SUMMARY
Disrupted epidermal differentiation characterizes numerous diseases that impact >25% of the population. In a search for dominant mediators of differentiation, we defined a requirement for ZNF750 in terminal epidermal differentiation. ZNF750 controlled genes mutated in numerous human skin diseases, including FLG, LOR, LCE3B, ALOXE3, and SPINK5. ZNF750 induced progenitor differentiation via an evolutionarily conserved C2H2 zinc finger motif. The epidermal master regulator, p63, bound the ZNF750 promoter and was necessary for its induction. ZNF750 restored differentiation to p63-deficient tissue, suggesting it acts downstream of p63. A search for functionally important ZNF750 targets via analysis of ZNF750-regulated genes identified KLF4, a transcription factor that activates late epidermal differentiation. ZNF750 binds to KLF4 at multiple sites flanking the transcriptional start site and controls its expression. ZNF750 thus directly links a tissue-specifying factor, p63, to an effector of terminal differentiation, KLF4, and represents a potential future target for disorders of this process.
Keywords: Differentiation, Stem Cell, ZNF750, p63, KLF4
INTRODUCTION
Transcription factors mediating somatic tissue development, such as p63 in epidermis, can also act in homeostasis to enable differentiation. In the case of epidermis, terminal differentiation is essential for the acquisition of normal skin barrier function and homeostasis, as demonstrated by both common and rare human skin disorders, including atopic dermatitis, psoriasis, ichthyosis vulgaris, and epidermal cancers, which in lifetime aggregate afflict a large portion of the U.S. population(de Cid et al., 2009; Leech and Moss, 2007; Nestle et al., 2009; Rogers et al., 2010; Smith et al., 2006). Cells leaving the epidermal progenitor layer first express early differentiation proteins, such as keratins 1 and 10, then, upon subsequently migrating to outer epidermal layers, induce terminal differentiation proteins, such as filaggrin, loricrin and the late cornified envelope proteins (LCEs). p63 is required both for the development(Mills et al., 1999; Yang et al., 1999) and homeostasis of stratified epithelial tissues. p63 acts in the latter setting to sustain stem cell function,(Senoo et al., 2007) to maintain stratified epithelial tissue fate by preventing reversion to simple epithelium as well as, paradoxically, to enable terminal differentiation(Truong and Khavari, 2007; Truong et al., 2006). Several transcription factors, including KLF4(Patel et al., 2006; Segre et al., 1999), PRDM1/Blimp1(Magnusdottir et al., 2007), and OVOL1(Teng et al., 2007) have been implicated in the induction of terminal differentiation, however, the mechanisms controlling their expression and how they are linked to p63 is unclear. Here we demonstrate that ZNF750 is a nuclear factor that is upregulated during epidermal differentiation by p63. ZNF750 is necessary to turn on the terminal epidermal differentiation gene program, which is due in part to ZNF750 regulation of the differentiation specific transcription factor, KLF4.
RESULTS
Induction of ZNF750 is Necessary for Terminal Epidermal Differentiation
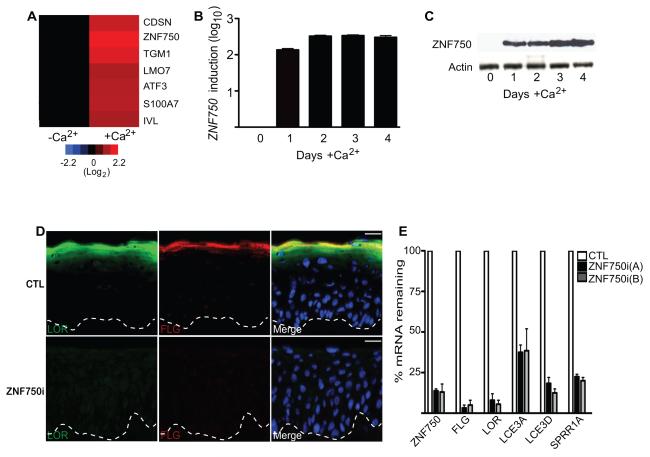
To identify potential effectors of terminal epidermal differentiation, we searched for genes whose transcript levels were temporally increased prior to this process. ZNF750, which encodes a 723 amino acid protein of unknown function, was increased >300 fold in epidermal cells by day 2 of differentiation in calcium-containing media in vitro (Figures 1A-1C). In tissue, ZNF750 protein was absent from the undifferentiated cell compartment adjacent to the basement membrane, but displayed nuclear localization in spinous layer cells and above (Figure S1A). ZNF750 is thus induced immediately prior to engagement of the terminal differentiation gene expression program. To test for a potential functional role for ZNF750 in this setting, ZNF750 expression was depleted in regenerated organotypic human epidermal tissue, a setting which recapitulates the structure and differentiation gene expression program of normal skin(Sen et al., 2010; Truong et al., 2006). Although leaving early differentiation gene markers keratin 1 and 10 unaffected (Figure S1B-S1C), ZNF750 loss abolished induction of late differentiation genes, including FLG, LOR, LCE3A, LCE3D, and SPRR1A (Figures 1D-1E). Loss of ZNF750 did not result in increased apoptosis or affect the percentage of proliferative cells (Figure S1D-S1F). Although the percentage of proliferative cells did not significantly change, there was an increased number of proliferative cells in the normally differentiated suprabasal layer suggesting that withdrawal from the cell cycle did not properly occur in ZNF750i epidermis (Figure S1E and S1G). ZNF750 is thus necessary for terminal epidermal differentiation.
Figure 1.
ZNF570 is required for terminal differentiation. (A) Differentiation gene induction heatmap on day 3 of calcium treated keratinocytes (+Ca2+) to induce differentiation as compared to undifferentiated sub confluent control keratinocytes (-Ca2+). (B) ZNF750 mRNA induction during calcium-induced differentiation in vitro [ordinate is log10 scale]. Data show standard error of the mean (SEM). (C) ZNF750 protein induction during calcium-induced differentiation in vitro. (D) Effects of siRNA-mediated ZNF750 depletion (ZNF750i) versus scrambled control (CTL) on terminal differentiation proteins in regenerated organotypic human epidermal tissue; loricrin [LOR; green] and filaggrin [FLG; red], epidermal basement membrane is noted with dashed lines (scale bar=25μm). (E) Terminal differentiation gene mRNA quantitation in epidermal tissue with two independent ZNF750 siRNAs vs. scrambled control. Data show standard error of the mean (SEM). N=3. See also Figure S1.
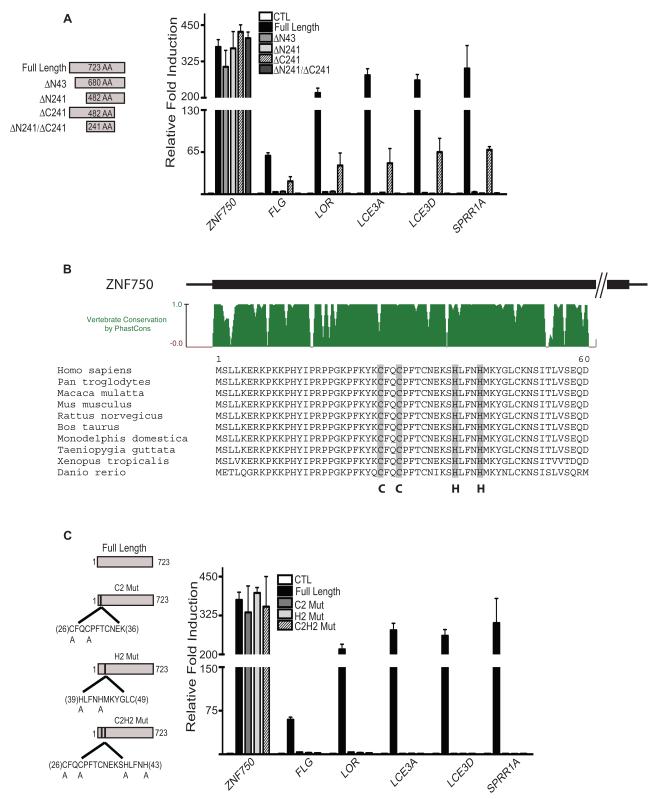
The Highly Conserved C2H2 Domain is Necessary for ZNF750 Function
Expression of ZNF750 in undifferentiated cells induced terminal differentiation genes (Figure 2A), indicating that ZNF750 is not only required for this process but is also sufficient to engage it. Deletion analysis demonstrated that even partial ZNF750-driven differentiation required amino-terminal but not carboxyl-terminal ZNF750 sequences (Figure 2A). Loss of differentiation induction ability of amino terminal mutants was not due to lack of expression since all mutants were comparably expressed (Figure S2A and S2B). The amino terminal region of ZNF750 encompasses an evolutionarily conserved block of 60 amino acids containing a predicted C2H2 zinc finger sequence (Figure 2B). Point mutagenesis of the C2H2 sequence abolished the capacity of ZNF750 to induce terminal differentiation despite being expressed to wild-type levels (Figure 2C; Figure S2A). ZNF750 differentiation effects thus depend upon a conserved amino terminal zinc finger motif. Mutations in ZNF750 have also been linked to abnormal terminal differentiation and enhanced keratinocyte proliferation in a Jewish Moroccan family with seborrheic dermatitis-like and psoriasiform elements(Birnbaum et al., 2006). This was found to be caused by a frameshift mutation in ZNF750 that results in a truncated protein of 43 amino acids with only the first 19 amino acids being identical to wildtype ZNF750. To determine if this mutant, truncated ZNF750 (ZNF750 56_57 dupCC) is responsible for abnormal proliferation and differentiation, the mutant construct was generated and expressed in keratinocytes induced to differentiate. Expression of ZNF750 56_57 dupCC did not have any significant impacts on epidermal differentiation or proliferation suggesting that the phenotype in patients may be due to a haploinsufficiency of ZNF750 (Figure S2C and S2D).
Figure 2.
An evolutionarily conserved amino terminal C2H2 zinc finger motif is required for ZNF750-driven terminal differentiation. (A) Induction of terminal differentiation genes in undifferentiated sub confluent cells requires an intact ZNF750 amino terminus. Fold mRNA induction quantified over undifferentiated control cells receiving vector control (CTL). Data shown SEM, n=3. (B) A C2H2 sequence in the evolutionarily conserved ZNF750 amino-terminus. (C) C2H2 sequence requirement in ZNF750-driven differentiation. Differentiation gene expression is shown with expression of full-length ZNF750 with point mutation in the amino-terminal C2H2 motif of either both cysteines (C2 Mut), both histidines (H2 Mut), all four residues (C2H2 Mut) or wild-type full-length control in undifferentiated human keratinocytes. Data shown SEM, n=3. See also Figure S2.
p63 is Necessary for ZNF750 Induction During Differentiation
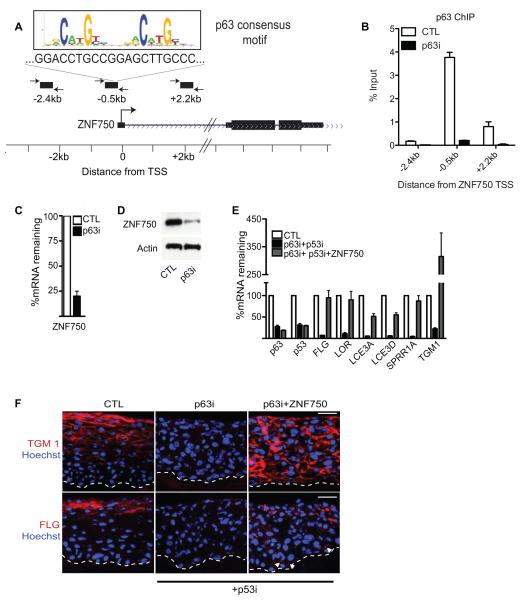
To study mechanisms of ZNF750 gene activation during epidermal differentiation, we examined the promoter of ZNF750 for binding sites of transcription factors implicated in epidermal gene regulation and found a region, highly conserved by PhastCons, 500bp upstream of the ZNF750 promoter (Figure S3A). This region also contains a sequence(Kouwenhoven et al.) matching the canonical p63 binding site (Figure 3A and Figure S3A). p63, which regulates epidermal development, stem cell function and differentiation(Crum and McKeon, 2010; Koster, 2010; Truong and Khavari, 2007), is expressed in all living layers of human epidermal tissue (www.proteinatlas.org and Figure S3B), where it is required for induction of both early and late epidermal differentiation genes(Truong et al., 2006). In differentiated cells, chromatin immunoprecipitation (ChIP) using a p63 antibody that recognizes all isoforms of p63 confirmed p63 binding to the putative p63 binding site in the ZNF750 promoter but not to adjacent upstream and downstream sequences (Figure 3B). A recent ChIP-Seq study(Kouwenhoven et al.) on genome-wide p63 binding also shows enrichment of p63 binding 500bp upstream of the promoter of ZNF750 (Figure S3A). Pan p63 depletion resulted in loss of p63 binding to the ZNF750 promoter and diminished ZNF750 expression during differentiation at both the mRNA and protein levels (Figures 3B-3D). To explore which isoform of p63 is responsible for binding the ZNF750 promoter, ChIP was performed using a TAp63 or p63α specific antibody. p63α was found to bind to the ZNF750 promoter to the same levels as pan p63, whereas no binding could be detected with the TAp63 antibody (Figure S3C). Knockdown of ΔNp63 using siRNAs targeting only the ΔNp63 isoform confirmed the specificity of ΔNp63 binding (Figure S3C). Taken together, these data suggest that ΔNp63α, the major p63 isoform expressed in epidermis, is also the major p63 isoform that binds the ZNF750 gene.
Figure 3.
ZNF750 is a p63-dependent gene and rescues terminal differentiation gene expression in p63-deficient tissue. (A) ZNF750 gene locus with canonical p63 binding sequence noted in the promoter -0.5kb upstream of the transcriptional start site (TSS). (B) p63 chromatin immunoprecipitation at the ZNF750 promoter in differentiated keratinocytes with p63 depletion [p63i] versus scrambled siRNA control [CTL] to verify specificity. Data show mean with SEM. (C) Reduction in ZNF750 mRNA with p63 depletion at day 3 of calcium-induced differentiation in vitro; siRNA to p63 (p63i) and scrambled siRNA control (CTL). Data show SEM. (D) Reduction in ZNF750 protein with p63 depletion. (E) ZNF750 rescues loss of terminal differentiation gene expression due to p63 deficiency. mRNA quantification in tissue; p63i tissue is also depleted of p53 to bypass the growth arrest that occurs with loss of p63 alone. Data show SEM (F) Protein expression in tissue quantified in E. Note filaggrin protein expression in lower epidermis with ZNF750 (white arrowheads), scale bar= 30μm. See also Figure S3.
To gain insight into how p63 controls ZNF750 expression, ChIP was also performed on p63 in undifferentiated cells. Surprisingly, p63 binds to the promoter of ZNF750 in the presence or absence of differentiation (Figure S3C). This suggests that during differentiation p63 may interact with additional proteins to alter the chromatin state. Supporting this, epigenetic marks of active transcription (H3K4me3, H3K27ac, and RNA Pol II) were only found on the promoter of ZNF750 in differentiated cells whereas minimal binding was detected in undifferentiated cells (Figure S3D). This RNA Pol II binding and incorporation of H3K4me3 and H3K27ac marks during differentiation were dependant on p63 binding as knockdown of ΔNp63 prevents incorporation of these marks of active transcription (Figure S3D). To determine if the p63 binding site is necessary for activation of ZNF750 during differentiation, the ZNF750 promoter region containing the wildtype or mutated p63 binding site was cloned into the pGreenFire luciferase reporter. In differentiated cells, there was an induction of luciferase activity from the wildtype ZNF750 promoter as compared to undifferentiated cells, whereas there was no induction from the mutated promoter, confirming that the p63 binding site is necessary for ZNF750 induction during differentiation (Figure S3E). Thus, p63 binds directly to the ZNF750 promoter and is required for ZNF750 induction during differentiation.
Expression of ZNF750 Rescues Differentiation Loss Due to p63 Depletion
While mediators of p63 effects on proliferation and adhesion have been identified(Ihrie et al., 2005; Koster et al., 2007), effectors of p63 differentiation impacts have yet to be fully defined. To determine if ZNF750 could rescue differentiation loss due to p63 depletion, we expressed ZNF750 in the setting of p63 knockdown; simultaneous tissue depletion of p53 was used, as it has been previously confirmed to rescue proliferation and stratification due to p63 loss without affecting differentiation(Truong et al., 2006). ZNF750 restored terminal differentiation gene induction to p63 depleted cells and tissue, even driving differentiation gene expression ectopically into the normally undifferentiated basal layer when it was expressed throughout the epidermis (Figure 3E and 3F). These results also suggest that the basal layer remains undifferentiated due to an absence of ZNF750 since driving ZNF750 expression into the basal layer results in premature terminal differentiation. Expression of ZNF750 in the context of cultured p63 depleted cells placed in differentiation also restored the expression of 71% (12/17) of upregulated differentiation genes (Figure S3F). ZNF750 is therefore a p63-bound and regulated gene that can restore the expression of terminal differentiation genes in the setting of p63 loss.
ZNF750 Regulation of KLF4 Expression Mediates Terminal Epidermal Differentiation
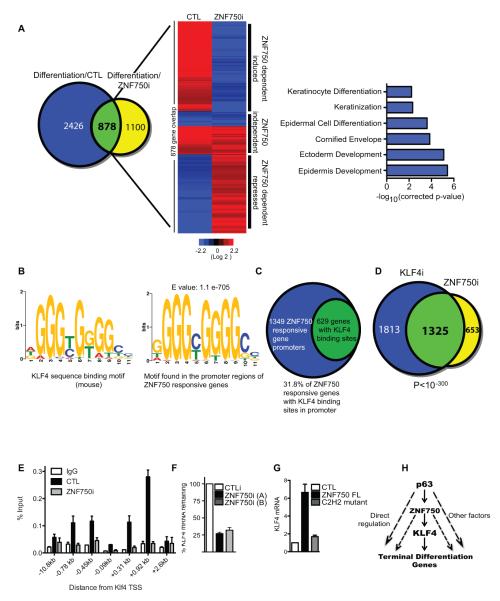
To identify potential effectors of ZNF750-mediated differentiation, ZNF750 was depleted in primary keratinocytes placed in differentiation conditions. 1,978 genes changed significantly upon ZNF750 knockdown as compared to control cells during differentiation (Figure 4A, yellow circle). To determine the number of genes regulated by ZNF750 that are differentiation regulated genes, the 1,978 genes were overlapped with the 3,138 genes that are differentially regulated during differentiation. A significant fraction of the genes regulated by ZNF750 (878/1,978) were differentiation genes (Figure 4A, overlap shown in green). In epidermal keratinocytes, ZNF750 controlled a subset of genes highly enriched for Gene Ontology terms related to terminal epidermal differentiation (Figure 4A and Tables S1 and S2). Among ZNF750-dependent genes were both structural and enzymatic genes implicated in terminal differentiation as well as KLF4, which is a transcription factor that has been previously identified as a regulator of late differentiation(Segre et al., 1999). Interestingly, a number of genes identified previously as KLF4-dependent using KLF4 knockout and over-expression mice(Patel et al., 2006), including ECM1, SPINK5, CDSN, FLG, and LCE3 were also ZNF750-dependent. Genomic binding sequences for KLF4, previously characterized by ChIP-seq(Yang et al., 2008), were strongly enriched in the ZNF750-dependent gene set [629 out of 1978 genes (31.8%) with an E-value=1.1×10−705], suggesting that ZNF750-regulated genes may be controlled in part through KLF4 action (Figures 4B and 4C and Table S3). Consistent with this, the gene expression signature of KLF4i and ZNF750i cells during differentiation strongly overlapped with each other [p<10−300] (Figure 4D, Figure S4A, and Tables S4), suggesting that ZNF750 regulated differentiation genes may be mediated to a major degree through the actions of KLF4.
Fig. 4.
ZNF750 regulates KLF4 to control terminal epidermal differentiation. (A) Venn diagram (left) illustrating overlap between ZNF750-dependent genes and differentiation genes. The blue circle (Differentiation/ CTL: 3,304 genes) represents all the genes that change significantly by >2-fold during epidermal differentiation. The yellow circle (Differentiation/ZNF750i: 1,978 genes) represents the genes that change significantly >2-fold when ZNF750 is knocked down during differentiation compared to control differentiated cells. The overlap (878 genes; green) shows ZNF750-regulated epidermal differentiation genes. Heat map (middle) of the overlap showing epidermal differentiation regulated genes affected by ZNF750 depletion during differentiation. CTL and ZNF750i represent heatmaps generated from control and ZNF750i cells induced to differentiate. Heatmaps were normalized to undifferentiated control cells (red [induced] and blue [repressed]), log2 based scale. Gene Ontology terms (right) of ZNF750-dependent genes (p-values represent Bonferroni- corrected EASE score). (B) Consensus mouse KLF4 binding motif (left panel) and consensus KLF4 binding motif (right panel) found informatically enriched in the promoter regions of ZNF750-dependent genes. (C) 629 of the 1978 ZNF750 dependent genes contain consensus KLF4 binding sites in their respective promoter regions. (D) Venn diagram illustrating overlap between changes identified with KLF4 and ZNF750 depletion in primary keratinocytes induced to differentiate with calcium. These data were derived by first determining the significantly changed genes between KLF4i and control cells during differentiation (3,138 genes) and overlapping them to the significantly changed genes between ZNF750i and control cells during differentiation (1,978 genes). (E) ChIP demonstrating ZNF750 binding to the regions of the KLF4 gene that flank the transcription start site (TSS) in differentiating epidermal cells. ZNF750i versus scrambled control (CTLi) knockdown represent specificity controls. Data show mean with SEM. (F) ZNF750 depletion impairs KLF4 induction in differentiating epidermal cells. Data show SEM (G) Ectopic expression of full length ZNF750 (ZNF750 FL), but not the C2H2 ZNF750 mutant or lacZ control (CTL), induces KLF4 in undifferentiated epidermal cells. Data show SEM. (H) Proposed model for epidermal differentiation. In this model, p63 induces the expression of ZNF750 during differentiation (solid arrow) as well as other factors necessary for differentiation (dashed arrow). ZNF750 induces differentiation in part by transcriptionally activating KLF4 (solid arrows). ZNF750 may also regulate other factors necessary for differentiation (dashed arrows). KLF4 can directly bind to differentiation gene promoters to activate terminal differentiation genes. See also Figure S4.
To explore if ZNF750 may directly regulate the expression of KLF4, ChIP was performed using a ZNF750 antibody in differentiated cells to determine if ZNF750 bound to KLF4 either downstream or upstream from the transcriptional start site (TSS). Binding was found at multiple sites flanking the TSS of KLF4, which was abrogated by ZNF750 depletion (Figure 4E) whereas no binding could be detected on the promoters of genes not regulated by ZNF750 (Figure S4B). To determine if ZNF750 acts as an activator or repressor of KLF4, regions -0.78, and +0.92kb from the TSS of KLF4 was cloned into the pGreenFire luciferase reporter. Luciferase expression was assayed for each reporter in both undifferentiated and differentiated cells. There was a >3-fold activation of luciferase during differentiation for the +0.92kb reporter construct (Figure S4C:left panel). The determine if this activation was a result of ZNF750 binding to the +0.92kb reporter construct, either wildtype or mutant C2H2 ZNF750 was coexpressed with the reporter construct in undifferentiated cells. Expression of wildtype but not C2H2 mutant ZNF750 resulted in activation of the reporter construct (Figure S4C:right panel).
Further demonstrating that ZNF750 activates KLF4 expression, knockdown of ZNF750 during differentiation prevented the induction of KLF4 gene expression, while enforced expression of wild-type but not the C2H2 ZNF750 mutant induced expression of KLF4 in undifferentiated cells (Figure 4F and 4G). Furthermore, enforced KLF4 expression partially rescued activation of ZNF750-dependent terminal differentiation genes in the context of ZNF750 depletion (Figure S4D).
DISCUSSION
Here we report that ZNF750 is differentiation p63 target gene that promotes terminal epidermal differentiation. Induction of ZNF750 expression is associated with ΔNp63α binding to the promoter region of ZNF750 during differentiation and mutation of the highly conserved p63 binding site prevents activation of a ZNF750 promoter reporter construct. Interestingly, p63 binds the promoter of ZNF750 in undifferentiated cells as well, suggesting that additional factors may cooperate with p63 during differentiation to turn on ZNF750 expression. p63 binding is associated with deposition of transcriptionally active marks of chromatin, such as H3K4me3 and H3K27 acetylation, as well as RNA polymerase II loading onto the ZNF750 promoter. Loss of p63 also prevents induction of ZNF750 expression during differentiation. Enforced expression of ZNF750 can restore the majority of p63 regulated differentiation genes studied that are lost with p63 knockdown. p63 may also regulate other as of yet unidentified factors to induce the differentiation program since ectopic ZNF750 expression does not rescue all of p63 dependent differentiation genes.
The function of ZNF750 is to promote terminal epidermal differentiation, as it is necessary for expression of late terminal differentiation genes but not early ones. ZNF750 promotes differentiation in part by inducing the expression of KLF4. ZNF750 binds multiple sites flanking the KLF4 TSS during epidermal differentiation. Wildtype but not C2H2 mutant ZNF750 is both necessary and sufficient for the induction of KLF4 expression. KLF4 and ZNF750 also regulate a common set of genes (1,325 genes P value <10−300). ZNF750 may also regulate the terminal differentiation program through regulating other transcription factors or directly binding to differentiation genes. We present a model where p63 induces ZNF750 as well as potentially other factors during differentiation to engage the terminal differentiation pathway. Then, ZNF750 can transcriptionally activate KLF4 to engage the terminal differentiation program (Figure 4H).
Functionally, ZNF750 had been previously unstudied, yet the only two prior publications to mention ZNF750 are consistent with the potential clinical relevance for this model. The first reported a single extended kindred with a ZNF750 mutation resulting in a truncated mutant protein of only 43 amino acids. The patients manifested clinically as a dominant generalized skin disease characterized by abnormal terminal epidermal differentiation with cutaneous inflammation, and defined ZNF750 mRNA as being predominantly expressed in stratified epithelia(Birnbaum et al., 2006). Expression of the 43 amino acid mutant had no impacts on epidermal differentiation or proliferation in our studies, suggesting that the phenotype may be due to a haploinsufficiency of ZNF750. This is supported by another familial genetic study that linked genetic mutations which resulted in lower levels of ZNF750 to psoriasis(Yang et al., 2008). ZNF750 thus connects the tissue-specifying factor p63 to the KLF4 transcription factor to enable terminal differentiation and may provide a future potential target in human disorders of this process.
EXPERIMENTAL PROCEDURES
The Supplemental Experimental Procedures section is available online which includes list of plasmids/retroviral constructs, siRNA, antibodies, and primer sequences.
Tissue culture
Primary human keratinocytes were derived from freshly isolated foreskin. Cells were grown in KCSFM (GIBCO-BRL) supplemented with epidermal growth factor and bovine pituitary extract. Cells were induced to differentiate by the addition of 1.2 mM calcium for 1 through 4 days in full confluence. Amphotrophic phoenix cells were maintained in DMEM and 10% fetal bovine serum.
Retroviral Gene transfer
Amphotrophic phoenix cells were transfected with 3 ug of each retroviral expression construct. Transfections were done in 6 well plates using Lipofectamine 2000 (Invitrogen). Viral supernatants were collected 48 hours post transfection and polybrene added (5ug/ml). These supernatants were placed on primary human keratinocytes and centrifuged for 1 hour. Cells were transduced and selected using puromycin (2ug/ml) after the last transduction.
Western blotting and immunofluorescence
50 ug of the cell lysates were used for immunoblotting and resolved on 10% SDS-PAGE and transferred to PVDF membranes. Membranes were incubated in primary and secondary for 1 hour each. For immunofluorescence experiments, 7 μm thick epidermal sections from adult human skin or organotypic cultures were fixed in 4% paraformaldehyde for 15 minutes followed by blocking in PBS with 2.5% normal goat serum, 0.3% triton X100, and 2% bovine serum albumin for 30 minutes. Sections were incubated in primary antibodies for 1 hour and secondary antibodies for another hour. TUNEL staining was done with the TMR Red In Situ Cell Death Detection Kit from Roche.
Genetically altered regenerated human skin
For organotypic cultures, 1 million genetically modified keratinocytes were seeded onto devitalized human dermis for 72 hours and then harvested for RNA or protein expression.
Quantitative reverse transcriptase-PCR analysis
Total RNA from cells was extracted using the Rneasy mini kit (Qiagen) and quantified by Nanodrop. One ug of total RNA was reverse transcribed using the 1st Strand cDNA Synthesis Kit for RT-PCR from Roche. Quantitative PCR was performed using the Mx3000P (Stratagene) thermocycler. Samples were run in triplicate and normalized to GAPDH.
Gene expression profiling
Microarray analysis was performed on biological duplicate samples. Labeling of cDNA and hybridization to Affymetrix HG-U133 2.0 plus arrays were performed at Stanford’s Pan facility. For gene expression analysis, arrays were RMA normalized and differential expression was defined using the following filters: Significance Analysis of Microarrays 3.0 with a false discovery rate less than 5%, an average fold change ≥ 2 at any time point, and an average raw expression intensity ≥ 100 at any time point. Hierarchical clustering and heat map generation were performed using GeneSpring GX software (Agilent). For future analyses, Affymetrix probe IDs were converted to unique Unigene IDs. P-values indicating the significance of the overlap between various gene sets were calculated using Fisher’s Exact test. GO term enrichment was performed using DAVID with the total set of genes on the appropriate microarray as the background, p-values represent a Bonferroni-corrected modified Fisher’s Exact test. Microarray data has been deposited into the GEO database with accession number: GSE32685
siRNAs
All siRNAs used were generated from Dharmacon. The control siRNA used is a nonfunctional oligo targeting p639. siRNAs targeting p63 were used as previously described9. siRNAs were generated against ZNF750 and KLF4. One nanomole of each siRNA was electroporated into 1 million keratinocytes using Amaxa nucleofection reagents.
Reporter Assays
The lentiviral constructs for reporter assays were made by cloning promoter regions of ZNF750 or KLF4 into pGreenFire (System Biosciences). 293T cells were transfected with 11ug of pGreenFire vector, 8.3ug of pCMVΔ8.91 (gag/pol) and 2.8ug pUC MD.G (VSVg envelope). Transfections were done in a 10cm dish with calcium phosphate (HBSS at pH 7.01). Virus supernatant was collected 48- and 72- post transfection and concentrated using Lenti-X Concentrator (System Biosciences). Primary human keratinocytes were transduced overnight with concentrated virus and selected with puromycin (1ug/mL). Firefly luciferase activity was measured using the Dual-luciferase reporter assay system from Promega, on a TD-20/20 Luminometer (Turner Designs) with a 10 second integration time. Luminescence was corrected for protein concentration and normalized to proviral integrants. Genomic DNA from infected keratinocytes was isolated using Qiagen DNeasy Blood & Tissue Kit. 10ng of genomic DNA was used for quantitative PCR using ABI 7900HT. Samples were done in triplicate and normalized to PLAC2 promoter.
Chip-Seq analysis of p63 binding to the promoter of ZNF750
Raw sequencing reads were downloaded from GEO dataset GSE17611(Kouwenhoven et al., 2010), mapped to hg18 and plotted in the UCSC genome browser. The predicted p63 binding site from this study was utilized.
Supplementary Material
Acknowledgements
We thank A.Oro, L. Attardi, and H. Chang for pre-submission review and helpful discussions. We thank J. Elder for helpful discussions. We thank J. Kim for pathology samples and S. Tao, J. Chow, and L. Morcom for technical support. This work is supported by the U.S. Department of Veterans Affairs Office of Research and Development and by NIH R01 AR45192 and AR49737 (P.A.K.) and by a K01AR057828-04 Award to G.L.S and by an NSF GRF Grant No. DGE-0645962 (L.D.B).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Notes
- Birnbaum RY, Zvulunov A, Hallel-Halevy D, Cagnano E, Finer G, Ofir R, Geiger D, Silberstein E, Feferman Y, Birk OS. Seborrhea-like dermatitis with psoriasiform elements caused by a mutation in ZNF750, encoding a putative C2H2 zinc finger protein. Nat Genet. 2006;38:749–751. doi: 10.1038/ng1813. [DOI] [PubMed] [Google Scholar]
- Crum CP, McKeon FD. p63 in epithelial survival, germ cell surveillance, and neoplasia. Annu Rev Pathol. 2010;5:349–371. doi: 10.1146/annurev-pathol-121808-102117. [DOI] [PubMed] [Google Scholar]
- de Cid R, Riveira-Munoz E, Zeeuwen PL, Robarge J, Liao W, Dannhauser EN, Giardina E, Stuart PE, Nair R, Helms C, et al. Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat Genet. 2009;41:211–215. doi: 10.1038/ng.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihrie RA, Marques MR, Nguyen BT, Horner JS, Papazoglu C, Bronson RT, Mills AA, Attardi LD. Perp is a p63-regulated gene essential for epithelial integrity. Cell. 2005;120:843–856. doi: 10.1016/j.cell.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Koster MI. p63 in skin development and ectodermal dysplasias. J Invest Dermatol. 2010;130:2352–2358. doi: 10.1038/jid.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MI, Dai D, Marinari B, Sano Y, Costanzo A, Karin M, Roop DR. p63 induces key target genes required for epidermal morphogenesis. Proc Natl Acad Sci U S A. 2007;104:3255–3260. doi: 10.1073/pnas.0611376104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouwenhoven EN, van Heeringen SJ, Tena JJ, Oti M, Dutilh BE, Alonso ME, de la Calle-Mustienes E, Smeenk L, Rinne T, Parsaulian L, et al. Genome-wide profiling of p63 DNA-binding sites identifies an element that regulates gene expression during limb development in the 7q21 SHFM1 locus. PLoS Genet. 2010;6:e1001065. doi: 10.1371/journal.pgen.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leech SN, Moss C. A current and online genodermatosis database. Br J Dermatol. 2007;156:1115–1148. doi: 10.1111/j.1365-2133.2007.07834.x. [DOI] [PubMed] [Google Scholar]
- Magnusdottir E, Kalachikov S, Mizukoshi K, Savitsky D, Ishida-Yamamoto A, Panteleyev AA, Calame K. Epidermal terminal differentiation depends on B lymphocyte-induced maturation protein-1. Proc Natl Acad Sci U S A. 2007;104:14988–14993. doi: 10.1073/pnas.0707323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Nestle FO, Kaplan DH, Barker J. Psoriasis. N Engl J Med. 2009;361:496–509. doi: 10.1056/NEJMra0804595. [DOI] [PubMed] [Google Scholar]
- Patel S, Xi ZF, Seo EY, McGaughey D, Segre JA. Klf4 and corticosteroids activate an overlapping set of transcriptional targets to accelerate in utero epidermal barrier acquisition. Proc Natl Acad Sci U S A. 2006;103:18668–18673. doi: 10.1073/pnas.0608658103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers HW, Weinstock MA, Harris AR, Hinckley MR, Feldman SR, Fleischer AB, Coldiron BM. Incidence estimate of nonmelanoma skin cancer in the United States, 2006. Arch Dermatol. 2010;146:283–287. doi: 10.1001/archdermatol.2010.19. [DOI] [PubMed] [Google Scholar]
- Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature. 2010;463:563–567. doi: 10.1038/nature08683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoo M, Pinto F, Crum CP, McKeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- Smith FJ, Irvine AD, Terron-Kwiatkowski A, Sandilands A, Campbell LE, Zhao Y, Liao H, Evans AT, Goudie DR, Lewis-Jones S, et al. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat Genet. 2006;38:337–342. doi: 10.1038/ng1743. [DOI] [PubMed] [Google Scholar]
- Teng A, Nair M, Wells J, Segre JA, Dai X. Strain-dependent perinatal lethality of Ovol1-deficient mice and identification of Ovol2 as a downstream target of Ovol1 in skin epidermis. Biochim Biophys Acta. 2007;1772:89–95. doi: 10.1016/j.bbadis.2006.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong AB, Khavari PA. Control of keratinocyte proliferation and differentiation by p63. Cell Cycle. 2007;6:295–299. doi: 10.4161/cc.6.3.3753. [DOI] [PubMed] [Google Scholar]
- Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 2006;20:3185–3197. doi: 10.1101/gad.1463206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- Yang CF, Hwu WL, Yang LC, Chung WH, Chien YH, Hung CF, Chen HC, Tsai PJ, Fann CS, Liao F, et al. A promoter sequence variant of ZNF750 is linked with familial psoriasis. J Invest Dermatol. 2008;128:1662–1668. doi: 10.1038/jid.2008.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.