defend body temperature upon cold exposure and had reduced cardiac ATP production and work output in response to physiological stimuli [108,109,112]. A skeletal muscle-specific PGC-1α knockout mouse exhibited multiple muscle defects including reduced exercise tolerance and abnormalities in the maintenance of normal muscle fiber composition [113]. PGC-1β knockout mice were viable and although the animals were not defective in energy intake or expenditure they also exhibited mitochondrial dysfunction under stress conditions [110]. Mitochondrial volume density was only marginally affected in brown fat and mRNAs encoding several nucleus-encoded respiratory proteins were diminished. COX4 and SDHB protein levels were also down and the animals exhibited defects in thermoregulation. Mitochondrial content was reduced in soleus muscle and heart along with modest reductions in respiratory gene expression. However, the mitochondria retained normal internal structure and the respiratory function of isolated mitochondria was indistinguishable between the wild type and knockout animals. Decreased respiratory mRNA expression was confirmed in an independent experiment although this was not linked to any global systemic changes in mitochondrial biogenesis or morphology [111]. These results demonstrate that either coactivator alone is not required for normal development or for the global biogenesis or maintenance of mitochondria.
The potential for functional complementation in the individual PGC-1 knockouts was addressed by the construction and analysis of PGC-1α/β double knockout mice [114]. The viability of these mice at birth indicated that the absence of both coactivators was compatible with a full course of prenatal development. However, the majority died of cardiac failure within 24 hours with none surviving beyond 14 days. Unlike the individual knockouts, the mitochondrial volume density in the brown fat of the PGC-1α/β null mice was reduced significantly along with diminished cristae density coinciding with reduced expression of several respiratory genes. Moreover, cardiac failure was accompanied by heart mitochondrial defects marked by a reduction in mitochondrial number and size as well as the presence of internal vacuoles with reduced cristae. These differences were ascribed to a defect in mitochondrial maturation which normally occurs during the late fetal and early postnatal stages [115,116]. The double knockout also exhibited lower expression of fatty acid oxidation and oxidative phosphorylation genes including a number of ERRα and ERRγ target genes suggesting that impairment of the PGC-1-ERR pathway contributes to the mitochondrial maturation defect [114]. The results argue that the two coactivators provide complementary functions in supporting postnatal cardiac differentiation.
Although PRC knockout mice have not been generated, stable lentiviral PRC shRNA transductants of U2OS cells having either complete or partial PRC silencing have been analyzed [117]. Complete PRC silencing led to marked inhibition of respiratory growth on galactose, diminished expression of both nuclear and mitochondrial respiratory chain subunits, lower levels of respiratory complexes I and IV and reduced production of mitochondrial ATP. Moreover, this transductant had abundant, morphologically defective mitochondria, a phenotype similar to those observed in the tissue-specific disruption of Tfam, Tfb1m, Mterf3 and Mterf4 whose products are localized to mitochondria and are required by the mitochondrial genetic system [13,16-18]. In addition, a gene array revealed many mitochondria-related genes whose expression was altered significantly upon PRC silencing [117]. These included respiratory chain subunits, mitochondrial protein import and assembly factors, and mitochondrial ribosomal proteins and tRNA synthetases, among others. Notably, stable expression of a PRC subfragment that binds both NRF-1 and CREB inhibits respiratory growth on galactose [72] suggesting that the interaction between PRC and its target transcription factors contributes to mitochondrial respiratory function. In addition, transient PRC silencing by siRNA also led to diminished respiratory chain expression and was most pronounced for complexes I, III and IV [118], providing independent confirmation that PRC acts as a positive regulator of nuclear genes required for expression of the mitochondrial respiratory complexes. A second, independent lentiviral transductant with partial PRC silencing to a level of about 15 percent of control cells had a milder galactose growth defect, but had no significant deficit in respiratory chain expression and ATP production [117]. Apparently, relatively low PRC levels are sufficient to maintain a nearly normal mitochondrial phenotype in this system. These results suggest that PRC expression may account for the absence of a more severe mitochondrial defect in the PGC-1 knockout mice by maintaining mitochondrial biogenesis and respiratory function.
7. Nutrient sensing and metabolic stress
It is well established that the PGC-1 coactivators are regulated at the level of mRNA expression through signaling pathways modulating growth, differentiation and metabolism [119,120]. These include pathways that converge on the CREB-dependent induction of PGC-1α in response to cold exposure, nutrient deprivation and exercise [121-123]. Metabolic signaling through PGC-1α also occurs through post-translational modifications (Fig. 2). For example, nutrient sensing through the NAD+ -dependent deacetylase, SirT1, results in the deacetylation of multiple lysine residues in PGC-1α thereby promoting mitochondrial fatty acid oxidation in response to low glucose [124]. Moreover, energy depletion in muscle triggers the activation of AMP-activated protein kinase (AMPK) which phosphorylates PGC-1α on specific serine and threonine residues [125]. The subsequent enhancement of mitochondrial gene expression is consistent with the notion that AMPK can mediate at least some of its effects through PGC-1α [126]. The SIRT and AMPK pathways can also cooperate in promoting calcium dependent mitochondrial biogenesis in myocytes [127,128].
Recent findings support a role for PGC-1α in linking nutrient deprivation to mitochondrial oxidant production through its control over SirT3 expression (Fig. 2). SirT3 is a member of the sirtuin family of deacetylases that is localized to mitochondria where it modifies key enzymatic activities in optimizing metabolic function [129]. In addition, SirT3 can exert a negative effect on oxidant stress by precipitating a series of reactions beginning with the activation of isocitrate dehydrogenase 2 and culminating in the detoxification of reactive oxygen species by glutathione peroxidase [130]. It can also deacetylate manganese superoxide dismutase in the mitochondrial matrix leading to increased activity and enhanced oxidant scavenging [131,132]. SirT3 can deacetylate specific respiratory complex subunits leading to diminished complex I, II and III activity in cells devoid of the SirT3 gene [133-135]. Complex III is thought to be a major site of cytosolic ROS production [136] potentially linking SirT3 to the control of cytosolic ROS levels. Thus, multiple SirT3-dependent mechanisms are implicated in mediating antioxidant effects associated with increased longevity [137].
PGC-1α coactivates the expression of SirT3 through ERRα which binds the SirT3 proximal promoter [138,139]. Occupancy of the Sirt3 promoter region by both PGC-1α and ERRα occurs in vivo and silencing of ERRα had a negative effect on the induction of Sirt3 by PGC-1α. Importantly, SirT3 was required for the induction of several components of the ROS detoxifying machinery by PGC-1α. Since PGC-1α is activated by SirT1 deacetylation in response to nutrient deprivation [140], PGC-1α can potentially serve as a link between nutrient sensing pathways and the oxidant scavenging mechanisms facilitated by SirT3 (Fig. 2). This relationship may have important implications for understanding the positive effects on longevity associated with the restriction of caloric intake [137].
A recent study has implicated PRC as a sensor of metabolic stress in cultured U2OS cells [141]. Several metabolic insults including treatment with the respiratory chain uncoupler, CCCP, expression of a dominant negative allele of NRF-1 or glucose deprivation all resulted in the marked induction of PRC protein levels (Fig. 3A). A microarray screen comparing PRC-expressing and non-expressing cells identified at least 45 PRC-responsive genes, the majority of which are involved in inflammation, cell stress and proliferation. A major category of PRC-dependent inflammatory/stress genes included pro-inflammatory molecules (e.g. IL-1α, IL-8, cyclooxygenase 2) as well as members of the proline-rich protein family (SPRR2D and SPRR2F). The latter are associated with the response to DNA damage and exit from the cell cycle [142,143]. Several of these PRC-dependent stress genes have been previously linked to the inflammatory microenvironment in a number of human cancers [144,145] and may also be associated with cellular senescence [146,147]. This is consistent with the observations that c-myc is induced along with PRC by chemical uncoupling [141] and that PRC is markedly elevated in several human tumor specimens (Fig. 3B). PRC protein levels are substantially increased in breast, lung and liver tumors compared to modest expression in the surrounding normal tissue (Fig. 3B). It is unclear why PRC from breast tumor migrated at a higher molecular mass. Tfam was expressed at much reduced levels in colon, breast and lung tumor specimens compared to the surrounding tissue, possibly reflecting the decrease in mitochondrial mass associated with the glycolytic nature of many tumors (Fig. 3B). These preliminary findings, along with the observation that PRC is elevated in human thyroid oncocytoma [148], suggests that PRC may contribute to the chronic inflammation associated with certain cancers.
PRC may have a dual role in cell growth control. Under normal conditions it is transiently expressed upon initiation of the proliferative cycle [72]. However, under conditions of metabolic stress, PRC departs from its usual transient expression pattern and becomes constitutively elevated [141]. In this capacity it may promote an inflammatory/stress response that triggers adaptive changes in cell structure, metabolism and growth (Fig. 3A). Interestingly, the induction of PRC by respiratory chain uncoupler and the associated inflammatory/stress response is highly sensitive to antioxidant [141], suggesting that this pathway is optimized in a pro-oxidant environment. This contrasts with the PGC-1α/SirT3 pathway which, along with ERRα, links nutrient deprivation to oxidant scavenging through induction of the ROS detoxifying machinery [138,139]. Thus, these seemingly opposing pathways, directed by different members of the PGC-1 coactivator family, may provide a balance between oxidant detoxification linked to longevity and the chronic inflammation associated with cellular senescence and disease. In this scenario, PGC-1α promotes an antioxidant environment through SirT3 in support of the increased mitochondrial fatty acid oxidation precipitated by nutrient limitation [124]. This may exert positive effects in promoting growth and longevity (Fig. 2). However, under conditions of severe stress, possibly resulting from genetic mutation, environmental toxins or oncogene activation, PRC may contribute to the induction of an inflammatory/stress response that accompanies cellular senescence (Fig. 3A). It is tempting to suggest that balance between the PGC-1α/SirT3 and PRC pathways is reflected in the diminished levels of SirT3 in human cancer cells [135,149] as opposed to the elevated PRC protein levels (Fig. 3B).
8. Summary and perspective
Nucleus-encoded regulatory proteins are essential to the control of mitochondrial biogenesis and oxidative function. Those factors that act exclusively within the mitochondrion (Tfam,Tfb1m, Mterf3, Mterf4) are absolutely required to maintain the morphology, biogenesis and function of the organelle and their loss in mouse knockouts has severe consequences for embryonic development and tissue function. Knockouts of several nuclear transcription factors acting on nuclear respiratory genes (NRF-1, NRF-2, YY1) display early embryonic lethality whereas others (ERRα, PPARα) yield relatively mild phenotypes. The latter may appear non-essential because they exist in families of multiple isoforms that may compensate for the loss of a single member. This is illustrated in the PGC-1 coactivator family where the individual PGC-1α or β knockout mice are viable with a mild mitochondrial phenotype compared to the severe defect in postnatal cardiac mitochondrial maturation observed in the double PGC-1α/β knockout. In addition to its contributions to the energy generating functions of mitochondria, PGC-1α may help maintain a pro-oxidant environment in response to nutrient limitation by inducing the SirT3-dependent detoxification of reactive oxygen species. Thus, this coactivator may contribute to the effects of nutrient limitation on longevity by promoting efficient metabolic function coupled to oxidant detoxification. A third member of the PGC-1 family, PRC, appears to function as an immediate-early gene during cell cycle entry but can also orchestrate an anti-oxidant-sensitive inflammatory response to severe metabolic stress. The nutrient sensing pathways directed by these members of the PGC-1 coactivator family may help balance the cellular responses to nutrient availability and may serve as essential links between the energy generating functions of mitochondria and the cellular REDOX environment associated with longevity, senescence and disease.
Highlights.
Nucleus-encoded factors control the expression mitochondrial respiratory function
Nucleus-encoded factors acting within mitochondria are essential for embryogenesis
Ablation of factors acting on nuclear respiratory genes results in a range of phenotypes
Nuclear coactivators governing respiratory gene expression respond to metabolic stress
Acknowledgments
Work in the author's laboratory was supported by National Institute of General Medical Sciences Grant GM 32525-29.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hatefi Y. The mitochondrial electron transport chain and oxidative phosphorylation system. Annu Rev Biochem. 1985;54:1015–1069. doi: 10.1146/annurev.bi.54.070185.005055. [DOI] [PubMed] [Google Scholar]
- 2.Balaban RS. Regulation of oxidative phosphorylation in the mammalian cell. Am J Physiol Cell Physiol. 1990;258:C377–C389. doi: 10.1152/ajpcell.1990.258.3.C377. [DOI] [PubMed] [Google Scholar]
- 3.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 4.Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Taanman JW. The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta Bio-Energetics. 1999;1410:103–123. doi: 10.1016/s0005-2728(98)00161-3. [DOI] [PubMed] [Google Scholar]
- 6.Wallace DC. A Mitochondrial Paradigm of Metabolic and Degenerative Diseases, Aging, and Cancer: A Dawn for Evolutionary Medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costanzo MC, Fox TD. Control of mitochondrial gene expression in Saccharomyces cerevisiae. Annu Rev Genet. 1990;24:91–113. doi: 10.1146/annurev.ge.24.120190.000515. [DOI] [PubMed] [Google Scholar]
- 8.Poyton RO, McEwen JE. Crosstalk between nuclear and mitochondrial genomes. Annu Rev Biochem. 1996;65:563–607. doi: 10.1146/annurev.bi.65.070196.003023. [DOI] [PubMed] [Google Scholar]
- 9.Fisher RP, Clayton DA. Purification and characterization of human mitochondrial transcription factor 1. Mol Cell Biol. 1988;8:3496–3509. doi: 10.1128/mcb.8.8.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bogenhagen DF, Wang Y, Shen EL, Kobayashi R. Protein components of mitochondrial DNA nucleoids in higher eukaryotes. Mol Cell Proteomics. 2003;2:1205–1216. doi: 10.1074/mcp.M300035-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Bogenhagen DF. Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J Biol Chem. 2006;281:25791–25802. doi: 10.1074/jbc.M604501200. [DOI] [PubMed] [Google Scholar]
- 12.Kaufman BA, Durisic N, Mativetsky JM, Costantino S, Hancock MA, Grutter P, Shoubridge EA. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol Biol Cell. 2007;18:3225–3236. doi: 10.1091/mbc.E07-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsson NG, Wang JM, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nature Genetics. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Wilhelmsson H, Graff C, Li H, Oldfors A, Rustin P, Bruning JC, Kahn CR, Clayton DA, Barsh GS, Thoren P, Larsson NG. Dilated cardiomyopathy and atrioventricular conduction blocks induced by heart-specific inactivation of mitochondrial DNA gene expression. Nature Genet. 1999;21:133–137. doi: 10.1038/5089. [DOI] [PubMed] [Google Scholar]
- 15.Wredenberg A, Wibom R, Wilhelmsson H, Graff C, Wiener HH, Burden SJ, Oldfors A, Westerblad H, Larsson NG. Increased mitochondrial mass in mitochondrial myopathy mice. Proc Natl Acad Sci USA. 2002;99:15066–15071. doi: 10.1073/pnas.232591499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park CB, sin-Cayuela J, Camara Y, Shi Y, Pellegrini M, Gaspari M, Wibom R, Hultenby K, Erdjument-Bromage H, Tempst P, Falkenberg M, Gustafsson CM, Larsson NG. MTERF3 is a negative regulator of mammalian mtDNA transcription. Cell. 2007;130:273–285. doi: 10.1016/j.cell.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 17.Metodiev MD, Lesko N, Park CB, Camara Y, Shi Y, Wibom R, Hultenby K, Gustafsson CM, Larsson NG. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 2009;9:386–397. doi: 10.1016/j.cmet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 18.Camara Y, sin-Cayuela J, Park CB, Metodiev MD, Shi Y, Ruzzenente B, Kukat C, Habermann B, Wibom R, Hultenby K, Franz T, Erdjument-Bromage H, Tempst P, Hallberg BM, Gustafsson CM, Larsson NG. MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab. 2011;13:527–539. doi: 10.1016/j.cmet.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Graham BH, Waymire KG, Cottrell B, Trounce IA, MacGregor GR, Wallace DC. A mouse model for mitochondrial myopathy and cardiomyopathy resulting from a deficiency in the heart/muscle isoform of the adenine nucleotide translocator. Nature Genetics. 1997;16:226–234. doi: 10.1038/ng0797-226. [DOI] [PubMed] [Google Scholar]
- 20.Hance N, Ekstrand MI, Trifunovic A. Mitochondrial DNA polymerase γ is essential for mammalian embryogenesis. Hum Mol Genet. 2005;14:1775–1783. doi: 10.1093/hmg/ddi184. [DOI] [PubMed] [Google Scholar]
- 21.Evans MJ, Scarpulla RC. Interaction of nuclear factors with multiple sites in the somatic cytochrome c promoter. Characterization of upstream NRF-1, ATF and intron Sp1 recognition sites. J Biol Chem. 1989;264:14361–14368. [PubMed] [Google Scholar]
- 22.Evans MJ, Scarpulla RC. NRF-1:a trans-activator of nuclear-encoded respiratory genes in animal cells. Genes Dev. 1990;4:1023–1034. doi: 10.1101/gad.4.6.1023. [DOI] [PubMed] [Google Scholar]
- 23.Virbasius CA, Virbasius JV, Scarpulla RC. NRF-1, an activator involved in nuclear-mitochondrial interactions, utilizes a new DNA-binding domain conserved in a family of developmental regulators. Genes Dev. 1993;7:2431–2445. doi: 10.1101/gad.7.12a.2431. [DOI] [PubMed] [Google Scholar]
- 24.Gugneja S, Scarpulla RC. Serine phosphorylation within a concise amino-terminal domain in nuclear respiratory factor 1 enhances DNA binding. J Biol Chem. 1997;272:18732–18739. doi: 10.1074/jbc.272.30.18732. [DOI] [PubMed] [Google Scholar]
- 25.Herzig RP, Scacco S, Scarpulla RC. Sequential serum-dependent activation of CREB and NRF-1 leads to enhanced mitochondrial respiration through the induction of cytochrome c. J Biol Chem. 2000;275:13134–13141. doi: 10.1074/jbc.275.17.13134. [DOI] [PubMed] [Google Scholar]
- 26.Scarpulla RC. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochem Biophys Acta. 2002;1576:1–14. doi: 10.1016/s0167-4781(02)00343-3. [DOI] [PubMed] [Google Scholar]
- 27.Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- 28.Cam H, Balciunaite E, Blias A, Spektor A, Scarpulla RC, Young R, Kluger Y, Dynlacht BD. A common set of gene regulatory networks links metabolism and growth inhibition. Mol Cell. 2004;16:399–411. doi: 10.1016/j.molcel.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 29.Virbasius JV, Scarpulla RC. Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: A potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc Natl Acad Sci USA. 1994;91:1309–1313. doi: 10.1073/pnas.91.4.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gleyzer N, Vercauteren K, Scarpulla RC. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Mol Cell Biol. 2005;25:1354–1366. doi: 10.1128/MCB.25.4.1354-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braidotti G, Borthwick IA, May BK. Identification of regulatory sequences in the gene for 5-aminolevulinate synthase from rat. J Biol Chem. 1993;268:1109–1117. [PubMed] [Google Scholar]
- 32.Aizencang GI, Bishop DF, Forrest D, Astrin KH, Desnick RJ. Uroporphyrinogen III synthase. An alternative promoter controls erythroid-specific expression in the murine gene. J Biol Chem. 2000;275:2295–2304. doi: 10.1074/jbc.275.4.2295. [DOI] [PubMed] [Google Scholar]
- 33.Takahashi Y, Kako K, Arai H, Oishi T, Inada Y, Tkehara A, Fukamizu A, Munekata E. Characterization and identification of promoter elements in the mouse COX17 gene. Biochem Biophys Acta. 2002;1574:359–364. doi: 10.1016/s0167-4781(01)00374-8. [DOI] [PubMed] [Google Scholar]
- 34.Blesa JR, Prieto-Ruiz JA, Hernandez JM, Hernandez-Yago J. NRF-2 transcription factor is required for human TOMM20 gene expression. Gene. 2007;391:198–208. doi: 10.1016/j.gene.2006.12.024. [DOI] [PubMed] [Google Scholar]
- 35.Blesa JR, Prieto-Ruiz JA, Abraham BA, Harrison BL, Hegde AA, Hernandez-Yago J. NRF-1 is the major transcription factor regulating the expression of the human TOMM34 gene. Biochem Cell Biol. 2008;86:46–56. doi: 10.1139/o07-151. [DOI] [PubMed] [Google Scholar]
- 36.FitzGerald PC, Shlyakhtenko A, Mir AA, Vinson C. Clustering of DNA sequences in human promoters. Genome Res. 2004;14:1562–1574. doi: 10.1101/gr.1953904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baar K, Wende AR, Jones TE, Marison M, Nolte LA, Chen M, Kelly DP, Holloszy JO. Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1. FASEB J. 2002;16:1879–1886. doi: 10.1096/fj.02-0367com. [DOI] [PubMed] [Google Scholar]
- 38.Murakami T, Shimomura Y, Yosimura A, Sokabe M, Fujitsuka N. Induction of nuclear respiratory factor-1 expression by an acute bout of exercise in rat muscle. Biochem Biophys Acta. 1998;1381:113–122. doi: 10.1016/s0304-4165(98)00018-x. [DOI] [PubMed] [Google Scholar]
- 39.Ojuka EO, Jones TE, Han DH, Chen M, Holloszy JO. Raising Ca2+ in L6 myotubes mimics effects of exercise on mitochondrial biogenesis in muscle. FASEB J. 2003;17:675–681. doi: 10.1096/fj.02-0951com. [DOI] [PubMed] [Google Scholar]
- 40.Bergeron R, Ren JM, Cadman KS, Moore IK, Perret P, Pypaert M, Young LH, Semenkovich CF, Shulman GI. Chronic activation of AMP kinase results in NRF-1 activation and mitochondrial biogenesis. Am J Physiol Endocrinol Metab. 2001;281:E1340–E1346. doi: 10.1152/ajpendo.2001.281.6.E1340. [DOI] [PubMed] [Google Scholar]
- 41.Wan B, Moreadith RW. Structural characterization and regulatory element analysis of the heart isoform of cytochrome c oxidase VIa. J Biol Chem. 1995;270:26433–26440. doi: 10.1074/jbc.270.44.26433. [DOI] [PubMed] [Google Scholar]
- 42.Ramachandran B, Yu G, Gulick T. Nuclear respiratory factor 1 controls myocyte enhancer factor 2A transcription to provide a mechanism for coordinate expression of respiratory chain subunits. J Biol Chem. 2008;283:11935–11946. doi: 10.1074/jbc.M707389200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mattingly KA, Ivanova MM, Riggs KA, Wickramasinghe NS, Barch MJ, Klinge CM. Estradiol stimulates transcription of nuclear respiratory factor-1 and increases mitochondrial biogenesis. Mol Endocrinol. 2008;22:609–622. doi: 10.1210/me.2007-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suliman HB, Sweeney TE, Withers CM, Piantadosi CA. Co-regulation of nuclear respiratory factor-1 by NFkappaB and CREB links LPS-induced inflammation to mitochondrial biogenesis. J Cell Sci. 2010;123:2565–2575. doi: 10.1242/jcs.064089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moriuchi M, Moriuchi H, Fauci AS. HTLV type I Tax activation of the CXCR4 promoter by association with nuclear respiratory factor 1. Aids Res Hum Retrov. 1999;15:821–827. doi: 10.1089/088922299310728. [DOI] [PubMed] [Google Scholar]
- 46.Tarnowski M, Grymula K, Reca R, Jankowski K, Maksym R, Tarnowska J, Przybylski G, Barr FG, Kucia M, Ratajczak MZ. Regulation of expression of stromal-derived factor-1 receptors: CXCR4 and CXCR7 in human rhabdomyosarcomas. Mol Cancer Res. 2010;8:1–14. doi: 10.1158/1541-7786.MCR-09-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Virbasius JV, Scarpulla RC. Transcriptional activation through ETS domain binding sites in the cytochrome c oxidase subunit IV gene. Mol Cell Biol. 1991;11:5631–5638. doi: 10.1128/mcb.11.11.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Virbasius JV, Virbasius CA, Scarpulla RC. Identity of GABP with NRF-2, a multisubunit activator of cytochrome oxidase expression, reveals a cellular role for an ETS domain activator of viral promoters. Genes Dev. 1993;7:380–392. doi: 10.1101/gad.7.3.380. [DOI] [PubMed] [Google Scholar]
- 49.Gugneja S, Virbasius JV, Scarpulla RC. Four structurally distinct, non-DNA-binding subunits of human nuclear respiratory factor 2 share a conserved transcriptional activation domain. Mol Cell Biol. 1995;15:102–111. doi: 10.1128/mcb.15.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.LaMarco KL, McKnight SL. Purification of a set of cellular polypeptides that bind to the purine-rich cis-regulatory element of herpes simplex virus immediate early genes. Genes Dev. 1989;3:1372–1383. doi: 10.1101/gad.3.9.1372. [DOI] [PubMed] [Google Scholar]
- 51.Ongwijitwat S, Wong-Riley MT. Is nuclear respiratory factor 2 a master transcriptional coordinator for all ten nuclear-encoded cytochrome c oxidase subunits in neurons? Gene. 2005;360:65–77. doi: 10.1016/j.gene.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 52.Ongwijitwat S, Liang HL, Graboyes EM, Wong-Riley MT. Nuclear respiratory factor 2 senses changing cellular energy demands and its silencing down-regulates cytochrome oxidase and other target gene mRNAs. Gene. 2006;374:39–49. doi: 10.1016/j.gene.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 53.Dhar SS, Ongwijitwat S, Wong-Riley MT. Nuclear respiratory factor 1 regulates all ten nuclear-encoded subunits of cytochrome c oxidase in neurons. J Biol Chem. 2008;283:3120–3129. doi: 10.1074/jbc.M707587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Au HC, Scheffler IE. Promoter analysis of the human succinate dehydrogenase iron-protein gene. Both nuclear respiratory factors NRF-1 and NRF-2 are required. Eur J Biochem. 1998;251:164–174. doi: 10.1046/j.1432-1327.1998.2510164.x. [DOI] [PubMed] [Google Scholar]
- 55.Elbehti-Green A, Au HC, Mascarello JT, Ream-Robinson D, Scheffler IE. Characterization of the human SDHC gene encoding one of the integral membrane proteins of succinate-quinone oxidoreductase in mitochondria. Gene. 1998;213:133–140. doi: 10.1016/s0378-1119(98)00186-3. [DOI] [PubMed] [Google Scholar]
- 56.Hirawake H, Taniwaki M, Tamura A, Amino H, Tomitsuka E, Kita K. Characterization of the human SDHD gene encoding the small subunit of cytochrome b (cybS) in mitochondrial succinate-ubiquinone oxidoreductase. Biochim Biophys Acta. 1999;1412:295–300. doi: 10.1016/s0005-2728(99)00071-7. [DOI] [PubMed] [Google Scholar]
- 57.Blesa JR, Hernandez JM, Hernandez-Yago J. NRF-2 transcription factor is essential in promoting human Tomm70 gene expression. Mitochondrion. 2004;3:251–259. doi: 10.1016/j.mito.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 58.Blesa JR, Hegde AA, Hernandez-Yago J. In vitro methylation of nuclear respiratory factor-2 binding sites suppresses the promoter activity of the human TOMM70 gene. Gene. 2008;427:58–64. doi: 10.1016/j.gene.2008.09.016. [DOI] [PubMed] [Google Scholar]
- 59.Hayashi R, Ueda T, Farwell MA, Takeuchi N. Nuclear respiratory factor 2 activates transcription of human mitochondrial translation initiation factor 2 gene. Mitochondrion. 2007;7:195–203. doi: 10.1016/j.mito.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 60.Wagatsuma A, Kotake N, Mabuchi K, Yamada S. Expression of nuclear-encoded genes involved in mitochondrial biogenesis and dynamics in experimentally denervated muscle. J Physiol Biochem. 2011 doi: 10.1007/s13105-011-0083-5. [DOI] [PubMed] [Google Scholar]
- 61.Giguere V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr Rev. 2008;29:677–696. doi: 10.1210/er.2008-0017. [DOI] [PubMed] [Google Scholar]
- 62.Eichner LJ, Giguere V. Estrogen related receptors (ERRs): a new dawn in transcriptional control of mitochondrial gene networks. Mitochondrion. 2011;11:544–552. doi: 10.1016/j.mito.2011.03.121. [DOI] [PubMed] [Google Scholar]
- 63.Deblois G, Giguere V. Functional and physiological genomics of estrogen-related receptors (ERRs) in health and disease. Biochim Biophys Acta. 2011;1812:1032–1040. doi: 10.1016/j.bbadis.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 64.Vega RB, Kelly DP. A role for estrogen-related receptor α in the control of mitochondrial fatty acid β-oxidation during brown adipocyte differentiation. J Biol Chem. 1997;272:31693–31699. doi: 10.1074/jbc.272.50.31693. [DOI] [PubMed] [Google Scholar]
- 65.Sladek R, Bader J, Giguere V. The orphan nuclear receptor estrogen-related receptor α is a transcriptional regulator of the human medium-chain acyl coenzyme A dehydrogenase gene. Mol Cell Biol. 1997;17:5400–5409. doi: 10.1128/mcb.17.9.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huss JM, Kelly DP. Nuclear receptor signaling and cardiac energetics. Circ Res. 2004;95:568–578. doi: 10.1161/01.RES.0000141774.29937.e3. [DOI] [PubMed] [Google Scholar]
- 67.Schreiber SN, Emter R, Hock MB, Knutti D, Cardenas J, Podvinec M, Oakeley EJ, Kralli A. The estrogen-related receptor α (ERRα) functions in PPARgamma coactivator 1α (PGC-1α)-induced mitochondrial biogenesis. Proc Natl Acad Sci USA. 2004;101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mootha VK, Handschin C, Arlow D, Xie XH, St Pierre J, Sihag S, Yang WL, Altshuler D, Puigserver P, Patterson N, Willy PJ, Schulman IG, Heyman RA, Lander ES, Spiegelman BM. Errα and Gabpa/b specify PGC-1α-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci USA. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dufour CR, Wilson BJ, Huss JM, Kelly DP, Alaynick WA, Downes M, Evans RM, Blanchette M, Giguere V. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRalpha and gamma. Cell Metab. 2007;5:345–356. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 70.Shao D, Liu Y, Liu X, Zhu L, Cui Y, Cui A, Qiao A, Kong X, Liu Y, Chen Q, Gupta N, Fang F, Chang Y. PGC-1 beta-regulated mitochondrial biogenesis and function in myotubes is mediated by NRF-1 and ERR alpha. Mitochondrion. 2010;10:516–527. doi: 10.1016/j.mito.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 71.Gopalakrishnan L, Scarpulla RC. Differential regulation of respiratory chain subunits by a CREB-dependent signal transduction pathway. Role of cyclic AMP in cytochrome c and COXIV gene expression. J Biol Chem. 1994;269:105–113. [PubMed] [Google Scholar]
- 72.Vercauteren K, Pasko RA, Gleyzer N, Marino VM, Scarpulla RC. PGC-1-related coactivator (PRC): immediate early expression and characterization of a CREB/NRF-1 binding domain associated with cytochrome c promoter occupancy and respiratory growth. Mol Cell Biol. 2006;26:7409–7419. doi: 10.1128/MCB.00585-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Basu A, Lenka N, Mullick J, Avadhani NG. Regulation of murine cytochrome oxidase Vb gene expression in different tissues and during myogenesis - Role of a YY-1 factor- binding negative enhancer. J Biol Chem. 1997;272:5899–5908. doi: 10.1074/jbc.272.9.5899. [DOI] [PubMed] [Google Scholar]
- 74.Seelan RS, Grossman LI. Structural organization and promoter analysis of the bovine cytochrome c oxidase subunit VIIc gene - A functional role for YY1. J Biol Chem. 1997;272:10175–10181. doi: 10.1074/jbc.272.15.10175. [DOI] [PubMed] [Google Scholar]
- 75.van WC, Moraes CT. Transcriptional co-expression and co-regulation of genes coding for components of the oxidative phosphorylation system. BMC Genomics. 2008;9:18. doi: 10.1186/1471-2164-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 77.Morrish F, Giedt C, Hockenbery D. C-MYC apoptotic function is mediated by NRF-1 target genes. Genes Dev. 2003;17:240–255. doi: 10.1101/gad.1032503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O'Donnell KA, Kim JW, Yustein JT, Lee LA, Dang CV. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005;25:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, Dang CV, Semenza GL. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 80.Huo L, Scarpulla RC. Mitochondrial DNA instability and peri-implantation lethality associated with targeted disruption of nuclear respiratory factor 1 in mice. Mol Cell Biol. 2001;21:644–654. doi: 10.1128/MCB.21.2.644-654.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ristevski S, O'Leary DA, Thornell AP, Owen MJ, Kola I, Hertzog PJ. The ETS transcription factor GABPα is essential for early embryogenesis. Mol Cell Biol. 2004;24:5844–5849. doi: 10.1128/MCB.24.13.5844-5849.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Donohoe ME, Zhang X, McGinnis L, Biggers J, Li E, Shi Y. Targeted disruption of mouse yin yang 1 transcription factor results in peri-implantation lethality. Mol Cell Biol. 1999;19:7237–7244. doi: 10.1128/mcb.19.10.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang ZF, Mott S, Rosmarin AG. The Ets transcription factor GABP is required for cell-cycle progression. Nat Cell Biol. 2007;9:339–346. doi: 10.1038/ncb1548. [DOI] [PubMed] [Google Scholar]
- 84.Baltzer C, Tiefenbock SK, Marti M, Frei C. Nutrition controls mitochondrial biogenesis in the Drosophila adipose tissue through Delg and cyclin D/Cdk4. PLoS One. 2009;4:e6935. doi: 10.1371/journal.pone.0006935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Davis AC, Wims M, Spotts GD, Hann SR, Bradley A. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 1993;7:671–682. doi: 10.1101/gad.7.4.671. [DOI] [PubMed] [Google Scholar]
- 86.Morrish F, Neretti N, Sedivy JM, Hockenbery DM. The oncogene c-Myc coordinates regulation of metabolic networks to enable rapid cell cycle entry. Cell Cycle. 2008;7:1054–1066. doi: 10.4161/cc.7.8.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee SS, Pineau T, Drago J, Lee EJ, Owens JW, Kroetz DL, Fernandez-Salguero PM, Westphal H, Gonzalez FJ. Targeted disruption of the alpha isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol Cell Biol. 1995;15:3012–3022. doi: 10.1128/mcb.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Luo J, Sladek R, Carrier J, Bader J, Richard D, Giguere V. Reduced fat mass in mice lacking orphan nuclear receptor estrogen-related receptor α. Mol Bio Cell. 2003;23:7947–7956. doi: 10.1128/MCB.23.22.7947-7956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Alaynick WA, Kondo RP, Xie W, He W, Dufour CR, Downes M, Jonker JW, Giles W, Naviaux RK, Giguere V, Evans RM. ERRgamma directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell Metab. 2007;6:13–24. doi: 10.1016/j.cmet.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 90.Rossignol R, Malgat M, Mazat JP, Letellier T. Threshold effect and tissue specificity. Implication for mitochondrial cytopathies. J Biol Chem. 1999;274:33426–33432. doi: 10.1074/jbc.274.47.33426. [DOI] [PubMed] [Google Scholar]
- 91.Villena JA, Hock MB, Chang WY, Barcas JE, Giguere V, Kralli A. Orphan nuclear receptor estrogen-related receptor alpha is essential for adaptive thermogenesis. Proc Natl Acad Sci U S A. 2007;104:1418–1423. doi: 10.1073/pnas.0607696104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huss JM, Imahashi K, Dufour CR, Weinheimer CJ, Courtois M, Kovacs A, Giguere V, Murphy E, Kelly DP. The nuclear receptor ERRα is required for the bioenergetic and functional adaptation to cardiac pressure overload. Cell Metab. 2007;6:25–37. doi: 10.1016/j.cmet.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 93.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 94.Rodgers JT, Lerin C, Gerhart-Hines Z, Puigserver P. Metabolic adaptations through the PGC-1 alpha and SIRT1 pathways. FEBS Lett. 2008;582:46–53. doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lin JD. Minireview: the PGC-1 coactivator networks: chromatin-remodeling and mitochondrial energy metabolism. Mol Endocrinol. 2009;23:2–10. doi: 10.1210/me.2008-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lin J, Puigserver P, Donovan J, Tarr P, Spiegelman BM. PGC-1β: A novel PGC-1-related transcription coactivator associated with host cell factor. J Biol Chem. 2002;277:1645–1648. doi: 10.1074/jbc.C100631200. [DOI] [PubMed] [Google Scholar]
- 97.Kressler D, Schreiber SN, Knutti D, Kralli A. The PGC-1-related protein PERC is a selective coactivator of estrogen receptor α. J Biol Chem. 2002;277:13918–13925. doi: 10.1074/jbc.M201134200. [DOI] [PubMed] [Google Scholar]
- 98.Andersson U, Scarpulla RC. PGC-1-related coactivator, a novel, serum-inducible coactivator of nuclear respiratory factor 1-dependent transcription in mammalian cells. Mol Cell Biol. 2001;21:3738–3749. doi: 10.1128/MCB.21.11.3738-3749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and function through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 100.Meirhaeghe A, Crowley V, Lenaghan C, Lelliott C, Green K, Stewart A, Hart K, Schinner S, Sethi JK, Yeo G, Brand MD, Cortright RN, O'Rahilly S, Montague C, Vidal-Puig AJ. Characterization of the human, mouse and rat PGC1β (peroxisome-proliferator-activated receptor-gamma co-activator 1β) gene in vitro and in vivo. Biochem J. 2003;373:155–165. doi: 10.1042/BJ20030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lin JD, Tarr PT, Yang RJ, Rhee J, Puigserver P, Newgard CB, Spiegelman BM. PGC-1β in the regulation of hepatic glucose and energy metabolism. J Biol Chem. 2003;278:30843–30848. doi: 10.1074/jbc.M303643200. [DOI] [PubMed] [Google Scholar]
- 102.Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. Peroxisome proliferator-activated receptor γ coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vercauteren K, Gleyzer N, Scarpulla RC. PGC-1-related Coactivator Complexes with HCF-1 and NRF-2{beta} in Mediating NRF-2(GABP)-dependent Respiratory Gene Expression. J Biol Chem. 2008;283:12102–12111. doi: 10.1074/jbc.M710150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Herschman HR. Primary response genes induced by growth factors and tumor promoters. Annu Rev Biochem. 1991;60:281–319. doi: 10.1146/annurev.bi.60.070191.001433. [DOI] [PubMed] [Google Scholar]
- 105.Winkles JA. Serum- and polypeptide growth factor-inducible gene expression in mouse fibroblasts. Prog Nucleic Acid Res Mol Biol. 1998;58:41–78. doi: 10.1016/s0079-6603(08)60033-1. [DOI] [PubMed] [Google Scholar]
- 106.Mirebeau-Prunier D, Le PS, Jacques C, Gueguen N, Poirier J, Malthiery Y, Savagner F. Estrogen-related receptor alpha and PGC-1-related coactivator constitute a novel complex mediating the biogenesis of functional mitochondria. FEBS J. 2010;277:713–725. doi: 10.1111/j.1742-4658.2009.07516.x. [DOI] [PubMed] [Google Scholar]
- 107.Arany Z, Lebrasseur N, Morris C, Smith E, Yang W, Ma Y, Chin S, Spiegelman BM. The transcriptional coactivator PGC-1β drives the formation of oxidative type IIX fibers in skeletal muscle. Cell Metab. 2007;5:35–46. doi: 10.1016/j.cmet.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 108.Lin J, Wu P, Tarr PT, Lindenberg KS, St-Pierre J, Zhang C, Mootha VK, Jäger S, Vianna CR, Reznick RM, Cui L, Manieri M, Donovan MX, Wu Z, Cooper MP, Fan ML, Rohas LM, Zavacki AM, Cinti S, Shulman GI, Lowell BB, Krainc D, Spiegelman BM. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1α null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 109.Leone TC, Lehman JJ, Finck BN, Schaeffer PJ, Wende AR, Boudina S, Courtois M, Wozniak DF, Sambandam N, Bernal-Mizrachi C, Chen Z, Holloszy JO, Medeiros DM, Schmidt RE, Saffitz JE, Abel ED, Semenkovich CF, Kelly DP. PGC-1α deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:0672–0687. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lelliott CJ, Medina-Gomez G, Petrovic N, Kis A, Feldmann HM, Bjursell M, Parker N, Curtis K, Campbell M, Hu P, Zhang D, Litwin SE, Zaha VG, Fountain KT, Boudina S, Jimenez-Linan M, Blount M, Lopez M, Meirhaeghe A, Bohlooly Y, Storlien L, Stromstedt M, Snaith M, Oresic M, Abel ED, Cannon B, Vidal-Puig A. Ablation of PGC-1β results in defective mitochondrial activity, thermogenesis, hepatic function, and cardiac performance. PLoS Biol. 2006;4:2042–2056. doi: 10.1371/journal.pbio.0040369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sonoda J, Mehl IR, Chong LW, Nofsinger RR, Evans RM. PGC-1beta controls mitochondrial metabolism to modulate circadian activity, adaptive thermogenesis, and hepatic steatosis. Proc Natl Acad Sci U S A. 2007;104:5223–5228. doi: 10.1073/pnas.0611623104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Arany Z, He H, Lin J, Hoyer K, Handschin C, Toka O, Ahmad F, Matsui T, Chin S, Wu PH, Rybkin II, Shelton JM, Manieri M, Cinti S, Schoen FJ, Bassel-Duby R, Rosenzweig A, Ingwall JS, Spiegelman BM. Transcriptional coactivator PGC-1α controls the energy state and contractile function of cardiac muscle. Cell Metab. 2005;1:259–271. doi: 10.1016/j.cmet.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 113.Handschin C, Chin S, Li P, Liu F, Maratos-Flier E, Lebrasseur NK, Yan Z, Spiegelman BM. Skeletal muscle fiber-type switching, exercise intolerance and myopathy in PGC-1alpha muscle-specific knockout animals. J Biol Chem. 2007;282:30014–30021. doi: 10.1074/jbc.M704817200. [DOI] [PubMed] [Google Scholar]
- 114.Lai L, Leone TC, Zechner C, Schaeffer PJ, Kelly SM, Flanagan DP, Medeiros DM, Kovacs A, Kelly DP. Transcriptional coactivators PGC-1alpha and PGC-lbeta control overlapping programs required for perinatal maturation of the heart. Genes Dev. 2010;22:1948–1961. doi: 10.1101/gad.1661708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Valcarce C, Navarrete RM, Encabo P, Loeches E, Satrustegui J, Cuezva JM. Postnatal development of rat liver mitochondrial functions. J Biol Chem. 1988;263:7767–7775. [PubMed] [Google Scholar]
- 116.Marin-Garcia J, Ananthakrishnan R, Goldenthal MJ. Heart mitochondrial DNA and enzyme changes during early human development. Mol Cell Biochem. 2000;210:47–52. doi: 10.1023/a:1007031919298. [DOI] [PubMed] [Google Scholar]
- 117.Vercauteren K, Gleyzer N, Scarpulla RC. Short hairpin RNA-mediated silencing of PRC (PGC-1-related coactivator) results in a severe respiratory chain deficiency associated with the proliferation of aberrant mitochondria. J Biol Chem. 2009;284:2307–2319. doi: 10.1074/jbc.M806434200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Raharijaona M, Le PS, Poirier J, Mirebeau-Prunier D, Rouxel C, Jacques C, Fontaine JF, Malthiery Y, Houlgatte R, Savagner F. PGC-1-related coactivator modulates mitochondrial-nuclear crosstalk through endogenous nitric oxide in a cellular model of oncocytic thyroid tumours. PLoS One. 2009;4:e7964. doi: 10.1371/journal.pone.0007964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Scarpulla RC. Transcriptional paradigms in Mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 120.Fernandez-Marcos PJ, Auwerx J. Regulation of PGC-1alpha, a nodal regulator of mitochondrial biogenesis. Am J Clin Nutr. 2011;93:884S–8890. doi: 10.3945/ajcn.110.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Herzig S, Long FX, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, Spiegelman B, Montminy M. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 122.Wu H, Kanatous SB, Thurmond FA, Gallardo T, Isotani E, Bassel-Duby R, Williams RS. Regulation of mitochondrial biogenesis in skeletal muscle by CaMK. Science. 2002;296:349–352. doi: 10.1126/science.1071163. [DOI] [PubMed] [Google Scholar]
- 123.Handschin C, Rhee J, Lin JD, Tarr PT, Spiegelman BM. An autoregulatory loop controls peroxisome proliferator-activated receptor gamma coactivator 1α expression in muscle. Proc Natl Acad Sci USA. 2003;100:7111–7116. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Steinberg GR, Kemp BE. AMPK in Health and Disease. Physiol Rev. 2009;89:1025–1078. doi: 10.1152/physrev.00011.2008. [DOI] [PubMed] [Google Scholar]
- 126.Jager S, Handschin C, St-Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci U S A. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Iwabu M, Yamauchi T, Okada-Iwabu M, Sato K, Nakagawa T, Funata M, Yamaguchi M, Namiki S, Nakayama R, Tabata M, Ogata H, Kubota N, Takamoto I, Hayashi YK, Yamauchi N, Waki H, Fukayama M, Nishino I, Tokuyama K, Ueki K, Oike Y, Ishii S, Hirose K, Shimizu T, Touhara K, Kadowaki T. Adiponectin and AdipoR1 regulate PGC-1alpha and mitochondria by Ca(2+) and AMPK/SIRT1. Nature. 2010;464:1313–1319. doi: 10.1038/nature08991. [DOI] [PubMed] [Google Scholar]
- 129.Verdin E, Hirschey MD, Finley LW, Haigis MC. Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends Biochem Sci. 2010;35:669–675. doi: 10.1016/j.tibs.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, Tanokura M, Denu JM, Prolla TA. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143:802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010;12:662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 132.Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, Jiang H, Kim HS, Flynn CR, Hill S, Hayes MW, Olivier AK, Spitz DR, Gius D. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell. 2010;40:893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, Finkel T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci U S A. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cimen H, Han MJ, Yang Y, Tong Q, Koc H, Koc EC. Regulation of succinate dehydrogenase activity by SIRT3 in mammalian mitochondria. Biochemistry. 2010;49:304–311. doi: 10.1021/bi901627u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, ykin-Burns N, Pennington JD, van der MR, Nguyen P, Savage J, Owens KM, Vassilopoulos A, Ozden O, Park SH, Singh KK, Abdulkadir SA, Spitz DR, Deng CX, Gius D. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17:41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bell EL, Klimova TA, Eisenbart J, Moraes CT, Murphy MP, Budinger GR, Chandel NS. The Qo site of the mitochondrial complex III is required for the transduction of hypoxic signaling via reactive oxygen species production. J Cell Biol. 2007;177:1029–1036. doi: 10.1083/jcb.200609074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bell EL, Guarente L. The SirT3 Divining Rod Points to Oxidative Stress. Mol Cell. 2011;42:561–568. doi: 10.1016/j.molcel.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Giralt A, Hondares E, Villena JA, Ribas F, az-Delfin J, Giralt M, Iglesias R, Villarroya F. Peroxisome proliferator-activated receptor-gamma coactivator-1alpha controls transcription of the Sirt3 gene, an essential component of the thermogenic brown adipocyte phenotype. J Biol Chem. 2011;286:16958–16966. doi: 10.1074/jbc.M110.202390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kong X, Wang R, Xue Y, Liu X, Zhang H, Chen Y, Fang F, Chang Y. Sirtuin 3, a new target of PGC-1alpha, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS One. 2010;5:e11707. doi: 10.1371/journal.pone.0011707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dominy JE, Jr, Lee Y, Gerhart-Hines Z, Puigserver P. Nutrient-dependent regulation of PGC-1alpha's acetylation state and metabolic function through the enzymatic activities of Sirt1/GCN5. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbapap.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gleyzer N, Scarpulla RC. PGC-1-Related Coactivator (PRC), a Sensor of Metabolic Stress, Orchestrates a Redox-Sensitive Program of Inflammatory Gene Expression. J Biol Chem. 2011 doi: 10.1074/jbc.M111.291575. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lohman FP, Medema JK, Gibbs S, Ponec M, van de PP, Backendorf C. Expression of the SPRR cornification genes is differentially affected by carcinogenic transformation. Exp Cell Res. 1997;231:141–148. doi: 10.1006/excr.1996.3458. [DOI] [PubMed] [Google Scholar]
- 143.Tesfaigzi J, Carlson DM. Expression, regulation, and function of the SPR family of proteins. A review. Cell Biochem Biophys. 1999;30:243–265. doi: 10.1007/BF02738069. [DOI] [PubMed] [Google Scholar]
- 144.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mantovani A, Garlanda C, Allavena P. Molecular pathways and targets in cancer-related inflammation. Ann Med. 2010;42:161–170. doi: 10.3109/07853890903405753. [DOI] [PubMed] [Google Scholar]
- 146.Orjalo AV, Bhaumik D, Gengler BK, Scott GK, Campisi J. Cell surface-bound IL-1alpha is an upstream regulator of the senescence-associated IL-6/IL-8 cytokine network. Proc Natl Acad Sci U S A. 2009;106:17031–17036. doi: 10.1073/pnas.0905299106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Davalos AR, Coppe JP, Campisi J, Desprez PY. Senescent cells as a source of inflammatory factors for tumor progression. Cancer Metastasis Rev. 2010;29:273–283. doi: 10.1007/s10555-010-9220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Savagner F, Mirebeau D, Jacques C, Guyetant S, Morgan C, Franc B, Reynier P, Malthièry Y. PGC-1-related coactivator and targets are upregulated in thyroid oncocytoma. Biochem Biophys Res Commun. 2003;310:779–784. doi: 10.1016/j.bbrc.2003.09.076. [DOI] [PubMed] [Google Scholar]
- 149.Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, Clish CB, Pandolfi PP, Haigis MC. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell. 2011;19:416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]


