Abstract
Because chronic intestinal inflammation is a risk factor for colorectal cancer, we hypothesized that genetic variants of inflammatory mediators, such as mannose-binding lectin 2 (MBL2), are associated with colon cancer susceptibility. Here we report the association of 24 MBL2 single nucleotide polymorphisms (SNPs) and corresponding haplotypes with colon cancer risk in a case-control study. Four SNPs in the 3′-UTR region of the gene (rs10082466, rs2120132, rs2099902, and rs10450310) were associated with an increased risk of colon cancer in African Americans. Odds ratios (OR) for homozygous variants vs. wild-type ranged from 3.17 (95% CI, 1.57–6.40) to 4.51 (95% CI, 1.94–10.50), whereas the 3′-UTR region haplotype consisting of these four variants had an OR of 2.10 (95% CI, 1.42–3.12). The C allele of rs10082466 exhibited a binding affinity of miR-27a and this allele was associated with both lower MBL plasma levels and activity. We found that 5′ secretor haplotypes known to correlate with moderate and low MBL serum levels exhibited associations with increased risk of colon cancer in African Americans, specifically as driven by two haplotypes LYPA and LYQC relative to the referent HYPA haplotype (LYPA: OR 2.60; 95% CI 1.33–5.08 and LYQC: OR 2.28; 95% CI 1.20–4.30). Similar associations were not displayed in Caucasians. Together, our results support the hypothesis that genetic variations in MBL2 increase colon cancer susceptibility in African Americans.
Keywords: colon cancer, single nucleotide polymorphism, mannose-binding lectin 2, innate immunity, African American
Introduction
Cancer of the colon and rectum is estimated to be the third most common cause of cancer deaths in the United States in 2010 for men and women (1). SEER age-adjusted colon and rectal cancer incidence and mortality rates show a decrease in both Caucasians and African Americans from 1992 to 2008 and 2007, respectively. Although these rates are decreasing in both races, they continue to be higher among African Americans when compared with Caucasians (2).
Chronic intestinal inflammation has been identified as a risk factor for colorectal cancer (3). Additionally, inflammatory bowel disease, such as ulcerative colitis or Crohn’s disease, is a risk factor for colon neoplasia, further supporting the role of chronic inflammation in the development of colon cancer (4). Therefore, it is possible that functionally important genetic variants of inflammatory mediators, such as mannose-binding lectin 2 (MBL2), are also associated with susceptibility to colon cancer. MBL2, a gene on chromosome 10 (5), codes for a 32kDa plasma glycoprotein called mannose-binding lectin (MBL) which is a pattern recognition molecule of the innate immune system (6). Functionally, MBL binds to a spectrum of sugars on pathogens, leading to their destruction by opsonization and phagocytosis and/or the activation of complement (6), which can be activated by three distinct pathways: 1) the classical pathway, which recognizes via charge and hydrophobic clusters; 2) the alternative pathway, which recognizes via sugar and charge; and 3) the lectin pathway (7). The lectin pathway recognizes neutral sugar clusters and is activated via binding of MBL to carbohydrate structures present on micro-organisms. This activation is mediated by the serine protease MASP-2.
MBL deficiency is common and present in approximately 5% of the population (7). This deficiency predisposes individuals to bacterial, viral, and fungal infections. Additionally, altered serum levels have been found in the presence of six single nucleotide polymorphisms (SNPs) in both exon 1 and the promoter of the MBL2 gene; these SNPs are in linkage disequilibrium and comprise seven different haplotypes known as the secretor haplotypes (6,8–10). Functionally, these SNPs have been shown to interfere with the formation of MBL oligomers leading to decreased compliment activation potential and possibly enhanced turnover (11). Recent reports in the literature suggest that altered MBL2 genetic background is associated with risk of cancer. Baccarelli, et al. reported that one of the variant MBL2 secretor haplotypes is associated with 3.5-fold increased risk of stomach cancer when compared to the most common haplotype (12); whereas Wang et al. showed an association of a polymorphism in MBL2 codon 54 with advanced phenotypes of gastric cancer and increased risk of gastric cancer in Japanese patients 65 years old or younger (13). Olivo-Marston, et al. showed that secondhand smoke exposure during childhood is associated with increased risk of lung cancer among never smokers, specifically in those carrying a haplotype associated with high serum MBL levels (14); and Bernig et al. reported that an MBL2 3′UTR SNP was associated with decreased risk of breast cancer in African American women (15).
MicroRNAs (miRNAs) are short non-coding RNAs that regulate gene expression through binding to cognate sequences, preferentially 3′-UTR regions, of mRNAs. Such binding has been shown to reduce protein translation as well as hasten mRNA degradation (16). The degree of complementarity around nucleotides 2–7 of a miRNA, called the “seed” region, is the most important known determinant of recognition of an mRNA by a targeting miRNA (17). These interactions, although restricted to a few nucleotides, can have profound effects on gene expression. Therefore, SNPs located in the 3′-UTR regions of genes, which traditionally have been considered less informative than coding or promoter SNPs, are now being more closely evaluated. Such single variations, if occurring within the cognate seed region could create, destroy or change the affinity of a miRNA binding site (18).
Given the idea that functionally important genetic variants of inflammatory mediators are associated with susceptibility to colon cancer, we hypothesized that altered MBL2 genetic background may be a factor associated with colon cancer risk. Additionally, we hypothesized that SNPs in the 3′-UTR region of MBL2 associated with colon cancer risk may affect miRNA binding. To test these hypotheses, we used a candidate gene approach to examine 24 MBL2 SNPs in The NCI-Maryland Colon Cancer Case-Control Study, consisting of 261 colon cancer cases and 537 controls from the greater Baltimore, Maryland area and performed functional studies to test the miRNA hypothesis.
Materials and Methods
Study Population
The study population consists of 798 subjects, including 261 cases and 537 controls, accrued from 1992–2003 and 1998–2003, respectively, from the greater Baltimore, Maryland area. The inclusion and exclusion criteria have been previously described (19). In brief, the inclusion criteria were self-reported Caucasian or African American race born in the U.S., while subjects were excluded if they had a self-reported history of cancer other than colon, HIV, HBV or HCV infections, IV drug use, were institutionalized or had a mental impairment. Of the 436 colon cancer patients at the University of Maryland Medical Center and the Baltimore VA Medical Center from 1992–2003, 261 were found eligible and agreed to participate in the study. The controls, which were not matched to cases, were accrued from both a hospital setting (n = 230) and a community setting (n = 307). Specifically, the hospital controls were cancer-free patients recruited from the same hospitals as the cancer cases, specifically from Family/Internal Medicine, Thoracic Surgery and Pulmonary Clinics, and the population controls were identified through Maryland Motor Vehicle Administration (MVA) records, contacted by mail, and then by telephone. The overall participation rates for controls as of December 31, 2003 are as follows: (a) hospital controls: among 1,598 screened, 1,399 completed eligibility screening, 308 were eligible, and 268 (87%) participated in the study; and (b) population controls: among 3,289 screened, 1,801 completed eligibility screening, 388 were eligible, and 350 (90%) participated in the study. Informed consent was obtained from all subjects, epidemiology questionnaires were administered and blood and/or tissue were obtained. The epidemiology questionnaire included personal history, family medical history, medical history, smoking/tobacco history, dietary information, and information on work environment. This study was approved by the Institutional Review Boards of the participating institutions.
Genotyping
DNA was isolated from buffy coat or colon tissue, using the Qiagen FlexiGene DNA Kit or the DNeasy Tissue Kit, respectively (Qiagen, Valencia, CA). Blood was used to obtain DNA from the cases; however when blood was not available, DNA was extracted from colon tissue (n = 127). We performed a sensitivity analysis removing the 127 cases with tissue-derived DNA and determined that the observed associations using only blood-derived DNA (n = 134) were consistent with the observations using all cases. Cases and controls were genotyped for 24 MBL2 SNPs (Supplementary Table 1) at the National Cancer Institute Core Genotyping Facility (CGF), using validated assays that can be found on their website (20). The case, control, and 10% duplicate samples were randomly distributed for order of processing, and samples were assigned a unique identification number that lacked identifiers. Staff at the CGF was blinded to the case, control, and duplicate statuses. Samples that failed to genotype were scored as missing. Duplicates for ten percent of all samples were genotyped for quality control, and concordance was >98% for the majority of SNPs. SNPs rs7096206, rs930508, and rs10450310 had a concordance of 97%; whereas rs930507, rs10082466, and rs2099902 had a concordance of 96%. Samples found not to be in concordance were scored as missing and removed from the analysis. All SNPs were in Hardy-Weinberg equilibrium; see Supplemental Table 2 for SNPs found to be associated with colon cancer risk.
Enzyme-Linked Immunosorbent Assays
Plasma samples from a subset (n = 450) of cases (n = 108) and controls (n = 342) and a minimum of 5% duplicates were assayed. These samples were selected for a parallel study being conducted in our laboratory. Ultimately, we determined this subset to be appropriate for use in this study because the demographic characteristics and haplotype frequencies for the subset of samples were similar to the overall study (Tables 1 – 3, 5, 6). The case, control, and 5% duplicate samples were randomly distributed across plates and the samples were blinded. Plasma mannan-binding capacity was determined by a ligand-lectin solid-phase enzyme-linked immunoassay (Cell Sciences, Inc., Canton, MA). MBL C4-complement activation was measured by determining the ability of MBL/MASP complexes to initiate C4 cleavage when bound to mannan using a solid-phase enzyme-linked immunosorbent assay (Cell Sciences, Inc., Canton, MA). For the MBL C4-complement activation assay, only 449 samples were assayed due to depletion of one sample (n = 108 cases; 341 controls).
Table 1.
Demographic characteristics of the NCI-Maryland Colon Cancer Case-Control Study subjects.
| Characteristics | Cases | Population Controls | Hospital Controls | Total Controls |
|---|---|---|---|---|
| n | 261 | 307 | 230 | 537 |
| Age; mean±sd | 64.8 ± 11.6 | 65.9 ± 9.6 | 62.7 ± 12.1 | 64.5 ± 10.9 |
| BMI; mean±sd | 27.7± 6.5 | 29.0 ± 9.8 | 27.9 ± 6.5 | 28.5 ± 8.6 |
| Gender; n (%) | ||||
| Male | 197 (75)a | 149 (49) | 106 (46) | 255 (47) |
| Female | 64 (25) | 158 (51) | 124 (54) | 282 (53) |
| Race; n (%) | ||||
| African American | 103 (39) | 127 (41) | 74 (32) | 201 (37) |
| Caucasian | 158 (61) | 180 (59) | 156 (68) | 336 (63) |
Statistically significant; p-value < 0.05 vs. each control group.
Table 3.
Association of MBL plasma levels (A) and activity (B) with the MBL2 secretor haplotypes in a subset of 450 Caucasian and African American cases and controls.
| A. | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Haplotypea,b | MBL Plasma Level (ng/ml) | ||||||||
|
| |||||||||
| CAUCASIANS | AFRICAN AMERICANS | ||||||||
|
| |||||||||
| Cases (n = 63) | Cases (n = 45) | ||||||||
| Frequency | Mean±SE | Median | IQ Rangec | Frequency | Mean±SE | Median | IQ Rangec | p-valued | |
| HYPA | 0.262 | 2421±361 | 1975 | 1031, 2913 | 0.111 | 2291±1065 | 1026 | 195, 3398 | 0.206 |
| LYQA, LYPA | 0.302 | 2262±289 | 1910 | 1035, 2534 | 0.456 | 2082±411 | 1190 | 425, 2129 | 0.084 |
| LXPA, LYPB, HYPD, LYQC | 0.436 | 1000±147 | 561 | 135, 1680 | 0.433 | 468±107 | 195 | 23, 559 | 0.016 |
|
| |||||||||
| p-valuee 0.0001 | p-valuee 0.0001 | ||||||||
| trendf <0.0005 | trendf 0.0009 | ||||||||
|
| |||||||||
| Controls (n = 191) | Controls (n = 151) | ||||||||
|
| |||||||||
| HYPA | 0.270 | 1672±207 | 1160 | 359, 2032 | 0.156 | 1384±188 | 1158 | 496, 1941 | 0.654 |
| LYQA, LYPA | 0.260 | 1622±206 | 1145 | 462, 1922 | 0.414 | 1346±136 | 885 | 312, 1941 | 0.150 |
| LXPA, LYPB, HYPD, LYQC | 0.470 | 637±113 | 266 | 25, 765 | 0.430 | 469±66 | 200 | 26, 475 | 0.348 |
|
| |||||||||
| p-valuee 0.0001 | p-valuee 0.0001 | ||||||||
| trendf <0.0005 | trendf <0.0005 | ||||||||
| B. | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Haplotypea,b | MBL Plasma Activity (U/ml) | ||||||||
|
| |||||||||
| CAUCASIANS | AFRICAN AMERICANS | ||||||||
|
| |||||||||
| Cases (n = 63) | Cases (n = 45) | ||||||||
| Frequency | Mean±SE | Median | IQ Rangec | Frequency | Mean±SE | Median | IQ Rangec | p-valued | |
| HYPA | 0.262 | 1481±175 | 1224 | 636, 2551 | 0.111 | 1052±297 | 952 | 363, 1134 | 0.275 |
| LYQA, LYPA | 0.302 | 1091±118 | 854 | 624, 1332 | 0.456 | 1062±177 | 534 | 256, 1480 | 0.081 |
| LXPA, LYPB, HYPD, LYQC | 0.436 | 750±111 | 384 | 124, 1103 | 0.433 | 449±101 | 226 | 75, 547 | 0.072 |
|
| |||||||||
| p-valuee 0.0001 | p-valuee 0.0004 | ||||||||
| trendf 0.0001 | trendf 0.007 | ||||||||
|
| |||||||||
| Controls (n = 190) | Controls (n = 151) | ||||||||
|
| |||||||||
| HYPA | 0.270 | 1025±103 | 604 | 329, 1417 | 0.156 | 955±152 | 492 | 274, 1431 | 0.433 |
| LYQA, LYPA | 0.260 | 931±193 | 640 | 262, 1094 | 0.414 | 804±66 | 512 | 245, 1276 | 0.274 |
| LXPA, LYPB, HYPD, LYQC | 0.470 | 393±38 | 229 | 62, 459 | 0.430 | 382±61 | 159 | 44, 416 | 0.141 |
|
| |||||||||
| p-valuee 0.0001 | p-valuee 0.0001 | ||||||||
| trendf <0.0005 | trendf <0.0005 | ||||||||
Haplotype defined as: H/L = rs11003125 (-618G>C); Y/X = rs7096206 (-289G>C); P/Q = rs7095891 (-65T>C); A/D = rs503737 (Ex1-34C>T); A/B = rs1800450 (Ex1-27G>A); A/C = rs1800451 (Ex1-18G>A).
SNPs categorized by serum MBL levels (high, moderate, low).
IQ Range: Interquartile Range
Mann-Whitney U test comparing MBL plasma level between Caucasians and African Americans.
Kruskal Wallis test comparing MBL plasma level by haplotype group.
Proportional odds model
Table 5.
MBL2 3′ UTR region haplotypes are associated with increased susceptibility to colon cancer in African Americans.
| Caucasians | African Americans | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Haplotypea | Cases Freqb | Controls Freqb | OR (95% CI)c,d | p-value | Cases Freqb | Controls Freqb | OR (95% CI)c,d | p-valuee |
| TAAC | 0.776 | 0.774 | 1.00 (Ref) | 0.359 | 0.506 | 1.00 (Ref) | ||
| TAGC | -- | -- | -- | -- | 0.111 | 0.116 | 1.29 (0.70–2.35) | 0.42 |
| CGGT | 0.224 | 0.226 | 1.00 (0.71–1.42) | 0.98 | 0.530 | 0.378 | 2.10 (1.42–3.12) | <0.0005 |
| ptrendf | 0.0004 | |||||||
Haplotype defined as: rs10082466 (Ex4-1483T>C); rs2120132 (Ex4-901A>G); rs2099902 (Ex4-710A>G); rs10450310 (3283bp 3′ STP).
Freq = frequency
OR: odds ratio; CI: confidence interval
Adjusted for age and gender.
Statistically significant; p-value <0.05.
ptrend = Cochran-Armitage chi-square test for trend
Table 6.
Association of MBL plasma levels (A) and activity (B) with the MBL2 3′ UTR region haplotypes in a subset of 450 Caucasian and African American cases and controls.
| A. | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Haplotypea | MBL Plasma Level (ng/ml) | ||||||||
|
| |||||||||
| CAUCASIANS | AFRICAN AMERICANS | ||||||||
|
| |||||||||
| Cases (n = 63) | Cases (n = 45) | ||||||||
| Frequency | Mean±SE | Median | IQ Rangeb | Frequency | Mean±SE | Median | IQ Rangeb | p-valuec | |
| TAAC | 0.776 | 1848±175 | 1465 | 561, 2353 | 0.356 | 2270±482 | 1330 | 559, 3398 | 0.987 |
| TAGC | -- | -- | -- | -- | 0.035 | 974±270 | 253 | 119, 1056 | -- |
| CGGT | 0.224 | 1403±336 | 536 | 87, 2140 | 0.609 | 886±613 | 425 | 134, 2100 | 0.273 |
|
| |||||||||
| p-valued 0.053 | p-valued 0.002 | ||||||||
| trende 0.01 | |||||||||
|
| |||||||||
| Controls (n = 191) | Controls (n = 151) | ||||||||
|
| |||||||||
| TAAC | 0.772 | 1464±143 | 868 | 268, 1797 | 0.510 | 1421±133 | 989 | 414, 1887 | 0.19 |
| TAGC | -- | -- | -- | -- | 0.051 | 553±100 | 184 | 25, 457 | -- |
| CGGT | 0.228 | 474±98 | 215 | 14, 514 | 0.439 | 564±295 | 129 | 26, 344 | 0.994 |
|
| |||||||||
| p-valued 0.0001 | p-valued 0.0001 | ||||||||
| p-trende <0.0005 | |||||||||
| B. | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Haplotypea | MBL Plasma Activity (U/ml) | ||||||||
|
| |||||||||
| CAUCASIANS | AFRICAN AMERICANS | ||||||||
|
| |||||||||
| Cases (n = 63) | Cases (n =45) | ||||||||
| Frequency | Mean±SE | Median | IQ Rangeb | Frequency | Mean±SE | Median | IQ Rangeb | p-valuec | |
| TAAC | 0.776 | 1144±93 | 833 | 414, 1862 | 0.356 | 891±174 | 531 | 317, 952 | 0.032 |
| TAGC | -- | -- | -- | -- | 0.035 | 656±154 | 236 | 116, 674 | -- |
| CGGT | 0.224 | 713±160 | 417 | 110, 1061 | 0.609 | 939±291 | 832 | 118, 1279 | 0.82 |
|
| |||||||||
| p-valued 0.007 | p-valued 0.001 | ||||||||
| trende 0.01 | |||||||||
|
| |||||||||
| Controls (n = 190) | Controls (n = 151) | ||||||||
|
| |||||||||
| TAAC | 0.773 | 789±59 | 461 | 222, 995 | 0.510 | 725±91 | 403 | 135, 961 | 0.205 |
| TAGC | -- | -- | -- | -- | 0.051 | 571±73 | 279 | 127, 579 | -- |
| CGGT | 0.227 | 340±51 | 215 | 54, 459 | 0.439 | 368±79 | 243 | 77, 511 | 0.217 |
|
| |||||||||
| p-valued 0.0001 | p-valued 0.0001 | ||||||||
| trende <0.0005 | |||||||||
Haplotype defined as: rs10082466 (Ex4-1483T>C); rs2120132 (Ex4-901A>G); rs2099902 (Ex4-710A>G); rs10450310 (3283bp 3′ STP).
IQ Range: Interquartile Range
Mann-Whitney U test comparing MBL plasma activity between Caucasians and African Americans.
Kruskal Wallis test comparing MBL plasma activity by haplotype group.
Proportional odds model
Luciferase reporter assays
Genomic DNA from two cases, each representing one of the two main 3′-UTR haplotypes of MBL2: TAAC and CGGT, was subjected to whole MBL2 3′-UTR amplification followed by cloning into a psiCHECK-2 vector (Promega, Madison, WI) that was modified using Gateway technology (Invitrogen, Carlsbad, CA). This vector contains a SV40 promoter driving Renilla Luciferase, followed by a Gateway cloning site for introduction of a 3′-UTR, a polyA sequence, and another operon consisting of the TK promoter driving Firefly Luciferase as an internal control. Plasmid DNA and pre-miR-27a or negative control #1 pre-miR (Ambion, Austin, TX) were introduced into 293T cells by reverse transfection onto 96-well plates, using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Briefly, 293T cells grown in DMEM supplemented with 10% Fetal Bovine Serum and 1mM Glutamine were trypsinized on the day of the assay. Plasmid DNA (20 ng/well) and pre-miR (30 nM) were premixed with Lipofectamine 2000 and then added individually to the culture plate in a total volume of 50 μl. Each combination of plasmid DNA and pre-miR was plated in sextuplicates. Cells were then added to the wells at 45,000 cells/well in a volume of 150 μl. Cells were lysed and dual Luciferase measurements taken 24 hours after transfection using Dual-Luciferase Reporter Assay System (Promega, Madison, WI), according to instructions from the manufacturer, using a plate-reading luminometer. Individual Renilla Luciferase readings were normalized to Firefly Luciferase, and the mean of normalized values for each combination of plasmid DNA and pre-miR were used to calculate the effect of pre-miR-27a on Luciferase, compared to negative control pre-miR.
Statistical Analyses
Linkage disequilibrium of the secretor haplotype and 3′-UTR region haplotype SNPs have been previously demonstrated (15). Linkage disequilibrium was verified by the Haploview software (21) with implementation of the default modus for the haplotype block definition. Both the 3′- and 5′-region haplotype blocks were inferred separately for Caucasian and African American subjects. Estimates of haplotypes were obtained using Phase software, version 2.1 (22), which uses a Bayesian method for haplotype reconstruction. Statistical analyses were performed using Stata 11.2 (College Station, TX). Departures from Hardy-Weinberg equilibrium for each SNP were determined using a χ2 test. Frequency distributions of the SNPs between African American and Caucasian controls and population and hospital controls were compared using a χ2 test. Odds ratios (OR) and 95% confidence intervals (CI) were calculated using logistic regression adjusted for age (continuous) and gender. We adjusted for multiple comparisons using the Benjamini–Hochberg method (23). To test trends across genotypes, the Cochran-Armitage test for trend was performed.
Mannan-binding capacity and C4-complement activation were compared between haplotypes associated with high, moderate, and low serum MBL levels using a Kruskal-Wallis test and tests for trend were performed using proportional odds models. These tests were also performed when comparing mannan-binding capacity and C4-complement activation between the 3′-UTR haplotypes. Tests for trend were not conducted in Caucasians for the 3′-UTR haploytpes because they only carry two haplotypes with a frequency >0.05. Mannan-binding capacity and C4-complement activation were compared between African Americans and Caucasians using a Mann-Whitney U test. For ELISA assays, correlations between duplicates were determined using Spearman’s correlation. Spearman’s rho = 0.97 for intraplate mannan-binding capacity (p-value <0.001); 0.95 for interplate mannan-binding capacity (p-value < 0.001); 0.98 for intraplate C4-complement activation (p-value <0.001); 0.58 for interplate C4-complement activation (p-value = 0.0073).
Comparison of MBL2 plasma levels and activity, associated with variants of rs10082466, was conducted using a Kruskal Wallis test with a post hoc Mann-Whitney U test. Relative luciferase levels generated by binding of hsa-miR-27a to the C or T allele of rs10082466 in the 3′-UTR of MBL2, were compared using an unpaired t-test with Welch’s correction.
A p-value of less than 0.05 was used as the criterion of statistical significance, and all statistical tests were two-sided.
Results
Study Population and MBL2 Genotype Analysis
We investigated the relationship between 24 SNPs (Supplemental Table 1) across the MBL2 gene and risk of colon cancer among 261 colon cancer cases and 537 controls. The relevant demographic characteristics are given in Table 1. The cases did not differ from any of the control groups (population, hospital, or total controls) in age, body mass index (BMI), or race; however, cases did significantly differ from each control group in gender. Cases had significantly more males than each of the control groups. Furthermore, the frequencies of genotypes for SNPs in which an association was detected significantly differed between Caucasian and African American controls (Supplemental Table 3). Therefore, all further genotype and haplotype analyses were performed stratified by race. Of note, the genotype frequencies we observed in the controls are similar to the genotype frequencies for these SNPs seen in the dbSNP (24) and variant GPS databases (20) for those of European and African descent, and therefore reflective of the general population. Additionally, the frequencies of genotypes for SNPs in which an association was detected did not differ between hospital and population controls for either Caucasians or African Americans; therefore, the results are presented using the total control group (Supplemental Table 4). The results of all non-significant genotype analyses are shown in Supplemental Table 5.
MBL2 Secretor Haplotypes and Colon Cancer Risk
We examined the secretor haplotypes with a frequency >0.01, which captured >99% of all inferred haplotypes in both Caucasians and African Americans. The secretor haplotypes previously shown to correlate with moderate MBL serum levels (LYQA, LYPA) and low MBL serum levels (LXPA, LYPB, HYPD, LYQC) (6,8–10) are associated with increased risk of colon cancer in African Americans (OR 1.80; 95% CI 0.99–3.25 and OR 2.00; 95% CI 1.11–3.62, respectively) when compared with the referent haplotype HYPA [previously shown to be associated with high serum levels (6,8–10)] (Table 2). These associations are driven by LYPA and LYQC as exhibited when each haplotype is independently compared with the referent HYPA haplotype (LYPA: OR 2.60; 95% CI 1.33–5.08 and LYQC: OR 2.28; 95% CI 1.20–4.30) (Table 2); and the adjusted p-values for these associations are significant (p-value = 0.03 for each haplotype). Similar associations were not seen in Caucasians. LYPA and LYQC are associated with moderate and low serum levels of MBL, respectively (6,8–10). Of the individual SNPs within the secretor haplotype, only rs1800451 showed a borderline association with colon cancer (OR 3.14; 95% CI 1.03–9.62) (Supplemental Table 5); however the adjusted p-value for this association was not significant. The other five SNPs in the secretor haplotype showed no association.
Table 2.
MBL2 secretor haplotypes are associated with increased susceptibility to colon cancer in African Americans.
| Caucasians | African Americans | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Haplotypea | Cases Freqb | Controls Freqb | OR (95% CI)c,d | p-value | Cases Freqb | Controls Freqb | OR (95% CI)c,d | p-valuee |
| High | ||||||||
| HYPA | 0.289 | 0.277 | 1.00 | 0.093 | 0.151 | 1.00 | ||
| Moderatef | 0.279 | 0.257 | 0.99 (0.67–1.44) | 0.94 | 0.441 | 0.434 | 1.80 (0.99–3.25) | 0.05 |
| LYQA | 0.218 | 0.204 | 0.96 (0.64–1.45) | 0.87 | 0.225 | 0.289 | 1.39 (0.73–2.63) | 0.31 |
| LYPA | 0.062 | 0.053 | 1.05 (0.55–2.01) | 0.87 | 0.215 | 0.145 | 2.60 (1.33–5.08) | 0.005 |
| Lowf | 0.432 | 0.466 | 0.84 (0.59–1.18) | 0.33 | 0.466 | 0.415 | 2.00 (1.11–3.62) | 0.02 |
| LXPA | 0.221 | 0.226 | 0.87 (0.59–1.31) | 0.52 | 0.162 | 0.148 | 1.87 (0.94–3.74) | 0.08 |
| LYPB | 0.117 | 0.155 | 0.72 (0.45–1.17) | 0.19 | 0.015 | 0.033 | 0.73 (0.18–2.91) | 0.65 |
| HYPD | 0.081 | 0.065 | 1.01 (0.57–1.81) | 0.97 | 0.015 | 0.010 | 2.51 (0.49–12.92) | 0.27 |
| LYQC | 0.012 | 0.020 | 0.74 (0.22–2.51) | 0.64 | 0.275 | 0.224 | 2.28 (1.20–4.30) | 0.01 |
| ptrendg | 0.42 | ptrendg | 0.07 | |||||
Haplotype defined as: H/L = rs11003125 (-618G>C); Y/X = rs7096206 (-289G>C); P/Q = rs7095891 (-65T>C); A/D = rs503737 (Ex1-34C>T); A/B = rs1800450 (Ex1-27G>A); A/C = rs1800451 (Ex1-18G>A).
Freq = frequency
OR: odds ratio; CI: confidence interval
Adjusted for age and gender.
Statistically significant; p-value <0.05.
Data shown for haplotypes categorized by serum MBL levels (moderate, low) and individually. Moderate = combined data from haplotypes LYQA and LYPA. Low = combined data from haplotypes LXPA, LYPB, HYPD, and LYQC.
ptrend = Cochran-Armitage chi-square test for trend
MBL2 Secretor Haplotypes and MLB Plasma Levels and Activity
We further investigated the MBL2 associations in our study using plasma from a subset of Caucasian and African American cases and controls (n = 450). Similar to the full dataset, cases (n = 108) did not differ from the total control group (n = 342) in age, body mass index (BMI), or race; however, cases did significantly differ from the total control group in gender (data not shown). Median levels of plasma MBL differed significantly between haplotypes previously shown to be associated with high (HYPA), moderate (LYPA, LYQA) and low (HYPD, LYQC, LYPB, LXPA) serum levels in both Caucasian and African American cases and controls [Kruskal-Wallis test, p-value = 0.0001 (cases and controls); test for trend, p-value <0.0005 (Caucasian and African American cases and Caucasian controls), 0.0009 (African American controls] (Table 3). The decreasing trend in plasma levels associated with the secretor haplotypes correlates with what was previously shown for serum levels. However, in African American cases the median MBL plasma level for haplotypes LYPA and LYQA is greater than the median level for haplotype HYPA, while the mean plasma level of HYPA is greater than the mean level for LYPA and LYQA. Furthermore, we determined that the median MBL activity levels also differ significantly between the haplotypes that represent high, moderate, and low serum levels in Caucasian cases and controls [Kruskal-Wallis test, p-value = 0.0001 (cases and controls); test for trend, p-value 0.0001 (cases), <0.0005 (controls)] and African American cases and controls [Kruskal-Wallis test, p-value = 0.0004 (cases), 0.0001 (controls); test for trend, p-value 0.007 (cases), <0.0005 (controls)] (Table 3). The decreasing trend in plasma activity associated with the secretor haplotypes also corresponds to what was previously shown for serum levels. Median MBL levels were significantly lower in African American cases (195 ng/ml) when compared with Caucasian cases (561 ng/ml) for haplotypes LXPA, LYPB, HYPD, LYQC (associated with low serum levels) with a p-value of 0.016 (Mann-Whitney U test) (Table 3). Furthermore, we determined that median MBL activity levels do not differ between African American cases when compared with Caucasian cases (Table 3). We did not observe these associations in either Caucasian or African American controls.
MBL2 3′-UTR Region SNPs and Colon Cancer Risk
Four MBL2-specific allele variants in linkage disequilibrium located in the 3′-UTR region of the gene are associated with a higher risk of colon cancer in African Americans: odd ratios (OR) for homozygous variants vs. homozygous wild-types ranged from 3.17 (95% CI, 1.57–6.40) to 4.51 (95% CI, 1.94–10.50) when adjusting for age and gender (Table 4); and the adjusted p-values for the homozygous variants are significant (p-value = 0.005 for rs10082466, rs2120132, rs2099902, and rs10450310). The four SNPs are located in the same haplotype block; therefore, we analyzed the risk associated with the common haplotypes (frequency >0.05; capturing >99% of all inferred haplotypes in Caucasians and 97% in African Americans). We found that haplotype CGGT, which contains all variant alleles, was associated with increased risk of colon cancer in African Americans (OR 2.10; 95% CI 1.42–3.12) when compared with the referent haplotype TAAC (the most frequent haplotype; contains all wild-type alleles) (Table 5). Similar associations were not seen in Caucasians.
Table 4.
Four 3′ UTR region MBL2 single nucleotide polymorphisms are associated with increased risk of colon cancer in African Americans.
| Caucasians | African Americans | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| SNP | Cases n (%) | Controls n (%) | OR (95% CI)a,b | p-value | Cases n (%) | Controls n (%) | OR (95% CI)a,b | p-valuec |
| rs10082466 | ||||||||
| TT | 89 (59) | 193 (61) | 1.00 | 24 (24) | 73 (39) | 1.00 | ||
| TC | 53 (36) | 109 (34) | 1.01 (0.71–1.70) | 0.67 | 45 (45) | 80 (43) | 1.75 (0.95–3.22) | 0.07 |
| CC | 7 (5) | 16 (5) | 0.91 (0.35–2.39) | 0.85 | 31 (31) | 33 (29) | 3.17 (1.57–6.40) | 0.001 |
| ptrendd | 0.91 | ptrendd | 0.002 | |||||
| rs2120132 | ||||||||
| AA | 88 (60) | 186 (61) | 1.00 | 21 (23) | 73 (40) | 1.00 | ||
| AG | 52 (35) | 102 (33) | 1.10 (0.71–1.71) | 0.68 | 46 (49) | 85 (46) | 1.92 (1.03–3.57) | 0.04 |
| GG | 7 (5) | 17 (6) | 0.74 (0.33–2.22) | 0.74 | 26 (28) | 26 (14) | 3.95 (1.83–8.42) | <0.0005 |
| ptrendd | 0.96 | ptrendd | 0.0006 | |||||
| rs2099902 | ||||||||
| AA | 89 (59) | 186 (61) | 1.00 | 9 (9) | 51 (27) | 1.00 | ||
| AG | 55 (37) | 106 (34) | 1.13 (0.73–1.74) | 0.58 | 51 (52) | 86 (46) | 3.31 (1.48–7.41) | 0.004 |
| GG | 6 (4) | 16 (5) | 0.76 (0.27–2.09) | 0.59 | 39 (39) | 50 (27) | 4.51 (1.94–10.50) | <0.0005 |
| ptrendd | 0.98 | ptrendd | 0.0005 | |||||
| rs10450310 | ||||||||
| CC | 86 (59) | 185 (60) | 1.00 | 22 (22) | 72 (40) | 1.00 | ||
| CT | 54 (37) | 108 (35) | 1.17 (0.75–1.81) | 0.49 | 49 (50) | 82 (45) | 1.91 (1.03–3.53) | 0.04 |
| TT | 7 (5) | 17 (5) | 0.83 (0.32–2.16) | 0.70 | 27 (28) | 28 (15) | 3.59 (1.70–7.58) | 0.001 |
| ptrendd | 0.93 | ptrendd | 0.001 | |||||
OR: odds ratio; CI: confidence interval
Adjusted for age and gender.
Statistically significant; p-value <0.05.
ptrend = Cochran-Armitage chi-square test for trend
MBL2 3′-UTR Region Haplotypes and MLB Plasma Levels and Activity
We investigated the plasma levels and activity associated with the 3′-UTR region haplotype. Consistent with the full dataset, cases (n = 108) did not differ from the total control group (n = 341) in age, body mass index (BMI), or race; however, cases did significantly differ from the control group in gender (data not shown). Overall, the results were less consistent than those observed for the 5′ secretor haplotype; however significant associations were observed. Plasma levels differed significantly between the 3′UTR region haplotypes for Caucasian controls and African American cases and controls with a decreasing trend associated with the homozygous variant (TAAC) when compared with the TAGC or CGGT (wild-type) haplotypes [Kruskal-Wallis test, p-value = 0.0001 (Caucasian controls), 0.002 (African American cases), 0.0001 (African American controls); test for trend, p-value 0.01 (African American cases) and <0.0005 (African American controls)] (Table 6). Tests for trend were not performed in Caucasians because they only carry two haplotypes with a frequency >0.05; however the homozygous variant was associated with significantly lower plasma levels in controls. In Caucasian cases, lower plasma levels were observed to be associated the CGGT haplotype, although significance was borderline (Kruskal-Wallis test, p-value = 0.052). Whereas, the plasma levels for haplotype CGGT in African American cases had a higher median than the TAGC haplotype. Plasma activity levels differed significantly between the 3′ UTR region haplotypes for Caucasian and African American cases and controls [Kruskal-Wallis test, p-value = 0.0007 (Caucasian cases), 0.001 (African American cases), 0.0001 (Caucasian and African American controls); test for trend in African Americans, p-value 0.01 (cases), <0.0005 (controls)] (Table 6). Similar to plasma levels, a decreasing trend in plasma activity was associated with the homozygous variant (TAAC) when compared with the TAGC or CGGT haplotypes. Additionally, we compared the plasma levels and activity between African Americans and Caucasians and found no significant differences in cases or controls with the exception of plasma activity in cases for haplotype TAAC (Mann-Whitney U test, p-value = 0.032) (Table 6).
MBL2 3′-UTR Region Harbors a Polymorphic miRNA Site
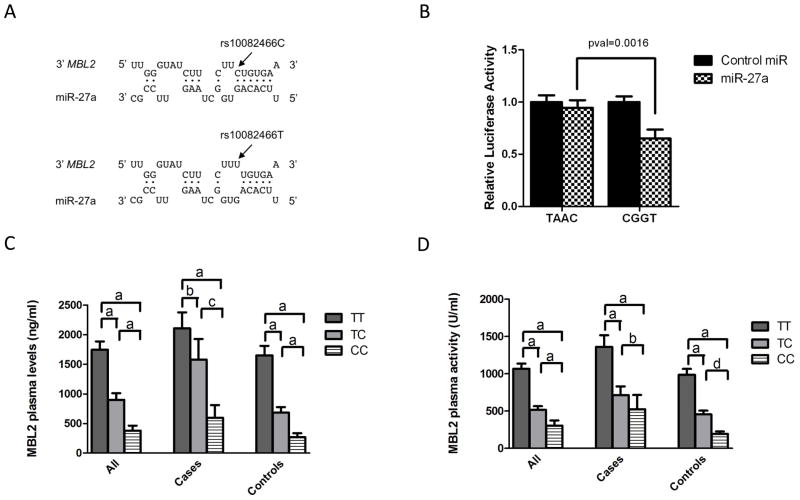
We investigated whether the SNPs in this haplotype block fell within putative miRNA binding sites due to the observed association of 3′UTR region haplotypes and plasma MBL levels. Two publicly available web-tools: Patrocles (25) and PolymiRTS (26) that compile information on 3′-UTR sequences, SNPs, and miRNA binding site predictions were used to identify SNPs that may alter predicted miRNA binding. Both databases predicted that the C allele of rs10082466 would likely create a binding target for hsa-miR-27a and hsa-miR-27b by generating a site with full complementarity to the 2–7 seed region (Figure 1A). Both hsa-miR-27a and hsa-miR-27b share the same seed region and spectrum of predicted targets; however, hsa-miR-27a shares a greater degree of complementarity outside the seed sequence. Moreover, when examining colon cancer tissue, the fold increase of hsa-miR-27a precursor is significantly higher in African Americans when compared with Caucasians (data not shown). Therefore, functional studies were carried out using hsa-miR-27a. The interaction of hsa-miR-27a with the 3′-UTR region of MBL2 and its modulation by 3′-UTR haplotypes were tested in vitro using reporter constructs cloned from genomic DNA of two cases each representing one of the main two 3′-UTR haplotypes of MBL2: TAAC and CGGT. In this system, normalized luciferase activity is a reporter for binding of miRNA to the 3′-UTR cloned downstream. The increased binding affinity predicted for the C allele of rs10082466 was reflected by a significant decrease in normalized luciferase activity, compared with the negative control, in the presence of construct CGGT (Figure 1B). More importantly, the sensitive allele rs10082466C is associated with lower plasma MBL levels (Figure 1C) and activity (Figure 1D) in cases and controls, as would be expected for a regulatory interaction involving a germline polymorphism.
Figure 1.
(A) The C allele of rs10082466 is predicted to create a binding site with full complementarity to the 2–7 seed region of hsa-miR-27a. (B) Relative luciferase levels are decreased when the C allele of rs10082466 3′-UTR is cloned downstream of the luciferase reporter gene (construct CGGT), compared to TAAC. The sensitive allele rs10082466C is associated with lower plasma MBL levels (C) and activity (D) in total cases and controls (a <0.0005; b = 0.0001; c = 0.0004; d = 0.0007).
Discussion
The primary hypothesis of our study is that variations in MBL2 genetic background alter susceptibility to colon cancer. Our results suggest that variations in both the secretor haplotype and 3′UTR region of the gene increase colon cancer risk in African Americans; whereas similar observations were not seen in Caucasians. We investigated whether plasma MBL levels and activity were altered in relation to the variants associated with increased risk of colon cancer. Our results suggest that the secretor and 3′UTR region haplotypes are associated with altered MBL plasma levels; however the results observed for plasma activity were less consistent. Furthermore, we showed evidence that one SNP in the 3′-UTR (rs10082466) may alter predicted miRNA binding, thus accounting for the lower plasma MBL levels.
Our results for the 3′-UTR region and secretor haplotypes are consistent with previously published studies that show these haplotypes modify serum levels (9,10,27,28). Additionally, we did confirm that genetic variation in the 5′-region of the gene is associated with both altered MBL plasma levels and activity in both Caucasians and African Americans. Interestingly, the median MBL plasma levels for haplotypes LXPA, LYPB, HYPD, and LYQC are lower in African American cases when compared with Caucasian cases. This correlates with previous reports that low serum MBL levels are more prevalent in populations from Africa. It has been suggested that low serum MBL levels have been selected for in African countries due to the protection that the lower levels provide against the deleterious effects of certain infections by intracellular bacteria and parasites (9).
It is possible that higher MBL plasma levels enhance immunological surveillance against colon neoplasia and, in turn, provide protection against the onset of disease. It has been shown that MBL recognizes and binds to oligosaccharide ligands expressed on the surfaces of human tumor cells, including a human colorectal carcinoma cell line SW1116 (29). Furthermore, MBL has been shown to inhibit tumor progression by MBL-dependent cell-mediated cytotoxicity (30). Thus, those individuals carrying the HYPA haplotype, which is associated with the high serum concentration of the protein, may be less susceptible to the onset of colon cancer. Our study results are supported by a previous study which found an association between an MBL2 haplotype associated with lower serum concentrations and increased risk of stomach cancer in a population-based case-control study in Warsaw, Poland (12).
We only observed evidence for increased susceptibility to colon cancer in African Americans. It has been shown that MBL serum concentrations correlate with the MBL2 secretor haplotypes in distinct populations (31) and we observed a similar correlation with MBL plasma levels in both Caucasians and African Americans in our study. Additionally, we observed differential levels of MBL activity correlating with the secretor haplotypes in both groups. These results suggest that similar functional consequences of the serum/plasma levels should be observed between races. However, we also observed that those African American and Caucasian cases carrying the haplotypes associated with low MBL serum levels differed in MBL plasma levels. Furthermore, the African American cases had lower MBL plasma levels when compared with Caucasian cases for these specific haplotypes. We did not see similar associations when Caucasian controls were compared with African American controls. This may account in part for the increased risk we see associated with colon cancer in those African Americans carrying specific haplotypes. It should be noted that although the cases have overall higher MBL levels and activity when compared with controls, this does not negate the possibility that the lower levels seen in African Americans could be associated with risk of developing cancer. Higher MBL levels in cases have been previously reported in the literature (32,33); and it has been suggested that this observation may be because MBL is a marker of inflammatory response associated with having the disease.
It is also plausible to hypothesize that the serum/plasma MBL levels may interact with additional genetic and/or environmental risk factors in which African Americans are exposed that we have not identified in our study. It has been shown that Caucasian individuals carrying the HYD haplotype, which is a truncated version of the HYPD haplotype, and a variant of IL-1β at position -511 have an increased risk of stomach cancer (12). Furthermore, it is possible that the colonic microbiota differ between African Americans and Caucasians. Evidence suggests that diet directly influences the bacterial diversity of the microbiota (34), therefore differences in diet between Caucasian and African American populations could influence the colonic microbiota. Thus, the increased susceptibility to colon cancer seen in African Americans in this study may be due to the response this specific population has to the lower levels of MBL when compared with Caucasians.
We also investigated the potential functional implications of the observed association of the 3′UTR region genotypes with increased risk of colon cancer and found that one of the associated SNPs leads to differential miRNA binding. Specifically, we found that more efficient binding of hsa-miR-27a to the risk-associated allele of MBL2 is consistent with lower levels and activity of MBL found in rs10082466C carriers. Of note, it was indicated in an earlier report investigating the association of MBL2 3′-UTR haplotypes and breast cancer risk in African Americans that carriers of the risk haplotype had lower MBL serum levels, although the association was only borderline significant (15). In our study, we observed a similar trend in cases and controls, regardless of race; therefore, the question of how this functional association explains the robustly increased colon cancer risk in African Americans still remains. Interestingly, evidence for differential miRNA expression associated with race is starting to emerge (35,36). Therefore, it is possible that expression of hsa-miR-27a in liver tissues, which is where MBL is mainly expressed, differs in African Americans when compared with Caucasians. When colon cancer tissue was examined, we could not show differential expression of mature hsa-miR-27a between African Americans and Caucasians; however we were able to demonstrate the fold increase of hsa-miR-27a precursor to be significantly higher in African Americans when compared with Caucasians (data not shown). A limitation of our study is that we have been unable to reliably quantify MBL2 protein expression in cultured cells or conditioned media, thus the correlation between rs10082466 and MBL2 expression levels could not be readily tested in vitro.
In addition to their functional association with miRNAs, it cannot be ignored that the 3′-UTR SNPs may be in linkage disequilibrium with another functional SNP not identified in our study. Bernig, et al. have shown that the CGGG 3′-UTR haplotype is most frequently seen in linkage with the truncated secretor haplotype 0, which also includes the secretor haplotype LYQC (28). We have observed similar results in that the CGGT 3′-UTR haplotype is most frequently seen in linkage with the truncated secretor haplotype 0 (data not shown). Therefore, the associations of the secretor haplotypes (LYPA, LYQC) and at the 3′-end of the gene could also represent an epistatic effect.
The main limitation of this study is the small sample size; therefore the observed associations will need to be replicated in a larger study. Additionally, further studies to elucidate the underlying biological mechanisms associated with MBL levels and the differences between colon cancer susceptibility in African Americans and Caucasians are warranted.
In conclusion, our study supports the hypothesis that MBL2 genetic variation in both the 3′- and 5′-region of the gene increase susceptibility of colon cancer, but only in African Americans. We also show that the genetic variation observed alters plasma MBL levels and activity. In the 3′UTR region, we showed that hsa-miR-27a more efficiently binds to rs10082466C which is consistent with the lower plasma MBL levels and activity observed. However, further studies are warranted to validate the associations we have observed in our study.
Supplementary Material
Acknowledgments
We thank Stefan Ambs, Sharon Pine, and Susan Olivo-Marston for valuable feedback, discussion, and constructive comments; Donna Perlmutter, Raymond Jones, Leoni Leonaridis, Audrey Salabes, John Cottrell, Glenwood Trivers, and the Surgery and Pathology Departments from the participating hospitals for their contribution to participant and biomaterial accrual; and Karen Yarrick for administrative assistance. This work was supported by funding from the Cancer Prevention Fellowship Program (2003–2007), Division of Cancer Prevention, National Cancer Institute to K.A.Z.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Surveillance Epidemiology and End Results [Internet] Bethesda: National Cancer Institute; [updated April 2011; based on the Nov 2010 submission; cited 2011 June 30]. Available from: http://seer.cancer.gov/ [Google Scholar]
- 3.Terzic J, Grivennikov S, Karin E, Karin M. Inflammation and colon cancer. Gastroenterology. 2010;138:2101–14. doi: 10.1053/j.gastro.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 4.Choi PM, Zelig MP. Similarity of colorectal cancer in Crohn’s disease and ulcerative colitis: implications for carcinogenesis and prevention. Gut. 1994;35:950–4. doi: 10.1136/gut.35.7.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hansen S, Holmskov U. Structural aspects of collectins and receptors for collectins. Immunobiology. 1998;199:165–89. doi: 10.1016/S0171-2985(98)80025-9. [DOI] [PubMed] [Google Scholar]
- 6.Turner MW. The role of mannose-binding lectin in health and disease. Mol Immunol. 2003;40:423–9. doi: 10.1016/s0161-5890(03)00155-x. [DOI] [PubMed] [Google Scholar]
- 7.Presanis JS, Kojima M, Sim RB. Biochemistry and genetics of mannan-binding lectin (MBL) Biochem Soc Trans. 2003;31:748–52. doi: 10.1042/bst0310748. [DOI] [PubMed] [Google Scholar]
- 8.Lee SG, Yum JS, Moon HM, Kim HJ, Yang YJ, Kim HL, et al. Analysis of mannose-binding lectin 2 (MBL2) genotype and the serum protein levels in the Korean population. Mol Immunol. 2005;42:969–77. doi: 10.1016/j.molimm.2004.09.036. [DOI] [PubMed] [Google Scholar]
- 9.Madsen HO, Satz ML, Hogh B, Svejgaard A, Garred P. Different molecular events result in low protein levels of mannan-binding lectin in populations from southeast Africa and South America. J Immunol. 1998;161:3169–75. [PubMed] [Google Scholar]
- 10.Madsen HO, Garred P, Thiel S, Kurtzhals JA, Lamm LU, Ryder LP, et al. Interplay between promoter and structural gene variants control basal serum level of mannan-binding protein. J Immunol. 1995;155:3013–20. [PubMed] [Google Scholar]
- 11.Petersen SV, Thiel S, Jensenius JC. The mannan-binding lectin pathway of complement activation: biology and disease association. Mol Immunol. 2001;38:133–49. doi: 10.1016/s0161-5890(01)00038-4. [DOI] [PubMed] [Google Scholar]
- 12.Baccarelli A, Hou L, Chen J, Lissowska J, El Omar EM, Grillo P, et al. Mannose-binding lectin-2 genetic variation and stomach cancer risk. Int J Cancer. 2006;119:1970–5. doi: 10.1002/ijc.22075. [DOI] [PubMed] [Google Scholar]
- 13.Wang FY, Tahara T, Arisawa T, Shibata T, Yamashita H, Nakamura M, et al. Mannan-binding lectin (MBL) polymorphism and gastric cancer risk in Japanese population. Dig Dis Sci. 2008;53:2904–8. doi: 10.1007/s10620-008-0249-3. [DOI] [PubMed] [Google Scholar]
- 14.Olivo-Marston SE, Yang P, Mechanic LE, Bowman ED, Pine SR, Loffredo CA, et al. Childhood exposure to secondhand smoke and functional mannose binding lectin polymorphisms are associated with increased lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2009;18:3375–83. doi: 10.1158/1055-9965.EPI-09-0986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernig T, Boersma BJ, Howe TM, Welch R, Yadavalli S, Staats B, et al. The mannose-binding lectin (MBL2) haplotype and breast cancer: an association study in African-American and Caucasian women. Carcinogenesis. 2007;28:828–36. doi: 10.1093/carcin/bgl198. [DOI] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 17.Brodersen P, Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Biol. 2009;10:141–8. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- 18.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodman JE, Bowman ED, Chanock SJ, Alberg AJ, Harris CC. Arachidonate lipoxygenase (ALOX) and cyclooxygenase (COX) polymorphisms and colon cancer risk. Carcinogenesis. 2004;25:2467–72. doi: 10.1093/carcin/bgh260. [DOI] [PubMed] [Google Scholar]
- 20.variant GPS [Internet] Bethesda: National Cancer Institute; [cited 2011 June 30]. Available from: http://variantgps.nci.nih.gov/cgfseq/pages/snp500.do. [Google Scholar]
- 21.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 22.Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 2005;76:449–62. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 24.National Center for Biotechnology Information [Internet] Bethesda: National Institutes of Health; [cited 2011 June 30]. Available from: http://www.ncbi.nlm.nih.gov/projects/SNP/ [Google Scholar]
- 25.Hiard S, Charlier C, Coppieters W, Georges M, Baurain D. Patrocles: a database of polymorphic miRNA-mediated gene regulation in vertebrates. Nucleic Acids Res. 2010;38:D640–D651. doi: 10.1093/nar/gkp926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ziebarth JD, Bhattacharya A, Chen A, Cui Y. PolymiRTS Database 2.0: linking polymorphisms in microRNA target sites with human diseases and complex traits. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr1026. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garred P, Larsen F, Madsen HO, Koch C. Mannose-binding lectin deficiency--revisited. Mol Immunol. 2003;40:73–84. doi: 10.1016/s0161-5890(03)00104-4. [DOI] [PubMed] [Google Scholar]
- 28.Bernig T, Breunis W, Brouwer N, Hutchinson A, Welch R, Roos D, et al. An analysis of genetic variation across the MBL2 locus in Dutch Caucasians indicates that 3′ haplotypes could modify circulating levels of mannose-binding lectin. Hum Genet. 2005;118:404–15. doi: 10.1007/s00439-005-0053-5. [DOI] [PubMed] [Google Scholar]
- 29.Nakagawa T, Kawasaki N, Ma Y, Uemura K, Kawasaki T. Antitumor activity of mannan-binding protein. Methods Enzymol. 2003;363:26–33. doi: 10.1016/S0076-6879(03)01041-3. [DOI] [PubMed] [Google Scholar]
- 30.Nakagawa T, Ma BY, Uemura K, Oka S, Kawasaki N, Kawasaki T. Role of mannan-binding protein, MBP, in innate immunity. Anticancer Res. 2003;23:4467–71. [PubMed] [Google Scholar]
- 31.Garred P, Larsen F, Seyfarth J, Fujita R, Madsen HO. Mannose-binding lectin and its genetic variants. Genes Immun. 2006;7:85–94. doi: 10.1038/sj.gene.6364283. [DOI] [PubMed] [Google Scholar]
- 32.Ytting H, Christensen IJ, Jensenius JC, Thiel S, Nielsen HJ. Preoperative mannan-binding lectin pathway and prognosis in colorectal cancer. Cancer Immunol Immunother. 2005;54:265–72. doi: 10.1007/s00262-004-0594-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ytting H, Jensenius JC, Christensen IJ, Thiel S, Nielsen HJ. Increased activity of the mannan-binding lectin complement activation pathway in patients with colorectal cancer. Scand J Gastroenterol. 2004;39:674–9. doi: 10.1080/00365520410005603. [DOI] [PubMed] [Google Scholar]
- 34.O’Keefe SJ. Nutrition and colonic health: the critical role of the microbiota. Curr Opin Gastroenterol. 2008;24:51–8. doi: 10.1097/MOG.0b013e3282f323f3. [DOI] [PubMed] [Google Scholar]
- 35.Zhao H, Shen J, Medico L, Wang D, Ambrosone CB, Liu S. A pilot study of circulating miRNAs as potential biomarkers of early stage breast cancer. PLoS ONE. 2010;5:e13735. doi: 10.1371/journal.pone.0013735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heegaard NH, Schetter AJ, Welsh JA, Yoneda M, Bowman ED, Harris CC. Circulating microRNA expression profiles in early stage non-small cell lung cancer. Int J Cancer. 2011 doi: 10.1002/ijc.26153. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.