Abstract
NO synthase enzymes (NOS) support unique single-electron transitions of a bound H4B cofactor during catalysis. Previous studies showed that both the pterin structure and surrounding protein residues impact H4B redox function during catalysis. A conserved Arg residue (Arg375 in iNOS) forms hydrogen bonds with the H4B ring. In order to understand the role of this residue in modulating the function of H4B and overall NO synthesis of the enzyme, we generated and characterized three mutants R375D, R375K and R375N of the oxygenase domain of inducible NOS (iNOSoxy). The mutations affected the dimer stability of iNOSoxy and its binding affinity towards substrates and H4B to varying degrees. Optical spectra of the ferric, ferrous, ferrous dioxy, ferrous-NO, ferric-NO, and ferrous-CO forms of each mutant were similar to the wild-type. However, mutants displayed somewhat lower heme midpoint potentials and faster ferrous heme-NO complex reactivity with O2. Unlike the wild-type protein, mutants could not oxidize NOHA to nitrite in a H2O2-driven reaction. Mutation could potentially change the ferrous dioxy decay rate, H4B radical formation rate, and the amount of the Arg hydroxylation during single turnover Arg hydroxylation reaction. All mutants were able to form heterodimers with the iNOS G450A full-length protein and displayed lower NO synthesis activities and uncoupled NADPH consumption. We conclude that the conserved residue Arg375 (1) regulates the tempo and extent of the electron transfer between H4B and ferrous dioxy species and (2) controls the reactivity of the heme-based oxidant formed after electron transfer from H4B during steady state NO synthesis and in H2O2-driven NOHA oxidation. Thus, Arg375 modulates the redox function of H4B and is important in controlling the catalytic function of NOS enzymes.
INTRODUCTION
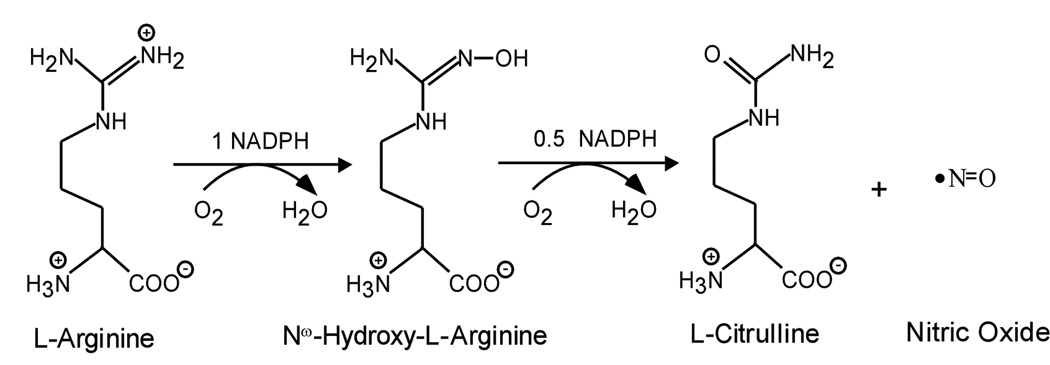
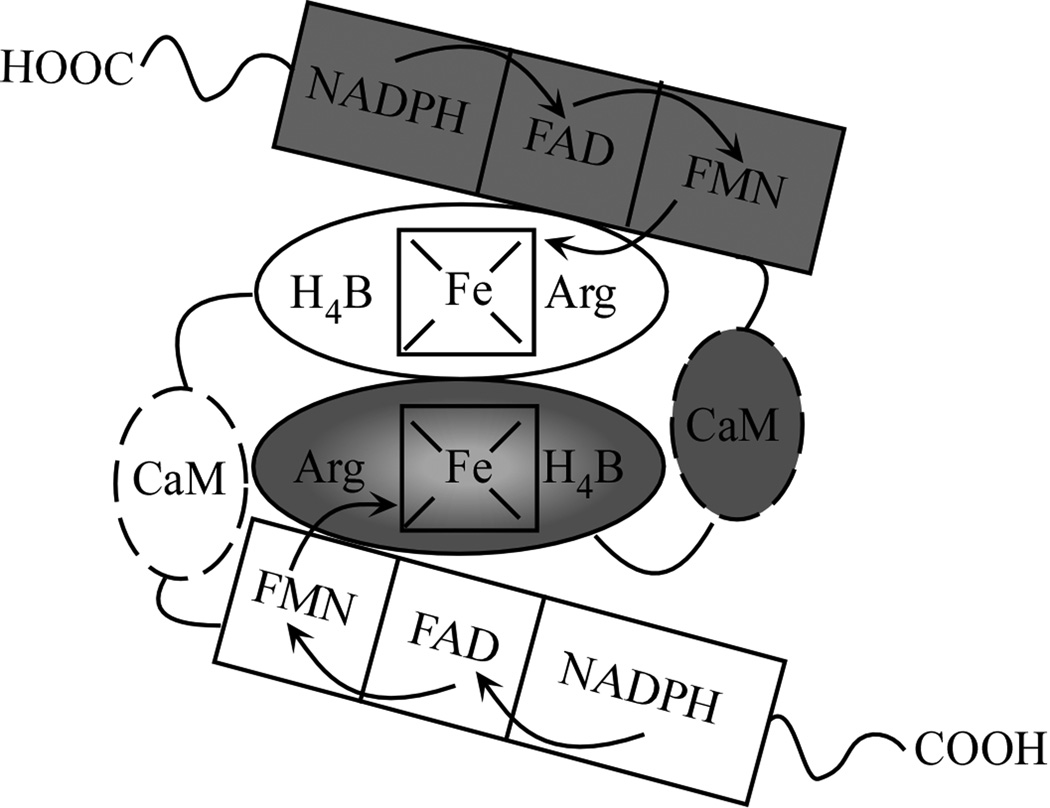
Nitric oxide (NO), an inorganic radical, plays an important role in physiology and pathology [1–3]. In mammals NO is largely produced by nitric oxide synthase (NOS) enzymes from L-arginine (Arg). NOSs are flavo-heme enzymes that catalyze a stepwise oxidation of L-Arg to form nitric oxide (NO) and L-citrulline [4–6]. The overall biosynthetic reaction consumes 1.5 NADPH and 2 O2 and involves two steps: the first being Arg hydroxylation to form N-hydroxy-L-Arg (NOHA) as a bound intermediate, and the second being NOHA oxidation to form citrulline and NO (Scheme 1). Three mammalian NOS isozymes have been characterized: neuronal NOS (nNOS, type I), macrophage inducible NOS (iNOS, type II) and endothelial NOS (eNOS, type III) [7–9]. Each NOS is only active as a homodimer [10;11], with each monomer being comprised of a N-terminal oxygenase domain that binds Fe-protoporphyrin IX (heme), the substrate Arg, and the cofactor 6R-tetrahydrobiopterin (H4B) [12;13], and a C-terminal flavoprotein domain that binds FAD, FMN, and NADPH [14–16]. A central calmodulin (CaM) binding motif links the two domains [10;15;16–18]. The heme is ligated to a cysteine thiolate and acts in conjunction with H4B to catalyze a reductive activation of molecular oxygen in both steps of NO synthesis [19–21]. During catalysis, NADPH-derived electrons are transferred to FAD and FMN in the NOS flavoprotein domain. CaM binding activates NO synthesis by triggering inter-domain electron transfer between the reduced FMN and ferric heme groups in the NOS homodimer (Fig. 1) [22–26]. Once ferrous heme forms, it can bind O2, obtain an electron from H4B, generate reactive heme dioxy species, and catalyze substrate oxidation in each NOSoxy domain [22–24;27].
Scheme 1.
Fig. 1. Model of NOS dimer structure and electron transfer pathway.
Inter-domain electron transfer occurs between FMN and the heme located on the adjacent subunit.
H4B was found to play novel structural and electronic roles in NOS [12;20;28;29]. It promotes dimer assembly of iNOS heme-containing monomers, increases Arg binding affinity, modifies heme midpoint potential and induces a high spin shift of the heme iron [30–33]. Moreover, H4B performs a critical redox function in NOS: it reduces the heme ferrous dioxy intermediate (FeIIO2) that is formed during oxygen activation and becomes an enzyme-bound H4B radical [20;21;34–40]. NOS flavoprotein domain then catalyzes reduction of the biopterin radical to H4B which remains bound in the enzyme to be used again in the second step of NO synthesis. The one-electron redox cycling of bound H4B in NOS enzymes is unique and differs from other enzymes like tryptophan/phenylalanine/tyrosine hydroxylases which also use H4B as a cofactor but only catalyze two-electron oxidation of H4B to generate H2B, and need a steady supply of fresh H4B during catalysis [27;41].
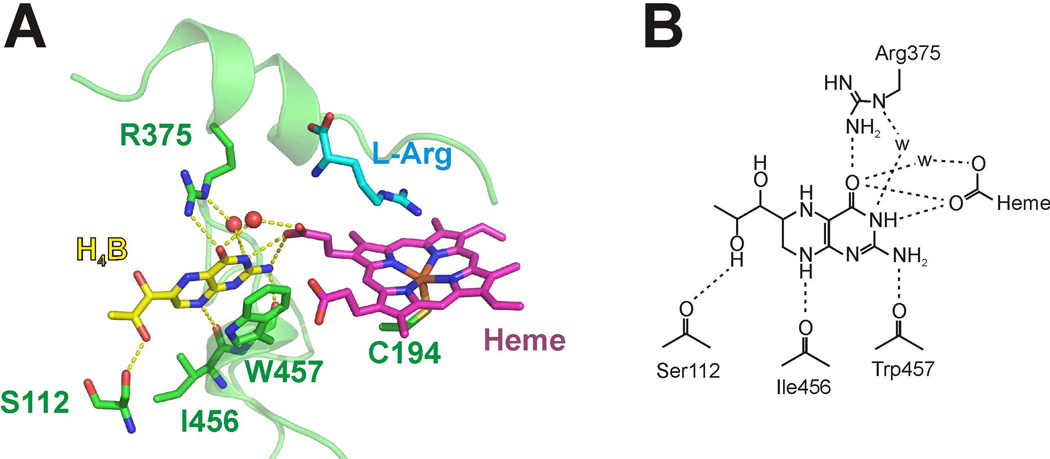
Crystal structures of nNOS, iNOS, and eNOS oxygenase domain dimers (i.e., NOSoxy) [12;13;15;42] reveal that H4B binds near the heme with its 2-amino-4-hydroxypyrimidine ring hydrogen bonded with a heme propionate group [43;44]. The H4B ring also makes π-stacking and/or hydrogen-bonding interactions with several conserved residues that are provided by both subunits of the NOSoxy dimer (Fig. 2). Point mutagenesis has been utilized to probe the roles of these residues in modulating the functions of H4B and catalytic properties of NO synthesis [37;38;45–48]. For instance, the conserved residue Trp457 in iNOS and its corresponding amino residue Trp678 in nNOS were found to regulate delivery of both electrons required for O2 activation (i.e., kinetics of ferric heme reduction by the NOS flavoprotein domain and reduction of the hemedioxy intermediate by H4B) [37;38;45]. Here we are particularly interested in a conserved Arg residue (Arg375 in iNOS), which is situated on the substrate binding helix and forms hydrogen bonds with O4 and N5 of the H4B ring (Fig. 2). To investigate the role of Arg375 in regulating the redox function of H4B and overall NO synthesis of the enzyme, we used point mutagenesis to generate the R375D, R375K and R375N in iNOSoxy. We then characterized each mutant with regard to their dimer content, H4B and substrate Arg binding, heme midpoint potential, extent of H4B radical formation, and a range of catalytic properties. Our results indicate that conserved residue Arg375 modulates the redox function of H4B and is important in controlling the catalytic function of NOS enzymes.
Fig. 2. Protein environment of the H4B cofactor.
A, Interactions between H4B and iNOS. Several iNOS residues and the heme cofactor form a hydrogen bonding network with the H4B cofactor. Relevant iNOS residues are shown in green; H4B (yellow), Heme (pink) and the substrate L-Arg (blue) are shown as sticks. Two water molecules that mediate H-bonding interactions are shown as red spheres. The hydrogen bonding interactions are shown as yellow dashes. Note the π-stacking interaction between the Trp457 side chain and H4B. The figure was made using PyMOL (http://www.pymol.org/) with the crystal structure of the mouse iNOSoxy dimer (PDB entry 1NOD [12;79]). B; Hydrogen bond interactions (dashed lines) between the H4B cofactor and iNOS. Ser112, Ile456 and Trp457 make H-bonds through the main chain carbonyl groups; Arg375 interacts through its side chain.
MATERIALS AND METHODS
Materials
All reagents and materials were obtained from Sigma, Aldrich, Alexis, or sources described previously [37;38].
Mutagenesis
Site-directed mutagenesis of Δ65iNOSoxy domain (amino acid 66–498) was performed using the QuikChange XL Mutagenesis Kit (Agilent Technologies-Stratagene, La Jolla, CA). Mutations were confirmed by DNA sequencing at the Cleveland Clinic Genomics Core Facility. Mutation bases (bold and underlined) and a silent restriction site (Italic) were introduced to the primers:
| R375K_Fw: | 5’ ACT GGG TGG CGA TTG TGC CTC CCA TGT CGG GAT CCA TCA CCC CTG TCT TCC 3’ |
| R375K_Rev: | 5’ GGA AGA CAG GGG TGA TGG ATC CCG ACA TGG GAG GCA CAA TCG CCA CCC AGT 3’ |
| R375N_Fw: | 5’ ACT GGG TGT TCA TTG TGC CTC CCA TGT CGG GAT CCA TCA CCC CTG TCT TCC 3’ |
| R375N_Rev: | 5’ GGA AGA CAG GGG TGA TGG ATC CCG ACA TGG GAG GCA CAA TCG ACA CCC AGT 3’ |
| R375D_Fw: | 5’ GGA AGA CAG GGG TGA TGG ATC CCG ACA TGG GAG GCA CAA GA ACA CCC AGT 3’ |
| R375D_Rev: | 5’ GGA AGA CAG GGG TGA TGG ATC CCG ACA TGG GAG GCA CAA TCT TCA CCC AGT 3’ |
DNA isolation, restriction enzyme digestion, transformation etc. were carried out using standard protocols.
Protein Expression and Purification
Wild type and mutants with a six-histidine tag at C terminus were over expressed in BL21 using pCWori vector and purified as reported previously by Ni2+-nitrilotriacetate (NTA) chromatography [46]. Concentrations of NOS proteins were determined from the 444 nm absorbance of the ferrous-CO complex, using an extinction coefficient 76 mM−1 cm−1 [49].
Dimerization Assessment
Protein stock solutions were incubated overnight at 4 °C in the presence of 1 mM H4B and 10 mM Arg. The dimer contents of the proteins were estimated by chromatography on an Amersham Pharmacia Biotech Superdex-200 HR size-exclusion column equilibrated with 40 mM EPPS buffer (pH 7.4), containing 10% glycerol, 250 mM NaCl and 3 mM DTT. The dimer:monomer ratios were derived from measuring the integrated area of the corresponding elution peaks determined at 280 nm. Molecular weights of the protein peaks were estimated relative to protein molecular weight standards.
Peroxide Assay
H2O2-dependent NOHA oxidation assays were performed as described previously [46;50;51]. In short, enzymes were incubated at room temperature with NOHA, DTT and different concentrations of H4B in 96-well plates. Reactions were initiated by adding 30 mM H2O2 and stopped after 10 min by adding catalase. Griess reagent solution was then added to enable detection of nitrite production as determined by the absorbance change at 550 nm. Nitrite was quantitated based on NaNO2 standard solutions.
Midpoint Potential Titration of the Mutants
Midpoint potential measurements were performed as previously described with some modifications [52]. Spectroelectrochemical titrations were carried out at room temperature in the presence of mixed dye (40 µM neutral red, 20 µM benzyl viologen, and 100 µM methyl viologen). Proteins (~ 15 µM), which were incubated with 10 mM Arg and 1 mM H4B at 4 °C overnight, were added to 40 mM phosphate buffer (pH 7.0) including 200 µM H4B, 10 mM Arg, 0.1% Tween and the mixture of dyes described above. Protein samples and working electrode (Au) in the cell were made anaerobic by alternating cycles of vacuation and flushing with nitrogen. The anaerobic reference electrode (AgCl(s)/Ag(s)/Cl(aq)) and auxiliary electrode (Ag(s)/KCl(aq)) were promptly placed into the cell under N2 pressure. Potential titrations were performed by gradually adding small amount of currents to the system using a Radiometer PGP201 potentiostat/galvanostat. The spectra were then recorded when the potential in the system were stabilized after each addition. The midpoint potential was determined using the absorbance changes at 400 and 645 nm, which indicate the ferric to ferrous heme transition. The midpoint potential was calculated according to the Nernst equation and is reported relative to the standard hydrogen electrode.
Single-Turnover Arg hydroxylations
Arg hydroxylation single-turnover reactions were carried out in a Hi-Tech SF-61 stopped flow apparatus equipped for anaerobic work and coupled to a Hi-Tech MG-6000 diode array detector, as reported previously [20;37]. The R375 mutants were pre-incubated with Arg plus H4B or H2B on ice overnight before performing experiments because of their lower affinity toward substrate and cofactor. An anaerobic solution that contained dithionite-reduced enzyme were transferred into the stopped-flow instrument and rapidly mixed with air-saturated EPPS buffer at 10 °C. Ninety-six spectral scans were obtained after each mixing. Sequential spectral data were fit to different reaction models using the Specfit/32 global analysis software, Version 3.0 (Spectrum Software Associates), which could calculate the number of different enzyme species, their spectra, and their concentrations versus time during the single turnover reactions. Data from eight to ten sequential reactions were averaged to obtain the final traces.
EPR Spectra Measurement
EPR spectra were recorded on a Bruker ESP 300 electron paramagnetic resonance spectrometer equipped with an ER 035 NMR gauss meter and a Hewlett-Packard 5352B microwave frequency counter. All spectra were obtained at 150 K using a microwave power of 2 milliwatts, microwave frequency of 9.45 GHz, modulation amplitude of 5.0 G, and modulation frequency of 100 kHz. 10 scans per sample were accumulated to improve the signal to noise ratio. Spin quantitations were calculated by double integration. We observed some variations in the protein concentration (around 25%). Thus, in order to improve the measurement, protein samples were thawed after EPR data collection and protein concentration were calculated. Double integration values were divided by the final protein concentration. Radical concentrations versus time were fit to an A → B → C kinetic model, where B is the radical, using the Origin 7.5 software (OriginLab, Northampton, MA).
Product Stoichiometry Analysis for Arg hydroxylation Single-turnover Reaction
NOHA production from Arg was measured using the following method. Reaction samples collected from single turnover reactions run in anaerobic vessels were derivatized with o-phthalaldehyde, and then analyzed by reverse-phase HPLC with fluorescence detection as described previously [20;37]. In summary, after reactions were completed, sample solutions were filtered through an Amicon Centricon device (10,000 MW cut-off) prior to derivatization for 2 min. An aliquot was then immediately injected into a Hewlett-Packard ODS-Hypersil column and eluted with a gradient of buffer A (5 mM ammonium acetate, pH 6.0, 20% methanol) and buffer B (100% methanol). Retention times and concentrations of amino acids were calculated based on Arg and NOHA standard solutions.
Ferrous NO Oxidation
Reduced protein was generated by the method described previously [53;54]. Small amount of anaerobic NO-saturated buffer were added to the reduced protein to form ferrous NO complex. Such solution was transferred to the stopped-flow instrument by a gastight syringe and rapidly mixed with air saturated buffer. Sequential spectral data of 96 scans were fit to A → B reaction models using the Specfit global analysis software.
Heterodimer Studies of Arg375 Mutant Monomer and G450A iNOS Full Length Monomer
60 µM iNOS wild type or mutants were incubated with urea (5 M) for 2.5 h at 15 °C to form protein monomer. For the heterodimer formation, the resulting urea-treated monomer was incubated with G450A iNOS full length protein (ratio 7:1) at 30 °C for 1 h in 40 mM EPPS, 10% glycerol, 250 mM NaCl buffer containing 1 mg/ml BSA, 1 mM H4B, 3 mM DTT, 10 mM Arg and then further incubated at 4 °C overnight. Nitrite productions of heterodimer were assayed as described previously in 96 well plates [46]. In short, about 50 ~ 80 µl heterodimers were incubated with cofactor FAD, FMN, and other reagents including SOD and catalase in each well, and NADPH was added to initiate the reaction at 37 °C. After 1 hour, excess NADPH was consumed by adding lactate dehydrogenase (LDH) and sodium pyruvate. Absorbance difference at 550 and 650 nm were measured after adding Griess reagent to each well. Nitrite productions were then calculated according to a standard curve made with nitrite solutions of known concentration.
NO synthesis of heterodimers were determined by spectrophotometric oxyhemoglobin assay [38;55] using extinction coefficient of 38 mM−1 cm−1 for methemoglobin minus oxyhemoglobin. All sample solutions contained 100 µM H4B, 5 mM Arg, 0.3 mM DTT, 0.1 mg/ml BSA, 10 U/ml SOD, 346 U/ml catalase, 1 mM oxyhemoglobin and 4 µM FMN/FAD. Reactions were initiated by adding NADPH (100 µM final concentration), and the absorbance changes at 401 nm were recorded at room temperature. NADPH oxidation was monitored at 340 nm using an extinction coefficient of 6.22 mM−1 cm−1.
Antagonist experiments were done in a similar way except that 300 nM wild type iNOSoxy monomer was included during the heterodimer formation of the mutants.
RESULTS AND DISCUSSION
Spectral Property, Stability, Binding Affinity for H4B and Arg, and Dimer Content of Mutants
Mutations at Arg375 in iNOSoxy changed enzyme’s binding affinity towards substrate Arg and cofactor H4B significantly. When the proteins were purified in the absence of Arg and H4B, adding high concentration of Arg and H4B at room temperature afterwards or incubating the enzyme overnight at 4 °C could not cause a complete high-spin shift in the heme iron spin state, which is a characteristic of substrate and cofactor binding (data not shown). Mutants were also found to be less stable compare to the wild type: two mutants R375N and R375D displayed a P420 peak instead of a P450 peak after the addition of CO to their dithionite reduced forms [56]. To maximize mutants’ stability and their binding affinity for substrate and cofactor, we henceforth utilized mutant proteins that were purified in the presence of 1 mM Arg and 2 µM H4B for our studies, unless noted otherwise.
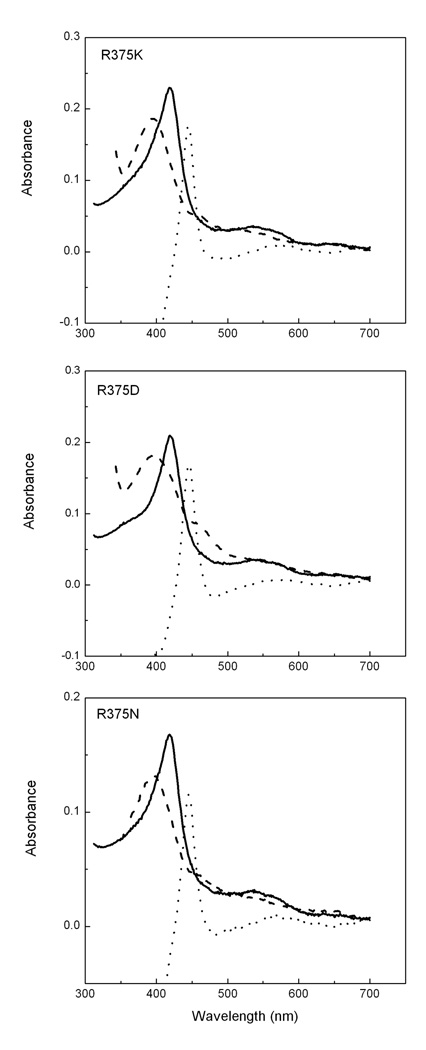
All three mutants displayed the Soret absorbance peaks at 418 nm when purified in the presence of H4B and Arg, indicating that they contained low-spin heme iron (Fig. 3) which is also observed in the wild type NOSoxy and other mutant like W457A iNOSoxy when purified in the absence of substrate and cofactor [46;57;58;59]. Overnight incubation of proteins at 4 °C with additional H4B and Arg induced Soret peak maxima shift to 395 nm for R375K and R375N, and 388 nm for R375D, which indicates that Arg and H4B could bind and stabilize the heme iron in a five-coordinate high-spin state as occurs in the wild type NOS (Table 1, Fig. 3). After addition of the reducing agent dithionite and CO gas to the solution, mutants generated stable ferrous-CO complexes with maximal Soret absorbance at 444 nm (Table 1, Fig. 3). Arg375 mutants were also able to form other heme species including ferric/ferrous NO and ferrous dioxy which displayed similar Soret band maxima as that of the corresponding wild type heme species. These spectral properties demonstrate that the mutation does not perturb the electronic properties of the heme significantly. This finding agrees well with the crystal structures of R375 iNOSoxy mutants which showed that there are very limited structural changes around the heme pocket compared to the wild-type iNOSoxy1. We also estimated the possible conformation changes after the mutations and observed that Lys residue in R375K can still maintain the interaction with the heme, as well as forming the hydrogen bonding with O4 of H4B without significant rearrangements of the protein structure. However, the side chain of Asn and Asp are too short to make direct interactions with H4B in the absence of noticeable structural changes (Supplementary Fig. S1).
Fig. 3. Spectral properties of the R375 mutants.
The spectra of proteins which were purified in the presence of 0.5 mM Arg and 3 µM H4B are shown in solid lines. Dashed lines show the spectra of mutants in the presence of 10 mM Arg and 200 µM H4B after incubation proteins with high concentration of Arg and H4B overnight at 4 °C. A difference spectrum created by subtracting the spectrum of the ferric enzyme from the spectrum of the enzyme ferrous-CO complex is shown in dotted line in each panel.
Table 1. Soret band peak absorbance values and dimeric contents of the wild-type and mutant enzymes.
UV-visible spectra were recorded at room temperature. The dimerization contents were estimated by gel filtration chromatography at 4 °C.
| WT | R375K | R375D | R375N | |
|---|---|---|---|---|
| FeIII (−Arg, −H4B) | 418 nm | 418 nm | 418 nm | 418 nm |
| FeIII (+Arg, +H4B) | 395 nm | 395 nm | 388 nm | 394 nm |
| FeII | 411 nm | 412 nm | 413 nm | 412 nm |
| Ferrous-CO | 444 nm | 444 nm | 445 nm | 444 nm |
| Ferrous-NO | 437, 567 nm | 437, 567 nm | 439, 568 nm | 437, 567 nm |
| Dimer content | 100% | 100% | 30% | 38% |
The dimer-monomer ratio of each Arg375 mutant was estimated by gel filtration chromatography [38;46;58]. After incubating the enzyme with sufficient amounts of substrate and cofactor, R375K mutant was approximately 100% dimeric. However, only approximately 38% and 30% dimer were obtained for R375N and R375D, respectively. Such decreases in dimer content were also observed in R375A and Trp678 mutants where Trp 678 is another conserved amino residue involved in the hydrogen bonding and π stacking with H4B ring in nNOSoxy [38].
Peroxide Assay
We next examined whether the mutants could oxidize NOHA to nitrite in a H2O2-driven reaction. Measurements of the reaction end product-nitrite were carried out photometrically using the Griess Assay in a 96-well microplate at 37 °C as described previously [46;50;51]. For this experiment we used mutants purified in the absence of Arg and with the presence of H4B. Protein solutions were first incubated with 1 mM H4B overnight at 4 °C, then were diluted to the desired concentration and further incubated with various concentration of NOHA (1 mM, 2 mM, 10 mM) or H4B (100 µM, 500 µM, 1 mM) at 30 °C for 30 min. H2O2 solution was added to initiate the reaction, then the reaction mixtures were incubated for 10 min at 30 °C.
Wild type iNOSoxy protein being as a positive control displayed normal activity comparable to our previous results [46]. However, none of the mutants showed activities under these experimental conditions regardless of increasing the enzyme concentration from 110 nM to 1.1 µM. To test whether the lower activity was attributed to the lower binding ability of substrate NOHA and H4B, we measured the spectra of these mutants in the presence of 1 mM NOHA and 0.5 mM H4B. About 80–90% of heme iron of R375K and R375N were in the high spin state suggesting normal binding of NOHA and H4B towards the enzyme under the assay conditions. However, heme iron of R375D mainly existed in the low spin state.
In peroxide assay system, H2O2 binds directly to the NOS ferric heme forming reactive heme-oxy species and further react with Arg or NOHA to produce nitrite in the presence or absence of H4B [46]. Since the NOS heme does not need to acquire electrons from the reductase domain or H4B [50;51;58]. This method can be used to test the reactivity of the heme pocket of the oxygenase domain for NOS enzymes. For R375K and R375N mutants, their lower activities compared to wild type iNOSoxy may be either due to the lower affinity of the enzyme for H2O2 or the reaction generating a heme-oxy species that is less reactive toward NOHA than wild type iNOSoxy. In the case of R375D, low binding affinity towards NOHA plus the two factors mentioned above contribute to its lower activity.
Heme Midpoint Potential
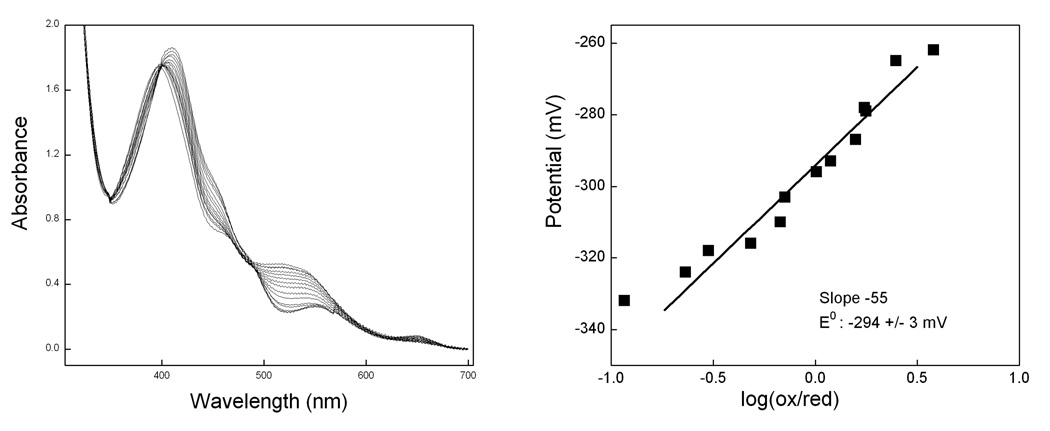
The NOS ferric heme midpoint potential is important in determining the thermodynamic driving force for electron transfer between the reductase domain flavins and the ferric heme. We next measured the heme midpoint potential for R375K and R375D. A spectro-electrochemical reductive titration was performed for the ferric enzyme in the presence of Arg, H4B, and mediator dyes [52]. Representative data were shown in Fig. 4. The heme midpoint potential were determined to be −294 ± 3 mV for R375K and −283 ± 3 mV for R375D, which are about 30 to 40 mV more negative than the wild type iNOSoxy when measured under similar conditions (−263 mV [52]; −253 mV 2).
Fig. 4. Potentiometric reductive titration of R375K iNOSoxy.
Left Panel shows spectra recorded during titration of the ferric enzyme in the presence of H4B, Arg, and mediator dyes. Right panel plots the measured potentials vs. the logarithmic ratio of ferric/ferrous heme.
In NOSs, the heme iron has a proximal cysteine thiolate ligand [12;13;42] (Fig. 2). Studies have shown that the strength of the heme-thiolate bond determines the heme midpoint potential of the enzyme. One conserved residue Trp409 in nNOS (Trp188 in iNOS) engages in π-stacking with the heme and forms hydrogen bond to the proximal cysteine ligand. Mutation on this residue turns out to alter the heme midpoint potential greatly by either weakening or strengthening the heme-thiolate bond [60]. Interestingly, our Arg375 mutation lower the heme midpoint potential, indicating that such mutation could fine tune the heme environment even though spatially Arg375 is far from the cysteine thiolate ligand. These effects are probably mediated through the direct interaction of the Arg375 side chain with the heme propionate. The decrease in the redox potential creates unfavorable thermodynamic change for the electron transfer from the flavins in the reductase domain and the ferric heme. In addition, the lower heme midpoint potential also implied the conceivable changes of the heme reactivity which is supported by NOHA oxidation reaction described above.
Oxidation Rate of the Ferrous Heme-NO (FeIINO) Complex (kox)
The differences in kox values between the wild type and mutants often indicate subtle changes of the heme environment. We next determined kox for the wild-type and Arg375 mutants by mixing their ferrous heme-NO complexes with an air-saturated buffer in the stopped-flow spectrophotometer, and following their subsequent conversion to ferric enzyme [61–64]. The starting FeIINO complex and ending ferric enzyme were observed for all proteins, and their conversion fit well to a monophasic transition (Fig. 5). The calculated kox values are listed in Table 2. All mutants showed higher kox rates, which were about 2.1 to 2.8 times faster than that of the wild-type iNOSoxy. In combination with the results of the heme midpoint potential measurements, mutation at Arg375 lowered the heme midpoint potential and increased the oxidation rate of the ferrous heme-NO complex. Such relationship was observed in other mutants as well. For example, W409F, W409Y nNOSoxy mutants and mesoheme-nNOSoxy also displayed lower heme midpoint potentials and high kox values relative to their wild-type enzyme [54;60;61;65–67]. For iNOSoxy, replacing Trp188 to His increases the heme midpoint potential by 80 mV due to the strengthened heme-thiolate bond, since His is able to form stronger hydrogen bonding with proximal Cys ligand, and its kox rate of W188H was found to decrease by 40% relative to wild type iNOSoxy correspondingly3.
Fig. 5. Spectra and kinetics of iNOSoxy FeIINO oxidation reactions.
Anaerobic ferrous iNOSoxy proteins in the presence of Arg (10 mM) and H4B (400 µM) were titrated by small amounts of anaerobic NO-saturated buffer to form the ferrous NO complex, which was then rapid-mixed in a stopped flow spectrophotometer with air-saturated buffer at 10 °C. Left panels contain spectra of the beginning ferrous-NO and ending ferric heme species as calculated by global analysis of the spectral data, while right panels show the concentration of both species versus time after mixing. Data are representative of three experiments.
Table 2. Rates of heme and H4B redox transitions and product yield during Arg single turnover reactions.
Ferrous wild type and mutant iNOSoxy proteins containing substrates Arg and H4B were rapid-mixed at 10° C with air-saturated buffer to start the reactions. Subsequent heme transitions were followed by stopped-flow rapid-scanning spectroscopy. Rates were calculated by Specfit global analysis of diode array spectral data. H4B radical formation and decay were monitored by EPR. NOHA productions were calculated based on HPLC results described in the Experimental section. Values are the means ± S.D. of three determinations.
| NOS | FeIIO2 formation (s−1) |
FeIIO2 decay (s−1) |
H4B radical formation (s−1) |
H4B radical decay (s−1) |
NOHA produced per heme |
|---|---|---|---|---|---|
| WT | 67.8±3.0 | 20.8±0.5 | 11.1a | 0.7±0.5 | 0.55±0.06 |
| R375K | 55.9±3.3 | 29.7±1.1 | 30.2±9.8 | 1.6±0.5 | 0.30±0.10 |
| R375N | 72.4±3.2 | 17.9±0.5 | 34.8±4.6 | 1.2±0.6 | 0.25±0.08 |
| R375D | NA | 0.13±0.01 | 8.8±1.6 | 0.3±0.1 | 0.11±0.02 |
values from ref [37].
The correlation between the kox rate and heme midpoint potential provides a good indication about the heme environment and the characteristics of the heme-thiolate bond. NOS heme FeII-NO complexes oxidize at much higher rates compare to any other heme protein including Hb or Mb [28;54;65;68;69]. Such difference is attributed in part by the heme-thiolate bond, causing the NOS heme FeIINO complex to have a lower midpoint potential. Our results on Arg375 further indicate that besides the difference between the heme proximal ligands (cysteine in NOSs versus histidine in Hb, Mb and other heme proteins), modifications of nearby amino residue in the heme pocket could also modulate the heme midpoint potentials and the kox rate of NOS enzymes.
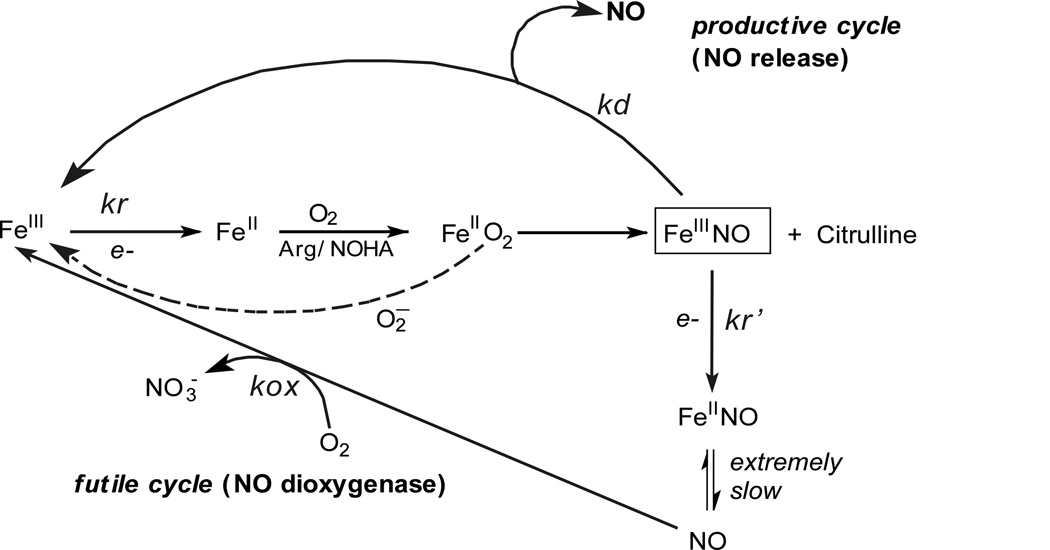
The rate of ferrous NO oxidation (kox) is one of the key parameters controlling the steady state NO synthesis in full length enzymes [63;70] (Fig. 6). During NO synthesis, practically all generated NO binds to the NOS ferric heme forming FeIIINO before exiting the enzyme. Part of the FeIIINO is reduced by the attached reductase domain and the resulting FeIINO complex must react with O2 (kox in Fig. 6) afterwards to return to the catalytic cycle [63;70]. Unlike other heme-thiolate proteins [54;71], NOS enzyme posses relatively high oxidization rate of FeIINO complex which is crucial to avoid the formation of a dead-end product during catalysis. The observed faster kox rates of Arg375 mutants possibly increase the steady-state activity of iNOS by speeding the return of enzyme molecules to the productive cycle (Fig. 6) [62;70;72].
Fig. 6. Global kinetic model for NO synthesis by NOS.
During steady state catalysis, the enzyme molecules engage in a productive cycle that releases free NO and in a futile cycle that releases a higher oxide of nitrogen (nitrate). Reduction of ferric enzyme to ferrous (kr) enables the heme to bind O2 and initiates catalytic reactions. After NO is made, an immediate product of catalysis is the ferric heme-NO complex (FeIII-NO), which can either release NO (kd) or become reduced (kr') to generate a ferrous heme-NO complex (FeII-NO). The ferrous heme-NO complex dissociates extremely slowly and instead regenerates the active ferric enzyme by reacting with O2 (kox). The FeIIO2 intermediate can also undergo an uncoupled reaction that generates ferric enzyme and superoxide.
Kinetics of Heme Transitions, H4B Radical Formation and Decay During Arg Hydroxylation
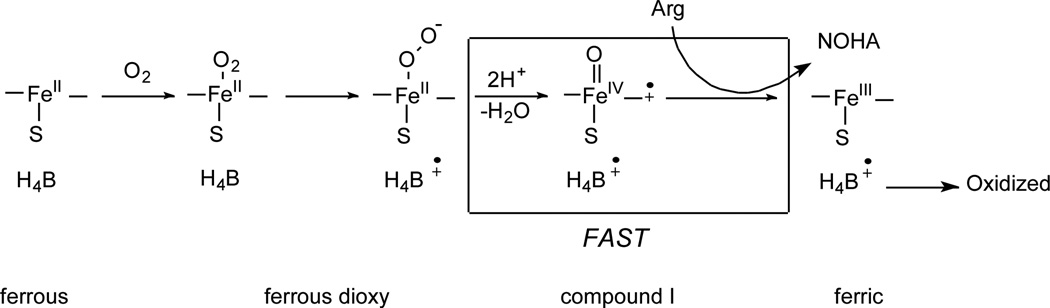
Previous studies show that H4B performs a novel redox function in NOS: it reduces the ferrous dioxy intermediate (FeIIO2) that forms during oxygen activation. This step is critical because it enables NOS to form the heme-oxy species that react with Arg or NOHA (Scheme 2). Since Arg375 forms hydrogen bond with the H4B ring, we investigated whether mutation affects the redox function of H4B during single catalytic turnover Arg hydroxylation.
Scheme 2.
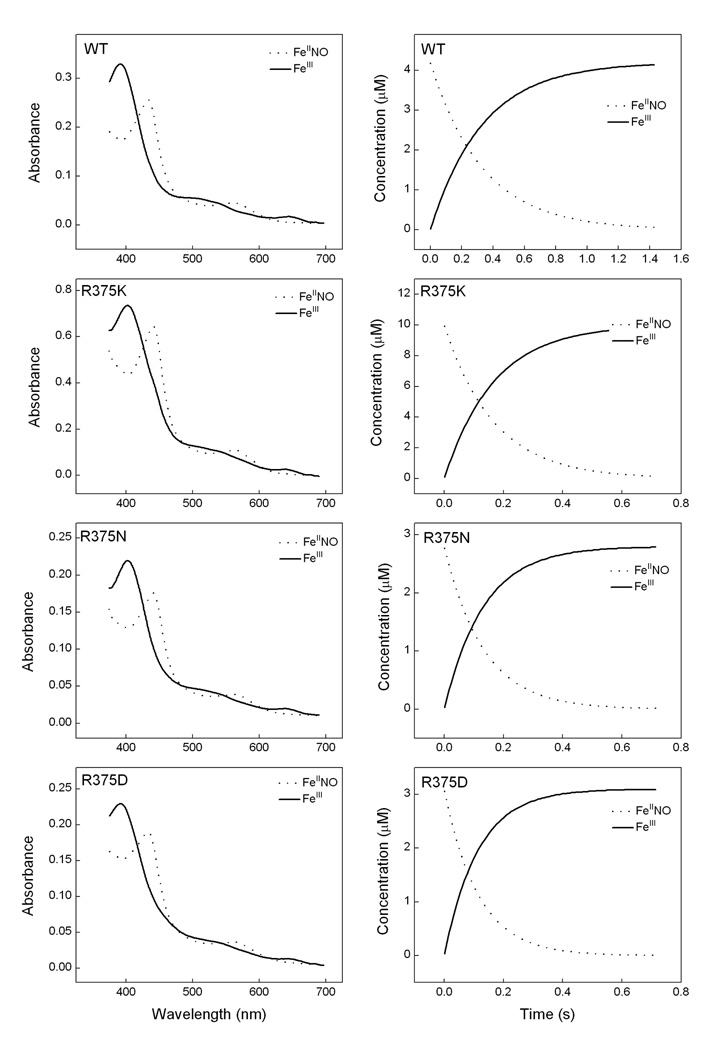
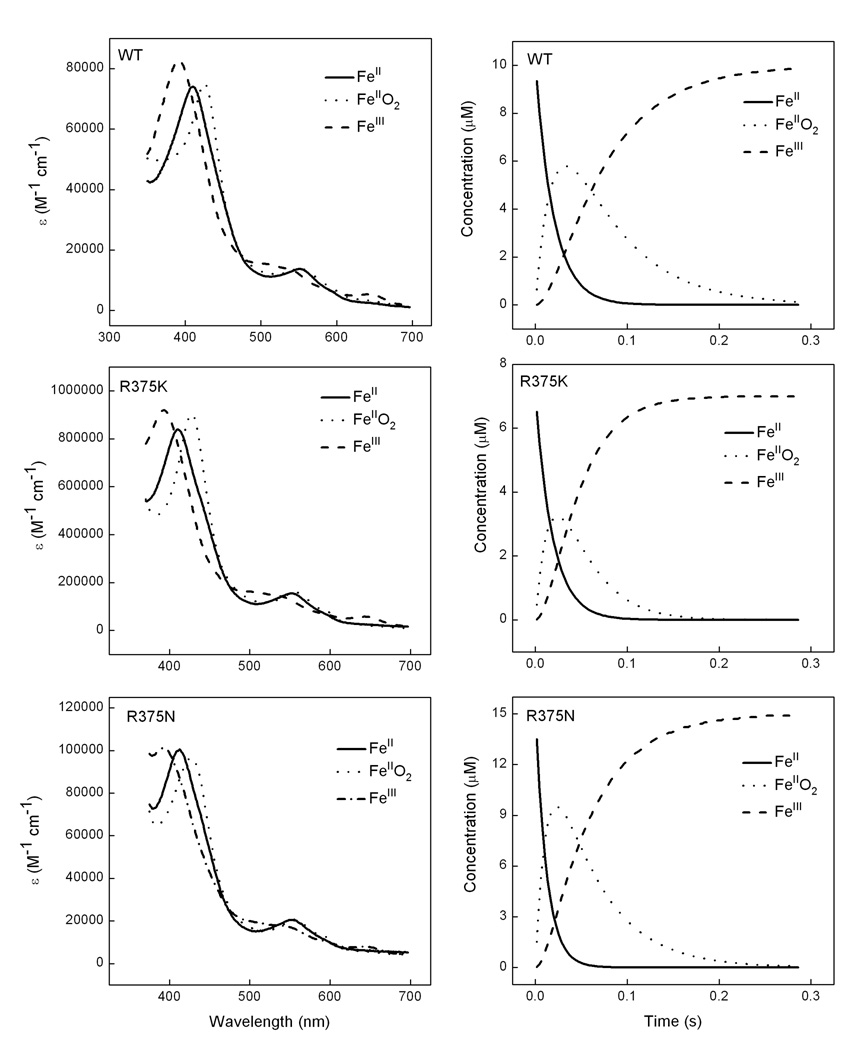
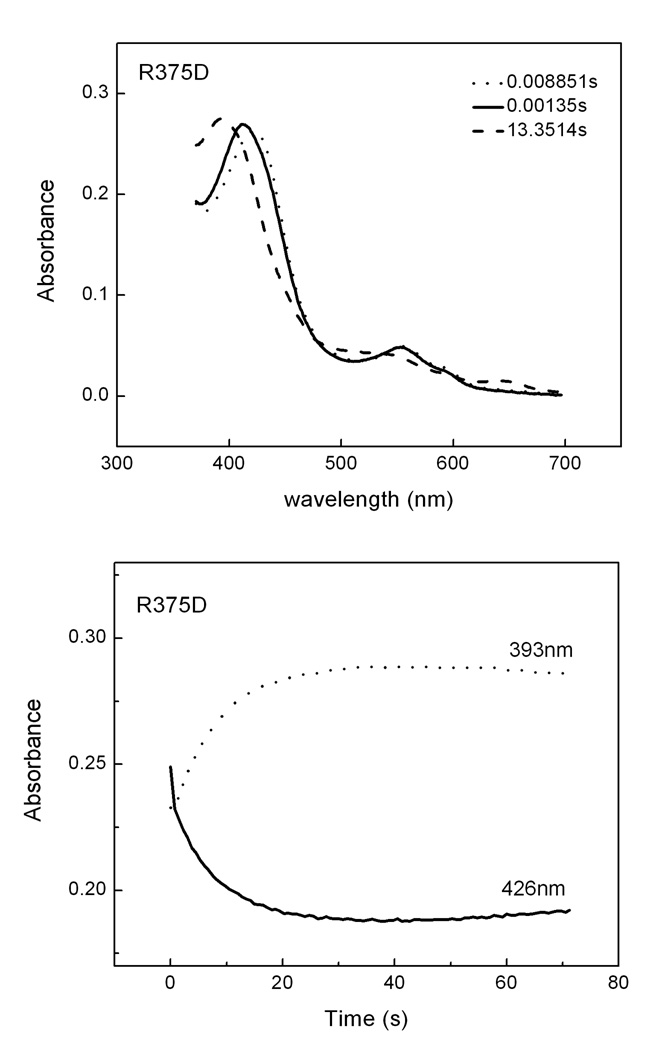
First, we monitored the heme transitions that occur during a single turnover Arg hydroxylation reaction run at 10 °C in the stopped-flow spectrophotometer. Air-saturated buffer was mixed with anaerobic ferrous iNOSoxy proteins that contained H4B and Arg and the subsequent heme transitions were followed by UV-visible scanning [20;37;73]. Fig. 7 and 8 contain representative data collected during reactions catalyzed by wild type, R375K, R375N and R375D iNOSoxy. In Fig. 7, the left panels show the light absorbance spectra of the three enzyme species that were detected during each reaction, while the right panels show the concentration of each species changed with time during the reaction. In all mutants we could identify a beginning ferrous species, a heme-dioxy (FeIIO2) intermediate, and an ending ferric species. Their Soret absorbance maxima for each heme species matched very well among wild-type and mutants. The ending ferric species for all mutants displayed a 390–393nm peak, which indicated that their heme groups were predominantly in high spin state at the end of the single turnover reactions. In Fig. 8, Panel A shows the scanning traces collected at different time after mixing anaerobic ferrous R375D proteins with air-saturated buffer at 10 °C. Panel B contains cross section analysis of the kinetic data at 426 nm for the decay of the heme ferrous dioxy and 393 nm for the formation of the ferric enzyme. These results showed that Arg375 mutations still permitted buildup of a FeIIO2 intermediate, and did not significantly alter its spectral characteristics or those of the ferrous or ferric species. Formation and disappearance of the FeIIO2 intermediate were best described by monophasic transitions, consistent with previous reports [20;37;73]. The mutants showed similar FeIIO2 formation rate as of the wild type iNOSoxy (Table 2). However, the transition of the FeIIO2 intermediate to ferric enzyme was found to be different among mutants: R375N and R375K mutants demonstrated similar or faster rate, respectively, compared to wild type, but much slower FeIIO2 decay rate was observed in the R375D mutant (Table 2). Our previous results show that the electron transfer from H4B to ferrous dioxy species in wild type iNOSoxy increases the ferrous dioxy decay rate by 40 fold compared to the H2B- bond protein (H2B cannot reduce the ferrous dioxy species). To elucidate the redox role of H4B in Arg375 mutants, we performed the single turnover Arg hydroxylation reactions using H2B-bound R375D mutant as well. Three heme species were observed, whose spectral features were identical to the ferrous, FeIIO2, and ferric species in the H4B-bound proteins (data not shown). However, conversion of the FeIIO2 intermediate to ferric enzyme was 5 times slower in the H2B-bound proteins, suggesting H4B is able to transfer an electron to the heme ferrous dioxy species of Arg375 mutants during Arg hydroxylation single turnover reactions.
Fig. 7. Stopped-flow analysis of heme transitions and kinetics during single-turnover Arg hydroxylation catalyzed by R375K, R375N and wild type iNOSoxy.
Anaerobic ferrous iNOSoxy proteins (4 µM) in the presence of Arg (10 mM) and H4B (400 µM) were mixed in a stopped flow instrument with air-saturated buffer at 10 °C and diode array spectra were collected. Left panels contain three heme species that were detected during each reaction as calculated by global analysis of the spectral data, while right panels show their concentration profiles versus time after mixing. Data are representative of three similar experiments.
Fig. 8. Heme transitions and kinetics of R375D iNOSoxy during Arg hydroxylation single turnover reactions.
Panel A contains spectral scans collected at 0.0089 s (…), 0.00135 s (––) and 13.35 s (---) after mixing anaerobic ferrous proteins with air-saturated buffer at 10 °C. Panel B contains cross section analysis of the kinetic data at 426 nm and 393 nm. Data are representative of three similar experiments.
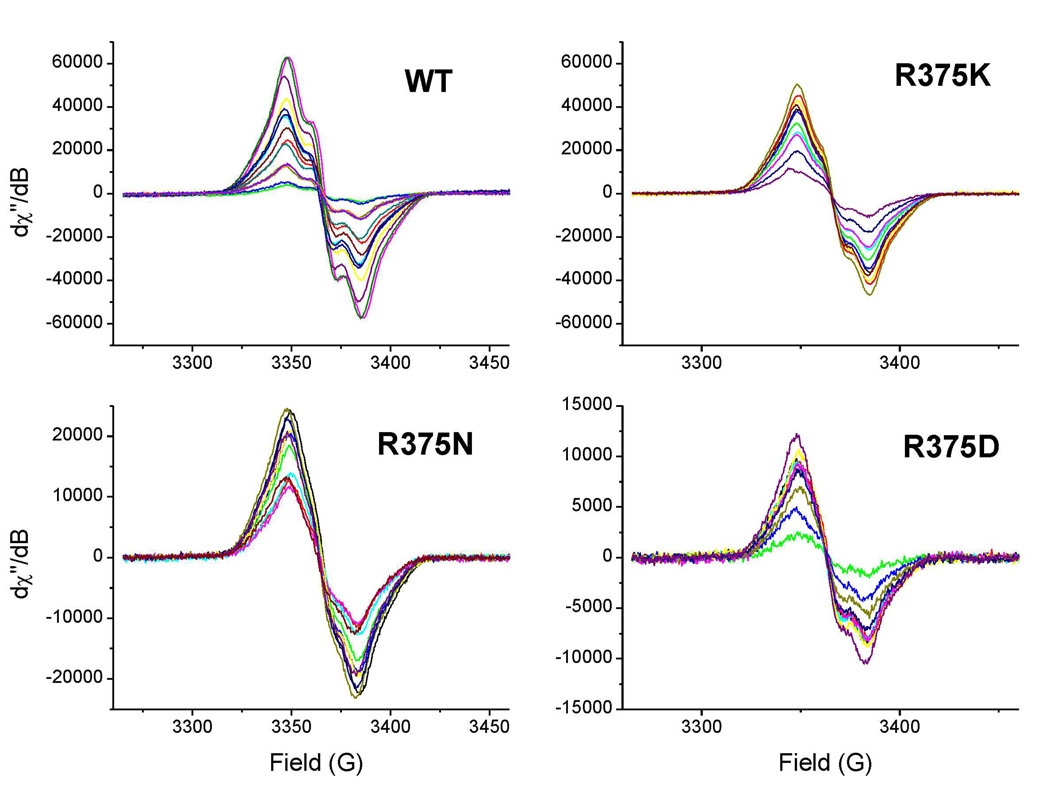
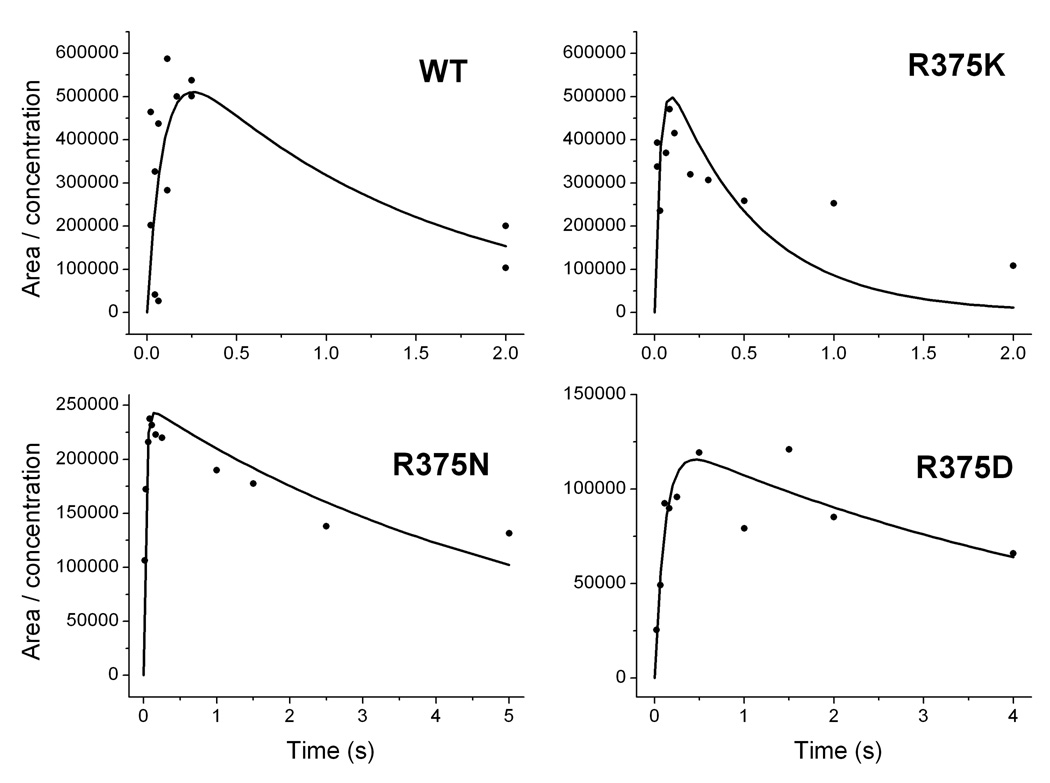
We next studied H4B radical formation and decay during Arg hydroxylation. Rapid-freeze experiments showed that a radical with g value of 2.0 built up in Arg375 mutants during the single turnover reactions (Fig. 9). The spectra and saturation properties of the mutant radicals were either identical or similar to those of the H4B radical formed in wild-type iNOSoxy [13;20;37;42;74]. Signal intensities were used to calculate the amount of H4B radical per heme that was present at each time point. The kinetic data were fit to a two step process including the radical formation and decay (Fig. 10). The relative rates of H4B radical formation are consistent with the different ferrous dioxy disappearance rates we observed in the mutants, and imply that there is faster ferrous dioxy reduction by H4B in the R375K and R375N mutants compared to the wild type. Apparently R375 mutations alter the tempo of electron transfer: in R375K and R375N mutants, H4B transfers an electron faster than in the wild type protein, while H4B transfers an electron slower in the R375D mutant. Our kinetic data also suggest the mutations do not greatly affect the stability of the H4B radical since all mutants displayed similar H4B decay rates compared to the wild type within experimental error.
Fig. 9. H4B radical formation and decay during Arg hydroxylation in Arg375 mutants and wild-type iNOSoxy.
Anerobic ferrous iNOSoxy proteins were rapidly mixed with an air-saturated solution at 10 °C to start the reaction. Reactions were aged at the various times (16 ms to 5 s), and then quenched by rapid freezing. EPR traces are shown for Arg375 mutants and wild-type iNOSoxy.
Fig. 10. Calculated concentrations of the H4B radical versus time during Arg hydroxylation in Arg375 mutants and wild-type iNOSoxy.
The relative concentration of H4B radical was calculated by double integration of the EPR signals shown in Figure 9 and divided by the protein concentration.
Our data analysis also shows that a normal level of H4B radical accumulated in the R375K mutant reaction (about 0.78 per heme) compared to wild type iNOSoxy [37]. However, there was less H4B radical accumulation (0.4 and 0.2 radical per heme, respectively) during the single turnover reactions of R375N and R375D iNOSoxy. Increasing the H4B concentration in the reactions from 1 to 5 mM did not alter the results in any way (data not shown), suggesting that the observed differences were not due to incomplete H4B binding. The poor radical buildup observed in these mutants might be due to their incomplete dimer formation, or possibly some unreacted H4B remaining in the enzyme.
We then studied the stoichiometry of Arg hydroxylation in the Arg375 mutants. NOHA was the only amino acid product in all cases and its formation depended completely on H4B (data not shown). The R375K, R375N and R375D mutants consistently catalyzed about 0.30, 0.25 and 0.11 of Arg hydroxylation on a per heme basis during the single turnover Arg hydroxylation reactions, respectively, which are all less extensive than in the wild type (0.55, Table 2).
Previous studies suggest that a complete and well coupled electron transfer between H4B and FeIIO2 occurred for wild type NOS proteins in Arg hydrxoylation single turnover reactions. Thus, one would expect that Arg hydroxylation should generate 1 NOHA/heme. However, we and others [20;73–75] typically observe substoichiometric NOHA formation in single turnover reactions ranging from 0.2 to 0.8 NOHA formed per heme, which implies that subsequent steps (i.e. conversion of the iron-peroxo intermediate to iron-oxo or its reaction with Arg) become uncoupled in the wild type iNOSoxy within this setting. We recently identified a heme-oxy reaction intermediate in the W188H iNOSoxy [60], further studies should be able to shed light on this issue. Here, the mutation at Arg375 even further decreased the product stoichiometry compared to the wild type protein. One possibility is that Arg375 mutations might diminish the reactivity of the heme-based oxidant toward Arg, since this effect will only occur after H4B reduces the FeIIO2 intermediate. Our H2O2-driven NOHA oxidation results supported this assumption since the heme-based oxidant formed when ferric NOS reacts with H2O2 is thought to be similar or identical to the species that hydroxylates NOHA during normal NOS catalysis [33;61]. For the R375N and D mutants, their incomplete enzyme dimerization would also diminish the product stoichiometry.
Activities of Mutant Heterodimers
To test whether mutation at Arg375 affect the NO synthesis activity, we made heterodimers by incubating the monomer of wild type iNOSoxy (or mutants) and full-length G450A iNOS. The point mutation Gly450 to Ala prevents itself forming homodimer, thus the reductase domain from G450A is only capable of transferring electron to the iNOSoxy which enables NO production from the oxygenase domain. [46;76;77]. We first converted oxygenase domains of wild type and mutants to monomer by adding urea, then incubated such monomer with full-length G450A protein in the presence of H4B and Arg to form heterodimer. We next measured the rates of NO synthesis and NADPH oxidation of these heterodimers in the presence of Arg and H4B at 25 °C using the spectrophotometric oxyhemoglobin assay. Results are summarized in Table 3.
Table 3. Catalytic activities of heterodimers formed by the Arg375 mutants or wild type iNOSoxy and G450A iNOS.
NO synthesis and NADPH oxidation were measured at room temperature and NO2− production measurement was performed at 30 °C as described under "Materials and Methods." The values are the mean ± S.D. of three measurements. WT, wild type.
| Heterodimer |
Native WT | ||||
|---|---|---|---|---|---|
| R375K | R375N | R375D | WT | iNOSfl | |
| NO synthesis (min−1) | 17.6±0.6 | 8.6±0.3 | 1.7±0.1 | 51.8±0.4 | 50.9±0.2 |
| NADPH oxidation (min−1) | 62.5±4.0 | 64.2±3.8 | 50.1±4.4 | 80.3±6.2 | 78.2±3.1 |
| NO2− production (µM) | 7.8±0.2 | 6.3±0.1 | 1.8±0.1 | 30.3±0.7 | NA |
| (NO2−/µM G450A/min) | |||||
| NO / NADPH | 3.6 | 7.5 | 29.4 | 1.6 | 1.5 |
| Antagonist Study | 29% | 30% | 13% | 100% | NA |
The NO synthesis activity of the heterodimer containing iNOSoxy wild type was very close to the native wild type full length iNOS enzyme, which validates the experimental approach. All mutants showed lower NO synthesis activities with only 35%, 17% and 7% of the wild type for R375K, N and D, respectively. However, their concurrent rates of NADPH oxidation were also reduced to a lesser degree. In general, NO synthesis from Arg displays a stoichiometry of 1.5 NADPH oxidized per NO formed for wild type full length NOSs [18;78]. This minimum value was observed for our wild-type heterodimer iNOS at 25 °C, while a stoichiometry of 3.5, 7.5, and 29 NADPH oxidized per NO formed were found for R375K, R375N and R375D, respectively (Table 3). This indicates that a majority of the NADPH oxidation is uncoupled from their NO synthesis in the Arg375 mutants.
Nitrite production of heterodimer measured by Griess assay also showed the lower activity of mutants compared to the wild type, which is consistent with the results from our NO measurements (Table 3). One could argue that the lower activity might be due to the low affinity of mutant monomer toward the G450A subunit. To test this, we measured the capability of each mutant monomer to “compete” or “antagonize” wild type iNOSoxy monomer in forming a heterodimer with full length G450A iNOS. First, 300 nM wild type iNOSoxy monomer formed heterodimer with G450A iNOS. If mutant monomers could compete with wild type effectively, then addition of excess of mutant monomer (~ 3 µM) to the protein solution should reduce the nitrite production to a level similar to that of the heterodimer we formed by mixing mutant monomer and G450 iNOS in the absence of wild type iNOSoxy monomer. Our results in Table 3 clearly indicate that Arg375 mutants compete successfully and completely with wild-type iNOSoxy monomer in forming the heterodimer, eliminating the possibility that lower activity is associated with a lower binding affinity of the mutant monomer to full length G450A iNOS. These results are also consistent with our previous heterodimer studies of R375A mutant [46].
The catalytic profiles we observed for heterodimer are consistent with our single turnover Arg hydroxylation results. Since all the heterodimers have the same reductase domain subunit, lower NO synthesis observed in the mutants solely come from the mutant oxygenase domain itself and/or the inter-domain electron transfer. Negative shift of the heme midpoint potential of Arg375 mutants disfavored the electron transfer from reductase domain to the oxygenase domain, which can decrease the NO synthesis activity. Also the less efficient electron transfer from H4B and/or reduced reactivity of heme-based oxidant further lower the NO synthesis rate in Arg375 mutants.
Conclusions
Mutation of the conserved residue Arg375, whose side chain H-bonds with heteroatoms in the ring of H4B, greatly changes iNOS properties and catalytic behavior. The decreased binding affinity towards substrate and cofactor can be overcome by purifying the enzyme in the presence of substrates and cofactor and later incubating the enzyme with additional amount of substrate and cofactor. However, the maximum dimer formation obtained in the R375N and D mutant are only 38% and 30%, respectively. Such a low percentage in these two mutants partially contributes to their lower Arg hydroxylation observed in the single turnover reactions and decreased NO synthesis rate. Mutation at Arg375 also perturbs the heme environment which leads to the negative shift of the heme midpoint potential, faster ferrous NO oxidation rate and vanished peroxide activity in H2O2-drived NOHA oxidation compare to the wild type protein. Although replacement of Arg375 by Lys (K) increases the tempo of electron transfer between H4B and the ferrous dioxy species formed during O2 activation, R375K still shared similar properties like other mutants, for instance, lower NOHA production in single turnover reactions and lower NO synthesis activities and uncoupled NADPH oxidation. We attribute these properties mainly to the reduced reactivity of heme-based oxidant formed after the electron transfer from H4B. Our results suggest that the conserved residue Arg375 modulates the redox and structural functions of H4B, and is thereby important in controlling the catalytic behavior of NOS enzymes.
Supplementary Material
Acknowledgements
We thank Manisha Sharma and John MacDonald for excellent technical support. This work was supported by National Institutes of Health Grants GM51491 and CA53914 (D. J. S.), American Heart Association Beginning Grant-in-aid 0565297B (Z-Q. W.), KSU Farris Innovation Award (Z-Q. W.) and KSU Tuscarawas Faculty Professional Development Release Time Award (Z-Q. W.).
Table of Abbreviations
- NOS
nitric oxide synthase
- iNOS
inducible nitric oxide synthase
- iNOSoxy
the oxygenase domain of inducible nitric oxide synthase
- Arg
L-arginine
- DTT
dithiothreitol
- NO
nitric oxide
- EPPS
4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid
- NOHA
Nω-hydroxy-L-arginine
- H4B
(6R)-5, 6, 7, 8-tetrahydro-L-biopterin
- H2B
7,8-dihydro-l-biopterin
- NED
N-1-naphthylethylenediamine dihydrochloride
- LDH
lactate dehydrogenase
- FeII
ferrous heme species
- FeIII
ferric heme species
- FeIIO2
ferrous dioxy species
- FeIIINO
ferric NO species
- FeIINO
ferrous NO species
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Z.-Q.Wang, M. Aoyagi, E. D. Getzoff and D. J. Stuehr, unpublished data.
J. Santolini and D. J. Stuehr, unpublished data.
Z.-Q.Wang, and D. J. Stuehr, unpublished data
Reference List
- 1.Nathan C, Shiloh MU. Proc.Natl.Acad.Sci.U.S.A. 2000;97:8841–8848. doi: 10.1073/pnas.97.16.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sasaki M, Gonzalez-Zulueta M, Huang H, Herring WJ, Ahn S, Ginty DD, Dawson VL, Dawson TM. Proc.Natl.Acad.Sci.U.S.A. 2000;97:8617–8622. doi: 10.1073/pnas.97.15.8617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stopper H, Moller M, Bommel HM, Schmidt HH. Toxicol.Lett. 1999;106:59–67. doi: 10.1016/s0378-4274(99)00019-3. [DOI] [PubMed] [Google Scholar]
- 4.Stuehr DJ, Ghosh S. Handbook of Experimental Pharmacol. 2000. pp. 33–70. [Google Scholar]
- 5.Alderton WK, Cooper CE, Knowles RG. Biochem.J. 2001;357:593–615. doi: 10.1042/0264-6021:3570593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gorren AC, List BM, Schrammel A, Pitters E, Hemmens B, Werner ER, Schmidt K, Mayer B. Biochemistry. 1996;35:16735–16745. doi: 10.1021/bi961931j. [DOI] [PubMed] [Google Scholar]
- 7.Bredt DS, Snyder SH. Proc.Natl.Acad.Sci.U.S.A. 1990;87:682–685. doi: 10.1073/pnas.87.2.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie QW, Cho HJ, Calaycay J, Mumford RA, Swiderek KM, Lee TD, Ding A, Troso T, Nathan C. Science. 1992;256:225–228. doi: 10.1126/science.1373522. [DOI] [PubMed] [Google Scholar]
- 9.Pollock JS, Forstermann U, Mitchell JA, Warner TD, Schmidt HH, Nakane M, Murad F. Proc.Natl.Acad.Sci.U.S.A. 1991;88:10480–10484. doi: 10.1073/pnas.88.23.10480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masters BS, McMillan K, Sheta EA, Nishimura JS, Roman LJ, Martasek P. FASEB J. 1996;10:552–558. doi: 10.1096/fasebj.10.5.8621055. [DOI] [PubMed] [Google Scholar]
- 11.Stuehr DJ. Annu.Rev.Pharmacol.Toxicol. 1997;37:339–359. doi: 10.1146/annurev.pharmtox.37.1.339. [DOI] [PubMed] [Google Scholar]
- 12.Crane BR, Arvai AS, Ghosh DK, Wu C, Getzoff ED, Stuehr DJ, Tainer JA. Science. 1998;279:2121–2126. doi: 10.1126/science.279.5359.2121. [DOI] [PubMed] [Google Scholar]
- 13.Fischmann TO, Hruza A, Niu XD, Fossetta JD, Lunn CA, Dolphin E, Prongay AJ, Reichert P, Lundell DJ, Narula SK, Weber PC. Nat.Struct.Biol. 1999;6:233–242. doi: 10.1038/6675. [DOI] [PubMed] [Google Scholar]
- 14.Sheta EA, McMillan K, Masters BS. J.Biol.Chem. 1994;269:15147–15153. [PubMed] [Google Scholar]
- 15.Gachhui R, Presta A, Bentley DF, Abu-Soud HM, McArthur R, Brudvig G, Ghosh DK, Stuehr DJ. J.Biol.Chem. 1996;271:20594–20602. doi: 10.1074/jbc.271.34.20594. [DOI] [PubMed] [Google Scholar]
- 16.Garcin ED, Bruns CM, Lloyd SJ, Hosfield DJ, Tiso M, Gachhui R, Stuehr DJ, Tainer JA, Getzoff ED. J.Biol.Chem. 2004;279:37918–37927. doi: 10.1074/jbc.M406204200. [DOI] [PubMed] [Google Scholar]
- 17.Mayer B, Hemmens B. Trends Biochem.Sci. 1997;22:477–481. doi: 10.1016/s0968-0004(97)01147-x. [DOI] [PubMed] [Google Scholar]
- 18.Stuehr DJ. Biochim.Biophys.Acta. 1999;1411:217–230. doi: 10.1016/s0005-2728(99)00016-x. [DOI] [PubMed] [Google Scholar]
- 19.Li H, Poulos TL. J.Inorg.Biochem. 2005;99:293–305. doi: 10.1016/j.jinorgbio.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 20.Wei CC, Wang ZQ, Wang Q, Meade AL, Hemann C, Hille R, Stuehr DJ. J.Biol.Chem. 2001;276:315–319. doi: 10.1074/jbc.M008441200. [DOI] [PubMed] [Google Scholar]
- 21.Wei CC, Wang ZQ, Hemann C, Hille R, Stuehr DJ. J.Biol.Chem. 2003;278:46668–46673. doi: 10.1074/jbc.M307682200. [DOI] [PubMed] [Google Scholar]
- 22.Griffith OW, Stuehr DJ. Annu.Rev.Physiol. 1995;57:707–736. doi: 10.1146/annurev.ph.57.030195.003423. [DOI] [PubMed] [Google Scholar]
- 23.Masters BS, McMillan K, Sheta EA, Nishimura JS, Roman LJ, Martasek P. FASEB J. 1996;10:552–558. doi: 10.1096/fasebj.10.5.8621055. [DOI] [PubMed] [Google Scholar]
- 24.Marletta MA. Cell. 1994;78:927–930. doi: 10.1016/0092-8674(94)90268-2. [DOI] [PubMed] [Google Scholar]
- 25.Abu-Soud HM, Yoho LL, Stuehr DJ. J.Biol.Chem. 1994;269:32047–32050. [PubMed] [Google Scholar]
- 26.Roman LJ, Miller RT, de La Garza MA, Kim JJ, Siler Masters BS. J.Biol.Chem. 2000;275:21914–21919. doi: 10.1074/jbc.M002449200. [DOI] [PubMed] [Google Scholar]
- 27.Wei CC, Crane BR, Stuehr DJ. Chem.Rev. 2003;103:2365–2383. doi: 10.1021/cr0204350. [DOI] [PubMed] [Google Scholar]
- 28.Abu-Soud HM, Gachhui R, Raushel FM, Stuehr DJ. J.Biol.Chem. 1997;272:17349–17353. doi: 10.1074/jbc.272.28.17349. [DOI] [PubMed] [Google Scholar]
- 29.Sorlie M, Gorren AC, Marchal S, Shimizu T, Lange R, Andersson KK, Mayer B. J.Biol.Chem. 2003;278:48602–48610. doi: 10.1074/jbc.M305682200. [DOI] [PubMed] [Google Scholar]
- 30.Gerber NC, Nishida CR, Ortiz de Montellano PR. Arch.Biochem.Biophys. 1997;343:249–253. doi: 10.1006/abbi.1997.0187. [DOI] [PubMed] [Google Scholar]
- 31.Mayer B, Wu C, Gorren AC, Pfeiffer S, Schmidt K, Clark P, Stuehr DJ, Werner ER. Biochemistry. 1997;36:8422–8427. doi: 10.1021/bi970144z. [DOI] [PubMed] [Google Scholar]
- 32.Abu-Soud HM, Loftus M, Stuehr DJ. Biochemistry. 1995;34:11167–11175. doi: 10.1021/bi00035a023. [DOI] [PubMed] [Google Scholar]
- 33.Rusche KM, Spiering MM, Marletta MA. Biochemistry. 1998;37:15503–15512. doi: 10.1021/bi9813936. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt PP, Lange R, Gorren AC, Werner ER, Mayer B, Andersson KK. J.Biol.Inorg.Chem. 2001;6:151–158. doi: 10.1007/s007750000185. [DOI] [PubMed] [Google Scholar]
- 35.Hurshman AR, Krebs C, Edmondson DE, Marletta MA. Biochemistry. 2003;42:13287–13303. doi: 10.1021/bi035491p. [DOI] [PubMed] [Google Scholar]
- 36.Berka V, Yeh HC, Gao D, Kiran F, Tsai AL. Biochemistry. 2004;43:13137–13148. doi: 10.1021/bi049026j. [DOI] [PubMed] [Google Scholar]
- 37.Wang ZQ, Wei CC, Ghosh S, Meade AL, Hemann C, Hille R, Stuehr DJ. Biochemistry. 2001;40:12819–12825. doi: 10.1021/bi011182s. [DOI] [PubMed] [Google Scholar]
- 38.Wang ZQ, Wei CC, Santolini J, Panda K, Wang Q, Stuehr DJ. Biochemistry. 2005;44:4676–4690. doi: 10.1021/bi047508p. [DOI] [PubMed] [Google Scholar]
- 39.Wei CC, Wang ZQ, Durra D, Hemann C, Hille R, Garcin ED, Getzoff ED, Stuehr DJ. J.Biol.Chem. 2005;280:8929–8935. doi: 10.1074/jbc.M409737200. [DOI] [PubMed] [Google Scholar]
- 40.Wei CC, Wang ZQ, Tejero J, Yang YP, Hemann C, Hille R, Stuehr DJ. J.Biol.Chem. 2008;283:11734–11742. doi: 10.1074/jbc.M709250200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagatsu T, Ichinose H. Mol.Neurobiol. 1999;19:79–96. doi: 10.1007/BF02741379. [DOI] [PubMed] [Google Scholar]
- 42.Raman CS, Li H, Martasek P, Kral V, Masters BS, Poulos TL. Cell. 1998;95:939–950. doi: 10.1016/s0092-8674(00)81718-3. [DOI] [PubMed] [Google Scholar]
- 43.Aoyagi M, Arvai AS, Ghosh S, Stuehr DJ, Tainer JA, Getzoff ED. Biochemistry. 2001;40:12826–12832. doi: 10.1021/bi011183k. [DOI] [PubMed] [Google Scholar]
- 44.Crane BR, Arvai AS, Ghosh S, Getzoff ED, Stuehr DJ, Tainer JA. Biochemistry. 2000;39:4608–4621. doi: 10.1021/bi992409a. [DOI] [PubMed] [Google Scholar]
- 45.Wang ZQ, Wei CC, Stuehr DJ. J.Biol.Chem. 2002;277:12830–12837. doi: 10.1074/jbc.M111967200. [DOI] [PubMed] [Google Scholar]
- 46.Ghosh S, Wolan D, Adak S, Crane BR, Kwon NS, Tainer JA, Getzoff ED, Stuehr DJ. J.Biol.Chem. 1999;274:24100–24112. doi: 10.1074/jbc.274.34.24100. [DOI] [PubMed] [Google Scholar]
- 47.Sagami I, Sato Y, Daff S, Shimizu T. J.Biol.Chem. 2000;275:26150–26157. doi: 10.1074/jbc.M000534200. [DOI] [PubMed] [Google Scholar]
- 48.Chen PF, Berka V, Wu KK. Arch.Biochem.Biophys. 2003;411:83–92. doi: 10.1016/s0003-9861(02)00729-4. [DOI] [PubMed] [Google Scholar]
- 49.Stuehr DJ, Ikeda-Saito M. J.Biol.Chem. 1992;267:20547–20550. [PubMed] [Google Scholar]
- 50.Gachhui R, Ghosh DK, Wu C, Parkinson J, Crane BR, Stuehr DJ. Biochemistry. 1997;36:5097–5103. doi: 10.1021/bi970331x. [DOI] [PubMed] [Google Scholar]
- 51.Pufahl RA, Wishnok JS, Marletta MA. Biochemistry. 1995;34:1930–1941. doi: 10.1021/bi00006a014. [DOI] [PubMed] [Google Scholar]
- 52.Presta A, Weber-Main AM, Stankovich MT, Stuehr DJ. J.Am.Chem.Soc. 1998;120:9460–9465. [Google Scholar]
- 53.Wang ZQ, Wei CC, Sharma M, Pant K, Crane BR, Stuehr DJ. J.Biol.Chem. 2004;279:19018–19025. doi: 10.1074/jbc.M311663200. [DOI] [PubMed] [Google Scholar]
- 54.Tejero J, Santolini J, Stuehr DJ. FEBS J. 2009;276:4505–4514. doi: 10.1111/j.1742-4658.2009.07157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adak S, Aulak KS, Stuehr DJ. J.Biol.Chem. 2001;276:23246–23252. doi: 10.1074/jbc.M102509200. [DOI] [PubMed] [Google Scholar]
- 56.Huang L, Abu-Soud HM, Hille R, Stuehr DJ. Biochemistry. 1999;38:1912–1920. doi: 10.1021/bi981954t. [DOI] [PubMed] [Google Scholar]
- 57.Abu-Soud HM, Wu C, Ghosh DK, Stuehr DJ. Biochemistry. 1998;37:3777–3786. doi: 10.1021/bi972398q. [DOI] [PubMed] [Google Scholar]
- 58.Ghosh DK, Wu C, Pitters E, Moloney M, Werner ER, Mayer B, Stuehr DJ. Biochemistry. 1997;36:10609–10619. doi: 10.1021/bi9702290. [DOI] [PubMed] [Google Scholar]
- 59.Abu-Soud HM, Wang J, Rousseau DL, Stuehr DJ. Biochemistry. 1999;38:12446–12451. doi: 10.1021/bi990698x. [DOI] [PubMed] [Google Scholar]
- 60.Tejero J, Biswas A, Wang ZQ, Page RC, Haque MM, Hemann C, Zweier JL, Misra S, Stuehr DJ. J.Biol.Chem. 2008;283:33498–33507. doi: 10.1074/jbc.M806122200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adak S, Wang Q, Stuehr DJ. J.Biol.Chem. 2000;275:17434–17439. doi: 10.1074/jbc.M000846200. [DOI] [PubMed] [Google Scholar]
- 62.Santolini J, Meade AL, Stuehr DJ. J.Biol.Chem. 2001;276:48887–48898. doi: 10.1074/jbc.M108666200. [DOI] [PubMed] [Google Scholar]
- 63.Wang ZQ, Wei CC, Stuehr DJ. J.Inorg.Biochem. 2010;104:349–356. doi: 10.1016/j.jinorgbio.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 64.Adak S, Wang Q, Stuehr DJ. J.Biol.Chem. 2000;275:33554–33561. doi: 10.1074/jbc.M004337200. [DOI] [PubMed] [Google Scholar]
- 65.Tejero J, Biswas A, Haque MM, Wang ZQ, Hemann C, Varnado CL, Novince Z, Hille R, Goodwin DC, Stuehr DJ. Biochem.J. 2010;433:163–174. doi: 10.1042/BJ20101353. [DOI] [PubMed] [Google Scholar]
- 66.Adak S, Crooks C, Wang Q, Crane BR, Tainer JA, Getzoff ED, Stuehr DJ. J.Biol.Chem. 1999;274:26907–26911. doi: 10.1074/jbc.274.38.26907. [DOI] [PubMed] [Google Scholar]
- 67.Adak S, Stuehr DJ. J.Inorg.Biochem. 2001;83:301–308. doi: 10.1016/s0162-0134(00)00176-8. [DOI] [PubMed] [Google Scholar]
- 68.Ost TW, Daff S. J.Biol.Chem. 2005;280:965–973. doi: 10.1074/jbc.M411191200. [DOI] [PubMed] [Google Scholar]
- 69.Cooper CE. Biochim.Biophys.Acta. 1999;1411:290–309. doi: 10.1016/s0005-2728(99)00021-3. [DOI] [PubMed] [Google Scholar]
- 70.Stuehr DJ, Santolini J, Wang ZQ, Wei CC, Adak S. J.Biol.Chem. 2004;279:36167–36170. doi: 10.1074/jbc.R400017200. [DOI] [PubMed] [Google Scholar]
- 71.Quaroni LG, Seward HE, McLean KJ, Girvan HM, Ost TW, Noble MA, Kelly SM, Price NC, Cheesman MR, Smith WE, Munro AW. Biochemistry. 2004;43:16416–16431. doi: 10.1021/bi049163g. [DOI] [PubMed] [Google Scholar]
- 72.Santolini J, Adak S, Curran CM, Stuehr DJ. J.Biol.Chem. 2001;276:1233–1243. doi: 10.1074/jbc.M006858200. [DOI] [PubMed] [Google Scholar]
- 73.Boggs S, Huang L, Stuehr DJ. Biochemistry. 2000;39:2332–2339. doi: 10.1021/bi9920228. [DOI] [PubMed] [Google Scholar]
- 74.Hurshman AR, Krebs C, Edmondson DE, Huynh BH, Marletta MA. Biochemistry. 1999;38:15689–15696. doi: 10.1021/bi992026c. [DOI] [PubMed] [Google Scholar]
- 75.Bec N, Gorren AC, Voelker C, Mayer B, Lange R. J.Biol.Chem. 1998;273:13502–13508. doi: 10.1074/jbc.273.22.13502. [DOI] [PubMed] [Google Scholar]
- 76.Siddhanta U, Wu C, Abu-Soud HM, Zhang J, Ghosh DK, Stuehr DJ. J.Biol.Chem. 1996;271:7309–7312. doi: 10.1074/jbc.271.13.7309. [DOI] [PubMed] [Google Scholar]
- 77.Siddhanta U, Presta A, Fan B, Wolan D, Rousseau DL, Stuehr DJ. J.Biol.Chem. 1998;273:18950–18958. doi: 10.1074/jbc.273.30.18950. [DOI] [PubMed] [Google Scholar]
- 78.Marletta MA, Hurshman AR, Rusche KM. Curr.Opin.Chem.Biol. 1998;2:656–663. doi: 10.1016/s1367-5931(98)80098-7. [DOI] [PubMed] [Google Scholar]
- 79.Crane BR, Arvai AS, Gachhui R, Wu C, Ghosh DK, Getzoff ED, Stuehr DJ, Tainer JA. Science. 1997;278:425–431. doi: 10.1126/science.278.5337.425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.