Abstract
Protein methyltransferases (PMTs) play various physiological and pathological roles through methylating histone and nonhistone targets. However, most PMTs including more than 60 human PMTs remain to be fully characterized. The current approaches to elucidate the functions of PMTs have been diversified by many emerging chemical biology technologies. This review focuses on progress in these aspects and is organized into four discussion modules (assays, substrates, cofactors and inhibitors) that are important to elucidate biological functions of PMTs. These modules are expected to provide general guidance and present emerging methods for researchers to select and combine suitable PMT-activity assays, well-defined substrates, novel SAM surrogates and PMT inhibitors to interrogate PMTs.
Keywords: epigenetic, cancer, posttranslational modification, transferase enzymes, PRMT, PKMT, protein methylation, bioorthorgonal, high throughput screening (HTS)
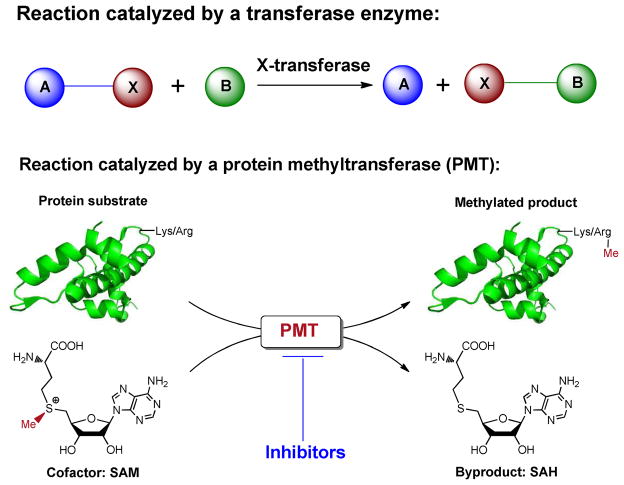
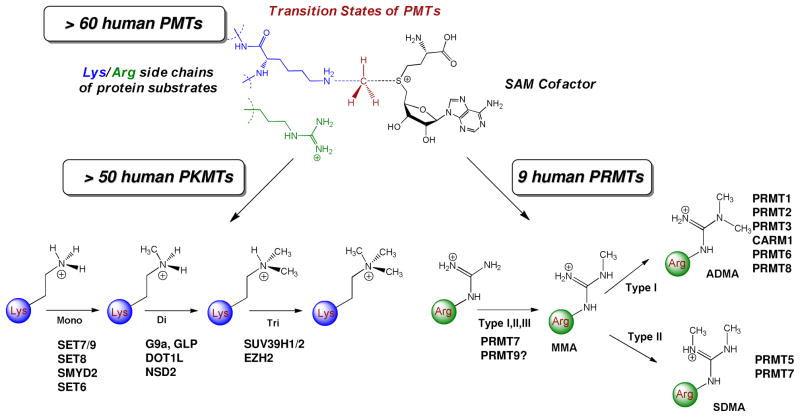
According to biochemical reactions and enzyme numerical classification (EC number), protein methyltransferases (PMTs), together with acetyltransferases, glycosyltransferases and kinases, belong to the family of transferase enzymes (EC 2). The common feature of these enzymes is to transfer a functional group from a donor (cofactor or coenzyme) to an acceptor (Fig. 1). For PMTs, the cofactor and acceptor are S-adenosylmethionine (SAM) and lysine or arginine side chains of protein substrates, respectively (Fig. 1). The human genome encodes more than 60 PMTs including 9 known protein arginine methyltransferases (PRMTs) and > 50 protein lysine methyltransferases (PKMTs) (Fig. 2).1
Figure 1.
Reactions catalyzed by transferase enzymes and protein methyltransferases (PMTs). A transferase enzyme (X-transferase) has the ability to transfer a functional moiety (X) from a cofactor or coenzyme (A–X) to its substrates (B) and generate modified products (B–X) and a cofactor metabolite (A). In the case of PMTs, the functional moiety, the cofactor, substrates, products and the byproduct are a methyl group, S-adenosylmethionine (SAM), specific Lys/Arg side chains of proteins, methylated products and S-adenosylhomocysteine, respectively.
Figure 2.
Human PMTs. The human genome encodes > 60 PMTs, which diverge into protein lysine methyltransferases (PKMTs, > 50) and protein arginine methyltransferases (PRMTs, around 9). PKMTs can mono-/di-/tri-methylate their substrates in a processive or distributive manner. According to the three forms of arginine methylation products (MMA, ADMA and SDMA for monomethylarginine, asymmetric dimethylarginine and symmetric dimethylarginine, respectively), PRMTs can be categorized into three subtypes: Type I (MMA-then-ADMA products for PRMT1, 2, 3, 4, 6, 8), Type II (MMA-then-SDMA products for PRMT5, 7) and Type III (MMA product for certain targets of PRMT7). The methylation pattern of PRMT9 remains to be characterized unambiguously.
The 9-member human PRMTs (PRMT1-9) share a set of four conserved motifs (I, post-I, II and III) and the characteristic THW loop for SAM binding.2 With SAM as the methyl donor, PRMTs modify arginine’s ω-guanidino nitrogen in a target-specific manner (Fig. 2).2 The three forms of arginine methylation products (MMA, ADMA and SDMA for monomethylarginine, asymmetric dimethylarginine and symmetric dimethylarginine, respectively) further distinguish PRMTs into three subtypes (Fig. 2): Type I (MMA-then-ADMA products for PRMT1, 2, 3, 4, 6, 8), Type II (MMA-then-SDMA products for PRMT5, 7) and Type III (MMA product for certain targets of PRMT7).2 The methylation pattern of PRMT9 remains to be characterized unambiguously.2 Except DOT1L, whose catalytic domain is similar to that of PRMTs, PKMTs harbor a canonical SET domain comprised of 130 amino acids for SAM binding and enzyme catalysis.3 PKMTs methylate lysine’s ε-amino group to specific degrees: mono-, di- and tri-methylation (Fig. 2).4,5
PRMTs and PKMTs methylate histone targets.4,5 For instance, PRMT1 and CARM1 methylate arginine 3 of histone H4 (H4R3) and arginines 2/17/26 of histone H3 (H3R2/17/26), respectively.2,4,5 These events have been linked to transcriptional activation.2,4,5 In contrast, PRMT5 and PRMT6 modify H4R3 and H3R2. These methylation events are associated with transcriptional repression.2,4,5 This yin-yang-type of switch has also been observed for PKMT-involved histone methylation. For instance, tri-methylation of H3 lysine 4 (H3K4me3) and trimethylation of H3 lysine 36 (H3K36me3) and lysine 79 (H3K79me3) are the marks for active genes, whereas H3 lysine 9 di-/tri-methylation (H3K9me2/3) and H4 lysine 20 methylation (H4K20me1/2/3) are the marks for silenced genes.2,4–6
Besides histones, PMTs also methylate diverse nonhistone targets.7 The majority of PRMT substrates are nonhistone targets including transcription factors STAT1, RUNX1 and FOXO1 (PRMT1 substrates);8–10 transcription coactivators p300 and CBP (CARM1 substrates);11,12 and RNA-binding proteins (substrates of PRMTs).13 Efforts over the past decade have led to the characterization of many PKMT nonhistone substrates as well (e.g. the tumor suppressor p53 as the substrate of SET7/9, SET8, SMYD2, G9a and GLP).14–19 PMT-mediated histone and nonhistone methylation, together with other posttranslational modifications (e.g. acetylation, phosphorylation, sumolyation and ubiquitination), can regulate binding partners (activators or repressors), localization or stability of the PMT substrates.2,4,5,7 These modifications alone or in combination can modulate downstream signals in an epigenetic manner and thus render meaningful biological readouts.2,4,5,7
Apart from PMTs’ roles in normal physiology, their dysregulation has been implicated in many diseases including cancer.20 For instance, oncogenic properties of PMTs (e.g. EZH2, G9a, PRMT5, SUV39H1 and SMYD2) can rely on target methylation that destabilize or downregulate tumor suppressors.20 PMTs can also be linked to cancer through aberrant upregulation of oncogenes.20 For example, the enzymatic activities of DOT1L and PRMT1 were shown to be essential for downstream signals of mixed lineage leukemia (MLL) transcriptional complex. The constitutive recruitment of DOT1L and PRMT1 by MLL-fusion protein stimulates hematopoietic transformation.21,22 Additionally, overexpression of PMTs such as GLP, SUV39H2, NSD2, NSD3, SMYD3 and PRDM14 has been reported in many primary tumors.20 These findings further underscore the cancer relevance of PMTs.
Most PMT substrates were identified through a conventional candidate-based approach. In this approach, a proposed PMT substrate is tested against a panel of PMTs in vitro with [Me-3H]SAM as a cofactor. The radioactive methyl group is expected to be delivered to a bona fide substrate only by matched PMTs. To map the site(s) of the methylation, truncated or site-specifically-mutated substrates are then examined for either gain or loss of the methylation signal. The confirmed enzyme-substrate pair can then be validated in cellular contexts with other biochemical and genetic methods. After the methylation activities of PMT-substrate pairs were validated in vitro and in cellular contexts, their upstream and downstream events can be further pursued with accurate disease or animal models.
Although the well-established candidate-based approach demonstrated the feasibility for identifying and validating individual PMT targets, their application to proteome-wide profiling of PMT substrates is questionable. As exemplified with SET7/9, a PKMT initially characterized as a H3K4 methyltransferase, the efforts over the past decade have led to identification of a dozen of SET7/9 nonhistone substrates, such as p53, TAF10, ERα, PCAF, NF-χB, DNMT1 and HIV transactivator Tat.17,23–25 However, new SET7/9 targets keep emerging and give no sign to end the decade-long endeavor in searching SET7/9 targets.26 In addition, target-recognizing patterns of PMTs cannot be readily rationalized because of the lack of consensus sequences. These challenges emphasize the need for new tools to elucidate how PMTs recognize structurally-diverse substrates. Given the biological relevance of PMTs, it is equally important to develop tools to elucidate and manipulate the functions of PMTs in normal and disease contexts.
As chemical biology methods emerge to study transferase enzymes such as glycosyltransferases,27 kinases28 and acetyltransferases,29,30 these approaches have been proven or show potential to be transformed for PMTs. Meanwhile, PMT-catalyzed reactions have been or can be investigated with PMT-specific methods.31,32 This review focuses on providing the present status and additional perspectives on how chemical biology methods can be applied to interrogate PMTs. Given the feature of the PMT-catalyzed transferase reaction, the review is organized into four discussion modules: assays, substrates, cofactors and inhibitors. To minimize redundancy of the topics that have been covered by other excellent reviews,33,34 this article mainly deals with a collection of recently-published literature and their chemical biology aspects. I apologize for the omission of many high-quality works because of space limitation.
PMT-activity Assays
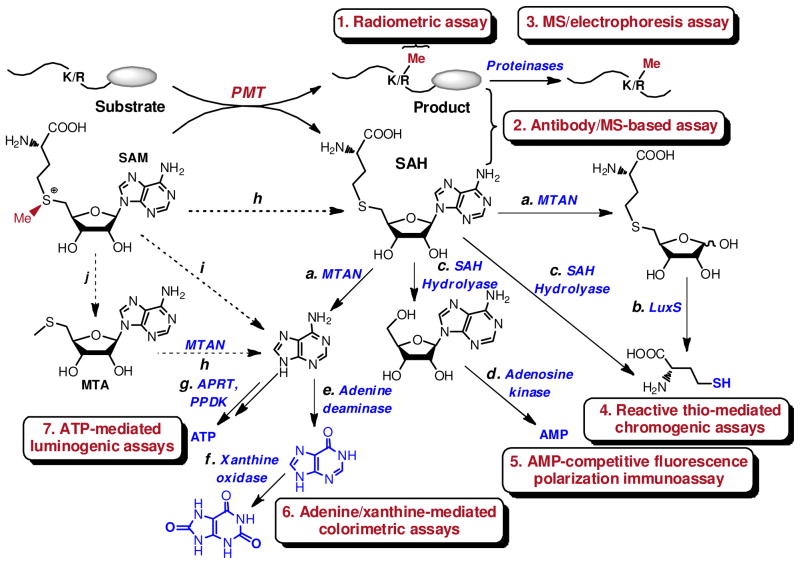
In a PMT-catalyzed methylation reaction, the substrate (peptide/protein/protein complex) and SAM will be enzymatically processed into the methylated product and the byproduct S-adenosylhomocysteine (SAH), respectively (Fig. 1). Because of PMTs’ slow enzymatic turnovers, most PMT-activity assays were developed by detecting reaction products rather than measuring depletion of starting materials. Methylated products and SAH can be quantified either directly (e.g. autoradiography, top/middle-down mass spectrometry (MS) and anti-methyllysine/arginine antibodies) or indirectly after processing them into various derivatives (e.g. enzyme-coupled colorimetric assays and shot-gun MS) (Fig. 3). The adaptability of these assays for high throughput screening (HTS) will also be discussed below.
Figure 3.
Schematic presentation of current PMT-activity assays and potential interfering factors. The current PMT-activity assays mainly rely on quantification of methylated protein products or the byproduct SAH. Methylated peptide/protein products can be quantified directly by radiometric and top-down mass spectrometric methods (Methods 1,2). The digested products of peptide/protein can be quantified by middle-down/shot-gun mass spectrometric or electrophoresis methods (Method 3). In contrast, the byproduct SAH can be quantified directly by anti-SAH antibody or MS (Methods 1,2) or indirectly by various colorimetric assays (Methods 4–7) with coupling enzymes (Pathways a–g): MTAN (5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase), LuxS (S-ribosylhomocysteine lyase), SAH hydrolyase, adenosine kinase, adenine deaminase, xanthine oxidase, APRT (adenine phosphoribosyl transferase) or PPDK (pyruvate orthophosphate dikinase). SAM can spontaneously decompose into SAH, adenine and MTA (Pathways h, i, j).
Radiometric quantification of substrate methylation (Method 1 in Fig. 3)
For PMT-catalyzed methylation, the radiolabeled methyl group, from either [Me-3H]- or [Me-14C]-SAM, can be enzymatically incorporated into PMT targets (Method 1 in Fig. 3). After removing the unreacted SAM, the enzymatically-incorporated radioactive moiety can be quantified by autoradiography or liquid scintillation counting. To separate radiolabeled products from residual SAM, the accepted practices are to use phosphocellulose filter paper to bind peptide or protein products, followed by washing and scintillation counting or SDS-PAGE separation, followed by autoradiography or gel extraction/scintillation counting.35 Although these methods are favored for their straightforward protocols as well as facile access to reagents and instruments, they are laborious (multi-step washing or transferring) and time-consuming (~ 6 h–7 d for a single run).35
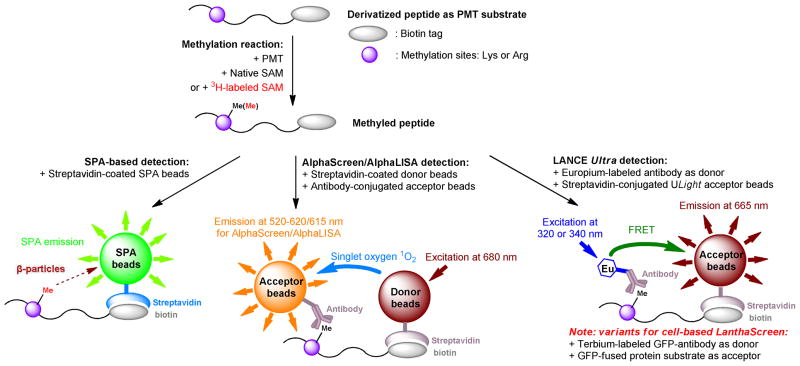
To accelerate the assay turnover, the Hevel group found that radiolabeled protein products can be readily separated from unreacted SAM with ZipTipc4 pipette tips.35 With the ZipTip protocol, the entire process can be completed within ~ 2 – 14 min. Alternatively, the Jeltsch and the Zheng laboratories adapted scintillation proximity assay (SPA) to rapidly quantify radiolabeled products.36–39 In the SPA-based assay, biotinylated peptides and [Me-3H]-labeled-SAM were used as PMT substrates and cofactor, respectively. After the [Me-3H]-labeled products were immobilized to avidin-conjugated plates or beads, the proximity between the β-particles from the immobilized 3H-labeled peptide and SPA-plate/bead-coated scintillation fluid triggered an emission of scintillation signal (Fig. 4). This SPA-based approach has been applied for measuring the activities of Dim5, G9a and PRMT1.36–39 In comparison with other radiometric methods, the homogenous SPA approach features no separation of residual radioactive SAM and is thus adaptable for a mix-and-measure HTS format (The HTS application of SPA will be discussed later).36
Figure 4.
Emerging SPA-based and antibody-based homogenous PMT-activity assays for HTS. The principles of SPA, AlphaScreen/LISA, LANCE Ultra and LanthaScreen are briefly described below. In the reported SPA-based PMT-activity assay,36 a biotinylated peptide substrate was radiolabeled by PMT with [Me-3H]-labeled-SAM and then immobilized to streptavidin-conjugated SPA beads. The proximity between the β-particles from the immobilized peptide and SPA-coated scintillation fluid led to light emission. For the reported AlphaScreen and AlphaLISA PMT-activity assays,40,41 the methylated, biotin-tagged peptide product caused the proximity between streptavidin-coated donor beads and anti-methyllysine antibody-conjugated acceptor beads (or primary antibody-immunized secondary-antibody-conjugated acceptor beads). Exciting the donor beads at 680 nm led to emitting singlet oxygen (1O2), which excited the acceptor beads to generate emission at 520–620 nm (AlphaScreen) or at 615 nm (AlphaLISA). For the reported LANCE Ultra and LanthaScreen PMT-activity assays,42,43 europium/terbium-labeled anti-methyllysine antibodies were used as FRET donors and streptavidin-conjugated Ulight (LANCE Ultra) or GFP-fused protein (LanthaScreen) as FRET acceptors. Exciting the donors at 320 nm or 340 nm led to FRET-mediated light emission of the acceptors.
Antibody-based detection of substrate methylation (Method 2 in Fig. 3)
Although radiometric assays are often used to study PMTs, their radioactive format is not environmentally friendly. In addition, positive radioactive signals only report the methylation activity, but not the degree of methylation (e.g. MMA/ADMA/SDMA for PRMTs or mono-/di-/tri-methylation for PKMTs). However, these limitations can be addressed by antibody-based PMT-activity assays. Diverse primary monoclonal or polyclonal antibodies are available to recognize specific methylation epitopes for Western blot, CHIP, CHIP-on-chip and CHIP-seq analysis.6
In conjunction with several recent technologies, such as AlphaScreen (PerkinElmer), AlphaLISA (PerkinElmer), LANCE Ultra (PerkinElmer) and LanthaScreen (Invitrogen), anti-methyllysine(arginine) antibodies have demonstrated their use in homogeneous PMT-activity assays (Fig. 4).40–43 These assays share a similar principle by pairing a PMT substrate and an anti-methyllysine antibody with donor and acceptor dyes (Fig. 4).40–43 The anticipated interaction between the methylated product and the antibody brings the donor and acceptor dyes in a proximity. The excitation of the donor dye then leads to emission of the acceptor dye through either singlet oxygen (1O2) (AlphaScreen and AlphaLISA) or time-resolved fluorescence resonance energy transfer (TR-FRET) (LANCE Ultra and LanthaScreen).40–43 As the first application of PMTs, Quinn et. al. reported chemiluminescence AlphaScreen immunoassay technology, combined with a polyclonal anti-methyl-H3K9 antibody, to examine G9a-catalyzed H3K9 methylation.40 Gauthier et. al. and Hauser et. al. then developed an antibody-based AlphaLISA approach to monitor SET7/9-catalyzed H3K4 methylation and PRMT1-catalyzed H4R3 methylation, respectively.41,42 Gauthier et. al. also demonstrated a similar application combining LANCE Ultra technology and a europium-labeled anti-methyllysine antibody.41 With terbium-labeled anti-methyl H3K9 antibody (the donor) and GFP-fused histone H3 (the acceptor), Machleidt et. al. for the first time developed a LanthaScreen TR-FRET approach to visualize H3K9 dimethylation in cellular contexts.43 The merit of these antibody-based homogeneous assays lies in their adaptability for HTS as discussed later.
Though the antibody-based approaches have the merit for the ready readouts, the specificity of the antibodies and the dynamic range of epitope concentrations need to be well defined prior to their use in PMT-activity assays. Given the general narrow range of the latter, the antibody-based PMT-activity assays are not suitable to measure quantitative data such as Km and kcat (the PMT-activity assays for this purpose will be discussed later).
MS-based detection of intact protein samples (Method 2 in Fig. 3)
When small peptides are used as PMT substrates, the reaction products can be analyzed by MS after simple workup.31,32 The level of methylation is directly reflected by corresponding mass shifts (e.g. +14 Da for mono- or + 28 Da for di-methylation). Since lysine/arginine methylation does not significantly alter the size and the charge of substrates, the peak ratio between unmodified and modified peptides is sometimes used for direct quantification.31,32,44 When small-to-medium-size proteins (e.g. histones) are examined as PMT substrates, top-down MS is often used to monitor the level of methylation (e.g. mono-/di-/tri) as well as map the site(s) of methylation. Combining top-down MS with metabolic labeling, Pesavento et. al. were able to monitor cell-cycle-dependent dynamics of H4K20 methylation. Their work revealed that H4K20 methylation progressively accumulates on newly-translated histones during G2, M and G1 phases, and reaches to a maximal degree within 2 ~ 3 cell cycles.45 Combining top-down MS with heavy methyl-SILAC labeling, the Garcia laboratory was able to analyze systematically in vivo dynamics of multiple histone lysine and argnine methylations and showed that active-gene-associated histones are methylated faster than silenced-gene-associated histones.46 A key advantage of using intact peptide/protein samples for MS analysis is the ability to unambiguously detect the methylation(s) together with other posttranslational modifications on a single target.
PMT-activity assays using digested protein samples (Method 3 in Fig. 3)
Top-down MS approach is largely limited to small-size, high-quality protein samples such as histones.46 In contrast, middle-down/shot-gun MS using digested protein samples is more generally applicable. One general application of middle-down/shot-gun MS analysis is to map protein methylation sites. For example, after confirming SMYD2’s activity on pRb with a radiometric assay, Addict et. al. were able to rely on the shotgun MS and tandem MS (MS/MS) approach to conclude readily that the methylation occurs at Lys 860 but not at adjacent Lys 844/847.47 The La Thangue laboratory was able to use the same approach to identify Lys 810 of pRb as the methylation site for SET7/9.48 Compared with the laborious radiometric approach with truncated or site-specifically-mutated proteins to map protein methylation, the shotgun proteomic approach avoids the need to test multiple samples and thus significantly simplifies the mapping process (Method 3 in Fig. 3).
Although peptide samples are generally subject to MS analysis without protease digestion, the Janzen laboratory reported a microfluidic capillary electrophoresis using endoproteinase-digested peptides to quantify PMT-catalyzed reactions (Method 3 in Fig. 3).49 The authors relied on a methylation-sensitive endoproteinase, which cleaves unmethylated peptide but not methylated peptide.49 The resultant digested (unmethylated substrate) peptide and undigested peptide (methylated product) were resolved by microfluidic capillary electrophoresis according to their different charge-to-mass ratios. With G9a as a model PMT, the authors demonstrated that the approach is highly quantitative and suitable for characterizing the kinetics of PMT-catalyzed reactions.49
PRMTs generate three types of arginine methylation products (MMA, ADMA and SDMA) (Fig. 2).2 To distinguish the three types of products, [Me-3H/14C]-SAM-labeled substrate samples can be subjected to acid hydrolysis to yield MMA, ADMA and SDMA amino acids, which can be further characterized by column/thin-layer chromatography or MS analysis. With the acid-hydrolysis approach, Branscombe et. al. and Lee et. al. were able to detect the SDMA products of PRMT5 and PRMT7, and categorized the two enzymes as Type II PRMTs.50,51 With the same approach, the Frankel laboratory was able to experimentally define PRMT2 as a Type I PRMT.52 The Wang laboratory further demonstrated a MALDI-TOF MS/MS approach to differentiate MMA, ADMA and SDMA at the peptidic level.53 The MMA-, ADMA- and SDMA-containing peptides showed characteristic neutral losses of (−56 Da, −31 Da), (−45 Da) and (−70 Da, −30 Da), respectively.53
Direct Quantification of SAH with MS or ANTI-SAH antibody (Method 2 in Fig. 3)
MS- and antibody-based approaches have also been used to measure the byproduct SAH in PMT-catalyzed reactions (Method 2 in Fig. 3). The Frankel lab reported a tandem MS/MS approach to quantify SAH. 54 With this assay, they were able to quantify the sources causing SAH background in PRMT1-catalyzed reactions and concluded that, besides the SAH from the contamination in commercial SAM and from SAM’s nonenzymatic decomposition (Pathway h in Fig. 3), automethylation of PRMT1 accounts for a portion of the observed SAH background.54
The byproduct SAH in PMT-catalyzed reactions can also be quantified by antibody-based competitive assays (Method 2 in Fig. 3). Capdevila et. al. first reported a competitive immunoassay using SAH-BSA conjugate and anti-SAH antibody (~ 150-fold preference to SAH over SAM) to quantify SAH in plasma.55 In this assay, SAH competes with microplate-coated SAH-BSA to bind anti-SAH antibody and thus reduces ELISA signal from the microplate-immobilized antibody. Graves et. al. developed a similar competitive assay with fluorescein-SAH and anti-SAH antibody.56 In Graves’s approach, SAH is quantified by competing fluorescein-SAH to bind the antibody and thus cause the loss of fluorescence polarization signal. The assay has demonstrated its feasibility for catechol-O-methyltransferase and is likely applicable to PMTs, given their shared byproduct SAH.56 However, one should be cautious to use the SAH-based fluorescence polarization because the readout is linear only in a narrow range of SAH concentration (10 nM – 500 nM).56
PMT-activity assays through SAH derivatives (Methods 4–7 in Fig. 3)
Many SAH-based quantification assays were developed for small-molecule methyltransferases such as salicylic acid methyltransferase57 and catechol-O-methyltransferase.58 The Zhou laboratory reported an enzyme-coupled chromogenic assay for salicylic acid methyltransferase.57 This assay relied on two coupling enzymes MTAN (5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase) and LuxS (S-ribosylhomocysteine lyase) to convert SAH into homocysteine (Method 4, Pathway a+b in Fig. 3). Homocysteine can then be quantified with Ellman’s reagent (UV change at 412 nm). The Hrycyna laboratory reported a comparable fluorogenic assay for catechol-O-methyltransferase (Method 4, Pathway c in Fig. 3).58 This assay relies on the coupling enzyme SAH hydrolase to process SAH into homocysteine, which is then quantified by a free-thiol-activated dye fluorescein-cystamine-methyl red. The Trievel laboratory developed the first SAH-based quantification assay for PMTs.59 Although Trievel’s assay also relied on SAH hydrolase as a coupling enzyme (Method 4, Pathway c in Fig. 3), it was improved by using a more sensitive free-thiol-reactive dye ThioGlo 1 for better signal and a cysteine-free SAH hydrolase for lower background.59 Our laboratory noticed that replacing ThioGlo 1 with another dye, 7-diethylamino-3-(4′-maleimidylphenyl)-4-methylcoumarin, further improves signal-to-noise separation.60 In comparison with the radiometric, antibody- or MS-based assays as reviewed above, most SAH-based chromogenic assays are valuable because of their capacity to tolerate a broad concentration range of PMT substrates and cofactors, and thus are more suitable for measuring the kinetics of PMTs (e.g. kcat and Km).59,60
To enhance the detection threshold of SAH-based quantification assays, our laboratory developed an ultrasensitive luminescence assay (Method 7, Pathway a+g in Fig. 3).60 In this assay, SAH is sequentially converted into adenine, adenosine monophosphate 61, and then adenosine 5′-triphosphate (ATP) by three coupling enzymes: MTAN, adenine phosphoribosyl transferase and pyruvate orthophosphate dikinase. The resultant ATP is quantified with a sensitive luciferin/luciferase kit. This assay is ultrasensitive and is able to detect 0.3 pmol of SAH (30-fold better than prior SAH-based colorimetric assays) and has been validated by measuring the kinetics of SET7/9.60 To adapt a SAH-based colorimetric assay in a continuous format, the Hevel laboratory used MTAN and adenine deaminase as coupling enzymes to convert SAH into hypoxanthine (Method 6, Pathway a+e in Fig. 3).62 The amount of SAH was then quantified by the change of the UV absorption at 265 nm. The authors demonstrated the merit of the continuous assay by determining the kinetic parameters of PRMT1. G-Biosciences commercialized a methyltransferase assay kit with three coupling enzymes: MTAN, adenine deaminase and xanthine oxidase to convert SAH into highly-chromogenic xanthine derivatives (Method 6, Pathway a+e+f in Fig. 3). This format is an extended version of Hevel’s continuous assay and is expected to be applicable to other PMTs, given that the byproduct SAH is shared by all SAM-dependent methyltransferases (Compare Pathway a+e with Pathway a+e+f for Method 6 in Fig. 3). Klink et. al. developed another generic PMT assay by converting SAH into adenosine and then AMP by two coupling enzymes SAH hydrolase and adenosine kinase (Method 5, Pathway c+d in Fig. 3).63 The resultant AMP can be quantified by Transcreener AMP/GMP assay kit (TranscBellBrook Labs). As will be discussed later, the assay was developed in a HTS format.
To compare SAH-dependent chromogenic PMT-activity assays, several interfering factors should be considered (Pathways h, i, j in Fig. 3). The cofactor SAM can decompose spontaneously through three main pathways (Pathways h, i, j in Fig. 3): hydrolysis of methyl-sulfonium bond to SAH, cleavage of N-ribosyl bond to adenine and intramolecular SN2 lactonization to methylthioadenosine (MTA).60 The SAM-to-SAH decomposition can interfere with all SAH-mediated PMT-activity assays (Pathways h, i in Fig. 3).54,60,64 The Frankel laboratory found that this degradation occurs at a slow rate and its effect can be mitigated by using Tris buffer rather than Hepes buffer and freshly-purified SAM. 54 SAM’s degradation also affects the PMT-activity assays that rely on MTAN as one coupling enzyme and adenine or its derivatives as readouts. Since MTAN is promiscuous toward SAH and MTA, all nonenzymatic SAM-degrading products (SAH, MTA and adenine) will contribute signal readouts as enzymatic adenine production (Pathways h+a, i, j+h in Fig. 3).64 With the ATP-mediated luminogenic assay as a model, our laboratory evaluated the effect of three SAM-degrading products and found that SAH, MTA and adenine together gave 2-fold higher background than SAH alone.64 The spontaneous decomposition of SAM to SAH, MTA and adenine therefore restricts the use of the SAH-dependent chromogenic assays for PMTs of low-activity.
In many SAH-based chromogenic assays, SAH is degraded in situ by coupling enzymes (Pathways a,c in Fig. 3). The lack of accumulation of SAH is expected to be beneficial by releasing potential SAH inhibition of PMTs. However, our laboratory showed that SAH-based chromogenic assays can be carried out in an uncoupled format by allowing SAH accumulation followed by SAH quantification.64 The potential SAH inhibition won’t be dominant if the examined PMTs have low affinity to SAH or a high concentration of SAM is used.64 In addition, reactive-thiol-based chromogenic PMT-activity assays should be carried out under conditions free of reducing reagents such as DTT and β-mercaptoethanol, because these reagents interfere with the assays by reacting with the dyes directly (Method 4 in Fig. 3). Cysteines of PMTs and coupling enzymes are another source of high background in reactive-thiol-based PMT-activity assays. This effect can be minimized by using cysteine-free coupling enzymes.59
HTS adaptability of PMT-activity assays
PMT-activity assays have caught increasing attention for their potential medium/high throughput screening of PMT inhibitors (these inhibitors will be discussed later). As an early effort toward HTS of PRMT inhibitors, the Bedford laboratory formulated an antibody-based ELISA PMT-activity assay and applied it to identify a suite of PRMT inhibitors (e.g. AMI1, 5, 6, 9, 18) from a 9,000-compound library;65 the Imhof laboratory applied a radiometric filter-binding assay to a pooled mixture of 2,976 compounds (eight compounds per pool) and identified an SU(VAR)3-9 inhibitor chaetocin;66 Purandare et. al. developed a similar radiometric filter-binding assay and identified a pyrazole-based CARM1 inhibitor.67 The medium throughput format of these assays, though feasible for a small library of compounds, is not efficient to handle current HTS compound libraries, which generally contain > 100K entities.
Kubicek et. al. developed the first HTS assay for PMTs (Fig. 4).68 In this dissociation enhanced lanthanide fluoroimmunoassay (DELFIA), N-terminal biotinylated H3 1–20 amino-acid peptide was dimethylated by G9a at H3K9 and then immobilized onto a neuroavidin-coated 384-well microtiter plane. After multiple-step washing, the microtiter-plate-immobilized H3Kme2-epitopes were probed by primary rabbit α-H3Kme2 antibody followed by secondary europium-labeled goat α-rabbit antibody, which has characteristic fluorescence emission at 620 nm. The hits were identified by observing the loss of the signals. After screening a library of 125K compounds, Kubicek et. al. identified seven G9a inhibitors including BIX-01294 (the inhibitor will be discussed later).68
The so-far reviewed medium-to-high throughput PMT assays, though feasible for compound screening, require multiple-step washing and therefore have certain limitations for a broader application. The PMT-activity assays in a homogenous mix-and-measure format have their merit in HTS automation (Fig. 4).36,40–43,63 The new technologies such as AlphaScreen, AlphaLISA, LANCE Ultra and LanthaScreen (discussed above) have been explored as potential HTS platforms for PMTs (Fig. 4).41–43 A key statistical parameter of their HTS adaptability is to evaluate signal-to-background separation by Z′ factors (Z′ = 1 − [(3δ+ + 3δ−)/(μ+ − μ−)], where δ+, δ−, μ+ and μ− are denoted for standard deviations (δ) and average values (μ) for the high (+, the maximal signal) and low (−, the background signal) controls, respectively).64 Assays with Z′ value greater than 0.5 are suitable for HTS. Gauthier et. al. and Machleidt et. al. evaluated the Z′ factors of AlphaLISA for in vitro SET7/9-catalyzed H3K4 monomethylation and LanthaScreen TR-FRET assay for cellular H3K9 dimethylation, respectively (discussed above).41,43 The excellent Z′ values (> 0.7) of both the assays demonstrated their HTS adaptability. Klink et. al. also measured the Z− of their AMP-competitive fluorescence polarization immunoassay (discussed above).63 Although it only has a modest Z− of 0.59, the assay has merit in being generic for multiple PMTs by quantifying SAH-derivatized AMP (Fig. 3). HTS adaptability of other PMT assays remains to be evaluated.
Another major consideration for HTS adaptability is a low false-positive hit rate. In the course of searching for SMYD2 inhibitors, Ferguson et. al. developed an AlphaScreen HTS PMT assay (Fig. 4).69 The authors point out that the AlphaScreen assay intrinsically has a high false-positive hit rate. The false-positive hit rates in enzyme-coupled PMT-activity assays are also expected to be high, given potential false inhibition of coupling enzymes. To rapidly triage false-positive hits, a secondary orthogonal assay is necessary. Ferguson et. al. described a radiometric SPA-based approach as a robust secondary assay to validate the hits of SMYD2 after the primary AlphaScreen.69 To identify PRMT1 inhibitors, the Zheng laboratory independently reported the feasibility of using the radiometric SPA approach as a primary HTS assay.36 The radiometric SPA HTS is expected to be robust because of its simple detection format by involving only radiolabeled SAM, biotinylated substrate, a PMT and streptavidin-coated SPA beads (Fig. 4). In terms of reagents, the SPA HTS approach is more generic in comparison with the antibody-based HTS assays because the latter require high-quality antibodies in individual assays (Fig. 4). However, the SPA approach, which generates radioactive wastes, can raise environmental concerns given the amount of radioactive SAM needed in any typical HTS of 100 ~ 500K compounds. The HTS merits of the radiometric SPA approach versus antibody-based or coupling-enzyme-based assays therefore need to be evaluated case by case.
General guidance in selecting PMT-activity assays
With so many PMT-activity assays available, general guidelines may help select PMT-activity assays for specific research purposes. Here I summarized the Rule of Six followed by our laboratory as a quick reference: (1) use filter-radiometric binding/scintillation counting or SDS-PAGE/autoradiography assays to demonstrate and validate new PMT activities; (2) apply top-down/middle-down/shotgun MS analysis to map methylation sites (straightforward). Otherwise use the radiometric assays for this purpose; (3) develop sequence-specific anti-methyllysine/arginine antibodies or quantitative MS approach to probe cell-based methylation events; (4) use SAH-based MS or colorimetric assays to measure kinetics of high-turnover PMTs; (5) use radiometric medium-throughput PMT-activity assays to measure kinetics of low-turnover PMTs; (6) apply mix-and-measure homogenous SPA or antibody-based assays for HTS.
Substrates of PMTs
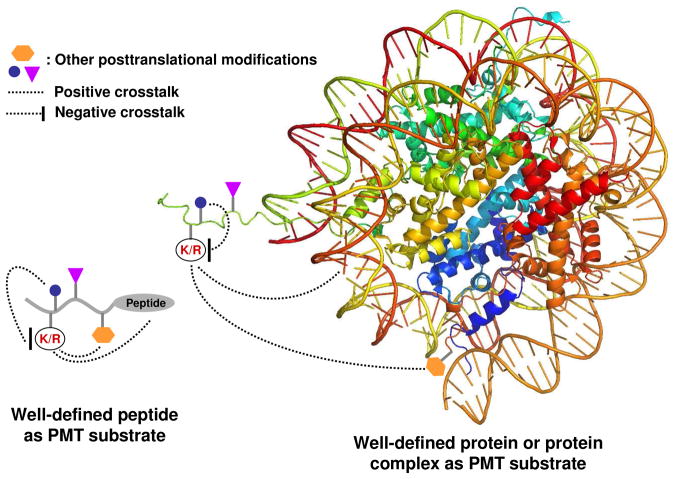
It remains challenging to identify substrates of designated PMTs and map their methylation sites solely based on their primary sequences. The adjacent or remote residues (or other posttranslational modifications) of a PMT target can positively or negatively modulate its reactivity as a substrate (Fig. 5). Current chemical biology approaches allow many PMT substrates to be synthesized or even arrayed with well-defined structures. The studies using these homogenous substrates and arrayed libraries have shed light on how PMTs recognize their targets.
Figure 5.
Well-defined homogenous peptides and proteins as PMT substrates. PMT-substrate specificity can be affected by amino acid sequences that are adjacent to or remote to methylation sites. In a similar manner, PMT-substrate specificity can be affected by other posttranslational modifications or allosteric factors that are adjacent to or remote to methylation sites. The crosstalk can only be examined with well-defined peptide or protein substrates.
Peptides as PMT substrates
Many PMTs can recognize protein substrates as well as the corresponding peptides (Fig. 5). Since peptides and their variants can be readily prepared through solid-phase peptide synthesis, they have been widely used as in vitro substrates to characterize PMTs. With PRMT1 as an example, the Thompson laboratory used various N-terminal H4 peptide to examine PRMT1’s substrate specificity.70 The detailed kinetic analysis on these peptide substrates revealed that, although PRMT1 has comparable H4R3 methylation activities (kcat/Km) on histone H4 and N-terminal H4 1–21 peptide, its activities on N-terminal H4 1–18 peptide and the corresponding R19A peptide drop 200-fold. This difference therefore indicated that a long-distance interaction between PRMT1 and a remote positively-charged region of the substrate is essential for substrate recognition (a step affecting Km). With the same N-terminal H4 1–21 peptide as well as its R3-methylated variant as substrates, the Thompson laboratory further demonstrated that PRMT1 catalyzes H4R3 dimethylation in a partially processive manner.71 Interestingly, when examining PRMT1 with a different substrate eIF4A1 R362 peptide, the Hevel laboratory found that PRMT1-mediated dimethylation occurs in a dissociative manner.72 The discrepancy argues the importance of the PMT substrates in the course of characterizing PMT-catalyzed methylation.
Examining crosstalk between methylation and other posttranslational modifications is also benefited from using well-defined homogenous peptides as PMT substrates (Fig. 5). With an N-terminal H3 peptide and its posttranslationally-modified variants as substrates, the Pradhan laboratory examined how Ser10 phosphorylation and Thr11 phosphorylation affect G9a-catalyzed H3K9 methylation.73 The kinetic analysis showed that S10 phosphorylation decreased kcat and Km of the methylation for more than 10-fold in comparison with only 2-fold decrease of kcat/Km by T11 phosphorylation. Yamagata et. al. demonstrated that PRMT1 methylates FOXO1 at R248 and R250.9 The two methylations inhibited Akt-mediated phosphorylation of S253, but the S253 phosphorylation doesn’t inhibit the methylation of R248/R250. Upon reviewing this work as well as other crosstalk involved with RXRXXS/T motif, Rust and Thompson proposed a dozen proteins including B-Raf, EZH2 and FOXG1 as highly probable PRMT1 substrates.74 This prediction is expected to be tested readily after obtaining the corresponding peptides.
The Zheng laboratory recently reported an approach using a fluorescent peptide as a chemical probe to study the transient kinetics of PMT catalysis.75,76 In Zheng’s work, Leu10 of a H4 N-terminal peptide was replaced by a fluorescein moiety. The resultant fluorescent H4 peptide showed comparable kinetics to native H4 peptide as a PRMT1 substrate. As reflected by fluorescence change, the fluorescein-labeled peptide displayed multiple-phase kinetics upon binding PRMT1. After dissecting the kinetics, the authors concluded that PRMT1 catalyzes H4 methylation via a multiple-step process including an ultra fast substrate-binding step, then a modestly fast formation of the ternary PRMT1-SAM-substrate complex, and lastly the rate-limiting methylation.75 This exemplifies an elegant utilization of substrate-type chemical probes to characterize PMTs.
Proteins or protein complexes as PMT substrates
The target specificity of PMTs can be altered dramatically depending on the nature of their substrates (Fig. 5). For instance, NSD2 methylates H3K36 if nucleosomes are provided as substrates but acts on H4K44 if histone octamers as the substrates.77 In these cases, full-length proteins or protein complexes are more relevant as in vitro substrates of PMTs. Using in vitro reconstituted chromatin templates as substrates of PRMT1, p300 and CARM1, the Roeder laboratory was able to study the p53-dependent crosstalk between the three (co)activators.78 The authors showed that PRMT1-involved H4R3 methylation, p300-involved H3/H4 acetylation and CARM1-involved H3R2/17/26 methylation can occur in a sequentially-stimulated manner. Daujat et. al. showed a similar crosstalk on the pS2 promoter, where CBP-mediated H3K14/18 acetylation stimulates the tight association of CARM1 with chromatin and the resultant H3R17 methylation.79 Besides the cis-crosstalk of posttranslational modifications, which occurs in the same peptide, trans-crosstalk of posttranslational modifications has also been implicated in multiple biological contexts. For example, the ubiqutination of H2K120 often precedes the methylation of H3K79 for transcriptional activation.61,80 These substrate-dependent target preferences and cis/trans-crosstalk therefore underscore the relevance of using proteins or protein complexes as substrates to elucidate PMTs’ functions.
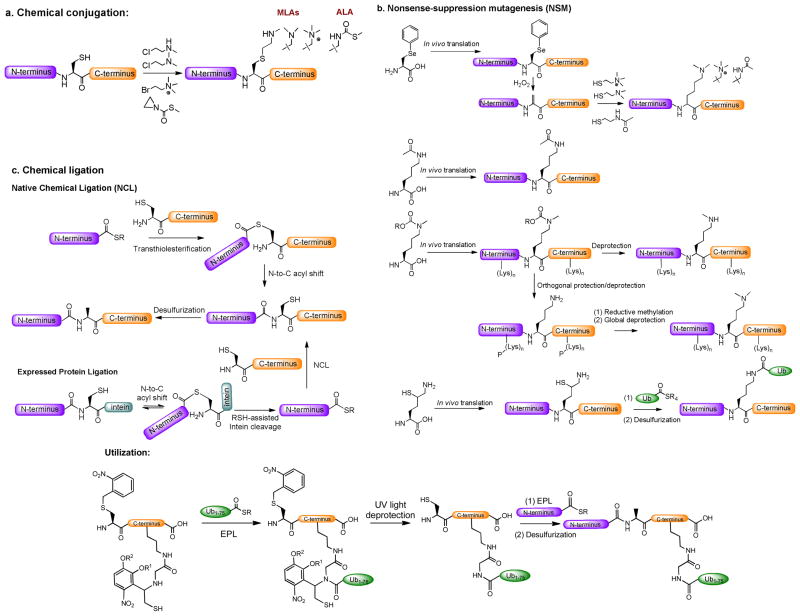
Homogenous proteins or protein complexes with well-defined posttranslational modifications cannot be prepared readily from cell lysates or via in vitro enzymatic reactions. In contrast, they can be accessed efficiently through emerging chemical biology approaches.33 This review will briefly highlight three such approaches (Fig. 6): (a) chemical conjugation, (b) nonsense-suppression mutagenesis (NSM) and (c) chemical ligation (see Chatterjee’s recent review for more details about the semisynthetic approaches).81 These approaches alone or their combination allow scientists to access various recombinant proteins containing well-defined posttranslational modifications (Fig. 6). This collection of recombinant proteins serves as an unprecedented substrate repertoire to study PMTs and their crosstalk with other posttranslational modifications.
Figure 6.
Emerging chemical biology approaches to prepare PMT protein substrates containing well-defined posttranslational modifications: (a) chemical conjugation, (b) nonsense-suppression mutagenesis (NSM) and (c) chemical ligation. These approaches alone or in combination have been applied to prepare histones containing mono/di/trimethyllysine, acetyllysine, ubiquitin or their mimics. A utilization of these approaches to prepare K120-ubiquitinated H2B is described as well.
a. Chemical conjugation
The free-thiol position of site-specifically-introduced cysteine is an ideal warhead for chemical conjugation. To exploit this chemistry, the Shokat laboratory first reported the approach to conjugate an N-methyl aminoethyl moiety to proteins (Fig. 6).82 The resultant N-methylated aminoethylcysteine proved to be an excellent methyllysine analogue (MLA), which can be recognized by α-methyllysine antibodies, methyllysine-binding protein HP1α and multiple PMTs.82 As one application, Margueron et. al. relied on this MLA approach to prepare a series of MLA-containing histones and used them as substrates to examine the crosstalk between PRC2-EZH2/EED (a PMT complex that methylate H3K27) and histone methylation marks (e.g. H3K27, H3K36 and H3K9).83 This work showed that the EED subunit of PRC2 complex strongly interacts with nucleosomes containing H3K27me3 and H3K9me1/2/3 MLAs but not H3K36me1/2/3 MLAs. Together with other biochemical evidence, the authors concluded that this interaction leads to the allosteric elevation of EZH2’s methyltransferase activity and suggested that PRC2 complex self-propagates to nearby chromatins by interacting with its own methylation product. To incorporate an acetyllysine analogue (ALA) into histones (Fig. 6), the Cole laboratory explored similar cysteine-S-alkylation chemistry using methylthiocarbonyl-aziridine as an electrophile.84 The chemical conjugation approach is restricted to incorporation of only one type of posttranslational modifications and has only been demonstrated with MLA and ALA on histones so far. There is thus a need to extend the approach to other posttranslational modifications as well as nonhistone targets.
b. Nonsense-suppression mutagenesis (NSM)
NSM allows unnatural amino acids to be introduced site-specifically into a recombinant protein (Fig. 6). Once orthogonally engineered tRNA/tRNA synthetase pairs are available, matched amino acid analogues can be introduced readily into proteins by supplying them to a cell-free translational system, or to E. coli., yeast, mammalian cells or animals.85 The incorporation of posttranslational modifications into recombinant proteins has been demonstrated in several recent NSM applications (Fig. 6). For instances, the Schultz laboratory was able to prepare recombinant proteins containing racemic methyllysine and acetyllysine mimics through site-specific phenylselenocysteine chemistry (Fig. 6).86 To access recombinant proteins containing enantiomerically pure methyllysine, Chin/Schutlz/Liu laboratories developed NSM by incorporating N-protected-methyllysine into a recombinant protein, followed by deprotection (Fig. 6).87–90 With a similar NSM, The Chin and Liu laboratories can also access enantiomerically pure acetyllysine in a high efficiency (Fig. 6).91–93. To use NSM to prepare recombinant proteins containing dimethyllysine, the Chin laboratory developed a multiple-step orthogonal protection/deprotection strategy (Fig. 6).87 The Chin group recently demonstrated an NSM approach for site-specific ubiquitination of recombinant proteins using δ-thiol-L-lysine as a building block, which was later used as an anchor for native chemical ligation followed by desulfurization (a strategy as discussed later) (Fig. 6). 94 The Chin and Liu laboratories also developed the strategies using a quadruplet-decoding ribosome (ribo-Q1) and the ochre stop codon UAA, respectively, to incorporate two amino acid analogues into multiple sites of a recombinant protein.95,96 The combined efforts of the Schultz/Chin/Liu laboratories therefore allowed the current NSM strategies to generate recombinant histone H3 containing mono/di/trimethyllysine, acetyllysine, ubiquitin or their mimics alone or in combination (Fig. 6).
c. Chemical ligation
In comparison with site-specific chemical conjugation and NSM, chemical ligation is featured by its ability to assemble a target protein from well-defined peptide fragments (Fig. 6). The approach is expected to be a powerful method for introducing complex patterns of posttranslational modifications to protein targets. Native chemical ligation (NCL) and expressed protein ligation (EPL) are by far the most widely-employed technologies in chemical ligation (Fig. 6).33,34 The residual cysteine in both NCL and EPL can be optionally converted into alanine through desulfurization. Multi-step sequential ligation, combined with chemical protection/deprotection and chemical conjugation, has also been developed to access targets that harbor distantly-separated posttranslational modifications or branched ubiquitination (Fig. 6).97–99
As an application of chemical ligation to PMTs, the Muir laboratory relied on the chemical ligation strategy to access H2BK120-ubiquitinated nucleosome (Fig. 6).33,98 Using the nucelosome as a substrate, they were able to study the crosstalk between H2BK120 ubiquitination and H3K79 methylation, which are catalyzed by RNF20 E3 ligase and DOT1L, respectively. The first step in Muir’s approach was to conjugate a short Cys117-protected, K120-modified H2B 117–125 peptide with a recombinant C-terminal intein-fused ubiquitin via an EPL-like auxiliary-facilitated chemical ligation. After removing the auxiliary and the Cys117-protecting group through UV irradiation, the resultant fragment was then connected to the N-terminal 1–116 fragment of H2B via NCL and the resultant cysteine was desulfurized. By combining chemical ligation and chemical conjugation, the Muir laboratory later developed a simplified strategy to access disulfide-linked analogues of H2BK120ub.33,99 With the aid of these ubiquitinated histones/nucleosomes as substrates, they were able to show that H2BK120ub is sufficient to stimulate DOT1L-mediated H3K79 methylation.97–99 This observation presented direct in vitro evidence that H2BK120-ubiquitination is an immediate upstream event of DOT1L-mediated H3K79 methylation.
Identifying PMT targets via consensus sequences and peptide array
Although efforts over the past decade have led to identification and characterization of hundreds of PMT targets, dissecting target profiles for individual PMTs is still a formidable task. For the conventional candidate-based approach, novel targets of designated PMTs were identified from the peptide library generated based on the known substrate sequences. As an example, to explore the substrates of PRMT1 beyond the classical RGG sequence, the Hevel laboratory used a focused peptide library (ca. 20 peptides) derived from the PRMT1 substrate fibrillarin.72 From this peptide collection, they were able to confirm eleven new PRMT1 substrate sequences.
To expand the candidate-based approach, the Jeltsch laboratory transformed a SPOT synthesis method to array peptide substrate candidates onto functionalized cellulose membrane (Fig. 7).26,100,101 With Dim5, G9a, and SET7/9 substrate peptides as lead sequences, the Jeltsch laboratory designed a peptide library by systematically replacing each amino acid with the other 19 amino acids. The resultant peptides were SPOT-synthesized and arrayed on cellulose membrane. The membrane was then incubated with recombinant PMTs and radiolabeled SAM, followed by autoradiography to map hot spots. With these peptide-array libraries, the authors were able to study the substrate-specificity of Dim-5, G9a, and SET7/9, and conclude that Dim-5 recognizes R8-G12 of H3 tail with T11 and G12 being most important for the substrate recognition, but Arg8 and Lys9 most important for G9a’s substrate recognition.26,100,101 Through proteome-wide search on the basis of the consensus sequences of active peptide substrates, the authors were able to report and validate a dozen of novel proteins including CDYL1, WIZ, ACINUS and G9a (automethylation) as G9a targets and AKA6, CENPC1, MeCP2, MINT, PPARBP, ZDH8, Cullin1, IRF1 as SET7/9 targets.26,100,101
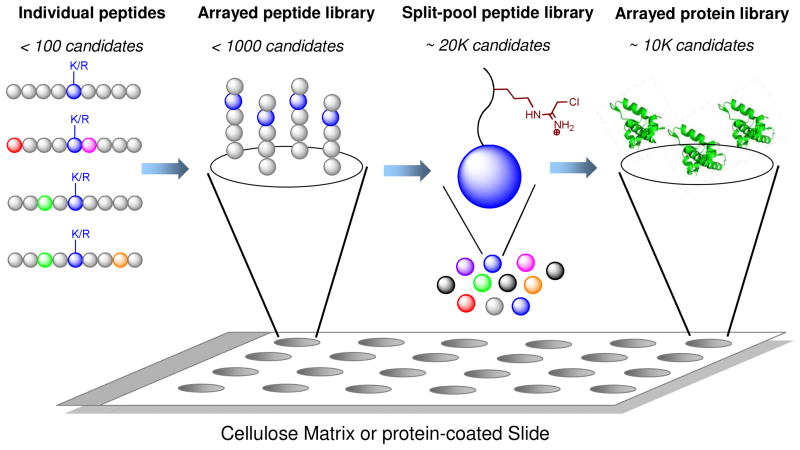
Figure 7.
Substrate candidates that can be used to identify novel PMT targets. The conventional approach relies on laborious screening of <100 individual peptides to identify novel PMT substrates. The peptide array approach allows several hundred distinct peptides to be examined in a single run. Split-pool peptide library allows several thousand candidates to be examined in a single run. The protein array approach maintains comparable capability of the split-pool library but present structurally-relevant proteins as substrate candidates.
To further enhance the throughput of the peptide-based approach for PMT target identification, the Thompson laboratory reported a randomized screening using a combinatorial peptide library (Fig. 7).102 The one-compound-one-bead split-pool peptide library utilizes a Cl-acetamidine warhead at the Arg site of the PRMT1 target. Prior to this work, the Thompson laboratory had demonstrated that the Cl-acetamidine moiety in the context of substrate covalently interacts with PRMT1.103 The active PRMT1 substrates containing the chemical moiety are expected to immobilize the enzyme onto the beads. Upon screening a +3 to −3 region of H4R3 using a pool of 21,000 peptides and with a fluorescein isothiocyanate labeled PRMT1 as a probe, the authors were able to identify 57 distinct hits as potential PRMT1 targets.
Although a few novel PMT targets were identified through the sequence-guided peptide-array strategy, these targets only account for a small portion of PMT substrates. Many PMT targets lack consensus sequences and there is no simple rule to generalize the substrate-recognition pattern of PMTs. These observations suggest that factors besides the sequences adjacent to methylation sites can be essential for PMTs’ substrate recognition.
Identify PMT targets with protein array libraries
In contrast to peptides, full-length proteins have more merit as PMT substrates, since certain PMTs function only in the context of full-length proteins (Fig. 7). The Gozani laboratory recently demonstrated the feasibility of using a protein-array approach to identify PMT substrates.104 In this study, the commercially available ProtoArray glass slide (coated with 9,500 proteins) was used for proteome-wide identification of SETD6 substrates. After the on-chip methyltransferase reaction, the hits were identified either by fluorescence signals when primary pan-anti-methyllysine antibody and secondary Alexa Fluor 647-conjugated antibody were used for readouts or through autoradiography when radiolabeled SAM was used as the cofactor. From 9,500 proteins arrayed on the glass slide, 118 proteins were identified as hits by the fluorescence method and 114 by the radiometric method with 26 proteins overlapped. Six proteins were cherry-picked for validation and were shown to be SET6 targets in vitro. Two of them were further validated as physiological substrates. In this work, however, detecting on-chip methylation with either antibody or autoradiography did not seem to be robust, because overlap analysis showed that each detection method favors a subset of targets with only 20% overlap. It is likely that the radiometric method is relatively robust but less sensitive and therefore can only detect more active substrates. In contrast, the antibody-based assay is more sensitive for slow substrates but may be restricted by the epitopes that the antibodies can recognize. The Gozani laboratory showed that the quality of commercial antibodies varies dramatically.104 In order to improve this protein-array approach, more effort can be made to increase the quantity of arrayed proteins as well as improve detection methods.
Profiling PMT targets from cellular proteomes
Although novel PMT targets can be identified from arrayed peptide or protein libraries, the in vitro assay conditions frequently do not reflect those occurring in cellular contexts or in vivo. PMTs often associate with other binding partners in vivo to form multimeric complexes78,83 and identification of authentic PMT targets may therefore rely on the native contacts. Some PMT-mediated methylations also depend on specific cellular or in vivo stimulation (e.g. methylation of Reptin by G9a only under hypoxic conditions and p65 by SET7/9 under TNFα stimulation).105,106 These observations therefore argue the importance to profile PMT targets in their native contexts.
To profile PMT targets in a cellular context, Frankel et. al. incubated recombinant enzymes with whole cell extracts in the presence of radiolabeled SAM, followed by autoradiography.107 The substrates can be labeled in the presence of matched PMTs. With this in vitro approach, the authors were able to radiolabel the targets of PRMT1, CARM1 and PRMT6. The different labeling patterns between the three closely-related PRMTs indicated their distinct substrate preference.107 To identify substrates of PRMT3 in a cellular context, the Bedford laboratory developed a comparable in vivo labeling approach by culturing cells in methionine-free medium and then supplying L-[methyl-3H]methionine.108 After the radiolabeled methionine was transported into the cells and processed into SAM (presumably by endogenous SAM synthetase), PMTs utilized the radiolabeled SAM to label substrates in the native cellular context. Because of the presence of protein synthesis inhibitors cycloheximide and chloramphenicol, radiolabeled methionine was not directly translated into proteins.108
Although the radiometric approach allows the PMT targets to be visualized by autoradiography, it does not provide direct information for target identification. As a complementary approach, the Richard laboratory generated ADMA- and SDMA-specific antibodies for proteome-wide profiling of PRMT targets.109 These antibodies allowed ADMA/SDMA-containing substrates to be pulled down from HeLa cell lysate. The reagents combined with shot-gun MS analysis enabled the Richard group to identify several hundreds of potential PRMT targets.109 However, this approach cannot assign the substrates to specific PRMTs (A bioorthogonal approach to label the substrates of individual PMTs in a cellular context will be discussed later).
Cofactors of PMTs
SAM ranks after ATP as the second most commonly used enzyme cofactor.110 The cofactor reactivity is harbored around the sulfonium center in most SAM-involved biochemical transformations. For instance, the sulfonium carbon bond in SAM’s thio-adenosyl moiety undergoes an enzyme-catalyzed homolytic cleavage to form a 5′-deoxyadenosyl radical, a key intermediate for canonical radical SAM enzymes.111 The sulfonium carbon bond in SAM’s homocysteine moiety can also undergo non-canonical homolytic cleavage to generate the 3-amino-3-carboxypropyl radical.112 The same sulfonium carbon bond can also be subject to intra- and intermolecular heterolylic cleavage, which provides the building blocks for biosynthesis of acylhomoserine and polyamine, respectively.60 Despite the diverse reactivity of SAM as a cofactor, the most ubiquitous role of SAM remains its use as a biological methyl donor for SAM-dependent methyltransferases. As reviewed below, several efforts have been made over the past decade to develop SAM analogues as cofactor surrogates or chemical probes for PMTs (Fig. 9).
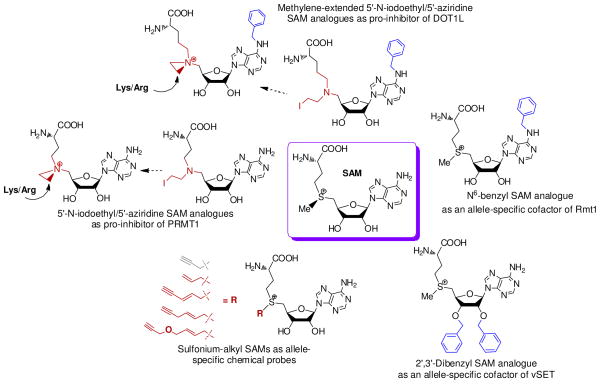
Figure 9.
SAM analogues as cofactor surrogates or chemical probes for PMTs. Clockwise: N6-benzyl SAM analogue as allele-specific cofactor and inhibitor of Rmt1; 2′,3′-dibenzyl SAM analogue as an allele-specific cofactor of vSET; sulfonium-alkyl SAM as cofactor surrogates and allele-specific chemical probes; 5′-N-iodoethyl/5′-aziridine SAM analogues as precursors of bisubstrate inhibitors of PRMT1 and DOT1L.
N6-benzyl SAM analogues as allele-specific cofactor and inhibitor of PRMTs (Fig. 9)
Lin et. al. designed a series of N6-substituted SAM analogues and examined their activity as cofactors of Rmt1 (a yeast PRMT) and its variants.113 Using a “bump-and-hole” approach guided by the structure of Rmt1 (the only available PRMT structure at that moment), the authors were able to identify an Rmt1 mutant (E117G) that can utilize N6-benzyl-SAM as a cofactor. This analogue is preferentially processed by E117G Rmt1 at the rate 67-fold faster than by native Rmt1. Following the same trend, N6-benzyl-SAH is an allele-specific inhibitor to the mutant with 20-fold increased selectivity versus the wild-type enzyme. The active enzyme-cofactor pair can be used for allele-specific labeling of Rmt1 targets. This was the first effort toward manipulating PMTs with SAM analogue cofactors.
2′,3′-Dibenzyl SAM analogue as an allele-specific cofactor of PKMT (Fig. 9)
Besides N6-substituted SAM analogues, the Zhou laboratory explored 2′- or 3′-substituted SAM analogues as potential SAM surrogates of engineered PMTs.114 The authors focused on vSET, a viral SET-domain-containing PKMT. Like human EZH2, the enzymatic component of PRC2, vSET methylates H3K27 in vivo. Guided by the structure of vSET, the Zhou laboratory located two residues that are expected to be sensitive to SAM’s 2′- or 3′-substitient. Upon mutating them followed by screening against 2′- or 3′-substituted SAM analogues, the Zhou laboratory were able to identify vSET L116A mutant and its matched 2′,3′-dibenzyl SAM cofactor. The enzyme-cofactor pair showed comparable kcat/Km to that of native vSET and SAM. Since the authors only examined a small number of SAM analogues and vSET mutants, more active mutant-cofactor pairs may exist. These active enzyme-cofactor pairs can be used for vSET-specific labeling.
5′-N-iodoethyl/5′-aziridine SAM analogues as precursors of bisubstrate inhibitors of PMTs (Fig. 9)
5′-N-adenosylaziridine and its SAM-like derivatives were reported to be active cofactors of bacterial DNA and small-molecule methyltransferases.110 The Thompson laboratory first examined whether PMTs can act on a 5′-aziridine SAM analogue.115 With PRMT1 as a model system, the authors demonstrated that the 5′-aziridine SAM analogue rapidly reacts with an N-terminal H4 peptide in an enzyme-dependent manner. H4R3 of the peptide (the methylation site of PRMT1) conjugates with the 5′-aziridine SAM analogue in situ to form a bisubstrate analogue inhibitor of PRMT1. This inhibitor showed a modest IC50 and 4.4-fold preference to PRMT1 over CARM1. The Song laboratory then examined the 5′-aziridine SAM analogue against DOT1L, G9a and SUV39H1. Only a modest IC50 against DOT1L was observed.116
In the course of developing DOT1L inhibitors, the Song laboratory noticed that, unlike PRMTs and other SET-domain-containing PKMTs, DOT1L has a relatively spacious binding site for SAM’s 6-NH2 group.116 By introducing the N6-benzyl-substituient to the 5′-aziridine SAM analogue (Fig. 9), the authors observed a 15-fold improvement of IC50 against DOT1L but not other PMTs (e.g. PRMT1, CARM1, G9a and SUV39H1). In addition, the authors reasoned that since C-N bonds (~ 1.47 Å) in the 5′-aziridine SAM analogue are slightly shorter than C-S bonds (~ 1.82 Å) in SAM and SAH, extending one more methylene in the 5′-aziridine SAM analogue would further improve the potency. The resultant methylene-extended 5′-aziridine-N6-benzyl SAM analogue (Fig. 9) showed an IC50 of 110 nM against DOT1L and > 1000-fold selectivity over PRMT1, CARM1, G9a and SUV391.116 Although the authors did not further characterize the mechanism of the inhibition, the DOT1L inhibitor is expected to behave much like the N-adenosylaziridine through the substrate-participating formation of a bisubstrate analogue inhibitor (Fig. 9).115,116 However, since aziridine SAM analogues are not stable under physiological pH, their broad application within biological contexts remains to be investigated.
Sulfonium-alkyl SAM as cofactor surrogates and allele-specific chemical probes (Fig. 9)
The Weinhold laboratory explored the use of sulfonium-β-sp2/sp1-doubled-activated SAM analogues as cofactors for bacterial DNA/RNA methyltransferases for target labeling (Fig. 9).110 However, the implementation of these SAM analogues to label PMT substrates had not been reported until recently. Peters et. al. developed (E)-pent-2-en-4-ynyl-SAM (EnYn-SAM) as an SAM surrogate and showed that the SAM analogue can be utilized by Dim-5 for target labeling under basic conditions (pH=9).117 The authors also demonstrated that the same SAM analogue can be utilized by native MLL4 and ASH2-MLL complex to some degree. Binda et. al. developed a propargyl-SAM analogue for PMT target labeling (Fig. 9).118 With a clickable FLAG probe coupled to a sensitive anti-FLAG antibody, Binda et. al. showed that SETDB1 but not SET7/9, SMYD2, PRMT1, CARM1, PRDM8, -10, and -16 can utilize the propargyl-SAM analogue. Interestingly, the Weinhold laboratory noticed that the propargyl-SAM analogue suffers a rapid decomposition at neutral and basic conditions.117 This discrepancy may be rationalized if SETDB1 can rapidly process the SAM analogue before decomposition.
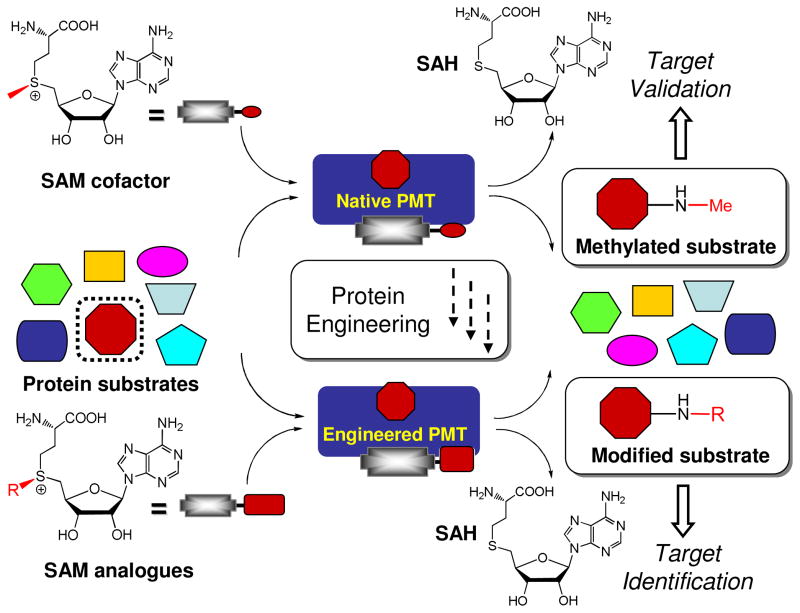
Although the prior cases demonstrated the feasibility of using the SAM analogue cofactors to label PMT substrates, the activities of native PMTs on these synthetic cofactors are generally low. A limitation in the prior approaches is that they cannot unambiguously assign the labeled targets to designed PMTs in cellular contexts because other promiscuous PMTs may be present to label their own substrates with these cofactors. To address these limitations, our laboratory aimed at developing SAM analogue cofactors that are inert toward native PMTs but can be recognized by engineered PMTs.31,64 We envisioned that this bioorthogonal approach would allow the labeled substrates to be assigned to engineered enzymes in an unambiguous manner (Fig. 8). Toward this goal, we developed (E)-hex-2-en-5-ynyl-SAM (Hey-SAM) and 4-propargyloxy-but-2-enyl-SAM (Pob-SAM), respectively, to profile the substrates of G9a and PRMT1 (Fig. 8).31,64 The two SAM analogues are inactive with native PMTs but can be processed efficiently by engineered G9a and PRMT1. Furthermore, Pob-SAM was demonstrated to be an excellent SAM surrogate for labeling PRMT1 substrates in a complex cellular milieu.
Figure 8.
Bioorthogonal profiling approach to label PMT substrates. Native PMTs use SAM as the cofactor to methylate their targets. In contrast, PMTs can be engineered to accommodate SAM derivatives as cofactors and label their substrates with distinct chemical groups. Since bulky SAM analogues are inert to native PMTs, the resultant distinctly-modified substrates can be assigned to a single, designated PMT.
With the aid of a reformulated fluorogenic assay, our laboratory systematically evaluated the activities of native PMTs (PRMT1, PRMT3, CARM1, SUV39H2, SET7/9, SET8, G9a and GLP) on a panel of SAM analogues (allyl-SAM, propargyl-SAM, (E)-pent-2-en-4-ynyl-SAM (EnYn-SAM), (E)-hex-2-en-5-ynyl-SAM (Hey-SAM) and 4-propargyloxy-but-2-enyl-SAM (Pob-SAM)).64 Among the examined 8×5 pairs of PMTs and SAM analogues, only native SUV39H2, G9a and GLP show slight activity toward allyl-SAM. The bulky SAM analogues, such as EnYn-, Hey- and Pob-SAM are inert toward the screened native PMTs. This finding is also consistent with the observed low activity of native MLL4 or ASH2-MLL on EnYn-SAM. These results therefore argue that the SAM-binding pocket of native PMTs needs to be tailored to accommodate bulky SAM analogues for efficient substrate labeling. The suitability of these SAM analogues to other engineered PMTs is being investigated in our laboratory.
Inhibitors of PMTs
Given that the methylation activities of PMTs associate with diverse cellular processes and their dysregulation is implicated in many diseases including cancer,20 many efforts have been made in academia and industry to develop PMT inhibitors as chemical probes and therapeutic reagents. However, the success in finding lead compounds is still limited and many of those have not been fully characterized. Because all PMTs have one of two types of highly-conserved SAM-binding pockets and utilize less-structured substrate-binding regions, it remains challenging to develop selective and potent PMT inhibitors for these enzymes. At present, rational design, HTS and in silico screening are three mainstream approaches in developing PMT inhibitors. The successful implementations and potential pitfalls of these approaches will be discussed in this section.
Principles to define high-quality PMT inhibitors
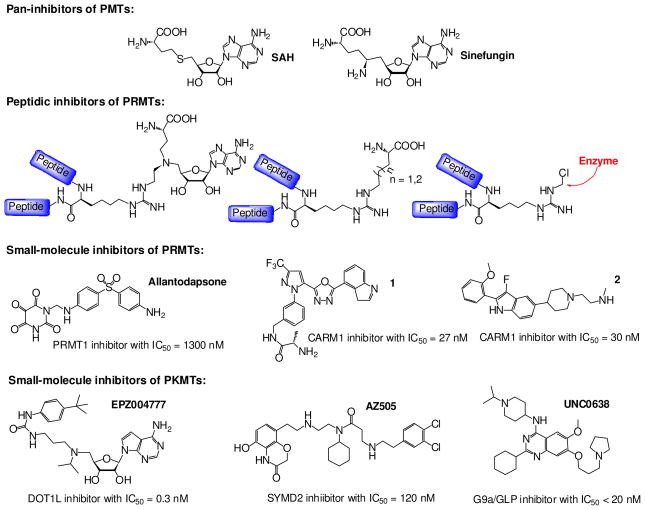
Sinefungin and SAH are SAM analogue inhibitors that have been claimed as pan-inhibitors of PMTs (Fig. 10). The former is a natural product available from Sigma. The latter is the metabolite byproduct of SAM-dependent methylation reactions. To achieve high intracellular concentrations of SAH, a common practice is to treat cells with adenosine dialdehyde,119 an irreversible SAH hydrolase inhibitor that blocks SAH hydrolase-mediated SAH degradation and thus causes its intracellular accumulation. However, caution should be taken with these SAM analogues as pan-inhibitors of PMTs because IC50 of these inhibitors can alter by two orders of magnitude for different PMTs (e.g. Ki > 25 μM for sinefungin against EZH2 versus 0.10 μM against CARM1).3 Therefore, the activity profile of the PMT pan-inhibitors needs to be defined carefully prior to their uses in biological contexts.
Figure 10.
Representative inhibitors of PMTs. SAH and sinefungin are the best characterized pan-inhibitors of PMTs. Peptidic inhibitors were designed on the basis of the sequenence of PMT substrates with their Arg residue conjugated with some moiety of the SAM or replaced with a functional group. Allantodapsone is a potential PRMT1 inhibitor with IC50 = 1300 nM. Compounds 1 and 2 are so far the most potent CARM1-selective inhibitors. EPZ004777 and UNC0639 are so far the most potent and best-characterized inhibitors of DOT1L and G9a/GLP, respectively. AZ505 is so far the most potent SMYD2 inhibitor.
PMT inhibitors can be developed either as chemical genetic probes to modulate the activities of PMTs or as potential drugs to treat patients. Although the rules applied to the former are less strict, several common principles still need to be fulfilled for high-quality PMT chemical genetic probes. In the article “the art of the chemical probe”, Frye introduced five simple principles for general development of chemical probes.120 Here I have rephrased these principles in the specific context of PMTs as the following: (1) a high-quality chemical genetic probe should show sufficient in vitro potency (inhibit one or several designated PMT(s)) and selectivity (does not inhibit irrelevant PMTs and other targets); (2) such PMT chemical genetic probes should show decent in vivo or at least cellular-level potency and selectivity that correlates with the in vitro data; (3) the inhibition mechanism should be clear and consistent in vitro and in vivo or in a cellular-level context (e.g. SAM-competitive, substrate-competitive, or irreversible inhibitors; identity of the active species as intact chemical probes versus well-characterized derivatives); (4) high-quality chemical genetic probes of designed PMTs should show at least one proved utilization (e.g. the treatment of a chemical genetic probe should recapitulate the biological readouts anticipated by genetically disrupting the corresponding PMT target); (5) As a bonus point, an ideal chemical genetic probe should be accessible either through commercial vendors or synthetically via well-described chemical methods. The recent advancement in PMT chemical genetic probes will be discussed according to these criteria (some of them listed in Fig. 10).
Rationally-designed PMT inhibitors
Based on the amino acid sequences of PMT substrates, several peptidic PMT inhibitors were reported recently (Fig. 10). As one example described above, the Thompson laboratory showed the H4R3 can react with the 5′-aziridine SAM analogue in a PRMT1-dependend manner. The resultant bisubstrate inhibitor of PRMT1 can be generated in situ with its IC50 in the range of single-digit μM.115 The Thompson laboratory also developed substrate-based, irreversible PRMT inhibitors containing the Cl-acetamidine warhead and implemented them as activity-based probes (described above, Figs. 7, 10).103,121 In contrast, the Martin and Frankel laboratory prepared partial-bisubstrate PRMT inhibitors that contain the peptidic fragments of PRMTs’ substrates and the α-amino carboxylic acid moiety of SAM.122 Though some of these peptidic PRMT inhibitors showed decent IC50 and were utilized as chemical probes in vitro, their utilization in vivo remain to be tested given general undesirable pharmacological properties of peptidic molecules. Since peptidic bisubstrate inhibitors have been only reported for PRMTs so far, examining whether a similar strategy can be applied to PKMTs can be interesting.
Thus far, known rationally-designed small-molecule PMT inhibitors were developed either by conjugating a moiety of PMT substrates with an azo-SAM analogue (bisubstrate-type inhibitors) or by exploring distinct SAM-binding pockets of specific PMTs. For example, the Ward laboratory reported efforts in developing PRMT-specific bisubstrate-type inhibitors by connecting a guanidium moiety with the azo-SAM analogue via various linkers.123,124 The series of compounds showed modest in vitro single-digit μM values of IC50 against PRMTs (only PRMT1 and CARM1 were tested) and > 10-fold selectivity over SET7/9. The Hirano laboratory reported similar efforts in developing bisubstrate-type inhibitors of PKMTs by linking the azo-SAM analogue with various N2-alkyl-aminoethyl moieties, which resemble the lysine side chain in a PKMT-catalyzed reaction.125 Surprisingly, their best inhibitors only showed modest in vitro IC50 values of 10 ~ 100 μM against SET7/9, the only PKMT that was tested. The in vitro IC50 of these PMT bisubstrate-type inhibitors against other PMTs remains to be measured. More mechanistic studies may help the design of bisubstrate-type PMT inhibitors to achieve better potency and selectivity.
An alternative approach to design rationally target-specific PMT inhibitors is to explore the difference of SAM-binding sites in PMTs. One of the most successful example is the DOT1L-specific inhibitor EPZ004777 (Fig. 10).22 Daigle et. al. reported EPZ004777 as a SAM-competitive inhibitor with an in vitro Ki of 0.3 nM, a cellular-level EC50 of sub-μM, and > 3000-fold selectivity over 9 other examined PMTs. Because DOT1L is an oncoprotein in several subtypes of mixed lineage leukemia (MLL), EPZ004777’s efficacy was also validated in the context of the relevant leukemia cells and with a mouse MLL xenograft model.22 In addition to this work, the Song laboratory reported a suite of 5′-N-iodoethyl (5′-aziridine)-based SAM analogues as potent DOT1L inhibitors (described above, Fig. 9).116 Although the Song laboratory did not perform biological validation of their DOT1L inhibitors, their work shed light on how EPZ004777 achieves high selectivity for DOT1L versus other PKMTs. They noticed that, because DOT1L-bound SAM (unlike the SAM bound by other PKMTs) adapts an open conformation, extending the 5′-region by a methylene moiety significantly enhanced the potency of their 5′-N-iodoethyl (5′-aziridine) SAM analogue inhibitors.116 The same rationale may be applicable to EPZ004777, whose 5′-linker may mimic the length and extended conformation of DOT1L-bound SAM. Although EPZ004777 was demonstrated to be a high-quality chemical genetic probe, its synthesis remains to be disclosed.
Recent structural and chemogenetic analysis on a dozen of human PMTs reveal that closely-related PMTs can bind to SAM, SAH or sinefungin preferentially.3,126 Many human PMTs have distinct SAM-recognizing motifs as well.3,126 The chemogenetic and structural information therefore present a general road map to further explore the differences between these SAM-binding sites for more potent and selective SAM analogue inhibitors of PMTs.
PMT inhibitors from HTS leads
HTS is so far the most powerful approach for identifying PMT inhibitors. As discussed in a previous section (PMT-activity assays), several HTS assays have been developed to facilitate the HTS-based identification of PMT inhibitors. From a collection of 9000 compounds, the Bedford laboratory identified a series of PRMT inhibitors including AMI-1 (will be discussed later), AMI-5 (eosin, a SAM-competitive nonspecific inhibitor), AMI6, AMI9 and AMI18.65 After optimizing AMI-5, the Bedford laboratory reported a CARM1-specific AMI-5 derivative, which shows a decent in vitro IC50 and a cellular-level EC50 of around 10 μM with >100-fold in vitro selectivity over PRMT1 and SET7/9.127 By focusing on the PRMT-specific inhibitor AMI-6 and non-specific PMT inhibitor AMI-9, Bonham et. al. merged their pharmacological components to invent the hybridized inhibitor.128 This compound showed a decent IC50 in vitro of 2 – 4 μM against PRMT1 and CARM1 and a cellular-level EC50 of 100 – 300 μM for CARM1-mediated H3R17 methylation. The authors showed that this compound modulates T-helper-cell function at a dose of > 50 μM, which turns out to be lower than their cellular-level EC50. Further studies are still needed to validate its use as a PRMT1/CARM1 chemical genetic probe and elucidate how the AMI inhibitors interact with their targets.
Purandare et. al. reported a pyrazole-based CARM1-specific inhibitor with an in vitro IC50 of 1.8 μM.67 Optimization of the lead compound led to a potent and selective CARM1 inhibitor (Compound 1 in Fig. 10) with an in vitro IC50 of 27 nM and >500-fold in vitro selectivity over PRMT1 and PRMT3 (Fig. 10).129 Sack et. al. recently released the structure of a new indole-type CARM1 inhibitor (Compound 2 in Fig. 10) with a potent in vitro IC50 of 30 nM (Fig. 10).129 Although the in vitro IC50 values of the two CARM1 inhibitors are more promising than those of the AMI-derived CARM1 inhibitors, no in vivo or cell-based efficacy of the two compounds has been reported (as discussed below).
From a collection of 2,976 compounds, the Imhof laboratory identified chaetocin as the first PKMT inhibitor, which has an in vitro IC50 and a cellular-level EC50 around 0.8 μM against Drosophila melanogaster SU(VAR)3-9.66 Unfortunately, the natural product lacks selectivity because it also inhibits G9a and DIM5 with in vitro IC50 of 2.5 and 3 μM, respectively. A following cell-based characterization showed that chaetocin can block histone H3K9 trimethylation (a target of SUV39H1, a human homologue of SU(VAR)3-9).130 However, given the complex synthesis of chaetocin and its derivatives,131 use of chaetocin as a general chemical probe may be limited.
From a 125K-compound library, Kubicek et. al. identified the first G9a inhibitor BIX-01294, which has an in vitro IC50 of 2.7 μM and doesn’t inhibit SUV39H1 and PRMT1.68 The following lead optimization led to a series of derivatives with improved potency and selectivity.132–135 At this point, the best characterized BIX-01294 derivative is UNC0638 (Fig. 10), a substrate-competitive inhibitor with ~ 20 nM in vitro and cellular-level IC50 values for G9a and GLP (closely-related homologue of G9a), > 3000-fold selectivity over other so-far-examined PMTs.132 Treatment with UNC0638 can reactivate silenced genes by reprogramming H3K9me2 and DNA methylation in mouse embryonic stem cells. This observation recapitulates the anticipated phenotype of genetic disruption of G9a and GLP. Other important properties of UNC0638 include no significant degradation in cellular contexts and low cellular toxicity. According to the five rules in Frye’s “the art of the chemical probe”,120 UNC0638, which is available from Sigma, is arguably a high-quality chemical genetic probe (Fig. 10). However, UNC0638 displays a fast clearance rate in animals, which may limit its use as a therapeutic reagent.
Using the AlphaScreen HTS assay, Ferguson et. al. reported AZ505, an inhibitor of SMYD2 with an in vitro IC50 of 0.12 μM and > 800-fold selectivity over other PMTs including the closely-related SMYD3 (Fig. 10).69 However, the compound was characterized to be a substrate-competitive, SAM-uncompetitive inhibitor, a mechanism that requires the formation of a SAM-inhibitor-enzyme ternary complex to satisfy the observed high potency (a Hook effect of SAM).69 Given the uncertainty of intracellular concentrations of SAM,136,137 the cellular-level inhibition of AZ505 remains to be tested.
PMT inhibitors identified through IN SILICO screening, intuition and serendipity
Besides rational design and HTS, virtual screening is another complementary approach to identify inhibitors of PMTs. As the first effort of in silico screening for PMT inhibitors, the Jung and Sippl laboratories docked the NCI diversity-set compound library into RmtA (a fungal homologue of human PRMT1) for the primary screening and then into PRMT1 for validation.138,139 The authors were able to identify and validate multiple PRMT1 inhibitors including allantodapsone (Fig. 10), C-7280948, RM65, and stilbamidine with in vitro IC50 values of 1.3 μM, 12.8 μM, 55.4 μM and 56.0 μM, respectively.138–141 In cellular contexts, a C-7280948 derivative, allantodapsone, RM65, and stilbamidine showed EC50 values around 25–50 μM.138–141 Mechanisms of these inhibitors against PRMT1 remain to be examined. Although the current in silico screening still focuses on PRMT1, this approach is expected to be transferable to other PMTs, given that around 20 distinct structures of human PMTs have been deposited into the PDB database.
The aforementioned HTS performed by the Bedford laboratory also led to the discovery of a set of polyphenol-type PRMT inhibitors such as AMI-18, which are structurally related to xenoestrogens.142 Driven by this intuition, Cheng and Bedford tested a few xenoestrogens and were able to identify tamoxifen as a CARM1-specific inhibitor with a modest in vitro and cellular-level EC50 of around 30–50 μM.142 In contrast to Cheng and Bedford’s intuition, pure serendipity led Selvi et. al. to identify a substrate-uncompetitive CARM1 inhibitor.143 In the course of purifying the active ingredients of pomegranate extract, Selvi et. al. found that one component, ellagic acid, inhibits CARM1 as well as p300. Ellagic acid was then characterized as a substrate-uncompetitive CARM1 inhibitor that depends on the substrate’s “KAPRK” motif at H3R17 region to interact with the enzyme.143 The formation of the dead enzyme-substrate-inhibitor ternary complex (the Hook effect of substrate) accounts for the observed inhibition of CARM1-mediated H3R17 methylation. The intuition- and serendipity-based findings surely enriched our tool box and contributed to the urgent need for PMT inhibitors.
Pitfalls of PMT inhibitors
Lessons learned from previous experiences are valuable to avoid the pitfalls of PMT inhibitors. AMI-1 was identified through HTS as a PRMT-specifc inhibitor.65 When examining the fluorescein-conjugated H4 N-terminus peptide (a PRMT1 substrate), the Zheng laboratory noticed that AIM-1 preferentially interacts with the histone peptide rather than the enzyme.144 This interaction with the peptide, likely native histones, accounts for the observed PRMT1 inhibition. This scenario resembles that of sanguinarine, which inhibits PMT-mediated histone methylations by interacting with core histones rather than enzymes themselves.145
Another pitfall of certain PMT inhibitors are SAM-, SAH- or substrate-uncompetitive inhibitors, as exemplified by the pyrazole- or indole-based CARM1 inhibitors (Compounds 1,2) and the SMYD2 inhibitor AZ505.69,129 Kinetic analysis and inhibitor-substrate-enzyme structures suggest that the three inhibitors are substrate-competitive, SAM/SAH-uncompetitive inhibitors.69,129 The tight binding of these inhibitors to their targets requires the presence of uncompetitive SAM or SAH to form the ternary enzyme-inhibitor-SAM/SAH dead complex (aforementioned Hook effect of SAM/SAH). Characterizing these inhibitors in cellular contexts and in vivo can be complicated by the uncertainty of concentrations of SAM and SAH in different cell types.136,137 Although using a low concentration of SAM in HTS assays can minimize the Hook effect of SAM or SAH, the issue seems to be unavoidable for SMYD2 because of its high affinity to SAM (Kd = < 1 nM).3 It is also possible to identify substrate-uncompetitive inhibitors (the Hook effect of substrate), such as Ellagic acid as exemplified above. To avoid the pitfall of substrate-uncompetitive inhibitors, Ferguson et. al. recommended using a low concentration of substrate to run HTS.69
With these experiences in mind, it is thus important to use enzymatic kinetics or other complementary tools to elucidate and validate the inhibition mechanisms of potential PMT inhibitors at the early stage. For instance, if it is known that a PMT inhibitor is substrate competitive, it is worth testing its potency against several PMT substrates to avoid a situation where the PMT inhibitor could only compete with weak-binding but not tight-binding substrates. In contrast, if a PMT inhibitor is SAM competitive, more efforts should be made to examine how intracellular concentrations of SAM affect the EC50 of the inhibitor and to define potential cross-activity against other methyltransferases. For any irreversible (covalently-interacting) inhibitor, lack of off-target effects should be addressed vigorously. Although the initial characterization consumes extra time and resources, the effort will be repaid by narrowing the focus on well-behaving leads for optimization. The key here is to be aware of Frye’s five principles of chemical probes.
Summary and Perspective
During the past decade, PMTs have caught significant attention because of their roles in epigenetics and diseases. Academic and industrial laboratories are highly engaged in developing tools to elucidate and manipulate PMT-involved methylation. This article has reviewed the current available chemical biology approaches for PMTs. These tools were further categorized into four modules: assays, substrates, cofactors and inhibitors. Herein I reviewed how the current chemical and biochemical assays can be applied to study PMTs. In particular, reliable HTS assays are still needed for identifying PMT inhibitors. In terms of PMT substrates, examining PMTs in the context of well-defined proteins and protein complexes will surely shed light on how PMTs behave in biological contexts. The current focus on this aspect still lies in histones or nuclesomes, however should be extended to nonhistone proteins. Emerging SAM analogues and PMT inhibitors surely diversify our tools to interrogate PMT functions. However, more efforts need to be put into characterizing these inhibitors in details, and in particular how they interact with PMT targets. Few efforts have been made over the past decade to experimentally characterize the transition state structures of PMT-catalyzed reactions. Elucidating the transition state structures of PMT-catalyzed reactions can provide meaningful guidance in designing novel PMT inhibitors. These chemical biology approaches have infiltrated many aspects of PMT-related research and will contribute to our understanding of PMT biology.
Acknowledgments
The author thanks the members in the Luo laboratory for their comments about the manuscript and financial support from the V Foundation for Cancer Research through 2009 V Foundation Scholar Award, March of Dimes Birth Defects Foundation through 2010 Basil O’Connor Starter Scholar, Alfred W. Bressler Scholars Endowment Fund Alfred through Alfred W. Bressler Scholar, NIH through NINDS (1R21NS071520-01), NIGMS (1R01GM096056-01) and the NIH Director’s New Innovator Award Program (1-DP2-OD007335-01), The Starr Foundation, W. H. Goodwin and A. Goodwin the Commonwealth Foundation for Cancer Research and The Experimental Therapeutics Center of Memorial Sloan-Kettering Cancer Center.
Abbreviations
- ADMA
asymmetric dimethylarginine
- ALA
acetyllysine analogue
- AMI
arginine methyltransferase inhibitor
- APRT
adenine phosphoribosyl transferase
- CARM1 or PRMT4
coactivator-associated arginine methyltransferase 1
- CBP
CREB (cAMP-response element-binding protein)-binding protein
- CHIP
chromatin immunoprecipitation
- DELFIA
dissociation enhanced lanthanide fluoroimmunoassay
- DNMT1
DNA methyltransferase 1
- ERα
estrogen receptor alpha
- ELISA
enzyme-linked immunosorbent assay
- EPL
expressed protein ligation
- FOXO1
Forkhead box protein O1
- HTS
high throughput screening
- LuxS
S-ribosylhomocysteine lyase
- MALDI-TOF
matrix assisted laser desorption/ionization time-of-flight
- MMA
monomethylarginine
- MLA
methyllysine analogue
- MLL
mixed lineage leukemia
- MS
mass spectrometry
- MTAN
5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase
- NCL
native chemical ligation
- NSM
Nonsense-suppression mutagenesis
- PCAF
p300/CBP associated factor
- PKMT(s)
protein lysine methyltransferases
- PMT(s)
protein methyltransferase(s)
- PPDK
pyruvate orthophosphate dikinase
- PRMT(s)
protein arginine methyltransferases
- RUNX1
Runt-related transcription factor 1
- SAM
S-adenosylmethionine
- SDMA
symmetric dimethylarginine
- SILAC
stable isotope labeling with amino acids
- SPA
scintillation proximity assay
- STAT1
signal transducer and activator of transcription 1
- TAF10
transcription initiation factor TFIID subunit 10
- THW loop
threonine-histidine- tryptophan loop
- TR-FRET
time-resolved fluorescence resonance energy transfer
References
- 1.Schapira M. Structural Chemistry of Human SET Domain Protein Methyltransferases. Curr Chem Genomics. 2011;5:85–94. doi: 10.2174/1875397301005010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bedford MT, Clarke SG. Protein Arginine Methylation in Mammals: Who, What, and Why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richon VM, Johnston D, Sneeringer CJ, Jin L, Majer CR, Elliston K, Jerva LF, Scott MP, Copeland RA. Chemogenetic analysis of human protein methyltransferases. Chem Biol Drug Des. 2011;78:199–210. doi: 10.1111/j.1747-0285.2011.01135.x. [DOI] [PubMed] [Google Scholar]
- 4.Kouzarides T. SnapShot: Histone-modifying enzymes. Cell. 2007;128:802–803. doi: 10.1016/j.cell.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 5.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Barski A, Cuddapah S, Cui KR, Roh TY, Schones DE, Wang ZB, Wei G, Chepelev I, Zhao KJ. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 7.Lee YH, Stallcup MR. Minireview: Protein Arginine Methylation of Nonhistone Proteins in Transcriptional Regulation. Mol Endocrinol. 2009;23:425–433. doi: 10.1210/me.2008-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mowen KA, Tang J, Zhu W, Schurter BT, Shuai K, Herschman HR, David M. Arginine methylation of STAT1 modulates IFNalpha/beta-induced transcription. Cell. 2001;104:731–741. doi: 10.1016/s0092-8674(01)00269-0. [DOI] [PubMed] [Google Scholar]
- 9.Yamagata K, Daitoku H, Takahashi Y, Namiki K, Hisatake K, Kako K, Mukai H, Kasuya Y, Fukamizu A. Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol Cell. 2008;32:221–231. doi: 10.1016/j.molcel.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 10.Zhao X, Jankovic V, Gural A, Huang G, Pardanani A, Menendez S, Zhang J, Dunne R, Xiao A, Erdjument-Bromage H, Allis CD, Tempst P, Nimer SD. Methylation of RUNX1 by PRMT1 abrogates SIN3A binding and potentiates its transcriptional activity. Genes Dev. 2008;22:640–653. doi: 10.1101/gad.1632608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu W, Chen HW, Du KY, Asahara H, Tini M, Emerson BM, Montminy M, Evans RM. A transcriptional switch mediated by cofactor methylation. Science. 2001;294:2507–2511. doi: 10.1126/science.1065961. [DOI] [PubMed] [Google Scholar]
- 12.Chevillard-Briet M, Trouche D, Vandel L. Control of CBP co-activating activity by arginine methylation. EMBO Journal. 2002;21:5457–5466. doi: 10.1093/emboj/cdf548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bedford MT, Richard S. Arginine methylation: An emerging regulator of protein function. Mol Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, Barlev NA, Reinberg D. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 15.Huang J, Dorsey J, Chuikov S, Zhang XY, Jenuwein T, Reinberg D, Berger SL. G9a and Glp Methylate Lysine 373 in the Tumor Suppressor p53. J Biol Chem. 2010;285:9636–9641. doi: 10.1074/jbc.M109.062588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, Kubicek S, Opravil S, Jenuwein T, Berger SL. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444:629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- 17.Kurash JK, Lei H, Shen Q, Marston WL, Granda BW, Fan H, Wall D, Li E, Gaudet F. Methylation of p53 by Set7/9 mediates p53 acetylation and activity in vivo. Mol Cell. 2008;29:392–400. doi: 10.1016/j.molcel.2007.12.025. [DOI] [PubMed] [Google Scholar]
- 18.Scoumanne A, Chen X. Protein methylation: a new mechanism of p53 tumor suppressor regulation. Histol Histopathol. 2008;23:1143–1149. doi: 10.14670/hh-23.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi XB, Kachirskaia L, Yamaguchi H, West LE, Wen H, Wang EW, Dutta S, Appella E, Gozani O. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell. 2007;27:636–646. doi: 10.1016/j.molcel.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Copeland RA, Solomon ME, Richon VM. Protein methyltransferases as a target class for drug discovery. Nat Rev Drug Discov. 2009;8:724–732. doi: 10.1038/nrd2974. [DOI] [PubMed] [Google Scholar]
- 21.Cheung N, Chan LC, Thompson A, Cleary ML, So CWE. Protein arginine-methyltransferase-dependent oncogenesis. Nat Cell Biol. 2007;9:1208–1215. doi: 10.1038/ncb1642. [DOI] [PubMed] [Google Scholar]
- 22.Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, Johnston LD, Scott MP, Smith JJ, Xiao Y, Jin L, Kuntz KW, Chesworth R, Moyer MP, Bernt KM, Tseng J-C, Kung AL, Armstrong SA, Copeland RA, Richon VM, Pollock RM. Selective Killing of Mixed Lineage Leukemia Cells by a Potent Small-Molecule DOT1L Inhibitor. Cancer Cell. 2011;20:53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munro S, Khaire N, Inche A, Carr S, La Thangue NB. Lysine methylation regulates the pRb tumour suppressor protein. Oncogene. 2010;29:2357–2367. doi: 10.1038/onc.2009.511. [DOI] [PubMed] [Google Scholar]
- 24.Pagans S, Kauder SE, Kaehlcke K, Sakane N, Schroeder S, Dormeyer W, Trievel RC, Verdin E, Schnolzer M, Ott M. The Cellular Lysine Methyltransferase Set7/9-KMT7 Binds HIV-1 TAR RNA, Monomethylates the Viral Transactivator Tat, and Enhances HIV Transcription. Cell Host Microbe. 2010;7:234–244. doi: 10.1016/j.chom.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pradhan S, Chin HG, Esteve PO, Jacobsen SE. SET7/9 mediated methylation of non-histone proteins in mammalian cells. Epigenetics. 2009;4:282–285. doi: 10.4161/epi.4.6.9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dhayalan A, Kudithipudi S, Rathert P, Jeltsch A. Specificity analysis-based identification of new methylation targets of the SET7/9 protein lysine methyltransferase. Chem Biol. 2011;18:111–120. doi: 10.1016/j.chembiol.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 27.Prescher JA, Bertozzi CR. Chemistry in living systems. Nat Chem Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 28.Bishop AC, Buzko O, Shokat KM. Magic bullets for protein kinases. Trends Cell Biol. 2001;11:167–172. doi: 10.1016/s0962-8924(01)01928-6. [DOI] [PubMed] [Google Scholar]
- 29.Cole PA. Chemical probes for histone-modifying enzymes. Nat Chem Biol. 2008;4:590–597. doi: 10.1038/nchembio.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang YY, Hang HC. Chemical approaches for the detection and synthesis of acetylated proteins. Chembiochem. 2011;12:314–322. doi: 10.1002/cbic.201000558. [DOI] [PubMed] [Google Scholar]
- 31.Islam K, Zheng W, Yu H, Deng H, Luo M. Expanding Cofactor Repertoire of Protein Lysine Methyltransferase for Substrate Labeling. ACS Chem Biol. 2011;6:679–684. doi: 10.1021/cb2000567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang R, Zheng W, Yu H, Deng H, Luo M. Labeling Substrates of Protein Arginine Methyltransferase with Engineered Enzymes and Matched S-Adenosyl-L-methionine Analogues. J Am Chem Soc. 2011;133:7648–7651. doi: 10.1021/ja2006719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voigt P, Reinberg D. Histone tails: ideal motifs for probing epigenetics through chemical biology approaches. Chembiochem. 2011;12:236–252. doi: 10.1002/cbic.201000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allis CD, Muir TW. Spreading chromatin into chemical biology. Chembiochem. 2011;12:264–279. doi: 10.1002/cbic.201000761. [DOI] [PubMed] [Google Scholar]
- 35.Suh-Lailam BB, Hevel JM. A fast and efficient method for quantitative measurement of S-adenosyl-L-methionine-dependent methyltransferase activity with protein substrates. Anal Biochem. 2010;398:218–224. doi: 10.1016/j.ab.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 36.Wu J, Xie N, Feng Y, Zheng YG. Scintillation Proximity Assay of Arginine Methylation. J Biomol Screen. 2012;2011 doi: 10.1177/1087057111414903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rathert P, Cheng X, Jeltsch A. Continuous enzymatic assay for histone lysine methyltransferases. Biotechniques. 2007;43:602–608. doi: 10.2144/000112623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gowher H, Zhang X, Cheng XD, Jeltsch A. Avidin plate assay system for enzymatic characterization of a histone lysine methyltransferase. Anal Biochem. 2005;342:287–291. doi: 10.1016/j.ab.2005.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dhayalan A, Dimitrova E, Rathert P, Jeltsch A. A continuous protein methyltransferase (G9a) assay for enzyme activity measurement and inhibitor screening. J Biomol Screen. 2009;14:1129–1133. doi: 10.1177/1087057109345528. [DOI] [PubMed] [Google Scholar]
- 40.Quinn AM, Allali-Hassani A, Vedadi M, Simeonov A. A chemiluminescence-based method for identification of histone lysine methyltransferase inhibitors. Mol BioSyst. 2010;6:782–788. doi: 10.1039/b921912a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gauthier N, Caron M, Pedro L, Arcand M, Blouin J, Labonte A, Normand C, Paquet V, Rodenbrock A, Roy M, Rouleau N, Beaudet L, Padros J, Rodriguez-Suarez R. Development of Homogeneous Nonradioactive Methyltransferase and Demethylase Assays Targeting Histone H3 Lysine 4. J Biomol Screen. 2012;2011 doi: 10.1177/1087057111416659. [DOI] [PubMed] [Google Scholar]
- 42.Hauser AT, Bissinger EM, Metzger E, Repenning A, Bauer UM, Mai A, Schule R, Jung M. Screening Assays for Epigenetic Targets Using Native Histones as Substrates. J Biomol Screen. 2012 doi: 10.1177/1087057111423968. [DOI] [PubMed] [Google Scholar]
- 43.Machleidt T, Robers MB, Hermanson SB, Dudek JM, Bi K. TR-FRET Cellular Assays for Interrogating Posttranslational Modifications of Histone H3. J Biomol Screen. 2012:4. doi: 10.1177/1087057111422943. [DOI] [PubMed] [Google Scholar]
- 44.Patel A, Dharmarajan V, Vought VE, Cosgrove MS. On the Mechanism of Multiple Lysine Methylation by the Human Mixed Lineage Leukemia Protein-1 (MLL1) Core Complex. J Biol Chem. 2009;284:24242–24256. doi: 10.1074/jbc.M109.014498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pesavento JJ, Yang H, Kelleher NL, Mizzen CA. Certain and progressive methylation of histone H4 at lysine 20 during the cell cycle. Mol Cell Biol. 2008;28:468–486. doi: 10.1128/MCB.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zee BM, Levin RS, Xu B, LeRoy G, Wingreen NS, Garcia BA. In Vivo Residue-specific Histone Methylation Dynamics. J Biol Chem. 2010;285:3341–3350. doi: 10.1074/jbc.M109.063784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abu-Farha M, Lanouette S, Elisma F, Tremblay V, Butson J, Figeys D, Couture JF. Proteomic analyses of the SMYD family interactomes identify HSP90 as a novel target for SMYD2. J Mol Cell Biol. 2011;3:301–308. doi: 10.1093/jmcb/mjr025. [DOI] [PubMed] [Google Scholar]
- 48.Carr SM, Munro S, Kessler B, Oppermann U, La Thangue NB. Interplay between lysine methylation and Cdk phosphorylation in growth control by the retinoblastoma protein. Embo J. 2011;30:317–327. doi: 10.1038/emboj.2010.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wigle TJ, Provencher LM, Norris JL, Jin JA, Brown PJ, Frye SV, Janzen WP. Accessing Protein Methyltransferase and Demethylase Enzymology Using Microfluidic Capillary Electrophoresis. Chem Biol. 2010;17:695–704. doi: 10.1016/j.chembiol.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee JH, Cook JR, Yang ZH, Mirochnitchenko O, Gunderson SI, Felix AM, Herth N, Hoffmann R, Pestka S. PRMT7, a new protein arginine methyltransferase that synthesizes symmetric dimethylarginine. J Biol Chem. 2005;280:3656–3664. doi: 10.1074/jbc.M405295200. [DOI] [PubMed] [Google Scholar]
- 51.Branscombe TL, Frankel A, Lee JH, Cook JR, Yang Z, Pestka S, Clarke S. PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J Biol Chem. 2001;276:32971–32976. doi: 10.1074/jbc.M105412200. [DOI] [PubMed] [Google Scholar]
- 52.Lakowski TM, Frankel A. Kinetic analysis of human protein arginine N-methyltransferase 2: formation of monomethyl- and asymmetric dimethyl-arginine residues on histone H4. Biochem J. 2009;421:253–261. doi: 10.1042/BJ20090268. [DOI] [PubMed] [Google Scholar]
- 53.Zou Y, Wang YS. Tandem mass spectrometry for the examination of the posttranslational modifications of high-mobility group A1 proteins: Symmetric and asymmetric dimethylation of Arg25 in HMGA1a protein. Biochemistry. 2005;44:6293–6301. doi: 10.1021/bi0475525. [DOI] [PubMed] [Google Scholar]
- 54.Lakowski TM, Frankel A. Sources of S-adenosyl-L-homocysteine background in measuring protein arginine N-methyltransferase activity using tandem mass spectrometry. Anal Biochem. 2010;396:158–160. doi: 10.1016/j.ab.2009.08.043. [DOI] [PubMed] [Google Scholar]
- 55.Capdevila A, Burk RF, Freedman J, Frantzen F, Alfbeim I, Wagner C. A simple rapid immunoassay for S-adenosylhomocysteine in plasma. J Nutr Biochem. 2007;18:827–831. doi: 10.1016/j.jnutbio.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Graves TL, Zhang Y, Scott JE. A universal competitive fluorescence polarization activity assay for S-adenosylmethionine utilizing methyltransferases. Anal Biochem. 2008;373:296–306. doi: 10.1016/j.ab.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hendricks CL, Ross JR, Pichersky E, Noel JP, Zhou ZS. An enzyme-coupled colorimetric assay for S-adenosylmethionine-dependent methyltransferases. Anal Biochem. 2004;326:100–105. doi: 10.1016/j.ab.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 58.Wang CH, Leffler S, Thompson DH, Hrycyna CA. A general fluorescence-based coupled assay for S-adenosylmethionine-dependent methyltransferases. Biochem Biophys Res Comm. 2005;331:351–356. doi: 10.1016/j.bbrc.2005.03.170. [DOI] [PubMed] [Google Scholar]
- 59.Collazo E, Couture JF, Bulfer S, Trievel RC. A coupled fluorescent assay for histone methyltransferases. Anal Biochem. 2005;342:86–92. doi: 10.1016/j.ab.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 60.Ibáñez G, McBean JL, Astudillo YM, Luo M. An Enzyme-coupled Ultrasensitive Luminescence Assay for Protein Methyltransferases. Anal Biochem. 2010;401:203–210. doi: 10.1016/j.ab.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 61.Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 62.Dorgan KM, Wooderchak WL, Wynn DP, Karschner EL, Alfaro JF, Cui YQ, Zhou ZS, Hevel JM. An enzyme-coupled continuous spectrophotometric assay for S-adenosylmethionine-dependent methyltransferases. Anal Biochem. 2006;350:249–255. doi: 10.1016/j.ab.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 63.Klink TA, Staeben M, Twesten K, Kopp AL, Kumar M, Schall Dunn R, Pinchard CA, Kleman-Leyer KM, Klumpp M, Lowery RG. Development and Validation of a Generic Fluorescent Methyltransferase Activity Assay Based on the Transcreener AMP/GMP Assay. J Biomol Screen. 2012:28. doi: 10.1177/1087057111421624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang R, Ibáñez G, Islam K, Zheng W, Blum G, Sengelaub C, Luo M. Formulating Fluorogenic Assay to Evaluate S-adenosyl-L-methionine Analogues as Protein Methyltransferase Cofactors. Mol Biosyst. 2011;11:2970–2981. doi: 10.1039/c1mb05230f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cheng DH, Yadav N, King RW, Swanson MS, Weinstein EJ, Bedford MT. Small molecule regulators of protein arginine methyltransferases. J Biol Chem. 2004;279:23892–23899. doi: 10.1074/jbc.M401853200. [DOI] [PubMed] [Google Scholar]
- 66.Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3–9. Nat Chem Biol. 2005;1:143–145. doi: 10.1038/nchembio721. [DOI] [PubMed] [Google Scholar]
- 67.Purandare AV, Chen Z, Huynh T, Pang S, Geng J, Vaccaro W, Poss MA, Oconnell J, Nowak K, Jayaraman L. Pyrazole inhibitors of coactivator associated arginine methyltransferase 1 (CARM1) Bioorg Med Chem Lett. 2008;18:4438–4441. doi: 10.1016/j.bmcl.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 68.Kubicek S, O’Sullivan RJ, August EM, Hickey ER, Zhan Q, Teodoro ML, Rea S, Mechtler K, Kowalski JA, Homon CA, Kelly TA, Jenuwein T. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell. 2007;25:473–481. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 69.Ferguson AD, Larsen NA, Howard T, Pollard H, Green I, Grande C, Cheung T, Garcia-Arenas R, Cowen S, Wu J, Godin R, Chen H, Keen N. Structural Basis of Substrate Methylation and Inhibition of SMYD2. Structure. 2011;19:1262–1273. doi: 10.1016/j.str.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 70.Osborne TC, Obianyo O, Zhang X, Cheng X, Thompson PR. Protein arginine methyltransferase 1: Positively charged residues in substrate peptides distal to the site of methylation are important for substrate binding and catalysis. Biochemistry. 2007;46:13370–13381. doi: 10.1021/bi701558t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Obianyo O, Osborne TC, Thompson PR. Kinetic mechanism of protein arginine methyltransferase 1. Biochemistry. 2008;47:10420–10427. doi: 10.1021/bi800904m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wooderchak WL, Zang TZ, Zhou ZS, Acuna M, Tahara SM, Hevel JM. Substrate profiling of PRMT1 reveals amino acid sequences that extend beyond the “RGG” paradigm. Biochemistry. 2008;47:9456–9466. doi: 10.1021/bi800984s. [DOI] [PubMed] [Google Scholar]
- 73.Chin HG, Pradhan M, Esteve PO, Patnaik D, Evans TC, Jr, Pradhan S. Sequence specificity and role of proximal amino acids of the histone H3 tail on catalysis of murine G9A lysine 9 histone H3 methyltransferase. Biochemistry. 2005;44:12998–13006. doi: 10.1021/bi0509907. [DOI] [PubMed] [Google Scholar]
- 74.Rust HL, Thompson PR. Kinase consensus sequences: a breeding ground for crosstalk. ACS Chem Biol. 2011;6:881–892. doi: 10.1021/cb200171d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Feng Y, Xie N, Jin M, Stahley MR, Stivers JT, Zheng YG. A Transient Kinetic Analysis of PRMT1 Catalysis. Biochemistry. 2011;50:7033–7044. doi: 10.1021/bi200456u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Feng Y, Xie N, Wu J, Yang C, Zheng YG. Inhibitory study of protein arginine methyltransferase 1 using a fluorescent approach. Biochem Biophys Res Comm. 2009;379:567–572. doi: 10.1016/j.bbrc.2008.12.119. [DOI] [PubMed] [Google Scholar]
- 77.Li Y, Trojer P, Xu CF, Cheung P, Kuo A, Drury WJ, 3rd, Qiao Q, Neubert TA, Xu RM, Gozani O, Reinberg D. The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. J Biol Chem. 2009;284:34283–34295. doi: 10.1074/jbc.M109.034462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 79.Daujat S, Bauer UM, Shah V, Turner B, Berger S, Kouzarides T. Crosstalk between CARM1 methylation and CBP acetylation on histone H3. Curr Biol. 2002;12:2090–2097. doi: 10.1016/s0960-9822(02)01387-8. [DOI] [PubMed] [Google Scholar]
- 80.Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 81.Dhall A, Chatterjee C. Chemical approaches to understand the language of histone modifications. ACS Chem Biol. 2011;6:987–999. doi: 10.1021/cb200142c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007;128:1003–1012. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, Pirrotta V, Reinberg D, Gamblin SJ. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang R, Holbert MA, Tarrant MK, Curtet S, Colquhoun DR, Dancy BM, Dancy BC, Hwang Y, Tang Y, Meeth K, Marmorstein R, Cole RN, Khochbin S, Cole PA. Site-specific introduction of an acetyl-lysine mimic into peptides and proteins by cysteine alkylation. J Am Chem Soc. 2010;132:9986–9987. doi: 10.1021/ja103954u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Greiss S, Chin JW. Expanding the Genetic Code of an Animal. J Am Chem Soc. 2011;133:14196–14199. doi: 10.1021/ja2054034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guo J, Wang J, Lee JS, Schultz PG. Site-specific incorporation of methyl- and acetyl-lysine analogues into recombinant proteins. Angew Chem Int Ed. 2008;47:6399–6401. doi: 10.1002/anie.200802336. [DOI] [PubMed] [Google Scholar]
- 87.Nguyen DP, Alai MMG, Virdee S, Chin JW. Genetically Directing epsilon-N, N-Dimethyl-L-Lysine in Recombinant Histones. Chem Biol. 2010;17:1072–1076. doi: 10.1016/j.chembiol.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 88.Wang YS, Wu B, Wang Z, Huang Y, Wan W, Russell WK, Pai PJ, Moe YN, Russell DH, Liu WR. A genetically encoded photocaged N(epsilon)-methyl-L-lysinewz. Mol BioSyst. 2010;6:1575–1578. doi: 10.1039/c002155e. [DOI] [PubMed] [Google Scholar]
- 89.Groff D, Chen PR, Peters FB, Schultz PG. A genetically encoded epsilon-N-methyl lysine in mammalian cells. Chembiochem. 2010;11:1066–1068. doi: 10.1002/cbic.200900690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ai HW, Lee JW, Schultz PG. A method to site-specifically introduce methyllysine into proteins in E. coli. Chem Commun. 2010;46:5506–5508. doi: 10.1039/c0cc00108b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW. A Method for Genetically Installing Site-Specific Acetylation in Recombinant Histories Defines the Effects of H3 K56 Acetylation. Mol Cell. 2009;36:153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Neumann H, Peak-Chew SY, Chin JW. Genetically encoding N-epsilon-acetyllysine in recombinant proteins. Nat Chem Biol. 2008;4:232–234. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- 93.Huang Y, Russell WK, Wan W, Pai PJ, Russell DH, Liu W. A convenient method for genetic incorporation of multiple noncanonical amino acids into one protein in Escherichia coli. Mol BioSyst. 2010;6:683–686. doi: 10.1039/b920120c. [DOI] [PubMed] [Google Scholar]
- 94.Virdee S, Kapadnis PB, Elliott T, Lang K, Madrzak J, Nguyen DP, Riechmann L, Chin JW. Traceless and Site-Specific Ubiquitination of Recombinant Proteins. J Am Chem Soc. 2011;133:10708–10711. doi: 10.1021/ja202799r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Neumann H, Wang K, Davis L, Garcia-Alai M, Chin JW. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature. 2010;464:441–444. doi: 10.1038/nature08817. [DOI] [PubMed] [Google Scholar]
- 96.Wan W, Huang Y, Wang Z, Russell WK, Pai PJ, Russell DH, Liu WR. A Facile System for Genetic Incorporation of Two Different Noncanonical Amino Acids into One Protein in Escherichia coli. Angew Chem Int Ed. 2010;49:3211–3214. doi: 10.1002/anie.201000465. [DOI] [PubMed] [Google Scholar]
- 97.McGinty RK, Kohn M, Chatterjee C, Chiang KP, Pratt MR, Muir TW. Structure-activity analysis of semisynthetic nucleosomes: mechanistic insights into the stimulation of Dot1L by ubiquitylated histone H2B. ACS Chem Biol. 2009;4:958–968. doi: 10.1021/cb9002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453:812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chatterjee C, McGinty RK, Fierz B, Muir TW. Disulfide-directed histone ubiquitylation reveals plasticity in hDot1L activation. Nat Chem Biol. 2010;6:267–269. doi: 10.1038/nchembio.315. [DOI] [PubMed] [Google Scholar]
- 100.Rathert P, Zhang X, Freund C, Cheng XD, Jeltsch A. Analysis of the substrate specificity of the dim-5 histone lysine methyltransferase using peptide arrays. Chem Biol. 2008;15:5–11. doi: 10.1016/j.chembiol.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rathert P, Dhayalan A, Murakami M, Zhang X, Tamas R, Jurkowska R, Komatsu Y, Shinkai Y, Cheng XD, Jeltsch A. Protein lysine methyltransferase G9a acts on non-histone targets. Nat Chem Biol. 2008;4:344–346. doi: 10.1038/nchembio.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bicker KL, Obianyo O, Rust HL, Thompson PR. A combinatorial approach to characterize the substrate specificity of protein arginine methyltransferase 1. Mol Biosyst. 2011;7:48–51. doi: 10.1039/c0mb00015a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Obianyo O, Causey CP, Jones JE, Thompson PR. Activity-Based Protein Profiling of Protein Arginine Methyltransferase 1. ACS Chem Biol. 2011;6:1127–1135. doi: 10.1021/cb2001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Levy D, Liu CL, Yang Z, Newman AM, Alizadeh AA, Utz PJ, Gozani O. A proteomic approach for the identification of novel lysine methyltransferase substrates. Epigenetics Chromatin. 2011;4:19. doi: 10.1186/1756-8935-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lee JS, Kim Y, Kim IS, Kim B, Choi HJ, Lee JM, Shin HJ, Kim JH, Kim JY, Seo SB, Lee H, Binda O, Gozani O, Semenza GL, Kim M, Kim KI, Hwang D, Baek SH. Negative regulation of hypoxic responses via induced Reptin methylation. Mol Cell. 2010;39:71–85. doi: 10.1016/j.molcel.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li Y, Reddy MA, Miao F, Shanmugam N, Yee JK, Hawkins D, Ren B, Natarajan R. Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-kappaB-dependent inflammatory genes. Relevance to diabetes and inflammation. J Biol Chem. 2008;283:26771–26781. doi: 10.1074/jbc.M802800200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Frankel A, Yadav N, Lee J, Branscombe TL, Clarke S, Bedford MT. The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J Biol Chem. 2002;277:3537–3543. doi: 10.1074/jbc.M108786200. [DOI] [PubMed] [Google Scholar]
- 108.Swiercz R, Person MD, Bedford MT. Ribosomal protein S2 is a substrate for mammalian PRMT3 (protein arginine methyltransferase 3) Biochem J. 2005;386:85–91. doi: 10.1042/BJ20041466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Boisvert FM, Cote J, Boulanger MC, Richard S. A proteomic analysis of arginine-methylated protein complexes. Mol Cell Proteomics. 2003;2:1319–1330. doi: 10.1074/mcp.M300088-MCP200. [DOI] [PubMed] [Google Scholar]
- 110.Klimasauskas S, Weinhold E. A new tool for biotechnology: AdoMet-dependent methyltransferases. Trends Biotechnol. 2007;25:99–104. doi: 10.1016/j.tibtech.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 111.Yan F, LaMarre JM, Rohrich R, Wiesner J, Jomaa H, Mankin AS, Fujimori DG. RlmN and Cfr are radical SAM enzymes involved in methylation of ribosomal RNA. J Am Chem Soc. 2010;132:3953–3964. doi: 10.1021/ja910850y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang Y, Zhu X, Torelli AT, Lee M, Dzikovski B, Koralewski RM, Wang E, Freed J, Krebs C, Ealick SE, Lin H. Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme. Nature. 2010;465:891–896. doi: 10.1038/nature09138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lin Q, Jiang FY, Schultz PG, Gray NS. Design of allele-specific protein methyltransferase inhibitors. J Am Chem Soc. 2001;123:11608–11613. doi: 10.1021/ja011423j. [DOI] [PubMed] [Google Scholar]
- 114.Li J, Wei H, Zhou MM. Structure-guided design of a methyl donor cofactor that controls a viral histone h3 lysine 27 methyltransferase activity. J Med Chem. 2011;54:7734–7738. doi: 10.1021/jm201000j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Osborne T, Roska RLW, Rajski SR, Thompson PR. In situ generation of a bisubstrate analogue for protein arginine methyltransferase 1. J Am Chem Soc. 2008;130:4574–4575. doi: 10.1021/ja077104v. [DOI] [PubMed] [Google Scholar]
- 116.Yao Y, Chen P, Diao J, Cheng G, Deng L, Anglin JL, Prasad BVV, Song Y. Selective Inhibitors of Histone Methyltransferase DOT1L: Design, Synthesis, and Crystallographic Studies. J Am Chem Soc. 2011;133:16746–16749. doi: 10.1021/ja206312b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Peters W, Willnow S, Duisken M, Kleine H, Macherey T, Duncan KE, Litchfield DW, Luscher B, Weinhold E. Enzymatic Site-Specific Functionalization of Protein Methyltransferase Substrates with Alkynes for Click Labeling. Angew Chem Int Ed. 2010;49:5170–5173. doi: 10.1002/anie.201001240. [DOI] [PubMed] [Google Scholar]
- 118.Binda O, Boyce M, Rush JS, Palaniappan KK, Bertozzi CR, Gozani O. A Chemical Method for Labeling Lysine Methyltransferase Substrates. Chembiochem. 2011;12:330–334. doi: 10.1002/cbic.201000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen DH, Wu KT, Hung CJ, Hsieh M, Li C. Effects of adenosine dialdehyde treatment on in vitro and in vivo stable protein methylation in HeLa cells. J Biochem. 2004;136:371–376. doi: 10.1093/jb/mvh131. [DOI] [PubMed] [Google Scholar]
- 120.Frye SV. The art of the chemical probe. Nat Chem Biol. 2010;6:159–161. doi: 10.1038/nchembio.296. [DOI] [PubMed] [Google Scholar]
- 121.Obianyo O, Causey CP, Osborne TC, Jones JE, Lee YH, Stallcup MR, Thompson PR. A chloroacetamidine-based inactivator of protein arginine methyltransferase 1: design, synthesis, and in vitro and in vivo evaluation. Chembiochem. 2010;11:1219–1223. doi: 10.1002/cbic.201000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.t Hart P, Lakowski TM, Thomas D, Frankel A, Martin NI. Peptidic Partial Bisubstrates as Inhibitors of the Protein Arginine N-Methyltransferases. Chembiochem. 2011;12:1427–1432. doi: 10.1002/cbic.201100074. [DOI] [PubMed] [Google Scholar]
- 123.Dowden J, Hong W, Parry RV, Pike RA, Ward SG. Toward the development of potent and selective bisubstrate inhibitors of protein arginine methyltransferases. Bioorg Med Chem Lett. 2010;20:2103–2105. doi: 10.1016/j.bmcl.2010.02.069. [DOI] [PubMed] [Google Scholar]
- 124.Dowden J, Pike RA, Parry RV, Hong W, Muhsen UA, Ward SG. Small molecule inhibitors that discriminate between protein arginine N-methyltransferases PRMT1 and CARM1. Org Biomol Chem. 2011;9:7814–7821. doi: 10.1039/c1ob06100c. [DOI] [PubMed] [Google Scholar]
- 125.Mori S, Iwase K, Iwanami N, Tanaka Y, Kagechika H, Hirano T. Development of novel bisubstrate-type inhibitors of histone methyltransferase SET7/9. Bioorg Med Chem Lett. 2010;18:8158–8166. doi: 10.1016/j.bmc.2010.10.022. [DOI] [PubMed] [Google Scholar]
- 126.Campagna-Slater V, Mok MW, Nguyen KT, Feher M, Najmanovich R, Schapira M. Structural chemistry of the histone methyltransferases cofactor binding site. J Chem Inf Model. 2011;51:612–623. doi: 10.1021/ci100479z. [DOI] [PubMed] [Google Scholar]
- 127.Cheng D, Valente S, Castellano S, Sbardella G, Di Santo R, Costi R, Bedford MT, Mai A. Novel 3,5-Bis(bromohydroxybenzylidene)piperidin-4-ones as Coactivator-Associated Arginine Methyltransferase 1 Inhibitors: Enzyme Selectivity and Cellular Activity. J Med Chem. 2011;54:4928–4932. doi: 10.1021/jm200453n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bonham K, Hemmers S, Lim Y-H, Hill DM, Finn MG, Mowen KA. Effects of a novel arginine methyltransferase inhibitor on T-helper cell cytokine production. FEBS J. 2010;277:2096–2108. doi: 10.1111/j.1742-4658.2010.07623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sack JS, Thieffine S, Bandiera T, Fasolini M, Duke GJ, Jayaraman L, Kish KF, Klei HE, Purandare AV, Rosettani P, Troiani S, Xie D, Bertrand JA. Structural basis for CARM1 inhibition by indole and pyrazole inhibitors. Biochem J. 2011;436:331–339. doi: 10.1042/BJ20102161. [DOI] [PubMed] [Google Scholar]
- 130.Bernhard W, Barreto K, Saunders A, Dahabieh MS, Johnson P, Sadowski I. The Suv39H1 methyltransferase inhibitor chaetocin causes induction of integrated HIV-1 without producing a T cell response. FEBS Lett. 2011;585:3549–3554. doi: 10.1016/j.febslet.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 131.Iwasa E, Hamashima Y, Fujishiro S, Hashizume D, Sodeoka M. Total syntheses of chaetocin and ent-chaetocin. Tetrahedron. 2011;67:6587–6599. [Google Scholar]
- 132.Vedadi M, Barsyte-Lovejoy D, Liu F, Rival-Gervier S, Allali-Hassani A, Labrie V, Wigle TJ, DiMaggio PA, Wasney GA, Siarheyeva A, Dong A, Tempel W, Wang SC, Chen X, Chau I, Mangano TJ, Huang X-p, Simpson CD, Pattenden SG, Norris JL, Kireev DB, Tripathy A, Edwards A, Roth BL, Janzen WP, Garcia BA, Petronis A, Ellis J, Brown PJ, Frye SV, Arrowsmith CH, Jin J. A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nat Chem Biol. 2011;7:648–648. doi: 10.1038/nchembio.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liu F, Barsyte-Lovejoy D, Allali-Hassani A, He Y, Herold JM, Chen X, Yates CM, Frye SV, Brown PJ, Huang J, Vedadi M, Arrowsmith CH, Jin J. Optimization of cellular activity of G9a inhibitors 7-aminoalkoxy-quinazolines. J Med Chem. 2011;54:6139–6150. doi: 10.1021/jm200903z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liu F, Chen X, Allali-Hassani A, Quinn AM, Wasney GA, Dong A, Barsyte D, Kozieradzki I, Senisterra G, Chau I, Siarheyeva A, Kireev DB, Jadhav A, Herold JM, Frye SV, Arrowsmith CH, Brown PJ, Simeonov A, Vedadi M, Jin J. Discovery of a 2,4-diamino-7-aminoalkoxyquinazoline as a potent and selective inhibitor of histone lysine methyltransferase G9a. J Med Chem. 2009;52:7950–7953. doi: 10.1021/jm901543m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Liu F, Chen X, Allali-Hassani A, Quinn AM, Wigle TJ, Wasney GA, Dong A, Senisterra G, Chau I, Siarheyeva A, Norris JL, Kireev DB, Jadhav A, Herold JM, Janzen WP, Arrowsmith CH, Frye SV, Brown PJ, Simeonov A, Vedadi M, Jin J. Protein lysine methyltransferase G9a inhibitors: design, synthesis, and structure activity relationships of 2,4-diamino-7-aminoalkoxy-quinazolines. J Med Chem. 2010;53:5844–5857. doi: 10.1021/jm100478y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.McMillan JM, Walle UK, Walle T. S-adenosyl-L-methionine: transcellular transport and uptake by Caco-2 cells and hepatocytes. J Pharm Pharmacol. 2005;57:599–605. doi: 10.1211/0022357056082. [DOI] [PubMed] [Google Scholar]
- 137.Rydberg B, Lindahl T. Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-L-methionine is a potentially mutagenic reaction. EMBO J. 1982;1:211–216. doi: 10.1002/j.1460-2075.1982.tb01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Spannhoff A, Machmur R, Heinke R, Trojer P, Bauer I, Brosch G, Schuele R, Hanefeld W, Sippl W, Jung M. A novel arginine methyltransferase inhibitor with cellular activity. Bioorg Med Chem Lett. 2007;17:4150–4153. doi: 10.1016/j.bmcl.2007.05.088. [DOI] [PubMed] [Google Scholar]
- 139.Spannhoff A, Heinke R, Bauer I, Trojer P, Metzger E, Gust R, Schuele R, Brosch G, Sippl W, Jung M. Target-based approach to inhibitors of histone arginine methyltransferases. J Med Chem. 2007;50:2319–2325. doi: 10.1021/jm061250e. [DOI] [PubMed] [Google Scholar]
- 140.Heinke R, Spannhoff A, Meier R, Trojer P, Bauer I, Jung M, Sippl W. Virtual Screening and Biological Characterization of Novel Histone Arginine Methyltransferase PRMT1 Inhibitors. Chemmedchem. 2009;4:69–77. doi: 10.1002/cmdc.200800301. [DOI] [PubMed] [Google Scholar]
- 141.Bissinger EM, Heinke R, Spannhoff A, Eberlin A, Metzger E, Cura V, Hassenboehler P, Cavarelli J, Schuele R, Bedford MT, Sippl W, Jung M. Acyl derivatives of p-aminosulfonamides and dapsone as new inhibitors of the arginine methyltransferase hPRMT1. Bioorg Med Chem Lett. 2011;19:3717–3731. doi: 10.1016/j.bmc.2011.02.032. [DOI] [PubMed] [Google Scholar]
- 142.Cheng D, Bedford MT. Xenoestrogens Regulate the Activity of Arginine Methyltransferases. Chembiochem. 2011;12:323–329. doi: 10.1002/cbic.201000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Selvi BR, Batta K, Kishore AH, Mantelingu K, Varier RA, Balasubramanyam K, Pradhan SK, Dasgupta D, Sriram S, Agrawal S, Kundu TK. Identification of a Novel Inhibitor of Coactivator-associated Arginine Methyltransferase 1 (CARM1)-mediated Methylation of Histone H3 Arg-17. J Biol Chem. 2010;285:7143–7152. doi: 10.1074/jbc.M109.063933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Feng Y, Li M, Wang B, Zheng YG. Discovery and Mechanistic Study of a Class of Protein Arginine Methylation Inhibitors. J Med Chem. 2010;53:6028–6039. doi: 10.1021/jm100416n. [DOI] [PubMed] [Google Scholar]
- 145.Selvi BR, Pradhan SK, Shandilya J, Das C, Sailaja BS, Shankar GN, Gadad SS, Reddy A, Dasgupta D, Kundu TK. Sanguinarine interacts with chromatin, modulates epigenetic modifications, and transcription in the context of chromatin. Chem Biol. 2009;16:203–216. doi: 10.1016/j.chembiol.2008.12.006. [DOI] [PubMed] [Google Scholar]