Summary
Changes of epigenetic status and chromatin structure have been shown to associate with aging in many organisms. Here, we report an RNAi screen of putative histone methyltransferases and demethylases in wild type C. elegans using reproduction inhibitor. We identified six genes that, when inactivated by RNAi, consistently extend lifespan. Five of these genes do not require germline proliferation to affect lifespan. We further characterized two of these genes, the highly homologous SET-domain containing genes, set-9 and set-26. They share redundant functions in maintaining normal lifespan, while exhibiting differential tissue expression patterns. Furthermore, we found that set-9 and set-26 partially act through the FOXO transcription factor, DAF-16, to modulate lifespan. Interestingly, inactivation of somatic SET-26 alone results in a robust lifespan extension, and alters the levels of histone H3 protein and the repressive histone marks, H3K9me3 and H3K27me3, in an age-dependent manner. We hypothesize that inactivation of SET-26 triggers compensation mechanisms to restore repressive chromatin structure, and hence affects chromatin stability to promote longevity.
Keywords: aging, epigenetics, histone modification, chromatin, SET domain, C. elegans
Introduction
Epigenetic controls include DNA methylation, post-translational modifications of histone peptides, and others. These modifications can have a direct and complex impact on the chromatin structure, gene expression, and other DNA biology (Bonasio et al. 2010). However, there are a few widely accepted characteristics of some modifications (Ruthenburg et al. 2007). For example, histone tail hyper-acetylation is generally associated with chromatin decondensation and active transcription (Hebbes et al. 1988). Tri-methylated histone H3 lysine-4 and lysine-36 (H3K4me3 and H3K36me3) are usually coupled with gene activation and elongation, while tri-methylated histone H3 lysine-9 and lysine-27 (H3K9me3 and H3K27me3) usually associate with gene silencing and highly compacted heterochromatin (Bonasio et al. 2010).
Many studies have pointed to the possibility that epigenetic changes have a direct and functional impact on aging (Calvanese et al. 2009). Vertebrate studies showed that older age is often associated with global decrease of DNA methylation, which opens up the heterochromatin (Singhal et al. 1987; Wilson et al. 1987). In addition to global changes, many individual histone modification or DNA methylation sites on specific gene promoters or chromosomal regions are also linked to aging and aging-related diseases, like cancer (Calvanese et al. 2009). Presumably, sophisticated changes of the balance among many modifiers are responsible for these age-associated chromatin structure alterations. Interestingly, unbiased screens have identified genes that are functionally implicated in chromatin modifications to affect aging in the roundworm Caenorhabditis elegans (Hamilton et al. 2005).
To investigate whether and how chromatin status is associated with aging in C. elegans and whether there are other specific chromatin modifiers directly linking epigenetic changes and lifespan, we carried out a small-scale RNAi screen of the majority of the putative histone methyltransferases and demethylases in worms. From the screen, we identified six longevity genes that, when inactivated by RNAi, can extend the worm lifespan. Among the six genes, we further characterized two highly homologous genes, set-9 and set-26, and found that they have differential expression patterns but overlapping functions in normal lifespan maintenance. We also observed that set-9 and set-26 influence longevity partially dependent on daf-16, a prominent longevity gene that encodes the worm ortholog of the highly conserved Forkhead transcription factor FOXO (Kenyon et al. 1993; Antebi 2007). Furthermore, inactivation of somatic set-26 alone is sufficient to extend worm lifespan and affects histone protein levels and two repressive histone modifications in an age-dependent manner.
Results
Six putative histone methyltransferases and demethylases identified as longevity determinants by an RNAi screen
Our previous genome-wide RNAi screen revealed several putative chromatin modifiers to be important for longevity determination in C. elegans. In particular, we identified proteins containing characteristic domains that are important for either histone methylation or methylated histone recognition (Hamilton et al. 2005). Since high-throughput RNAi screens are prone to false negatives (Ni & Lee 2010), we carried out a small-scale RNAi screen targeting putative histone methyltransferases and demethylases in order to search for additional epigenetic regulators involved in aging modulation. For the histone methyltransferases, we focused on genes predicted to contain a SET domain, a signature motif of most histone methyltransferases (Dillon et al. 2005). For the histone demethylases, we concentrated on both the LSD1/amine oxidase family and the jmjC-domain containing family (Klose et al. 2006). Through bioinformatics and literature search, we identified 38 SET-domain containing genes (Andersen & Horvitz 2007), 6 LSD1/amine oxidase genes, and 14 jmjC-domain containing genes in C. elegans (Klose et al. 2006). We were able to confirm 50 (out of 58) correct RNAi clones (Supporting Information).
To assay the lifespan phenotype associated with knockdown of each of the 50 genes, wild type N2 worms were fed individual RNAi bacteria starting at the L1 stage of development, and their adult lifespans were monitored in the presence of the mitotic inhibitor FUDR, which inhibits progeny production (Mitchell et al. 1979). Whereas the majority of the RNAi treatments resulted in shortening or no significant change of lifespan, we found that RNAi knockdown of six genes reproducibly extended lifespan of the animals (Table 1A, Table S1). Four of the six genes, set-9, set-26, rbr-2, and utx-1 have recently been identified as longevity-associated genes in worms through either high-throughput or targeted screening methods (Lee et al. 2003; Hamilton et al. 2005; Greer et al. 2010; Jin et al. 2011; Maures et al. 2011), thus validating the efficacy of our screen. It is important to note that set-9 and set-26 genes are >95% identical and their corresponding RNAi clones likely target both genes simultaneously (Figure 1A). We also discovered two new worm longevity genes, mes-2 and jmjd-2, that have not previously been implicated in worm aging. From the screen, we also found four RNAi clones that caused obvious developmental defects or slow growth. We retested these RNAi clones by exposing worms to RNAi treatment after they had reached adulthood but did not observe any lifespan increase (Table S1).
Table 1a. Lifespan effects by the six positive candidates from the RNAi screen.
A) Six positive RNAi clones from the lifespan screen extend wild type worm lifespan (20°C). Representative data from 3 or more independent experiments are shown.
| Positive candidates from RNAi lifespan screen on wild type (20°C) | ||||||
|---|---|---|---|---|---|---|
| RNAi | Mean LS ± SEM |
N | P-Value vs. EV |
% of EV LS |
Domain | Homolog (species) |
| EV | 19.05±0.27 | 88 | - | 100 | - | - |
| set-9/26 | 21.40±0.33 | 95 | <0.001 | 112 | SET | MLL5 (hs) |
| mes-2 | 20.29±0.29 | 49 | <0.002 | 106 | SET | E(Z) (hs, dm) |
| utx-1 | 21.63±0.37 | 104 | <0.001 | 113 | JmjC | UTX (hs, dm) |
| jmjd-2 | 21.55±0.29 | 94 | <0.001 | 113 | JmjC | KDM4 (hs, dm) |
| rbr-2 | 26.12±0.34 | 105 | <0.001 | 137 | JmjC | JARID1 (hs) |
| Lid (dm) | ||||||
All survival analyses were done with SPSS software using Kaplan Meier analysis and log-rank test to compute p- values. A p-value ≤0.05 is considered statistically significant.
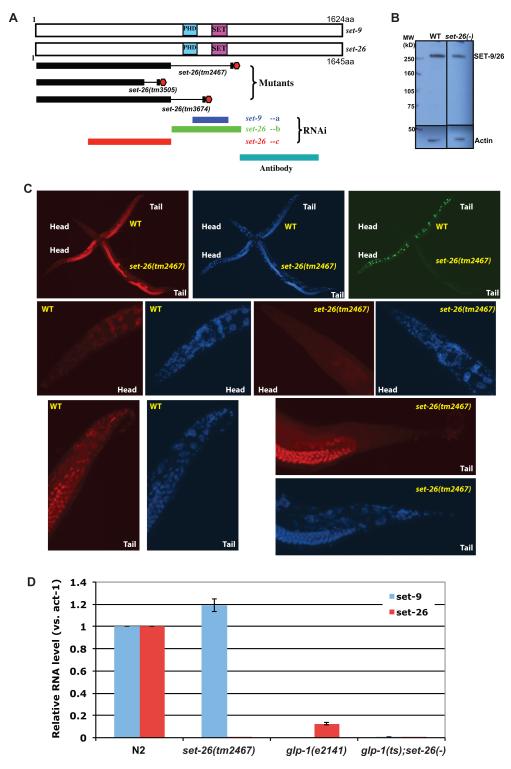
Figure 1. set-9 and set-26 genes share high homology but have differential expression patterns.
A) Schematic linear protein structures of SET-9 and SET-26. Deleted regions of the three set-26 mutant alleles were depicted as thin lines within thick black lines that indicate remaining coding regions. Premature stop codons caused by the deletions were depicted as “O”. Three RNAi constructs (a, b, and c) are shown aligned to their targeted sequences. The polypeptide used for antibody production is also shown. B) Western Blot of equal amount of whole worm lysate from wild type and the set-26(tm2467) mutant worms using SET-9/26 antibody. Actin is used as a loading control. C) SET-9 and SET-26 are nuclear proteins. The wild-type worms (labeled with an irrelevant GFP transgene sur-5::gfp, Green) and the set-26(tm2467) mutant worms (no GFP) were co-stained with a purified SET-26 antibody (Red) and Hoechst (Blue, DNA). Zoom-in frames of the heads and tails were shown at the bottom. D) Total RNA of young adult worms raised at 25°C were subject to reverse transcription and qPCR using primer sets and thermocycles that can distinguish set-9 and set-26 transcripts. The RNA levels of act-1 gene were used for normalization. Error bars show the difference between 2 independent experiments, and the pairwise two-tailed t-test were used to calculate the p-values.
While this work was underway, Greer et al published an RNAi screen of a similar set of putative histone methyltransferases with a major distinction being that no FUDR was used in their lifespan assays (Greer et al. 2010). As a result, they identified several genes that require germline proliferation and reproduction to affect lifespan, in addition to set-9 and set-26 that are the only common genes from the two screens. To test whether our positive candidates from the screen can affect lifespan independent of germline proliferation, we utilized glp-1(e2141) mutant worms, which lack a germline at non-permissive temperatures (Austin & Kimble 1987). We found that five of the six RNAi clones (except jmjd-2) continued to extend the lifespan of glp-1(e2141) worms to a similar extent as in wild type worms (Table 1B), suggesting that germline proliferation is not required for these genes to modulate lifespan. Our data suggest that jmjd-2 requires germline proliferation but not continuous progeny production as blocked by FUDR to affect lifespan (Table 1A and Table S1).
Table 1b. Lifespan effects by the six positive candidates from the RNAi screen.
B) Five of the six positive RNAi clones extend glp-1(e2141) mutant worm lifespan at the non-permissive temperature (25°C). Data are pooled from 2 independent experiments.
| RNAi lifespan on wild type (N2) and glp-1(e2141ts) worms (25°C) | ||||||
|---|---|---|---|---|---|---|
| Strain /RNAi |
Mean LS ± SEM |
N | P-Value vs. N2/EV |
% of N2/EV LS |
P-Value vs. glp-1/EV |
% of glp-1/EV LS |
| N2/EV | 16.16±0.18 | 193 | - | 100 | ||
|
N2
/set-9/26 |
19.65±0.25 | 176 | <0.001 | 122 | ||
| N2/mes-2 | 17.20±0.20 | 179 | <0.001 | 106 | ||
| N2/utx-1 | 19.25±0.25 | 175 | <0.001 | 119 | ||
| N2/jmjd-2 | 18.04±0.19 | 176 | <0.001 | 112 | ||
| N2/rbr-2 | 17.98±0.23 | 168 | <0.001 | 111 | ||
|
| ||||||
| glp-1/EV | 19.24±0.22 | 171 | <0.001 | 119 | - | 100 |
|
glp-1
/set-9/26 |
22.57±0.32 | 182 | <0.001 | 117 | ||
| glp-1/mes-2 | 20.61±0.30 | 129 | <0.001 | 107 | ||
| glp-1/utx-1 | 22.60±0.28 | 160 | <0.001 | 117 | ||
| glp-1/jmjd-2 | 19.09±0.18 | 160 | 0.201 | 99 | ||
| glp-1/rbr-2 | 21.07±0.41 | 108 | <0.001 | 110 | ||
All survival analyses were done with SPSS software using Kaplan Meier analysis and log-rank test to compute p- values. A p-value ≤0.05 is considered statistically significant.
SET-9 and SET-26 have similar gene structure but differential tissue expression patterns
As set-9 and set-26 knockdown exhibited strong effects on longevity independent of the reproductive status of the worms (Table 1), we sought to further characterize their functions. In addition to sharing very high identity at the level of nucleotide (97%) and amino acid (96%) sequences in the coding regions, set-9 and set-26 also have the same SL1 trans-splicing sites at their 5′ ends (Figure 1a and Supporting Information). Furthermore, they share high homology in the non-coding sequences flanking the coding regions (near 90% identity in the +/− 500bp regions). Although both SET-9 and SET-26 proteins contain a SET domain, the signature motif of most histone methyltransferases (Dillon et al. 2005), the position of the SET-domain within the polypeptides and the absence of several critical residues important for enzymatic activities suggest that SET-9/26 likely lack direct methyltransferase activities (Qian & Zhou 2006).
We examined the expression patterns of SET-9 and SET-26 using an antibody raised against a polypeptide downstream of the PHD and SET domains (Figure 1A). Because the two proteins are 96% identical, we predicted that the antibody would recognize both proteins. Consistent with this, immunoblotting results indicated that a ~250kD band (apparent MW of SET-9 and SET-26) showed diminished signal, but was not absent, in the set-26(tm2467) mutant that was expected to have no expression of the antigenic region (Figure 1B). The remaining signal likely represented SET-9 protein. Immunostaining in wild type worms using the same antibody showed nuclear signal in most tissues, except spermatids (Figure 1C, S1A, and data not shown). In contrast, signal was only found in the germline in the set-26(tm2467) mutant worms (Figure 1C). The same pattern was observed in all 4 larval stages (data not shown). These suggest that SET-9 protein is predominantly expressed in the germline with very little to no expression in the somatic tissues. However, we could not directly test this because no true set-9 loss-of-function mutant exists, as the only set-9 mutant available contains a tandem deletion/duplication (Wormbase). Similar as set-26(tm2467), two other set-26 mutants that have deletions of the PHD and SET domains and premature stop codons (Figure 1A and Supporting Information) exhibit the same expression pattern (data not shown).
To further distinguish the differential expression patterns between set-9 and set-26 genes, we designed primers and strategies to specifically amplify either set-9 or set-26 transcripts (Figure 1D). In the set-26(tm2467) mutant, set-26 transcript was undetectable, and the level of set-9 RNA remained largely the same as in wild type worms (p-value 0.184). In the germlineless glp-1(e2141) worms raised at a non-permissive temperature, consistent with the notion that SET-9 is likely to be a germline protein (Figure 1C), we detected very little set-9 RNA expression (Figure 1D). Interestingly, we also detected a dramatic decrease of the set-26 transcript level (down to ~12% of WT, p-value 0.01), suggesting that the majority of set-26 is expressed in the germline with a small but significant amount expressed in the somatic tissues, as was observed by immunostaining (Figure 1C). As a further corroboration, we observed that an RFP-tagged set-26 transgene was expressed in both the germline and somatic tissues (Figure S1B).
set-9 and set-26 genes have redundant functions in maintaining normal lifespan
To rule out possible off-target effects associated with the set-9/26 RNAi clones from the screen, we designed an additional RNAi construct (c) targeting a region of set-9/26 that is not overlapping with the two existing RNAi constructs (a and b) from the screen (Figure 1A). We found that all three constructs resulted in similar life extension effect in wild type worms (>20% extension, Figure 2A, Table S2). The RNAi-treated worms showed no obvious growth or developmental phenotypes (Figure S2E), suggesting the longevity effect of set-9/26 is not secondary to other developmental impairments.
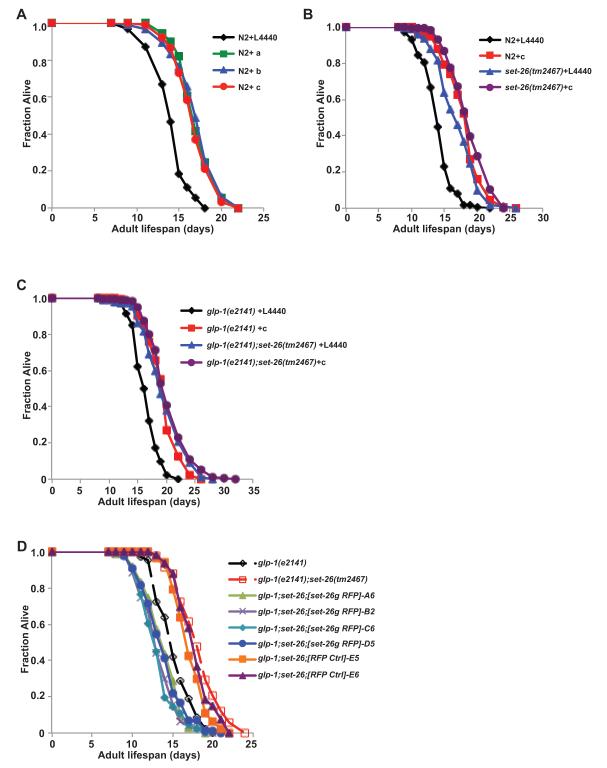
Figure 2. set-9 and set-26 genes have redundant functions in maintaining normal lifespan.
A) Knockdown of set-9 and set-26 by three RNAi constructs (as shown in Figure 1A) prolongs the lifespan of wild type worms to a similar extent at 25°C. B) set-26(tm2467) mutant lives moderately longer than wild type N2 worms when fed control RNAi bacteria (L4440), but has a similar lifespan as wild-type worms when fed set-9/26 RNAi bacteria. C) set-26(tm2467) mutation and set-26 RNAi similarly extend the lifespan of the germlineless glp-1(e2141) worms at the non-permissive temperature 25°C. D) Transgenic worms over-expressing the genomic set-26 gene in glp-1(e2141);set-26(tm2467) background live shorter than their corresponding transgenic controls. Multiple lines of transgenic worms are used. The set-26 over-expression lines are glp-1(e2141);set-26(tm2467);rwEx21[pJKL702-unc-119(mini)-set-26g mec-7::RFP pBSK] (lines A6, B2, C6, and D5) and the transgenic control lines are glp-1(e2141);set-26(tm2467);rwEx22[pJKL702-unc-119(mini) mec-7::RFP pBSK] (lines E5 and E6).
We next examined whether the set-26 mutation has a lifespan extension phenotype similar to what was seen with the set-9/26 RNAi treatment. We found that the three set-26(-) single mutant worms all live moderately longer than wild type worms (Figure 2B and Figure S2A-B). For the remainder of this study, we will focus on the set-26(tm2467) mutant. We note that the set-26(tm2467) single mutants did not live as long as wild type worms that were fed set-9/26 RNAi bacteria (21% vs. 34% in Figure 2B, 7% vs. 22% in Figure S2B), suggesting that set-9 likely has a redundant role in lifespan determination. Consistent with this prediction, we found that set-9/26 RNAi can further extend the lifespan of the set-26(tm2467) mutant, to a degree similar to that of wild type worms treated with set-9/26 RNAi (Figure 2B and S2B). Taken together, our results suggest that set-9 functions redundantly with set-26 in lifespan maintenance.
Considering both the expression and lifespan data discussed thus far, we conclude that deletion of set-26 alone in both the germline and soma has a small but significant effect on lifespan (Figure 2B and S2A-B) and that the germline-expressed set-9 also contributes to normal lifespan maintenance (Figure 2B). Because neither set-9 nor set-26 were detected in glp-1(e2141);set-26(tm2467) double mutants cultured at the non-permissive temperature (Figure 1D & S1D), we reasoned that this double mutant strain can serve as a mimic of double deletion of both set-9 and set-26, and that by comparing glp-1(e2141) versus glp-1(e2141);set-26(tm2467), we could deduce the functions of somatic SET-26. We found that deletion of set-26 in the glp-1(e2141) background substantially extended the lifespan of glp-1(e2141) worms (19%, Figure 2C). This finding is consistent with our earlier observation that set-9/26 RNAi could prolong the lifespan of germlineless mutants (Table 1b, Figure 2C) and suggests that somatic set-26 alone plays a significant role in modulating longevity. As a further validation of this notion that somatic set-26 has a direct role in longevity modulation, we found that somatic over-expression of a set-26 genomic fragment in the glp-1(e2141);set-26(tm2467) double mutant was able to reverse the prolonged lifespan of the double mutant as compared to the glp-1(e2141) single mutant (Figure 2D). Further experiments confirmed that this rescue of the lifespan phenotype is due to over-expression of wild-type SET-26 because RNAi knockdown of set-26 expression can abolish the rescue effects by the transgene (Figure S2C).
Because the lifespan extension effect by set-9/26 RNAi is not germline dependent, we wondered whether initiating such RNAi treatment after development might still exert a lifespan-extension phenotype. Interestingly, worms fed any of the three RNAi bacteria showed very little to no lifespan extension when RNAi treatment were started at the young adult stage (Figure S2D). This suggests that the functions of set-9/26 during development are important for normal adult lifespan maintenance.
SET-9 and SET-26 regulate lifespan in a partially daf-16-dependent manner
The Forkhead box O (FOXO) family of transcription factors are highly conserved key converging points of several longevity pathways (Antebi 2007). C. elegans has a single FOXO ortholog, daf-16, and mutants with daf-16 deficiency exhibit shortened lifespan compared to wild type animals (Kenyon et al. 1993; Antebi 2007). Very recent studies showed that daf-16 functions downstream of a histone modifier to influence lifespan (Jin et al. 2011; Maures et al. 2011). Thus, we examined whether set-9 and set-26 interact with daf-16 to modulate lifespan.
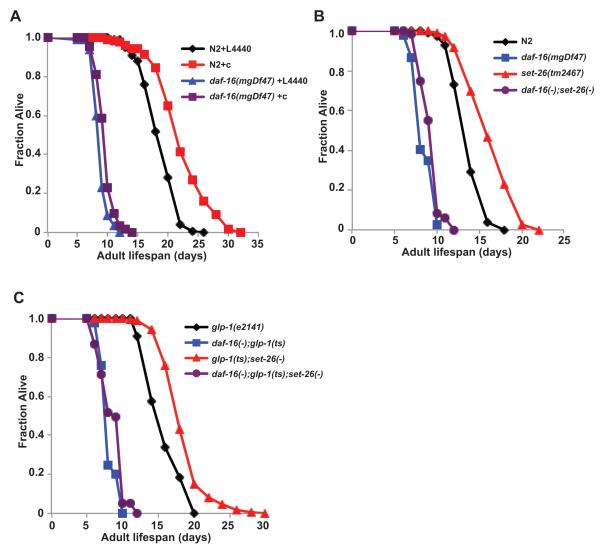
We found that daf-16(mgDf47) mutation could partially but not completely suppress the lifespan extension phenotype induced by set-9/26 RNAi (19% extension in WT vs. 10% in daf-16(mgDf47), p-value <0.001, Figure 3A), suggesting that SET-9/26 maintain normal lifespan partly by repressing DAF-16 activity. We note that our previous studies showed that set-9/26 RNAi cannot significantly extend the lifespan of daf-16(mgDf47) mutant worms in the RNAi enhancing mutant rrf-3(pk1426) background (Hamilton et al. 2005). Strain differences as well as the more frequent scoring and the increased statistical power gained by pooling multiple experiments in the current study likely explain the slight discrepancy. In addition, we observed that the set-26(tm2467) mutation also extended lifespan in a partially daf-16-dependent manner (19% extension in WT compared to 10% in daf-16(mgDf47) background, p-value <0.001, Figure 3B). Similarly, we found that somatic set-26 partially depends on daf-16 to affect lifespan as well (18% vs. 6% extension by set-26(tm2467) in glp-1(e2141) vs. glp-1(e2141);daf-16(mgDf47), p-value 0.001, Figure 3C; 24% vs. 20% extension by set-26(tm2467) in glp-1(e2141) treated with control RNAi compared to daf-16 RNAi, p-value <0.001, Figure S3A). Overall, our data support the model that set-9 and set-26 act through daf-16 to achieve part of its function in longevity modulation.
Figure 3. set-9 and set-26 partially function through daf-16 to influence lifespan.
A) daf-16(mgDf47) mutation partially suppresses the lifespan extension by set-9/26 RNAi in wild type N2 worms (20°C). B-C) daf-16(mgDf47) mutation partially suppresses the lifespan extension conferred by set-26(tm2467) single mutation in wild type animals (B) and in the germlineless glp-1(e2141) mutant worms (C) at 25°C.
Somatic SET-26 affects global levels of histone expression and repressive histone modifications in an age-dependent manner
To directly examine whether chromatin structure is affected when set-9/26 is inactivated, we monitored the global levels of several histone modifications that are well-characterized markers of chromatin structure. To avoid the complications by having hundreds of germ cells that are going through dramatic chromatin changes in wild type worms (Kimble & Crittenden 2005; Schaner & Kelly 2006), we used glp-1(e2141) mutants at a non-permissive temperature harboring either the wild type or tm2467 allele of set-26 gene to investigate the effect on histone modifications caused by loss of somatic SET-26. Extracts from synchronized worms at Day-1 (young adult) and Day-12 (old adult) were subjected to immunoblotting to compare the levels of SET-26, histone H3, and several specific modifications of histone H3 in both genetic backgrounds and age groups (Figure 4). We found that Actin protein levels do not significantly change with age or with the set-26 mutation and serves as an appropriate loading control. As shown in Figure S1, SET-26 protein level is undetectable in glp-1(e2141);set-26(tm2467) double mutant. Interestingly, we detected a large reduction in SET-26 protein level with age in glp-1(e2141) single mutant worms. Given that deletion of set-26 results in a robust lifespan extension in the glp-1 mutant background (Figure 2), this finding was somewhat unexpected. Further work is required to determine why SET-26 levels decrease with normal aging.
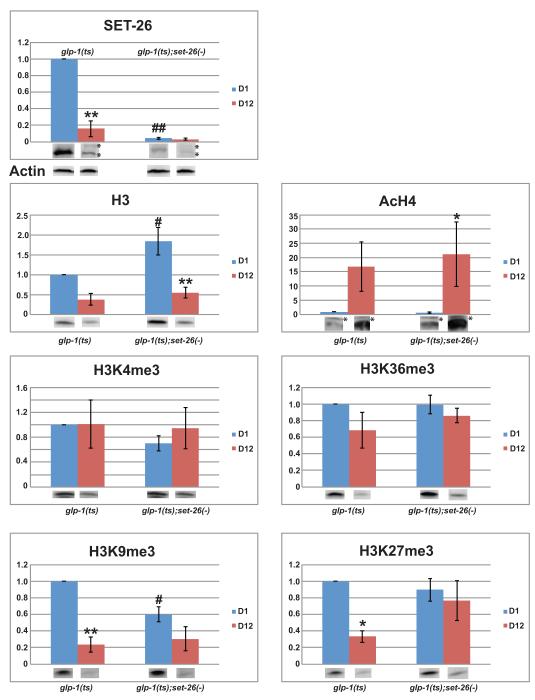
Figure 4. Deletion of somatic set-26 affects the levels of histone H3 protein and histone modifications in an age-dependent manner.
Synchronized glp-1(e2141) and glp-1(e2141ts);set-26(tm2467) worms were harvested at Day-1 (D1) and Day-12 (D12) adulthood at 25°C. Equal amount of worm total lysates were subjected to electrophoresis and western blot. A representative western blot is shown under each quantification chart, except that ACTIN blot is shown separately. Non-specific bands were labeled with “*”. SET-26 and histone H3 protein levels were normalized to Actin levels. AcH4, H3K4me3, H3K36me3, H3K9me3, and H3K27me3 were normalized to H3 levels (Supporting information). The amount of each epitope from the D1-glp-1(e2141) sample was set to 1. Error bars show standard error of mean (SEM) from 3-5 independent experiments. Two-way ANOVA was used to analyze the significance of the effect by genetic background (Mut), age (Age), and the interaction between Mutation and Age (Mut*Age). The set of p-values of (Mut, Age, Mut*Age) for each epitope is as following: SET-26 (<0.001, <0.001, <0.001), H3 (0.024, <0.001, 0.107), AcH4 (0.640, 0.001, 0.578), H3K4me3 (0.302, 0.462, 0.502), H3K36me3 (0.401, 0.042, 0.358), H3K9me3 (0.061, <0.001, 0.012), H3K27me3 (0.168, 0.005, 0.036). A p-value ≤0.05 is considered statistically significant. Tukey (HSD) pairwise comparison was also used. When comparing D1 and D12 samples of the same genetic background, we use “*” and “**” to denote p<0.05 and p <0.005, respectively. When comparing the two genetic mutants of the same age group, we use “#” and “##” to denote p <0.05 and p <0.005, respectively.
We found that total histone H3 protein level in somatic tissues decreased dramatically with age irrespective of the set-26(tm2467) mutation (2.63- and 3.33-fold in glp-1(e2141) and glp-1(e2141);set-26(tm2467) worms, respectively, Figure 4). Curiously, we found that deletion of set-26 moderately increased the histone H3 protein levels (1.83-fold for D1 adults), indicating that set-26(-) deletion causes accumulation of core histones.
Then we measured the abundance of several well-studied histone modifications (using total histone H3 level for normalization), including the open chromatin markers AcH4 (pan-acetylated histone H4), H3K4me3, and H3K36me3, as well as the repressive chromatin markers H3K9me3 and H3K27me3 (Bonasio et al. 2010). Statistical analysis showed that neither aging nor the set-26(tm2467) mutation had a significant effect on the global levels of the two active gene expression markers H3K4me3 and H3K36me3. We did observe a dramatic increase in the AcH4 levels with age, indicating an increase in open chromatin formation at older age. However, we did not detect any significant effect on AcH4 by the set-26(tm2467) mutation.
For the two repressive chromatin markers, H3K9me3 and H3K27me3, we detected strong age-dependent effects overall. In glp-1(e2141) worms, we observed a strong age-dependent decrease in both modifications (4.17- and 3-fold for H3K9me3 and H3K27me3, respectively), which suggests a loss of chromatin compaction during aging. In contrast, when set-26 is deleted, no significant age-dependent loss of either repressive histone mark was detected. This demonstrated a significant age-dependent set-26(tm2467) effect on the levels of H3K9me3 and H3K27me3 (Two-way ANOVA p-values are 0.012 and 0.036, respectively). It is of note that the H3K9me3 level was somewhat lower in young glp-1(e2141);set-26(tm2467) double mutant compared to young glp-1(e2141) single mutant, but the level of this histone mark did not significantly decrease with increasing age in the double mutants. Remarkably, the levels of H3K27me3 in young and old glp-1(e2141);set-26(tm2467) double mutants remained at a high level similar to young glp-1(e2141) single mutant animals. These data suggest that inactivation of somatic set-26 prevents the age-dependent loss of histone modifications that are normally associated with highly compacted heterochromatin.
Discussion
In our attempt to discover new histone modifiers that modulate longevity, we identified six putative histone methyltransferases and demethylases that, when inactivated by RNAi, extend wild type worm lifespan. The chromatin players we identified, with the exception of jmjd-2, do not require germline proliferation to affect normal lifespan. These results indicate that our screening method that uses FUDR creates a bias towards identification of germline-independent longevity genes, distinct from a similar chromatin gene screen previously done without FUDR (Greer et al. 2010). We further characterized the two highly homologous genes set-9 and set-26 that strongly extend the worm lifespan in both wild type and the germlineless glp-1(e2141) background. We found that set-9 and set-26 share redundant functions in maintaining normal lifespan despite their differential tissue expression patterns, and they function partially through the well-known longevity determinant daf-16. We found that inactivation of somatic set-26 alone is sufficient to extend lifespan, as well as to abrogate the age-dependent changes in the abundance of H3K9me3 and H3K27me3, two H3 modifications widely associated with compact chromatin.
Identification of histone modifiers that affect lifespan in a germline-independent pathway
Among the six longevity genes we found that rbr-2, utx-1, and jmjd-2 encode proteins with the JmjC-domain, a functional motif representative of a large class of histone demethylases (Klose et al. 2006). Intriguingly, previous reports showed that inactivation of rbr-2 results in a moderate lifespan extension when reproduction is inhibited by FUDR or glp-1 mutation, but shortens lifespan in wild type reproductive worms (Table 1) (Lee et al. 2003; Greer et al. 2010). Previous studies suggest that C. elegans RBR-2 (an H3K4me3 demethylase) antagonizes the activity of the methyltransferase SET-2/ASH-2 COMPASS complex in reproductive worms (Greer et al. 2010). Inactivation of rbr-2 is believed to deregulate H3K4me2/3 maintenance and impair germline stem cell proliferation (Li & Kelly 2011; Greer et al. 2010), thus likely contributing to the longevity phenotype associated with the rbr-2 inactivation. How RBR-2 inactivation in non-reproductive worms increases lifespan is unknown. We speculate that RBR-2 may antagonize other H3K4 methyltransferase and/or affect a different set of downstream target genes in the somatic tissues to restrict normal lifespan. The second jmjC gene we identified is utx-1, a member of the UTX/UTY family that demethylates H3K27me3 and H3K27me2 to H3K27me (Agger et al. 2007; Hong et al. 2007). We found that utx-1 RNAi substantially increases the lifespan of both wild type and germlineless worms in a daf-16-dependent manner (Table 1 and data not shown). This is consistent with two recent studies reporting that inactivation of utx-1 increases worm lifespan regardless of germline proliferation, and likely acts through repression of the components of IIS (Insulin/Insulin-like growth factor Signaling) pathway by increasing H3K27me3 levels on those genes and possibly other loci (Jin et al. 2011; Maures et al. 2011). The third gene we identified encodes JMJD-2 protein, a histone demethylase that affects global H3K9me3 levels and localized H3K36me3 levels in C. elegans (Whetstine et al. 2006). Nevertheless, the physiological role of JMJD-2 is poorly understood other than that it was found to regulate cell cycle progression by changing chromatin accessibility (Black et al. 2010). It is interesting to note that jmjd-2 is the only gene identified in this screen that requires germline proliferation to affect longevity but is not affected by FUDR, which inhibits mitosis therefore gradually inhibiting progeny production. This observation and some previous studies point to multi-layer signaling from the reproduction system to influence worm lifespan (Arantes-Oliveira et al. 2002, Greer 2010).
We also identified three SET-domain containing longevity genes, including set-9, set-26, and mes-2. MES-2 is a well-known Polycomb group protein that is essential for germline development and patterning. It functions in a repressive complex to maintain X-chromosome inactivation and somatic homeotic gene repression (Strome 2005; Schaner & Kelly 2006). Our mes-2 RNAi treatment did not yield apparent developmental defects but revealed a new function of this gene in maintaining normal worm lifespan in a germline-independent manner. Interestingly, heterozygous mutants of the E(z) gene (Drosophila homolog of mes-2) live longer than control flies and show derepression of some Polycomb target genes (Siebold et al. 2010). Our results suggest an intriguing possibility that MES-2 has conserved functions in longevity in diverse species.
In addition to the RNAi treatments that extended lifespan, our screen identified 36 RNAi (out of the 50) that resulted in significantly shortened lifespan (Table S1). Interestingly, among these 36 genes, 23 of them are known to have germline-enriched expression (Kim et al. 2001) and/or functions in normal development (Wormbase). Considering the known importance of these genes in development, it is not surprising that their RNAi knockdown leads to shortened lifespan.
SET-9 and SET-26 have differential tissue expression patterns but share redundant functions in lifespan modulation
Our lifespan assays suggest that germline-expressed SET-9 shares redundant functions with germline- and soma-expressed SET-26 to determine a normal lifespan, and the small fraction of SET-26 in the somatic tissues also plays a significant role in lifespan determination (Figure 2). Because the set-9 and set-26 genes are so similar in sequences and there is only one homologue gene in the close relative C. briggsae, it is likely that set-9 and set-26 are derived from a recent gene duplication event (Woollard 2005). It will be interesting to tease out whether the different expression patterns of set-9 and set-26 contribute to some specialized biological functions, how their functions in germline and soma affect aging differently, and how they function in comparison to their homologous genes in other species during evolution.
SET-9 and SET-26 are largely but not completely dependent on daf-16 to modulate lifespan
Our daf-16 lifespan epistasis analyses suggest that SET-9 and SET-26 work through DAF-16 to modulate lifespan (Figure 3). Since daf-16 transcript levels are not affected by set-26 status in glp-1(e2141) worms (Figure S3B) and that SET-26 regulates global histone modifications (Figure 4), an interesting possibility is that SET-26 acts as a cofactor of DAF-16 in target gene regulation via changes in local chromatin accessibility. To test this, we selected twelve known DAF-16 regulated genes (McElwee et al. 2003; Murphy et al. 2003) to test their transcript levels in response to set-26(tm2467) and daf-16(mgDf47) mutations in either wild type or glp-1(e2141) background (Figure S4). The results showed that five of the genes we tested showed substantial expression changes in set-26(tm2467) mutants compared to wild-type worms. However, their expression changes appeared to be independent of daf-16 status, as they showed similar down-regulation in both set-26(tm2467) and daf-16(mgDf47);set-26(tm2467) mutants. It is important to note that SET-26 may impact only a subset of DAF-16 targets and we may not have examined the appropriate DAF-16 target genes. Future genome-wide expression profiling will provide a clearer picture of whether and how SET-26 regulates DAF-16-mediated gene transcription.
It is also possible that, like UTX-1, SET-9 and SET-26 modulate lifespan by regulating expression of components of daf-16-dependent longevity pathways (Jin et al. 2011). Several well-known longevity pathways impinge on daf-16, including the daf-2/insulin pathway and the germline pathway (Kenyon et al. 1993; Hsin & Kenyon 1999). From our current study, the germline pathway is not likely to be involved as both set-9/26 RNAi and set-26 mutation are able to significantly extend worm lifespan in the germlineless mutant worms (Figure 2). It will be interesting to test whether set-9/26 genetically interacts with other known daf-16 dependent longevity pathways in the future.
Age-dependent decline of somatic SET-26
Even though SET-9 and SET-26 proteins may not have direct methyltransferase activity (Qian & Zhou 2006), their nuclear localization and effects on histone modifications suggest that they do have critical roles in affecting chromatin status. Indeed, they may act similarly to MLL5, the closest homolog of SET-9/26 in mammals, to affect chromatin structure through interactions with other chromatin modifiers (Sebastian et al. 2009). Interestingly, both worm SET-9/26 and mammalian MLL5 also contain a protein-protein interaction domain, PHD (plant homeodomain). Emerging studies have shown that PHD domains in some disease-linked chromatin remodeling factors can distinguish and bind certain forms of methylated histones to affect gene expression (Baker et al. 2008). It would be interesting to find out whether SET-9/26 and MLL5 can interpret some methylated histone status and associate with other regulators accordingly to affect local chromatin structure.
In our study, the levels of SET-26 protein in the soma decrease dramatically as worms age, reminiscent of the significant decline in abundance of some chromatin regulators (Pegoraro et al. 2009). We speculate that a dramatic age-dependent diminution in the levels of key chromatin modifiers leads to a more relaxed chromatin regulatory environment, which likely results in an increase in aging-associated genome instability. This is consistent with a previous study reporting that RNAi knockdown of set-9 and set-26 increased the rate of reporter gene frameshift (Pothof et al. 2003). We hypothesize that the genotoxic stress resulted by set-9/26 inactivation can trigger compensatory mechanisms that help promote chromatin health and therefore increase lifespan. This is similar to the hormesis responses from mild heat and oxidative stresses that can protect against later severe stresses to prolong worm lifespan (Lithgow et al. 1995; Cypser & Johnson 2002).
Inactivation of somatic SET-26 increases the histone H3 protein level
Similar to previous reports, we observed a clear age-dependent reduction of the levels of histone H3 protein (Maures et al. 2011). This is reminiscent of a recent report that histone protein levels decline during yeast replicative aging and that over-expression of some of the core histones increases the yeast replicative lifespan (Feser et al. 2010). Interestingly, we repeatedly observed a moderate increase in histone H3 protein level in the young glp-1(e2141);set-26(tm2467) versus glp-1(e2141) mutant worms. This suggests that loss of somatic SET-26 in non-dividing tissues directly or indirectly facilitates the accumulation of steady state levels of histone proteins, which might help promote chromatin maintenance and hence longevity. It will be informative to test whether over-expression of histone proteins in the adult worm soma can promote longer lifespan.
Somatic SET-26 affects the levels of repressive histone modifications in an age-dependent manner
We detected a strong increase in the levels of the open chromatin marker acetylated histone H4 in the aging adult worm soma. This finding suggests age-dependent opening of chromatin in worms, which might be related to the observation in vertebrates where DNA methylation, a repressive marker, decreases globally with age (Singhal et al. 1987; Wilson et al. 1987). On the same note, we also found clear decrease in the levels of two repressive histone marks, H3K9me3 and H3K27me3, during adult worm aging. Similar age-dependent trend of the two marks have been observed previously in worms and mammalian tissues (Sarg et al. 2002; Maures et al. 2011). These observations clearly indicate a global decrease of heterochromatin formation and gene repression with age, hence overall genome instability.
Interestingly, set-26(tm2467) alone also leads to a decrease in the levels of the repressive H3K9me3 mark in young worms, suggesting a decrease in heterochromatin formation (Figure 4). This observation might be related to earlier findings that inactivation of set-9/26 leads to increased genome instability (Pothof et al. 2003). However, in the glp-1(e2141);set-26(tm2467) double mutant, we observed no further age-dependent decrease in the levels of the two repressive gene marks, H3K9me3 and H3K27me3, when compared to glp-1(e2141) single mutant worms. Taking altogether, we hypothesize that set-26(-) mutation induces mild DNA damage stress early on to activate a hormesis mechanism that elicitsbeneficial responses, including elevation of histone H3 protein level and prevention/minimization of the deleterious loss of chromatin compaction through histone modifications, to restore genome stability and the youthfulness of the worms. It will be enlightening to find out what players are downstream of set-26(-) to slow down the aging process. In addition, we only assayed for global changes in histone H3 protein level and histone modifications in this study. In the future, it will be important and informative to perform genome-wide studies to map the localization of histone occupancy and redistribution of different histone modifications, so as to reveal locus-specific changes with age.
In summary, our studies revealed several new longevity genes that link various histone modifications to lifespan modulation. Additional characterization of two SET-domain containing genes, set-9 and set-26, further confirms the notion that global epigenetic status and genome stability affect organismal aging. These results have important implications for future development of interventions that can alter global chromatin structure to promote healthy aging.
Experimental procedure
Lifespan assays
Lifespan assays were performed similarly as described (Rizki et al. 2011). Details can be found in Supporting Information.
Immunostaining
Worm fixation and immunostaining were performed as previously described (Li et al. 2008) and see details in Supporting Information.
Immunoblotting and Quantification
Worm sample preparation was described in Supporting Information. The enhanced chemiluminescence was captured using ChemiDoc XRST (Bio-Rad, CA) and quantified using Image Lab Software.
Supplementary Material
Acknowledgments
We thank members of the Lee, Kemphues, and Liu Labs for insightful discussions and advice. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). We are grateful to Dr. Mitani (NBRP, Japan) and Dr. Ruvkun for providing some of the worm strains required for this study. We appreciate the discussion with Dr. Anderson about the set-9 mutant. We thank Ali Awan for help with SET-domain analysis. SSL is supported by the Senior Scholar Award in Aging from the Ellison Medical Foundation, the Glenn Award for Research in Biological Mechanisms of Aging and R01 grant AG024425 from the NIA. ZN was partially supported by a postdoctoral fellowship from American Heart Association.
Footnotes
Author contributions ZN and SSL conceived this project. ZN performed most of the experiments and data analyses. AE helped generate some of the transgenic worms and performed some lifespan experiments. EA helped with some strain crosses and performed some lifespan experiments. ZN and SSL wrote the paper.
Supporting information Additional supporting information may be found in the online version of this article:
References
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- Andersen EC, Horvitz HR. Two C. elegans histone methyltransferases repress lin-3 EGF transcription to inhibit vulval development. Development. 2007;134:2991–2999. doi: 10.1242/dev.009373. [DOI] [PubMed] [Google Scholar]
- Antebi A. Genetics of aging in Caenorhabditis elegans. PLoS Genet. 2007;3:1565–1571. doi: 10.1371/journal.pgen.0030129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin J, Kimble J. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell. 1987;51:589–599. doi: 10.1016/0092-8674(87)90128-0. [DOI] [PubMed] [Google Scholar]
- Baker LA, Allis CD, Wang GG. PHD fingers in human diseases: disorders arising from misinterpreting epigenetic marks. Mutat Res. 2008;647:3–12. doi: 10.1016/j.mrfmmm.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black JC, Allen A, Van Rechem C, Forbes E, Longworth M, Tschop K, Rinehart C, Quiton J, Walsh R, Smallwood A, Dyson NJ, Whetstine JR. Conserved antagonism between JMJD2A/KDM4A and HP1gamma during cell cycle progression. Mol Cell. 2010;40:736–748. doi: 10.1016/j.molcel.2010.11.008. [DOI] [PubMed] [Google Scholar]
- Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvanese V, Lara E, Kahn A, Fraga MF. The role of epigenetics in aging and age-related diseases. Ageing Res Rev. 2009;8:268–276. doi: 10.1016/j.arr.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Cypser JR, Johnson TE. Multiple stressors in Caenorhabditis elegans induce stress hormesis and extended longevity. J Gerontol A Biol Sci Med Sci. 2002;57:B109–114. doi: 10.1093/gerona/57.3.b109. [DOI] [PubMed] [Google Scholar]
- Dillon SC, Zhang X, Trievel RC, Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6:227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feser J, Truong D, Das C, Carson JJ, Kieft J, Harkness T, Tyler JK. Elevated histone expression promotes life span extension. Mol Cell. 2010;39:724–735. doi: 10.1016/j.molcel.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Maures TJ, Hauswirth AG, Green EM, Leeman DS, Maro GS, Han S, Banko MR, Gozani O, Brunet A. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature. 2010;466:383–387. doi: 10.1038/nature09195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton B, Dong Y, Shindo M, Liu W, Odell I, Ruvkun G, Lee SS. A systematic RNAi screen for longevity genes in C. elegans. Genes Dev. 2005;19:1544–1555. doi: 10.1101/gad.1308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbes TR, Thorne AW, Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci U S A. 2007;104:18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin H, Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature. 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- Jin C, Li J, Green CD, Yu X, Tang X, Han D, Xian B, Wang D, Huang X, Cao X, Yan Z, Hou L, Liu J, Shukeir N, Khaitovich P, Chen CD, Zhang H, Jenuwein T, Han JD. Histone demethylase UTX-1 regulates C. elegans life span by targeting the insulin/IGF-1 signaling pathway. Cell Metab. 2011;14:161–172. doi: 10.1016/j.cmet.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kim SK, Lund J, Kiraly M, Duke K, Jiang M, Stuart JM, Eizinger A, Wylie BN, Davidson GS. A gene expression map for Caenorhabditis elegans. Science. 2001;293:2087–2092. doi: 10.1126/science.1061603. [DOI] [PubMed] [Google Scholar]
- Kimble J, Crittenden SL. Germline proliferation and its control. WormBook. 2005:1–14. doi: 10.1895/wormbook.1.13.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- Li J, Ebata A, Dong Y, Rizki G, Iwata T, Lee SS. Caenorhabditis elegans HCF-1 functions in longevity maintenance as a DAF-16 regulator. PLoS Biol. 2008;6:e233. doi: 10.1371/journal.pbio.0060233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Kelly WG. A role for Set1/MLL-related components in epigenetic regulation of the Caenorhabditis elegans germ line. PLoS Genet. 2011;7:e1001349. doi: 10.1371/journal.pgen.1001349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lithgow GJ, White TM, Melov S, Johnson TE. Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc Natl Acad Sci U S A. 1995;92:7540–7544. doi: 10.1073/pnas.92.16.7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maures TJ, Greer EL, Hauswirth AG, Brunet A. The H3K27 demethylase UTX-1 regulates C. elegans lifespan in a germline-independent, insulin-dependent manner. Aging Cell. 2011 doi: 10.1111/j.1474-9726.2011.00738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee J, Bubb K, Thomas JH. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell. 2003;2:111–121. doi: 10.1046/j.1474-9728.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- Mitchell DH, Stiles JW, Santelli J, Sanadi DR. Synchronous growth and aging of Caenorhabditis elegans in the presence of fluorodeoxyuridine. J Gerontol. 1979;34:28–36. doi: 10.1093/geronj/34.1.28. [DOI] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Ni Z, Lee SS. RNAi screens to identify components of gene networks that modulate aging in Caenorhabditis elegans. Brief Funct Genomics. 2010;9:53–64. doi: 10.1093/bfgp/elp051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegoraro G, Kubben N, Wickert U, Gohler H, Hoffmann K, Misteli T. Ageing-related chromatin defects through loss of the NURD complex. Nat Cell Biol. 2009;11:1261–1267. doi: 10.1038/ncb1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pothof J, van Haaften G, Thijssen K, Kamath RS, Fraser AG, Ahringer J, Plasterk RH, Tijsterman M. Identification of genes that protect the C. elegans genome against mutations by genome-wide RNAi. Genes Dev. 2003;17:443–448. doi: 10.1101/gad.1060703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian C, Zhou MM. SET domain protein lysine methyltransferases: Structure, specificity and catalysis. Cell Mol Life Sci. 2006;63:2755–2763. doi: 10.1007/s00018-006-6274-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizki G, Iwata TN, Li J, Riedel CG, Picard CL, Jan M, Murphy CT, Lee SS. The Evolutionarily Conserved Longevity Determinants HCF-1 and SIR-2.1/SIRT1 Collaborate to Regulate DAF-16/FOXO. PLoS Genet. 2011;7:e1002235. doi: 10.1371/journal.pgen.1002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarg B, Koutzamani E, Helliger W, Rundquist I, Lindner HH. Postsynthetic trimethylation of histone H4 at lysine 20 in mammalian tissues is associated with aging. J Biol Chem. 2002;277:39195–39201. doi: 10.1074/jbc.M205166200. [DOI] [PubMed] [Google Scholar]
- Schaner CE, Kelly WG. Germline chromatin. WormBook. 2006:1–14. doi: 10.1895/wormbook.1.73.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian S, Sreenivas P, Sambasivan R, Cheedipudi S, Kandalla P, Pavlath GK, Dhawan J. MLL5, a trithorax homolog, indirectly regulates H3K4 methylation, represses cyclin A2 expression, and promotes myogenic differentiation. Proc Natl Acad Sci U S A. 2009;106:4719–4724. doi: 10.1073/pnas.0807136106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebold AP, Banerjee R, Tie F, Kiss DL, Moskowitz J, Harte PJ. Polycomb Repressive Complex 2 and Trithorax modulate Drosophila longevity and stress resistance. Proc Natl Acad Sci U S A. 2010;107:169–174. doi: 10.1073/pnas.0907739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal RP, Mays-Hoopes LL, Eichhorn GL. DNA methylation in aging of mice. Mech Ageing Dev. 1987;41:199–210. doi: 10.1016/0047-6374(87)90040-6. [DOI] [PubMed] [Google Scholar]
- Strome S. Specification of the germ line. WormBook. 2005:1–10. doi: 10.1895/wormbook.1.9.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, Shi Y. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Wilson VL, Smith RA, Ma S, Cutler RG. Genomic 5-methyldeoxycytidine decreases with age. J Biol Chem. 1987;262:9948–9951. [PubMed] [Google Scholar]
- Woollard A. Gene duplications and genetic redundancy in C. elegans. WormBook. 2005:1–6. doi: 10.1895/wormbook.1.2.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.