Abstract
Nitrogen heterocycles are the key functional and structural elements in both RNA and DNA, in half a dozen of the most important coenzymes, and in many synthetic drug scaffolds. On the other hand, only three of twenty proteinogenic amino acids have nitrogen heterocycles: proline, histidine and tryptophan. This inventory can be augmented in microbial proteins by posttranslational modifications downstream of leader peptide regions that convert up to ten serine, threonine and cysteine residues, side chains and peptide backbones, into oxazoles, thiazoles, and pyridine rings. Subsequent proteolysis releases these heterocyclic scaffolds in both linear and macrocyclic frameworks as bioactive small molecules.
Nature loves nitrogen heterocycles
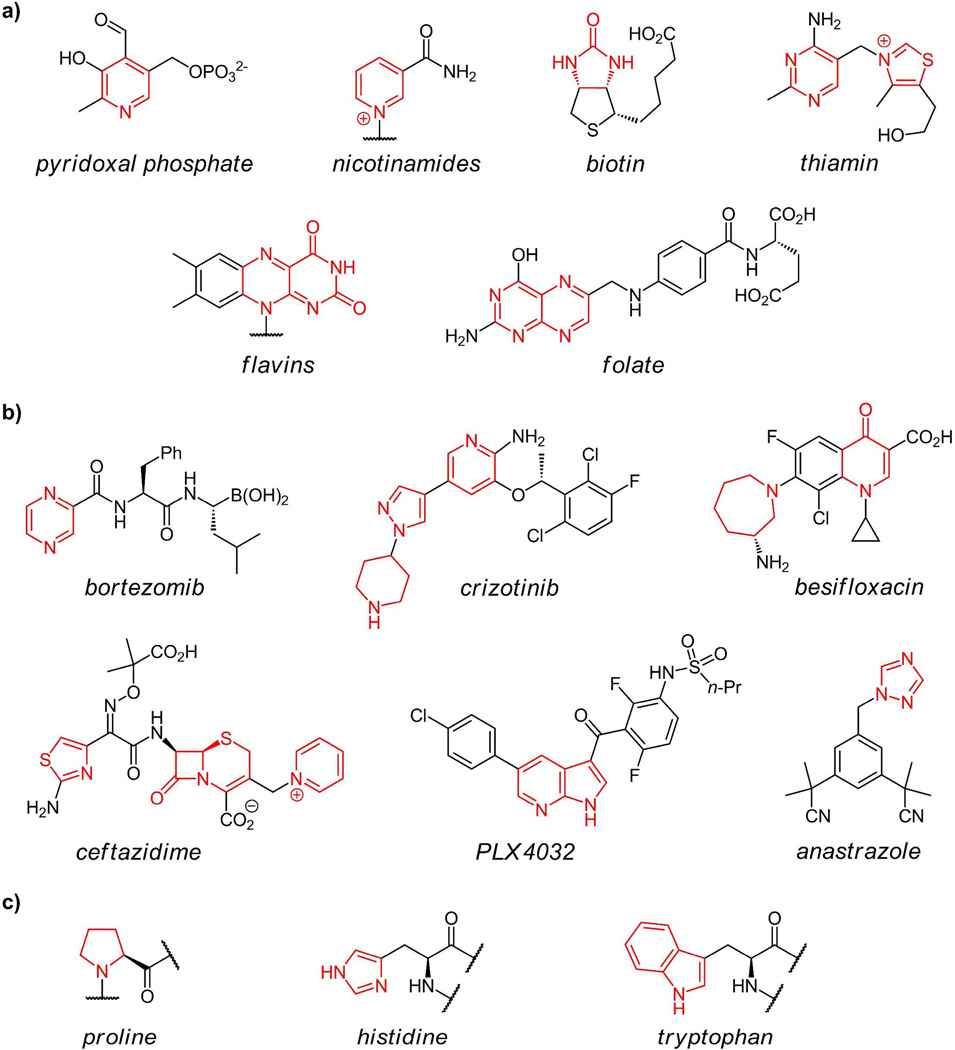
Nitrogen heterocycles are central structures for the chemistry of life. This is particularly evident in the heteroaromatic single ring pyrimidines and bicyclic purines that are the building blocks and molecular repositories for information in RNA and DNA. Also, more than a half dozen of the common coenzymes(1) (pyridoxal, biotin, folate, thiamin, nicotinamides, coenzyme B12, and flavins) have nitrogen heterocycles (figure 1a) that are essential for both structural and reactivity determinants of coenzyme function. The heterocyclic frameworks of such coenzymes suggest they may have had origins in an RNA world replete with the capacity to build two and three ring heteroaromatic nitrogen heterocycles (e.g. riboflavin, folates, and dimethylbenzimidazole).
Figure 1.
(a) 5- and 6-ring nitrogen heterocycles found in coenzymes with nitrogen heterocycles in red; (b) some approved drugs (anastrazole, bortezimib, crizotinib, besifloxacin, olanzapine, PLX4032) containing nitrogen heterocycles in their synthetic scaffolds; (c) nitrogen heterocycles found in proteinogenic amino acids proline, histidine, and tryptophan.
Medicinal Chemists also like Nitrogen Heterocycles
Medicinal chemists have also long been enamored with nitrogen heterocycles, both in aliphatic and heteroaromatic oxidation states, as ligands for specific biological targets. They recognize that nitrogen heterocycles offer hydrogen bonding possibilities, provide polarity and controllable basicity to particular regions of molecular scaffolds. When heteroaromatic, the rings are planar and can provide hydrophobic and π-stacking interactions. A survey of drugs approved in recent times or currently in clinical trials turns up many variants of five- and six-membered rings, often fused as bicyclic scaffolds, with multiple nitrogen atoms. As noted in the structures of six approved drugs(2) (figure 1b), chemists can utilize the same heterocyclic nitrogen building blocks as found in Nature while exploiting unnatural ones to bolster their chemical repertoire.
The biological utility of heterocyclic nitrogen-containing molecular frameworks is thus firmly established in both small molecule coenzymes and natural products, the RNA and DNA informational macromolecules, and also in synthetic drug chemistries. We now turn to the inventory of nitrogen heterocycles found in proteins to see what functionalities and strategies are at work in that context. The posttranslational morphing of protein residues into oxazoles, thiazoles, and pyridines and the release of these modified peptide fragments as the functional end products is the subject of this perspective.
Nitrogen heterocycles in proteins
Three nitrogen heterocycles are found in the 20 proteinogenic amino acids: proline, histidine, and tryptophan, the latter two containing heteroaromatic imidazole and indole rings respectively (figure 1c). The biosynthetic routes to these three building blocks are well established(1), with construction of both the pyrrole of Trp and the imidazole of His, involving some unusual chemistry. Proline is the only cyclic amino acid in proteins where the nitrogen heterocycle acts as a conformationally restrictive building block. The planar indole side chain of tryptophan can interact via π-stacking modes with other aromatic and hydrophobic side chains, generally in the interior of folded proteins, and can participate in long range electron transfers in DNA photolyase and in ribonucleotide reductase(3–4).
The special versatility of imidazole side chains
Histidine residues have a much more activist role in biological catalysis by virtue of the imidazole side chain which is optimized for several chemical manifolds at physiological pH. For example the large family of mononuclear nonheme iron oxygenases chelate FeII in the active site through a trio of amino acid side chains, involving the basic nitrogen atoms of two His residues and one Glu or Asp side chain carboxylate(5–6).
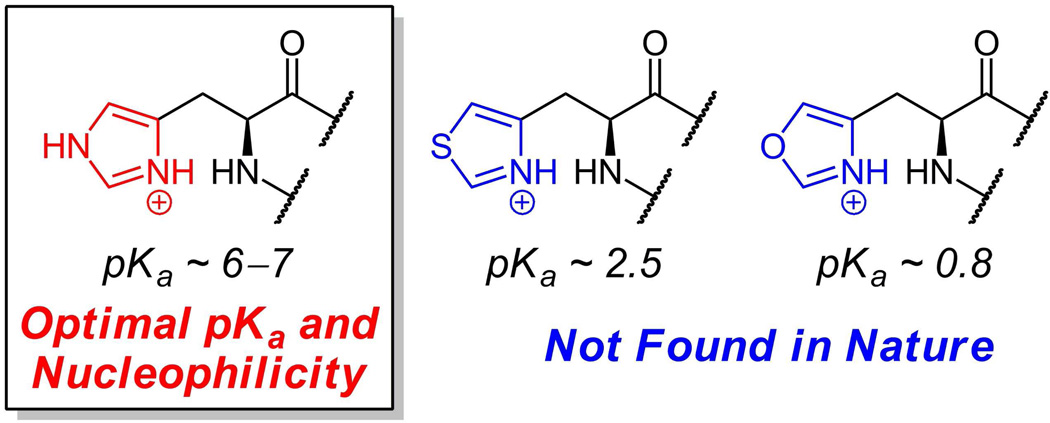
The more celebrated roles for imidazole side chains in enzymatic catalysis involve both general acid/base catalysis and nucleophilic catalysis(7) and are enabled by the pKa value for imidazole side chains which is between 6 and 7. Thus, a substantial fraction of the free base form of imidazole exists at neutral pH in a histidinyl side chain in an enzyme active site (figure 2). The superfamily of serine hydrolases use an equivalent of the serine residue alkoxide anion as nucleophile in the transition state. These enzymes almost invariably have a His residue positioned to act as a base (a proton transfer recipient) to convert the -Ser-O-H to a Ser-O-species in the transition state where a bound substrate carbonyl is attacked. The resultant tetrahedral adduct can break down to a covalent acyl-O-ser-enzyme intermediate when the imidazolium form of that His residue now acts as a general acid, donating a proton to the leaving group to lower the energy barrier for decomposition in the forward direction. In the second half reaction, deacylation of the acyl-O-Ser-enzyme intermediates in this enzyme superfamily, the His residue again first acts as a general base towards an attacking water molecule, and then as proton donor to regenerate the starting H-O-Ser-enzyme, ready for the next catalytic cycle(7).
Figure 2.
Oxazolyl alanine and thiazolyl alanine, arising from formal replacement of nitrogen in the imidazole side chain of histidine, are not found as natural amino acid metabolites. The pKa values indicate the lack of reactivity of the oxygen and sulfur atoms in those heterocycles as bases or nucleophiles.
The orthogonal reactivity of the imidazole side chain of histidine residues acting as nucleophile in proteins is exemplified in the large family of two component sensor kinase/response regulators in bacteria. Up to sixty such two component systems are found in Pseudomonas strains where they serve to report on organic and inorganic chemicals in the microenvironment and signal the conditional transcription of genes that elicit a specific metabolic response(8). When an external ligand is recognized, the transmembrane sensor kinases autophosphorylate a histidine residue in a cytoplasmic domain. The N-phosphohistidinyl enzymes arise by attack of one of the imidazole nitrogens (N1 or N3) of the active site His on the γ-phosphoryl group of bound Mg-ATP cosubstrate (it is the free base form of the imidazole side chain of the active site His in the sensor kinase superfamily that is the catalytic nucleophile)(9). The resultant N-P-His sensor kinase is thermodynamically activated for transfer of the PO32− group to a specific aspartyl side chain carboxylate in a partner response regulator protein. In turn the phosphoaspartyl form of the response regulator may now serve as a DNA binding protein at the promoter regions of specific target genes to allow transcription and production of proteins that carry out the new metabolic functions.
Imidazole is not interchangeable with Oxazole and Thiazole
As we will note below, the oxygen and sulfur congeners of the imidazole heterocycle – oxazole and thiazole – are found in a limited set of small proteins/peptides; however, they are always formed via posttranslational modifications of Cys and Ser residues, not by de novo incorporation of the corresponding oxazolyl alanine and thiazolyl alanine building blocks during protein synthesis (figure 3a). Replacement of N3 of the imidazole ring by oxygen would convert histidine into oxazolyl alanine while replacement of that nitrogen by sulfur would yield thiazolyl alanine. These have never been detected as free amino acids or in naturally occurring proteins. A rationale for this absence is that the lack of that second, basic nitrogen in the five ring oxazole and thiazole heterocycles removes both key facets of imidazole side chain functionality: the general acid/base role towards protons and the nucleophilic role towards carbon and phosphorus centers. One measure of the insufficient basicity of oxygen and sulfur compared to nitrogen in these heterocycles is the change in pKa for protonation shown in figure 2. Thus, it is likely the imidazole heterocycle represents optimal evolution for its central catalytic roles in protein catalysis.
Figure 3.
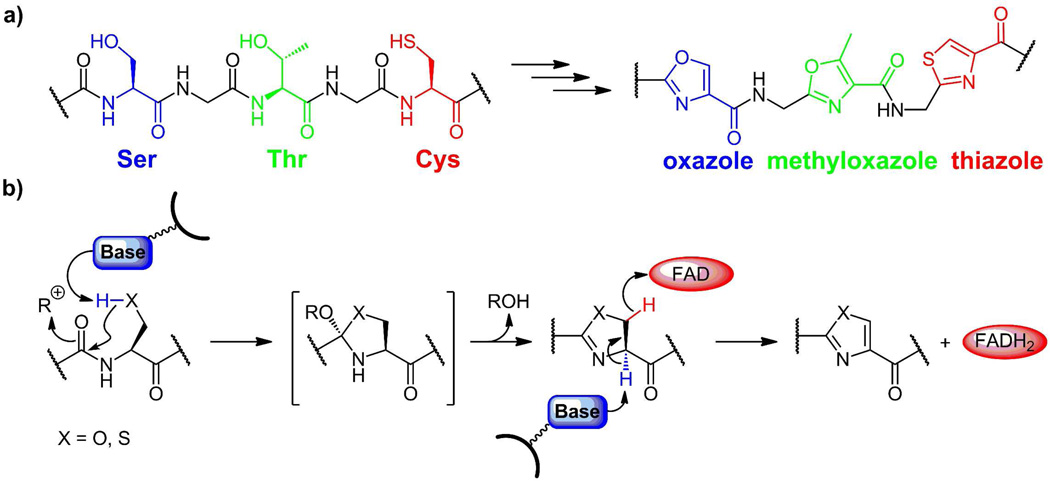
(a) Oxazoles (blue), methyloxazoles (green), and thiazoles (red) arise from posttranslational modification of -X-Ser, -X-Thr- and X-Cys dipeptide motifs in selected proteins. (b) Initial cyclodehydration on the carbonyl of the upstream amino acid yields the (methyl)oxazolines (X=O) and thiazolines (X=S), which can persist or be dehydrogenated to the (methyl)oxazoles and thiazoles by a subsequent enzymatic FAD dependent dehydrogenase.
Oxazoline/Oxazole and Thiazoline/Thiazole as Backbone Replacement for Peptide Bonds
Despite the lack of basicity and nucleophilicity of thiazoles and oxazoles, or perhaps because these reactivities have been factored out of the heterocycles, these rings are found as structural elements in a number of bacterial peptides that are derived from selective proteolysis of precursor proteins where they serve as rigidifying conformational elements. Also, as planar heteroaromatic scaffolds, they are peptide bond replacements that are resistant to hydrolysis and stable to proteases. Three amino acid residues can be modified posttranslationally to give these heterocycles: Ser to oxazole, Thr to methyloxazole, and Cys to thiazole (figure 3a). These conversions proceed through the dihydro intermediates, the oxazolines (from Ser, Thr) and the thiazolines (Cys)(10).
Cyclodehydration and Dehydrogenation to Yield Oxazole and Thiazole Backbones
The chemical mechanism and enzymatic machinery for conversion of Cys to thiazole and Ser and Thr to (methyl)oxazole residues was worked out in the microcin B17 system discussed specifically below (10–18). In general, one enzyme, in an ATP-dependent process, will cyclodehydrate both Cys and Ser residues that are downstream of an N-terminal leader sequence and produce the corresponding thiazolines and oxazolines as assayed by mass spectrometry. A companion FAD-dependent dehydrogenase then converts the dihydroheterocycles to the planar heteroaromatic thiazoles and oxazoles (figure 3b). Homologs of these two enzymes are found in all the subsequent systems that install oxazoline/oxazole and thiazoline/thiazole rings in peptide backbones. These enzyme families appear to recognize the leader peptide sequence, probably as an amphipathic helix, and then work on Cys, Ser, and Thr residues immediately downstream (19–20). The same enzyme works on both Cys and Ser (and presumably Thr) residues.
These remarkable morphings of peptide bonds into heterocycles are proposed to begin with an active site base removing the proton on the HS- or HO-side chain of a Cys or Ser residue to generate the thiolate or alkoxide ion equivalent in a transition state leading to attack on the upstream peptide carbonyl, creating the indicated cyclic tetrahedral adduct. The role of ATP in the cyclodehydration process is not yet clear but one suggestion is that the oxyanion in the tetrahedral adduct could act as nucleophile towards the γ-P of ATP to yield a phosphorylated adduct(21) Now loss of HOPO32− across the C-N bond would yield the C=N linkage in the thiazoline or oxazoline. The conversion of these cyclic imines to the heteroaromatic thiazoles and oxazoles is effected by an FAD-dependent dehydrogenase, presumably removing the C2-H of the thiazolines/oxazolines as a proton and the C3-H as a hydride equivalent sent to N5 of the FAD coenzyme. Reoxidation of the resultant FADH2 by oxygen completes the two enzyme catalytic cycle. In the microcin B17 case, it was observed that aromatizing dehydrogenation of thiazolines proceeds with 100-fold faster rate than of oxazolines(12). This kind of rate difference could explain the appearance of oxazolines co-existing with thiazoles in the patellamide and telomestatin frameworks as noted below.
Mechanistic Parallels to and Distinctions with Protein Splicing
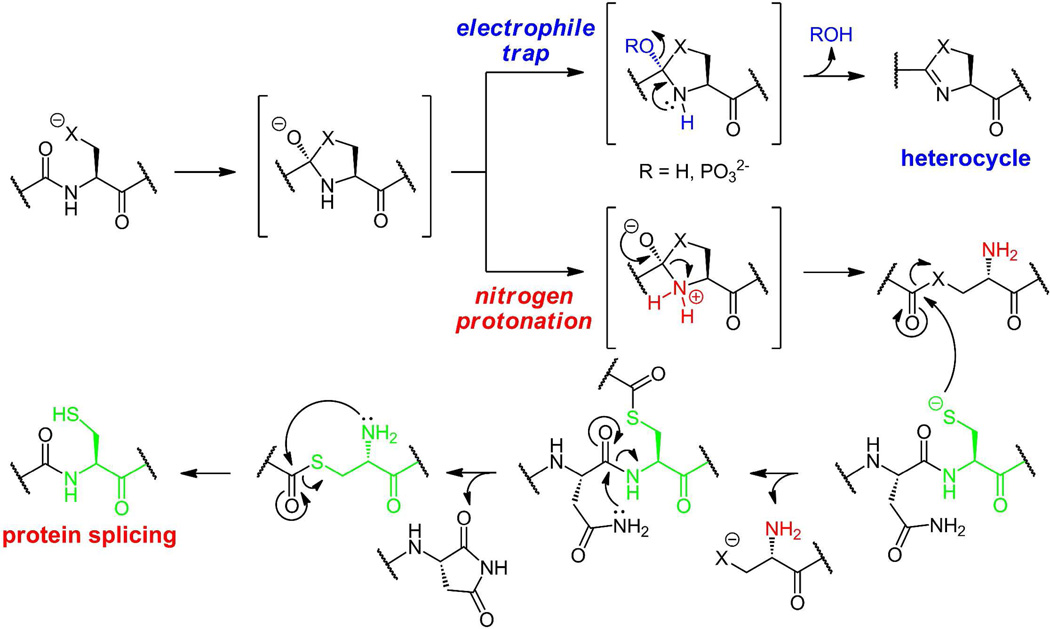
While the enzyme-catalyzed intramolecular attack of thiolate and alkoxide forms of Cys and Ser side chain to convert -X-Cys- and -X-Ser- dipeptide linkages into dihydroheterocycles is unusual chemistry, there are precedents in the first steps of protein splicing(9, 22). In self-spicing proteins, the same initial steps are posited: intramolecular attack of Cys and Ser side chains on the immediate upstream peptide carbonyl groups to yield the same tetrahedral adducts at extein/intein junctions (figure 4).
Figure 4.
Distinct outcome for a common tetrahedral adduct arising by attack of a Ser/Thr/Cys side chain on the upstream peptide carbonyl group: The adduct can proceed to the heterocycle (X=O oxazoline, X=S thiazoline, top scheme) or undergo C-N bond cleavage to yield an activated thioester (X=S) or an oxoester (X=O) (bottom scheme). Subsequent attack by a downstream cysteine residue followed by rearrangement yields the spliced protein.
The fate of these adducts is distinct. Rather than cleavage of the C-O bond as the thiazoline/oxazoline C=N bond forms, the elimination goes the other way. Protonation of the tetrahedral adduct on nitrogen, followed by collapse of the tetrahedral alkoxide back to a carbonyl breaks the C-N bond, with net cleavage of what had been the initial peptide bond as the covalent connectivity of the two parts of the protein are severed, leaving a free amine on the downstream side of the break. In the case of a Cys residue, a thioester is generated on the upstream side of the break, while an oxoester arises when a ser side chain has participated. Both the thioester and oxoester retain enough thermodynamic activation (and much more kinetic reactivity than the starting peptide) to be captured by thiolate side chains of Cys residues in the downstream extein. That thioester will eventually undergo an S to N shift to stitch two exteins together(9, 18).
It may be that the ATP consumed in the thiazoline/oxazoline intramolecular cyclizations(15, 21), by setting up the tetrahedral adduct oxygen as a phosphate ester, redirects that intermediate towards heterocyclization and away from net peptide bond cleavage. For both fates this may be quite ancient protein chemistry in terms of evolution of protein catalysts.
Posttranslational machinery and logic: heterocyclization and proteolysis
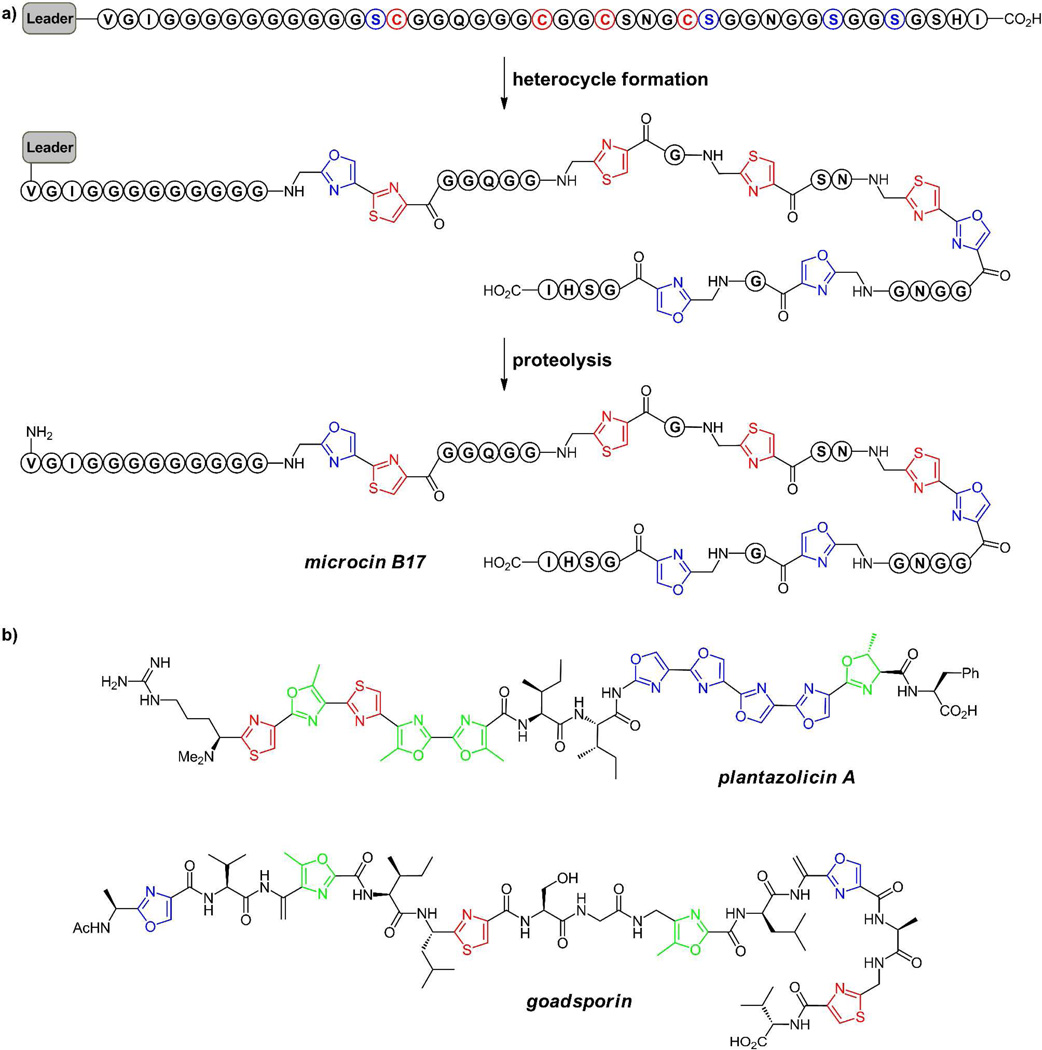
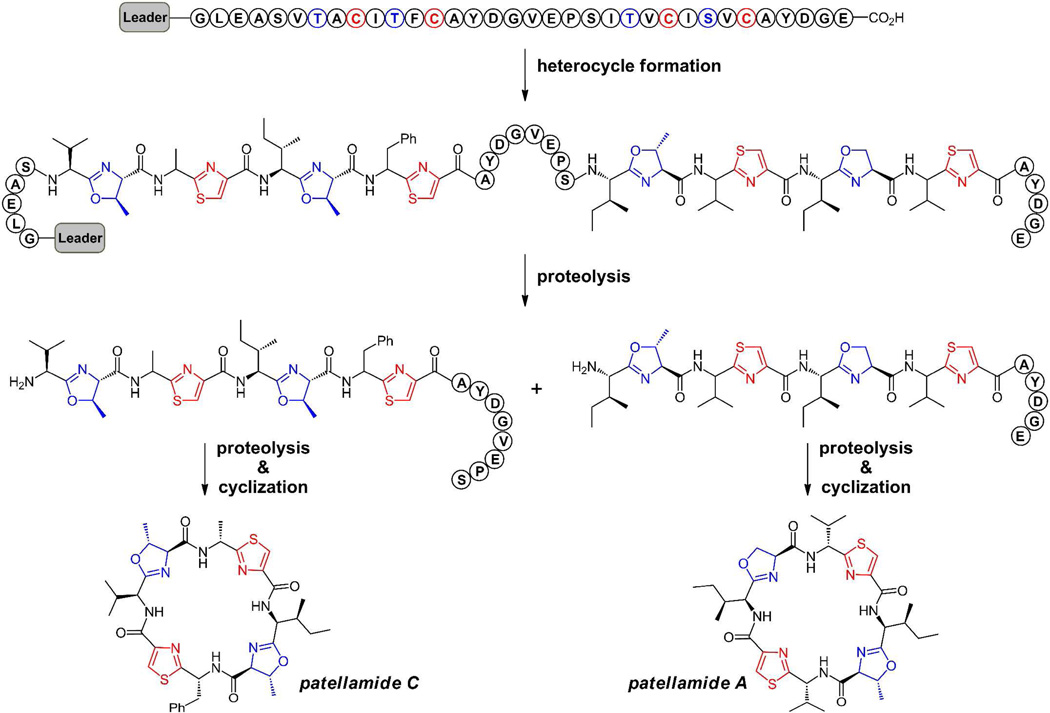
In the cases noted below for microcin B17, plantazolicin, and goadsporin, (figure 5) linear scaffolds are derived from nascent ribosomally generated small proteins of 50–75 residues, which serve as precursor for the posttranslational morphing of multiple Ser, Thr, or Cys residues clustered in the C-terminal region(11, 19–20). The N-terminal 35–50 residues serve as a helical leader sequence that acts as the recognition element for the tailoring enzymes, encoded adjacent to the preprotein structural gene. Once the heterocyclizations have occurred, the upstream leader peptide having performed its job of recruiting the tailoring enzymes, is chewed away by proteases in the producing cell (figure 5a). This constitutes a second posttranslational modification phase, returning the constituent amino acids to the cellular pool. The cyclic patellamides A and C, representatives of the large cyanobactin family discussed below, both derive from two distinct octapeptide sequences embedded within the same 71 residue PatE preprotein(23). The pat gene cluster encodes two proteases that excise and cyclize both patellamides A and C after the heterocycles have formed(24–25) (figure 6). Thus, all the oxazole and thiazole-modified peptide scaffolds are released as low molecular weight ligands for interaction with biologic targets.
Figure 5.
(a) Schematic for Microcin B17 processing. Eight Ser (blue) and Cys (red) residues to be morphed into heterocycles are clustered near the C-termini of the preprotein with N-terminal leader sequence. The leader region is likely helical and acts as a docking site for the tailoring enzymes encoded adjacent to the preprotein structural gene. Following heterocycle formation, proteolysis of the leader peptide releases microcin B17, a DNA gyrase-targeting antibiotic. (b) Plantazolicin has ten heterocycles, in two sets of five tandem rings. The 19 residue goadsporin with six heterocycles and also two dehydroalanine-derived residues, is a differentiation factor in streptomycetes.
Figure 6.
Schematic for conversion of octapeptide sequences within the PatE protein to cyclic peptides patellamide C and patellamide A via excision and intramolecular lactamization by dedicated peptidases. Patellamides A and C are members of the cyanobactin family of cytotoxic agents; the heterocycles alternate with proteinogenic amino acid side chains. Both the heterocyclization and macrocyclization events in these molecules provide highly constrained conformations of these morphed peptide natural products.
Linear Heterocyclic Peptide Scaffolds
Three examples of linear bacterial peptides that have multiple heterocyclic rings are microcin B17 from E.coli strains(19, 21, 26), plantazolicin from Bacillus amyloliquifaciens(27–28), and goadsporin from Streptomycetes(29–30) (figure 5). Microcin B17 arises from processing of a 69 residue protein, whose N-terminal 26 residues serve as a leader sequence that is ultimately removed by proteolytic processing. Of the remaining 43 residues, 14 are posttranslationally processed to generate four thiazoles and four oxazoles, including tandem oxazole-thiazole and thiazole-oxazole pairs. The antibacterial target of mature microcin B17 in enteric bacteria is DNA gyrase and the tandem bis-heterocycles are important activity determinants, perhaps by interaction with DNA in a DNA-gyrase complex(31).
Plantazolicin is a 14 residue peptide antibiotic active against gram positive pathogens, including B. anthracis, in which ten of the 14 residues have been heterocyclized: two thiazoles, three methyloxazoles, four oxazoles and one dihydro methyloxazoline. Remarkably the heterocycles come in two strings of five tandem rings (residues 2–6 and 9–13), indicating the efficiency of the posttranslational machinery to process the ten residues serially for heterocyclization. The last one, methyloxazline 13, has not undergone the aromatizing dehydrogenation, probably reflecting an N- to C-directional progression of the cyclodehydratase dehydrogenase pair from the upstream leader peptide(12–13) as well as rate-determining dehydrogenation of oxazolines vs thiazolines. The target of plantazolicin, which affects its antimicrobial activity, remains to be determined.
Goadsporin is a 19 residue modified peptide that similarly arises from posttranslational cleavage and morphing of a precursor protein in the producing streptomycetes. It functions as a differentiation signal although its streptomycete receptor is not yet determined. There are three methyloxazoles, one oxazole, and two thiazoles, indicating heterocyclization of three Thr, one Ser, and two Cys residues during posttranslational maturation. There are also two dehydroalanine (dhA) residues in goadsporin, reflecting a distinct mode of maturation of two additional Ser residues that will be addressed below. In both microcin B17 and presumably goadsporin, the multiple heterocycles (and the sp2 centers in the adjacent dhAs) impose conformational restrictions on otherwise mobile peptide backbones, populating conformers essential for high affinity recognition by their targets. The replacement of peptide bonds with heterocyclic backbones in small, otherwise unfolded peptides decreases entropic costs of binding to targets, dramatically decreases the lability to peptidases/proteases, and likely increases biological half lives for the antibiotic and differentiation functions.
Heterocycles in Macrocyclic Peptide Frameworks
Examples of cyclic peptides containing such five ring heterocycles are found in marine cyanobacteria(11, 23–24, 26, 32). The strategy and mechanism of maturation of patellamides, (figure 6) such as patellamide A and C, have been worked out in detail by Schmidt and colleagues, starting with genome sequencing of a marine cyanobacterial symbiont Prochloron didemni associated with the sponge Lissoclinum patella. These cyclic octapeptide cytotoxic agents have four heterocycles alternating with hydrophobic side chains. The two sulfur heterocycles are at the heteroaromatic thiazole oxidation states. The two oxygen rings are at the dihydroaromatic oxazoline (from Ser)/methyloxazoline (from Thr) oxidation states in both patellamides A and C.
The octapeptide sequences that give rise to both of these highly constrained low molecular weight heterocyclic and macrocyclic scaffolds are embedded within the same precursor protein sequence flanked by recognition sequences for the proteases(11, 24). This structural gene in turn is flanked by genes that encode both the heterocyclization enzymes and the proteases that carry out cyclization to the octapeptide lactam framework, a remarkable set of framework reconstructions from internal linear octapeptides. It is likely that the four heterocycles in patellamides A and C are generated before macrocyclization to create a conformation where residue 1 and 8 can be circularized after the first excision step.
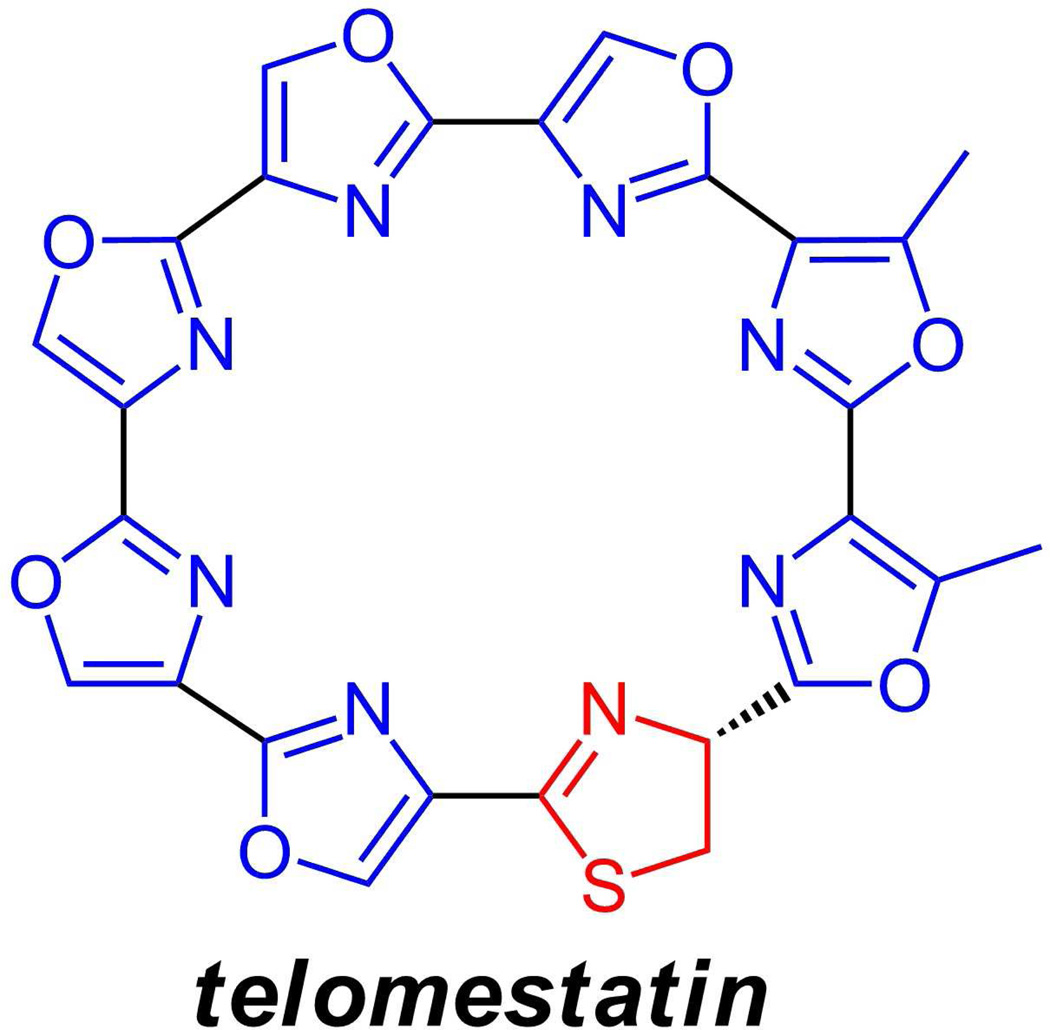
Also in this general family is telomestatin(33) (figure 7), a potent telomere binder from Streptomyces anulatus which is presumed to arise from an octapeptide motif of five serines, two threonines, and one cysteine (present as the thiazoline). Given the circularity of the eight ring macrocycle, until the precursor protein’s gene sequence has been determined, it is not clear whether the string of serines lies at the N-terminus, the middle, or the C-terminus of the linear precursor.
Figure 7.
Telomestatin from a streptomycete binds tightly to telomere regions of chromosomes; it has eight heterocycles, of which one, the thiazoline ring, is in the dihydroxoidation state.
Additional Posttranslational Modification Fates for Ser and Thr Side Chains: Phosphorylation Sequellae
Most of the posttranslational modifications at Ser and Thr residues in proteins are not the above cyclodehydrations or protein splicing reactions. They are instead phosphorylations by action of protein kinases, cleaving cosubstrate ATP into ADP while forming the P-Ser or P-Thr side chain of the protein substrate(s)(9, 34–35). These Ser and Thr residue phosphorylations occur across all kingdoms of life in diverse signaling pathways. The great bulk of these phosphate groups are removed hydrolytically by phosphoprotein phosphatases, releasing inorganic phosphate as the original Ser-OH/Thr-OH side chain is regenerated.
A small fraction of P-Ser and P-Thr side chains are subsequently targeted for enzymatic processing in a lyase reaction for elimination of the Cα-H along with the phosphate as the Cβ-O bond is cleaved(36–38). The result is a Cαβ double bond in the protein side chain: a dehydroalanine (dhA) from P-Ser and a dehydrobutyrine (dhB) from P-Thr residues (figure 8a). These PSer/P-thr lyases have now been found in several bacterial metabolic contexts. One involves secretion of P-Thr protein lyases by Shigella and Salmonella strains through type III secretion sytems into vertebrate host cells where they convert P-Thr residues in MAP kinases into dehydrobutryines(11). This is an irreversible inactivation of these host enzymes and blocks the signaling cascades in which they participate.
Figure 8.
Different fates for Phospho-Ser and Phospho-Thr residues in proteins. (a) Most Ser and Thr side chains that get phosphorylated by protein kinases are ultimately dephosphorylated hydrolytically by protein phosphatase, releasing Pi and regenerating the side chain-OH; some P-Ser/P-Thr intermediates can instead undergo a lyase-mediated reaction, initiated by loss of the Cα-H. Pi is again a product but now a double bond has been created between Cα and Cβ of the side chains to yield dehydroAla (dhA) and dehydrobutyrine (dhB) side chains; (b) In lantipeptide maturations some of those dhA and dhB residues can be captured intramolecularly by neighboring thiolate side chains of Cys residues to yield crosslinking lanthionine or methyllanthionine bridges. (c) Posttranslational processing of a –ser-thr-dipeptide moiety in the goadsporin precursor by kinase/lyase (ser) and cyclodehydratase/dehydrogenase (thr) replaces two tandem peptide bonds with a methylene-methyloxazole unit.
A more well studied context for net conversion of Ser and Thr to dhA and dhB residues is in the biosynthesis of lantibiotics(39–41). Enzymes in the Lan family are posttranslational modification catalysts, encoded adjacent to the structural gene that specifies a precursor form of the lantipeptide scaffold, which has an N-terminal signal peptide and then a downstream core peptide that undergoes the modifications. The Lan type enzymes transiently phosphorylate Ser and Thr side chains in the core region and catalyze the α,β lyase-mediated elimination of HPO42− to create the dehydroAla and dehydrobutyrine residues: the kinase and lyase activities can reside within the same enzyme(38). Some of these unsaturated side chains persist in lantipeptides but most are converted by an additional PTM enzyme, exemplified by NisC(40) in the nisin biosynthetic pathway. There, attack of the thiolate of Cys side chains on Cβ of the dehydro side chains yields a thioether linkage (figure 8b) between the two moieties. The adduct arising from Cys capture of dehydroAla is known as lanthionine (reflecting its original isolation from the proteins in wool) while the thioether with the β-methyl, from capture of dehydrobutyrine, is methyllanthionine. The presence of these double-headed lanthionine residues gives the lantibiotics/lantipeptides their name. This second PTM step highlights the electrophilic nature of Cβ of dhA (and dhB residues), relevant for the subsequent section on pyridine formation.
Returning to goadsporin(21, 25), we noted above that there are two dehydroalanine residues mixed in with two thiazoles and four (methyl)oxazoles, undoubtedly reflecting the phosphorylation/elimination of those two Ser side chains (at Ser4 and Ser15) in the goadsporin precursor. Indeed, examination of the goadsporin gene cluster shows not only the structural gene and the genes for cyclodehydration and dehydrogenation to form oxazoles and thiazoles posttranslationally but also homologs to the lantipeptide type Ser kinase/lyase orfs. In goadsporin processing, it is not yet clear how three threonines and one serine are sent down one pathway, to (methyl)oxazoles, while another two serines are processed to dehydroalanines. Tandem processing of S4/T5 and S15/T16 by the two distinct PTM pathways yields the methylene-methyloxazole part structures indicated in figure 8c, reflecting morphing of two consecutive peptide bonds into hydrolysis-resistant altered backbones by two distinct PTM pathways.
Nonribosomal assembly machinery also cyclodehydrates Cys, Ser and Thr
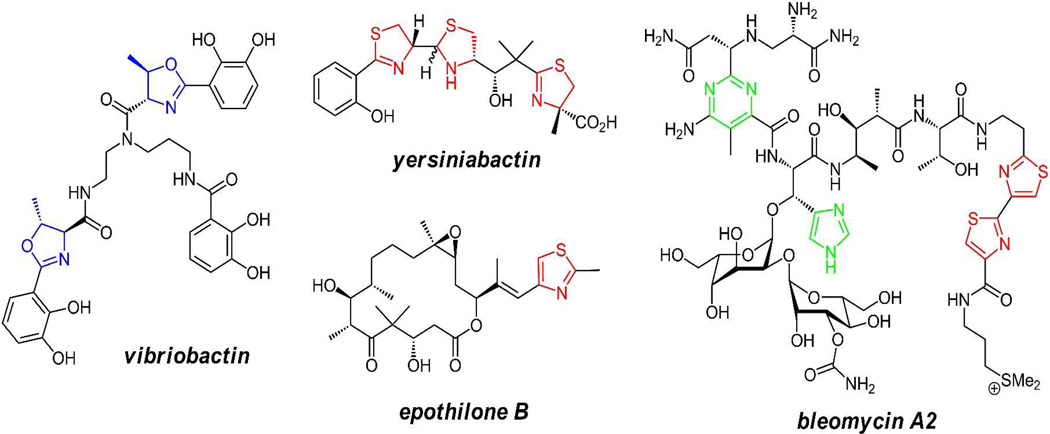
Although the focus here is on introduction of oxazoles and thiazoles within protein frameworks, these heterocycles are also put in place by nonribosomal peptide synthetase assembly lines, using similar cyclodehydration (and dehydrogenation where it occurs) enzyme chemistry(42) (figure 9). One such context is the assembly of thiazoline and oxazoline-containing siderophores(43–44) where the nitrogens in the dihydroheterocycles are part of the ligand set for binding ferric iron (yersiniabactin and vibriobactin). A parallel context is in the action of hybrid nonribosomal peptide synthetase/polyketide synthase assembly lines to make the methyl thiazole in the antitumor epothilones(45) and tandem bithiazoles in the DNA-targeting anticancer drug bleomycin(46–47).
Figure 9.
Yersiniabactin, vibriobactin, epothilone, and bleomycin A2 contain nitrogen heterocycles made by NRPS or NRPS/PKS enzymatic machinery rather than by posttranslational modifications of preproteins.
Adding pyridine rings to the posttranslational mix of Serine processing
A third heteroaromatic ring, pyridine or tetrahydro derivatives thereof, is found in the peptidic scaffold of a class of bacterial natural products with antibiotic activity, represented by thiostrepton(48), thiocillin(49), GE2270A(50) and berninamycin(51) as shown in figure 10. The founding member was isolated in 1948 and new examples continue to be isolated from actinomycetes by activity-guided fractionation schemes such that >80 molecules of this family have been reported(52). Most of these antibiotics target the 50S bacterial ribosome, although some with a 29-atom macrocyclic framework inhibit the chaperone protein EF-Tu from ferrying aminoacyl tRNAs to the ribosome for protein biosynthesis(53).
Figure 10.
Members of the thiazole-pyridine class of peptide antibiotics: thiostrepton, thiocillin, GE2270A, and berninamycin.
Although thiostrepton may be the best known of this antibiotic class because thiostrepton resistance (tsr) is a common genetic marker used in streptomyces molecular biology manipulations(54), it is unusual in having a tetrahydropyridine rather than the much more common pyridine oxidation state at the core of the framework. Over the past three years, several studies have shown these antibiotic peptides, albeit highly morphed, have a ribosomal origin in 50–60 residue long preproteins with 30–40 residues comprising N-terminal leader sequences(48, 55–57). The C-terminal 14–16 residues become dramatically altered by cascades of posttranslational modification. For example, inspection of the thiocillin framework shows six thiazoles, two dehydrobutyrines and a central pyridine ring. Three of the thiazoles are substituents on that pyridine such that the core can be considered to be a tetracyclic trithiazolylpyridine(49, 58).
The presence of thiazoles and dehydrobutyrines are reminiscent of the posttranslational enzyme machinery in microcins, goadsporin, and lantipeptides. So are the oxazoles and dehydroalanines seen, for example, in berninamycin. In accordance with these findings, the biosynthetic gene clusters indeed have the anticipated cyclodehydratase/dehydrogenase pair to make Cys, Ser, and Thr residues into thiazoles and (methyl)oxazoles as well as the kinase/lyase pair to make Ser and Thr into dehydroAla and dehydrobutyrine residues. Using gene replacement, the Cys, Ser, and Thr residues in the prethiocillin structural gene have been mutated to enable the producing B. cereus strain to make variants of thiocillin(58–59).
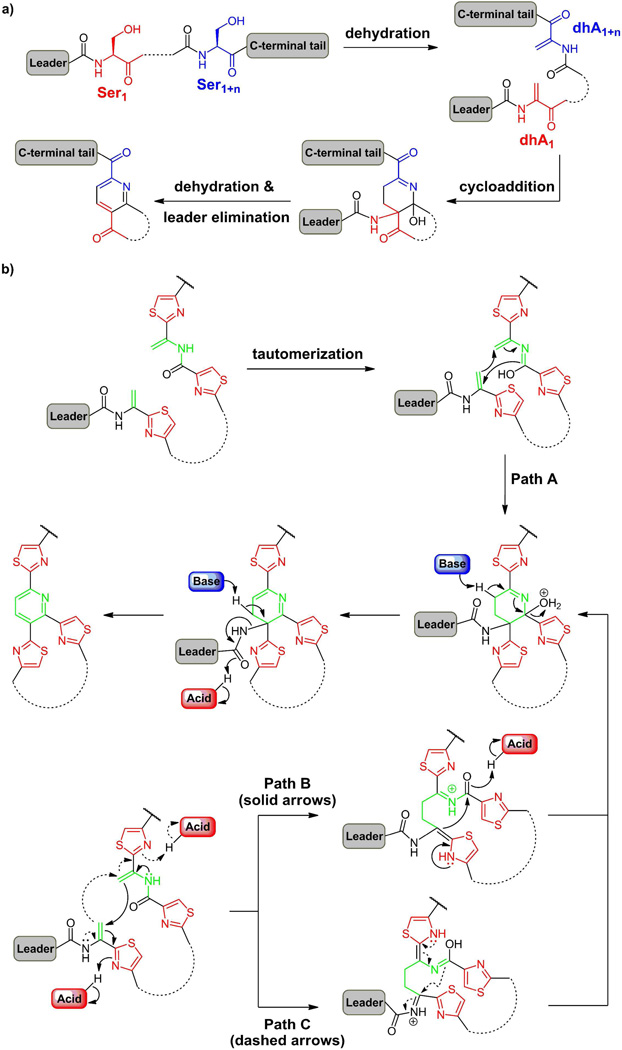
That leaves the question of how the pyridine ring is formed. The prime hypothesis(60–61) has been that two serine residues get converted to dehydroAlas and that pair undergoes a cycloaddition (figure 11a) to make two C-C bonds of the six-membered nitrogen ring. The oxidation state could then be adjusted to obtain either the tetrahydro- or the aromatized pyridine. Studies in the B. cereus strain that produces thiocillin show that a deletion of the tclM gene leads to accumulation of a linear precursor with all six thiazoles and the two dehydrobutyrines in place(62). In addition, Ser1 and Ser10 have been converted to dehydroAlas in the linear precursor. Complementation of a functional tclM back into this strain restores the ability to make the pyridine ring and thereby the mature thiocillin scaffolds. This indicates that pyridine ring formation is late, occurring after twelve posttranslational modifications have been processed, generating conformational restrictions (e.g. from three thiazoles at residues 2,9,11) to the 52 residue chain that may bring dhA1 and dhA10 into proximity for the pyridine formation.
Figure 11.
(a) The Bycroft-Floss hypothesis suggests pyridine ring formation from two dehydroalanine residues; (b) Construction of the central trithiazolyl pyridine core from two dhA residues at a late stage of antibiotic scaffold maturation after twin posttranslational modifications have already occurred; (b) possible concerted vs stepwise mechanisms for an aza-Diels-Alder cyclization, followed by dehydration to yield a dihydropyridine with the upstream leader peptide attached. Aromatizing elimination would yield the pyridine in the majority of cases while a two electron reduction would give the tetrahydropyridine core seen in thiostrepton with the leader peptide still in place.
As of yet, there is no mechanistic information on the timing of the two bonds that are formed as dhA1 and dhA10 are cyclized by TclM and homologs in the trithiazolylpyridine core construction. It is intriguing to speculate that this may be a biological version of an aza-Diels-Alder reaction(63) with both bonds formed in a single transition state through the intermediacy of the isoamide tautomer at dhA10 (figure 11b, Path A). Alternatively, the cycloaddition may proceed in a stepwise fashion, where one dhA acts as a nucleophile at Cβ while the other acts as an electrophile, with the thiazole ring serving as the electron sink (figure 11b, Paths B and C). In this latter scenario, the electron flow could in principle proceed in either direction. For instance, dhA10 might act as the nucleophile, undergoing Michael addition to Cβ of dhA1 (Path B); subsequently, attack of the α-carbon of dhA1 on the activated carboxamide adjacent to dhA10 would lead to six-membered ring formation. Instead, electrons may flow from dhA1 towards dhA10 (Path C), requiring the tautomer of dhA10 to function as the extended nucleophile in the cyclization event. All three plausible pathways outlined above intercept a common intermediate, dehydration of which leads to a dihydropyridine core. Now, loss of the upstream leader peptide (a 38mer carboxamide for thiocillin) would be driven by aromatization to the pyridine. The typical 2,3,6 pattern of trithiazole substitution on the pyridine in this antibiotic class is accommodated by this mechanism; the three thiazoles may position the dhA1 and dhA10 for this condensation (figure12). Instead of aromatizing by elimination of the leader peptide, the dihydropyridine intermediate could be intercepted for two electron reduction, generating the central ring at the tetrahydropyridine oxidation state. In this case, the upstream leader peptide would still be attached at the 2 position as found in thiostrepton (figure 10).
Figure 12.
(a) Simultaneous construction of the peptide macrocycle as pyridine ring formation occurs in the thiazolylpyridine class of peptide antibiotics. (b) Possible conformation of the linear intermediate poised for pyridine ring formation en route to the mature antibiotic (NMR solution structure of thiocillin(62)).
One additional distinguishing feature of the thiazolyl pyridine class of antibiotic peptides is the size of the macrocyclic ring. While thiocillins (and also thiostrepton) have a 26 atom macrocycle, in which the thiazolyl pyridine core is embedded, GE2270A has a 29 atom macrocycle and berninamycin a 35 atom version. For the 29 membered ring scaffolds, we have shown that instead of dhA10 being condensed with dhA1, it is dhA11(64) that gets selected by the TclM homologs in those assembly pathways. Also of note is that in every case the macrocycle is assembled at the same time and in the same step as the core pyridine – a remarkable piece of biosynthetic simultaneity (figure 12). The macrocyclic ring size and rigidifying constraints of the modified heterocycles and dehydroamino acids it contains in its backbone are crucial determinants of both antibiotic potency and target selectivity.
Formation of the pyridine, and thereby the tetracyclic trithiazolylpyridine core can be viewed as the biosynthetic device that builds the macrocyclic architecture into this antibiotic framework. To date this antibiotic family is the only one in which a pyridine ring is formed during posttranslational morphing of a nascent protein. Pyridine rings are found in other contexts in nature, including the N-ribosylated nicotinamide coenzymes and the pyridine aldehyde of pyridoxal-phosphate noted in figure 1a; however, those cofactors are biosynthesized by quite different routes and function as redox active heterocycles. By contrast, the pyridine in the tetracyclic core of the thiazolylpyridines appears to play an exclusively architectural role, creating the rigidified core required to display the macrocyclic loops for high affinity binding to the 50S ribosome or to EF-Tu. This strategy for conformational restriction of a modified peptide framework can be contrasted to the patellamides where the macrocyclization occurs by protease-mediated ligation rather than in a pyridine-forming step.
In sum, the thiazolylpyridine antibiotics are exemplars of how a protein scaffold can be morphed by a cascade of posttranslational modifications (at least thirteen modifications for the fourteen C-terminal residues of the thiocillin precursor protein). They also represent a confluence of several of the chemical transformations that can occur at Ser, Thr and Cys residues in proteins. X-Cys dipeptide moieties get converted to thiazoles, including a bithiazole (from Cys11 and Cys12 in thiocillins). Three distinct outcomes of X-Ser side chain processing are in play. One is the comparable conversion to oxazolines (GE2270A) and oxazoles (berninamycin). The other two outcomes follow from the initial formation of phosphoserine as the kinase/lyase pathway then yields dehydroAla residues. Cyclization of a pair of properly disposed dhA residues (where one may act as a formal nucleophile and the other as a formal electrophile) yields the pyridine ring, after aromatization.
Three Rings to Turn Peptides into Rigid Scaffolds
A small number of posttranslational chemical mechanisms with contextual variation (ser ⇒ P-ser ⇒ dhA ⇒ pyridine; ser ⇒ oxazole; cys ⇒ thiazole) result in the installation of three heterocycles into protein-derived backbones. Examples of morphed peptides containing two or all three, oxazole, thiazole and pyridine, in the same framework (as in berninamycin) show posttranslational modification virtuosity. The ring-forming enzymes, instructed by leader peptide recognition, act from one to thirty residues downstream. Strings of Ser, Thr, and Cys residues can be processed directionally but distributively(12). For the microcins, goadsporins and thiocillins, subsequent protease trimming away of the leader peptide yields the small molecule mature scaffold with (almost) no standard peptide bonds remaining.
For the patellamide cyanobactins double excision of the heterocyclic octapeptide moieties out of the precursor protein is the final maturation step. Complex three dimensional, rigid architectures are produced from linear 8–16 residue peptide precursors with gain of new biological functions. The net result is conversion of unstructured peptide backbones into constrained drug-like cyclic architectures.
Understanding the recognition and directional role of the leader peptide sequences may allow one to harness and deploy the Cys, Ser, and Thr posttranslational machineries noted in this review on other protein sequences, yielding the possibility to create designed architectural drug-like scaffolds from other protein precursors(65).
Acknowlegements
Work from the Walsh group noted in the references has been supported by NIH grant GM20011. Particular thanks to former coworkers M. Acker, A. Bowers, M. Fischbach, L. W. Brown on the thiocillins and to Y. Li, J. Milne, N. Kelleher, R. S. Roy, and P. Belshaw on microcin B17.
References
- 1.McMurry J, Begley T. The organic chemistry of biological pathways. Englewood Colorado: Roberts & Co; 2005. [Google Scholar]
- 2.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stuube J, Nocera D, Yee C, Chang M. Radical Initiation in the Class 1 Ribonucleotide Reductase: Long-Range Proton-Coupled Electron Transfer? Chemical Reviews. 2003;103:2167–2201. doi: 10.1021/cr020421u. [DOI] [PubMed] [Google Scholar]
- 4.Sancar A. Structure and Function of DNA Photolyase and Cryptochrome Blue-Light Photoreceptors. Chemical reviews. 2003;103:2203–2237. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- 5.Kohntrop K, Emerson JP, Que L. The 2-His-1carboxulatefacial triad: a versatile platform for dioxygen activation by mononuclear non-heme iron (II) enzymes. J Biol Inorg Chem. 2005;10:87–93. doi: 10.1007/s00775-005-0624-x. [DOI] [PubMed] [Google Scholar]
- 6.Krebs C, Galonic Fujimori D, Walsh CT, Bollinger JM., Jr. Non-heme Fe(IV)-oxo intermediates. Acc Chem Res. 2007;40:484–492. doi: 10.1021/ar700066p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walsh C. Enzymatic Reaction Mechanisms. San Francisco: Freeman; 1979. [Google Scholar]
- 8.Rodrigue A, Quentin Y, Lazdunski A, Mejean V, Foglino M. Two-component systems in Pseudomonas aeruginosa: why so many? Trends Microbiol. 2000;8:498–504. doi: 10.1016/s0966-842x(00)01833-3. [DOI] [PubMed] [Google Scholar]
- 9.Walsh C. Posttranslational Modification of Proteins. Englewood Colorado: Roberts & Co.; 2005. [Google Scholar]
- 10.Roy RS, Kim S, Baleja JD, Walsh CT. Role of the microcin B17 propeptide in substrate recognition: solution structure and mutational analysis of McbA1-26. Chem Biol. 1998;5:217–228. doi: 10.1016/s1074-5521(98)90635-4. [DOI] [PubMed] [Google Scholar]
- 11.Donia MS, Ravel J, Schmidt EW. A global assembly line for cyanobactins. Nat Chem Biol. 2008;4:341–343. doi: 10.1038/nchembio.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belshaw PJ, Roy RS, Kelleher NL, Walsh CT. Kinetics and regioselectivity of peptide-to-heterocycle conversions by microcin B17 synthetase. Chem Biol. 1998;5:373–384. doi: 10.1016/s1074-5521(98)90071-0. [DOI] [PubMed] [Google Scholar]
- 13.Kelleher NL, Hendrickson CL, Walsh CT. Posttranslational heterocyclization of cysteine and serine residues in the antibiotic microcin B17: distributivity and directionality. Biochemistry. 1999;38:15623–15630. doi: 10.1021/bi9913698. [DOI] [PubMed] [Google Scholar]
- 14.Milne JC, Roy RS, Eliot AC, Kelleher NL, Wokhlu A, Nickels B, Walsh CT. Cofactor requirements and reconstitution of microcin B17 synthetase: a multienzyme complex that catalyzes the formation of oxazoles and thiazoles in the antibiotic microcin B17. Biochemistry. 1999;38:4768–4781. doi: 10.1021/bi982975q. [DOI] [PubMed] [Google Scholar]
- 15.Milne JC, Eliot AC, Kelleher NL, Walsh CT. ATP/GTP hydrolysis is required for oxazole and thiazole biosynthesis in the peptide antibiotic microcin B17. Biochemistry. 1998;37:13250–13261. doi: 10.1021/bi980996e. [DOI] [PubMed] [Google Scholar]
- 16.Roy RS, Allen O, Walsh CT. Expressed protein ligation to probe regiospecificity of heterocyclization in the peptide antibiotic microcin B17. Chem Biol. 1999;6:789–799. doi: 10.1016/s1074-5521(99)80126-4. [DOI] [PubMed] [Google Scholar]
- 17.Sinha Roy R, Belshaw PJ, Walsh CT. Mutational analysis of posttranslational heterocycle biosynthesis in the gyrase inhibitor microcin B17: distance dependence from propeptide and tolerance for substitution in a GSCG cyclizable sequence. Biochemistry. 1998;37:4125–4136. doi: 10.1021/bi9728250. [DOI] [PubMed] [Google Scholar]
- 18.Madison LL, Vivas EI, Li YM, Walsh CT, Kolter R. The leader peptide is essential for the post-translational modification of the DNA-gyrase inhibitor microcin B17. Mol Microbiol. 1997;23:161–168. doi: 10.1046/j.1365-2958.1997.2041565.x. [DOI] [PubMed] [Google Scholar]
- 19.Roy RS, Gehring AM, Milne JC, Belshaw PJ, Walsh CT. Thiazole and oxazole peptides: biosynthesis and molecular machinery. Nat Prod Rep. 1999;16:249–263. doi: 10.1039/a806930a. [DOI] [PubMed] [Google Scholar]
- 20.Oman TJ, van der Donk WA. Follow the leader: the use of leader peptides to guide natural product biosynthesis. Nat Chem Biol. 2010;6:9–18. doi: 10.1038/nchembio.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li YM, Milne JC, Madison LL, Kolter R, Walsh CT. From peptide precursors to oxazole and thiazole-containing peptide antibiotics: microcin B17 synthase. Science. 1996;274:1188–1193. doi: 10.1126/science.274.5290.1188. [DOI] [PubMed] [Google Scholar]
- 22.Paulus H. Protein splicing and related forms of protein autoprocessing. Annu Rev Biochem. 2000;69:447–496. doi: 10.1146/annurev.biochem.69.1.447. [DOI] [PubMed] [Google Scholar]
- 23.Schmidt EW, Nelson JT, Rasko DA, Sudek S, Eisen JA, Haygood MG, Ravel J. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc Natl Acad Sci U S A. 2005;102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McIntosh JA, Schmidt EW. Marine molecular machines: heterocyclization in cyanobactin biosynthesis. Chembiochem. 2010;11:1413–1421. doi: 10.1002/cbic.201000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Severinov K, Semenova E, Kazakov A, Kazkov T, Gelfand M. Low molecular-weight posttranslationally modified microcins. Molecular Microbiology. 2007;65:1380–1394. doi: 10.1111/j.1365-2958.2007.05874.x. [DOI] [PubMed] [Google Scholar]
- 26.McIntosh JA, Robertson CR, Agarwal V, Nair SK, Bulaj GW, Schmidt EW. Circular logic: nonribosomal peptide-like macrocyclization with a ribosomal peptide catalyst. J Am Chem Soc. 2010;132:15499–15501. doi: 10.1021/ja1067806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalyon B, Helaly SE, Scholz R, Nachtigall J, Vater J, Borriss R, Sussmuth RD. Plantazolicin A and B: structure elucidation of ribosomally synthesized thiazole/oxazole peptides from Bacillus amyloliquefaciens FZB42. Org Lett. 2011;13:2996–2999. doi: 10.1021/ol200809m. [DOI] [PubMed] [Google Scholar]
- 28.Molohon KJ, Melby JO, Lee J, Evans BS, Dunbar KL, Bumpus SB, Kelleher NL, Mitchell DA. Structure Determination and Interception of Biosynthetic Intermediates for the Plantazolicin Class of Highly Discriminating Antibiotics. ACS Chem Biol. 2011 doi: 10.1021/cb200339d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Onaka H, Nakaho M, Hayashi K, Igarashi Y, Furumai T. Cloning and characterization of the goadsporin biosynthetic gene cluster from Streptomyces sp. TP-A0584. Microbiology. 2005;151:3923–3933. doi: 10.1099/mic.0.28420-0. [DOI] [PubMed] [Google Scholar]
- 30.Onaka H. Biosynthesis of indolocarbazole and goadsporin, two different heterocyclic antibiotics produced by actinomycetes. Biosci Biotechnol Biochem. 2009;73:2149–2155. doi: 10.1271/bbb.90263. [DOI] [PubMed] [Google Scholar]
- 31.Parks WM, Bottrill AR, Pierrat OA, Durrant MC, Maxwell A. The action of the bacterial toxin, microcin B17, on DNA gyrase. Biochimie. 2007;89:500–507. doi: 10.1016/j.biochi.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 32.Lee J, McIntosh J, Hathaway BJ, Schmidt EW. Using marine natural products to discover a protease that catalyzes peptide macrocyclization of diverse substrates. J Am Chem Soc. 2009;131:2122–2124. doi: 10.1021/ja8092168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shin-ya K, Wierzba K, Matsuo K, Ohtani T, Yamada Y, Furihata K, Hayakawa Y, Seto H. Telomestatin, a novel telomerase inhibitor from Streptomyces anulatus. J Am Chem Soc. 2001;123:1262–1263. doi: 10.1021/ja005780q. [DOI] [PubMed] [Google Scholar]
- 34.Pereira SF, Goss L, Dworkin J. Eukaryote-like serine/threonine kinases and phosphatases in bacteria. Microbiol Mol Biol Rev. 2011;75:192–212. doi: 10.1128/MMBR.00042-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Breitkreutz A, Choi H, Sharom JR, Boucher L, Neduva V, Larsen B, Lin ZY, Breitkreutz BJ, Stark C, Liu G, Ahn J, Dewar-Darch D, Reguly T, Tang X, Almeida R, Qin ZS, Pawson T, Gingras AC, Nesvizhskii AI, Tyers M. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen L, Wang H, Zhang J, Gu L, Huang N, Zhou JM, Chai J. Structural basis for the catalytic mechanism of phosphothreonine lyase. Nat Struct Mol Biol. 2008;15:101–102. doi: 10.1038/nsmb1329. [DOI] [PubMed] [Google Scholar]
- 37.Brennan DF, Barford D. Eliminylation: a post-translational modification catalyzed by phosphothreonine lyases. Trends Biochem Sci. 2009;34:108–114. doi: 10.1016/j.tibs.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 38.Goto Y, Okesli A, van der Donk WA. Mechanistic studies of Ser/Thr dehydration catalyzed by a member of the LanL lanthionine synthetase family. Biochemistry. 2011;50:891–898. doi: 10.1021/bi101750r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chatterjee C, Paul M, Xie L, van der Donk WA. Biosynthesis and mode of action of lantibiotics. Chem Rev. 2005;105:633–684. doi: 10.1021/cr030105v. [DOI] [PubMed] [Google Scholar]
- 40.Li B, Yu JP, Brunzelle JS, Moll GN, van der Donk WA, Nair SK. Structure and mechanism of the lantibiotic cyclase involved in nisin biosynthesis. Science. 2006;311:1464–1467. doi: 10.1126/science.1121422. [DOI] [PubMed] [Google Scholar]
- 41.Willey J, Van der Donk WA. Lantbiotics: Peptides of diverse structure and function. Annual Reviews of Microbiology. 2007;61:477–501. doi: 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]
- 42.McIntosh JA, Donia MS, Schmidt EW. Ribosomal peptide natural products: bridging the ribosomal and nonribosomal worlds. Nat Prod Rep. 2009;26:537–559. doi: 10.1039/b714132g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller DA, Luo L, Hillson N, Keating TA, Walsh CT. Yersiniabactin synthetase: a four-protein assembly line producing the nonribosomal peptide/polyketide hybrid siderophore of Yersinia pestis. Chem Biol. 2002;9:333–344. doi: 10.1016/s1074-5521(02)00115-1. [DOI] [PubMed] [Google Scholar]
- 44.Keating TA, Marshall CG, Walsh CT. Reconstitution and characterization of the Vibrio cholerae vibriobactin synthetase from VibB, VibE, VibF, and VibH. Biochemistry. 2000;39:15522–15530. doi: 10.1021/bi0016523. [DOI] [PubMed] [Google Scholar]
- 45.Chen H, O'Connor S, Cane DE, Walsh CT. Epothilone biosynthesis: assembly of the methylthiazolylcarboxy starter unit on the EpoB subunit. Chem Biol. 2001;8:899–912. doi: 10.1016/s1074-5521(01)00064-3. [DOI] [PubMed] [Google Scholar]
- 46.Schneider TL, Shen B, Walsh CT. Oxidase domains in epothilone and bleomycin biosynthesis: thiazoline to thiazole oxidation during chain elongation. Biochemistry. 2003;42:9722–9730. doi: 10.1021/bi034792w. [DOI] [PubMed] [Google Scholar]
- 47.Schneider TL, Walsh CT. Portability of oxidase domains in nonribosomal peptide synthetase modules. Biochemistry. 2004;43:15946–15955. doi: 10.1021/bi0481139. [DOI] [PubMed] [Google Scholar]
- 48.Kelly WL, Pan L, Li C. Thiostrepton biosynthesis: prototype for a new family of bacteriocins. J Am Chem Soc. 2009;131:4327–4334. doi: 10.1021/ja807890a. [DOI] [PubMed] [Google Scholar]
- 49.Walsh CT, Acker MG, Bowers AA. Thiazolyl peptide antibiotic biosynthesis: a cascade of post-translational modifications on ribosomal nascent proteins. J Biol Chem. 2010;285:27525–27531. doi: 10.1074/jbc.R110.135970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Selva E, Beretta G, Montanini N, Saddler GS, Gastaldo L, Ferrari P, Lorenzetti R, Landini P, Ripamonti F, Goldstein BP, et al. Antibiotic GE2270 a: a novel inhibitor of bacterial protein synthesis. I. Isolation and characterization. J Antibiot (Tokyo) 1991;44:693–701. doi: 10.7164/antibiotics.44.693. [DOI] [PubMed] [Google Scholar]
- 51.Lau RCM, Rinehart KL. BIOSYNTHESIS OF BERNINAMYCIN - INCORPORATION OF C-13-LABELED AMINO-ACIDS. Journal of the American Chemical Society. 1995;117:7606–7610. [Google Scholar]
- 52.Bagley MC, Dale JW, Merritt EA, Xiong X. Thiopeptide antibiotics. Chem Rev. 2005;105:685–714. doi: 10.1021/cr0300441. [DOI] [PubMed] [Google Scholar]
- 53.Anborgh PH, Parmeggiani A. New antibiotic that acts specifically on the GTP-bound form of elongation factor Tu. EMBO J. 1991;10:779–784. doi: 10.1002/j.1460-2075.1991.tb08009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu Z, Hopwood DA, Khosla C. Directed transfer of large DNA fragments between Streptomyces species. Appl Environ Microbiol. 2000;66:2274–2277. doi: 10.1128/aem.66.5.2274-2277.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morris RP, Leeds JA, Naegeli HU, Oberer L, Memmert K, Weber E, LaMarche MJ, Parker CN, Burrer N, Esterow S, Hein AE, Schmitt EK, Krastel P. Ribosomally synthesized thiopeptide antibiotics targeting elongation factor Tu. J Am Chem Soc. 2009;131:5946–5955. doi: 10.1021/ja900488a. [DOI] [PubMed] [Google Scholar]
- 56.Wieland Brown LC, Acker MG, Clardy J, Walsh CT, Fischbach MA. Thirteen posttranslational modifications convert a 14-residue peptide into the antibiotic thiocillin. Proc Natl Acad Sci U S A. 2009;106:2549–2553. doi: 10.1073/pnas.0900008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liao R, Duan L, Lei C, Pan H, Ding Y, Zhang Q, Chen D, Shen B, Yu Y, Liu W. Thiopeptide biosynthesis featuring ribosomally synthesized precursor peptides and conserved posttranslational modifications. Chem Biol. 2009;16:141–147. doi: 10.1016/j.chembiol.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Acker MG, Bowers AA, Walsh CT. Generation of thiocillin variants by prepeptide gene replacement and in vivo processing by Bacillus cereus. J Am Chem Soc. 2009;131:17563–17565. doi: 10.1021/ja908777t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bowers AA, Acker MG, Koglin A, Walsh CT. Manipulation of thiocillin variants by prepeptide gene replacement: structure, conformation, and activity of heterocycle substitution mutants. J Am Chem Soc. 2010;132:7519–7527. doi: 10.1021/ja102339q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mocek U, Zeng Z, O'Hagan D, Zhou P, Lai-Duen G, Beale J, Floss HG. Biosynthesis of the modified peptide antibiotic thiostrepton from Streptomyces azureus and Streptomyces laurentii. J Am Chem Soc. 1993;115:7992–8001. [Google Scholar]
- 61.Mocek UKAR, Tsuchiya R, Nguyne T, Beale J, Floss HG. Biosynthesis of the modified peptide antibiotic nosiheptide from Streptomyces actuosus. J Am Chem Soc. 1993;115:7557–7563. [Google Scholar]
- 62.Bowers AA, Walsh CT, Acker MG. Genetic interception and structural characterization of thiopeptide cyclization precursors from Bacillus cereus. J Am Chem Soc. 2010;132:12182–12184. doi: 10.1021/ja104524q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kelly WL. Intramolecular cyclizations of polyketide biosynthesis: mining for a "Diels-Alderase"? Org Biomol Chem. 2008;6:4483–4493. doi: 10.1039/b814552k. [DOI] [PubMed] [Google Scholar]
- 64.Young TS, Walsh CT. Identification of the thiazolyl peptide GE37468 gene cluster from Streptomyces ATCC 55365 and heterologous expression in Streptomyces lividans. Proc Natl Acad Sci U S A. 2011;108:13053–13058. doi: 10.1073/pnas.1110435108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.These three rings do not exhaust the capacity of proteins to fashion heterocycles out of peptide backbones. The remodeling of tripeptide motifs in proteins to make MIO (methylidene-imidazolone) cofactors in Phe and His ammonia lyases and the p-hydroxybenzylidene-imidazolone fluorophore in GFPs and congeneric chromoproteins are also remarkable but distinct from the transformations noted in this perspective in that they are autoctalytic, occur within the folded proteins and do not require partner proteins for the modification steps. These modified proteins stay intact rather than undergo proteolysis to release the chromophore as the functional end product.