Abstract
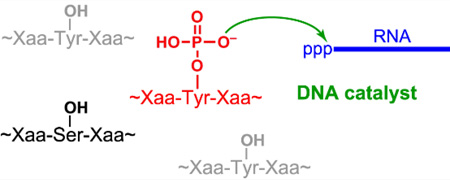
Phosphopeptides tagged: Phosphorylated tyrosine and serine residues in peptides are modified selectively by DNA catalysts (see the figure). The deoxyribozymes catalyze covalent attachment of an RNA tag to a range of peptide sequences, establishing proof-of-principle for a new approach to phosphopeptide analysis.
Keywords: deoxyribozymes, DNA, in vitro selection, phosphopeptide, peptides
Many natural peptides and proteins are phosphorylated on tyrosine (Tyr) or serine (Ser) residues. Phosphorylated peptides are important within neurochemistry (neuropeptides), immunology (cytokines), and endocrinology (hormones). For such peptides as well as for larger proteins, side chain phosphorylation is frequently associated with modulation of biological function.[1] Methods for analysis of phosphopeptides often depend upon their initial chromatographic separation from nonphosphorylated analogues using support-bound chelators or covalent binders of phosphate groups or products derived from them.[2] Alternatively, phosphotyrosine-specific antibodies can be generated, albeit with the attendant investments in cost and time.[2g,3] Here we describe proof of principle for an entirely different approach to phosphopeptide analysis in which DNA catalysts (deoxyribozymes) covalently tag phosphorylated amino acid side chains of peptides. In this approach, it is critical to ensure high selectivity for modification of phosphorylated amino acid side chains over their nonphosphorylated analogues.
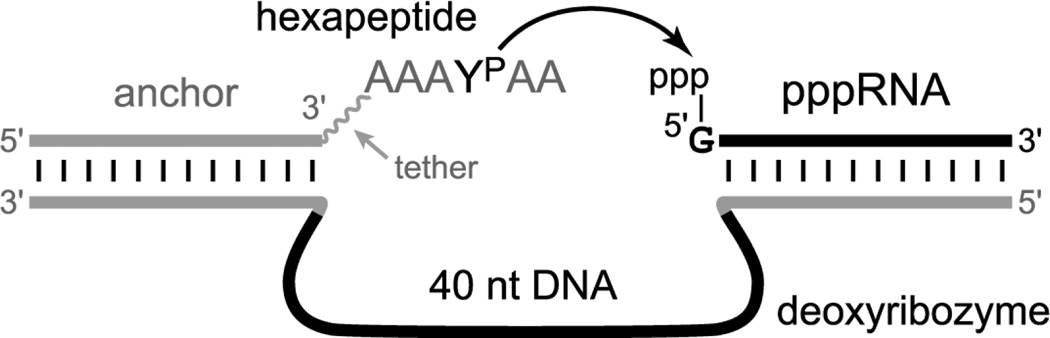
Deoxyribozymes were originally identified for the catalysis of RNA cleavage,[7] and their use has expanded to encompass a range of chemical reactions.[8] Our lab has reported a variety of deoxyribozymes for different chemical reactions,[9] including the covalent modification of amino acid side chains.[5,10] In particular, we have recently shown that tripeptide substrates can be covalently modified by the attachment of an RNA strand at nonphosphorylated Tyr or Ser.[6] Here we sought to identify deoxyribozymes that covalently modify phosphorylated TyrP (YP), using in vitro selection as shown in Figure 1. The hexapeptide substrate AAAYPAA was connected to a DNA anchor oligonucleotide via either a short or long tether [see structures in Supporting Information; the short tether connects the hexapeptide directly via its α-amino group to the DNA anchor, whereas the long tether includes an intervening hexa(ethylene glycol) moiety]. In vitro selection was used to identify deoxyribozymes that attach a 5′-triphosphorylated RNA tag to TyrP, with pyrophosphate as the leaving group.
Figure 1.
Strategy for selective covalent modification of phosphorylated Tyr (TyrP, YP) within a peptide substrate. In vitro selection identifies deoxyribozymes that function with a TyrP-containing hexapeptide substrate, catalyzing attachment of 5′-triphosphorylated RNA to the phosphorylated amino acid residue. The hexapeptide substrate is attached to an oligonucleotide anchor via a tether (see text for composition). The product has a pyrophosphate linkage between Tyr and RNA. Further information about the selection procedure, which followed our standard approach,[4–6] is provided in the Supporting Information.
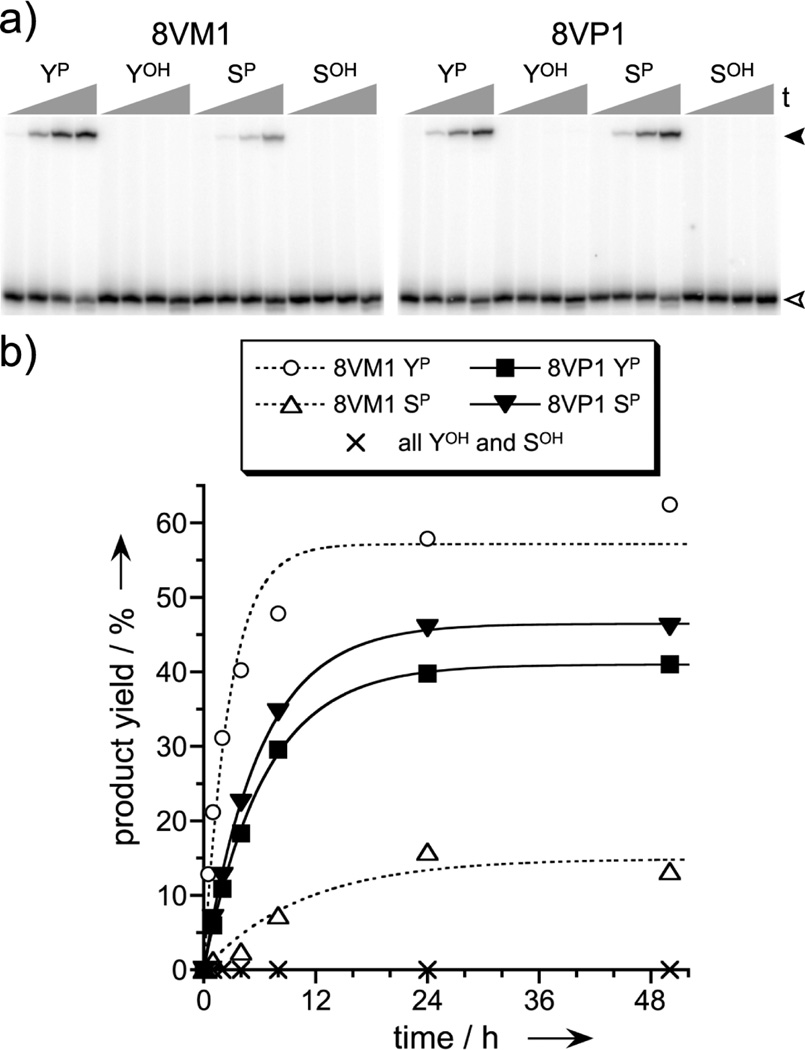
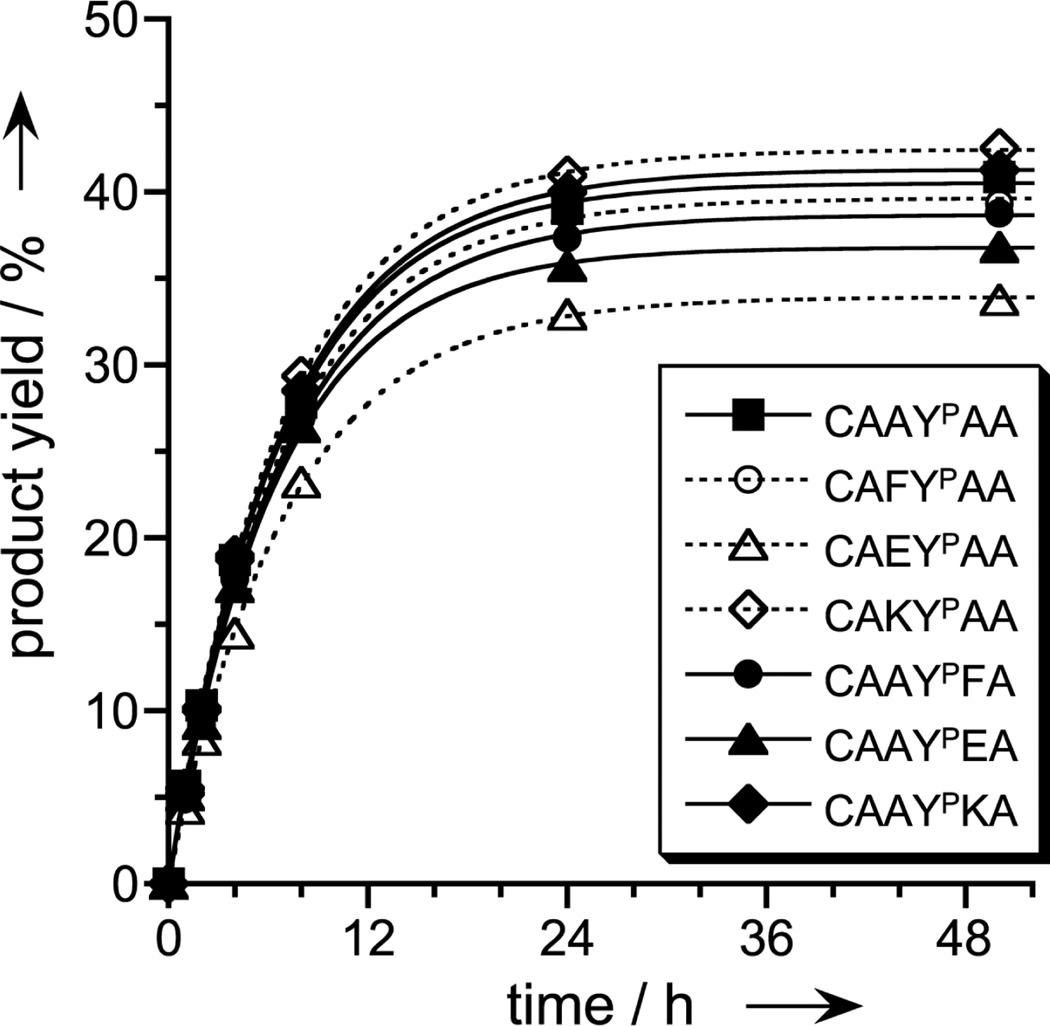
Two new deoxyribozymes from the selection process, 8VM1 and 8VP1 (one from each of the two selection experiments), were examined in more detail on the basis of their high catalytic activities with the TyrP- and analogous phosphoserine (SerP)-containing hexapeptides. Both deoxyribozymes were highly selective (>200:1) for each phosphorylated peptide over its nonphosphorylated analogue, with no detectable reaction at TyrOH or SerOH (<0.5%; Figure 2). 8VM1, which was identified by selection with the short tether, favored as its substrate the TyrP peptide over the SerP peptide by about 4- to 5-fold. In contrast, 8VP1, which was found via selection with the long tether, functioned equally well with TyrP or SerP peptides. 8VP1 was also found to accept a range of different amino acid identities—including hydrophobic and charged residues—flanking the TyrP that it covalently modifies (Figure 3), suggesting broad generality for different phosphopeptide sequences.
Figure 2.
The 8VM1 and 8VP1 deoxyribozymes covalently modify phosphotyrosine and phosphoserine. (a) PAGE image showing high selectivity for TyrP over TyrOH and for SerP over SerOH (50 mM HEPES, pH 7.5, 40 mM Mg2+, 20 mM Mn2+, 150 mM NaCl, 37 °C; single-turnover assays). Representative time points at 0, 0.5, 4, 24 h. Open arrowhead: 3′-32P-radiolabeled RNA substrate. Filled arrowhead: DNA-anchored hexapeptide attached to RNA. Each peptide was attached to the oligonucleotide anchor via its amino terminus and a short (8VM1) or long (8VP1) tether; see Supporting Information. Each product identity was verified by MALDI mass spectrometry (see Experimental Section). (b) Kinetic data for the tagging reactions. kobs (top to bottom for plots) = 0.37, 0.17, 0.15, 0.095 h−1. For 8VP1 (but not 8VM1), 2–4% product in 50 h is observed when the YP or SP hexapeptide is unattached to the DNA anchor, i.e., a free hexapeptide substrate (data not shown).
Figure 3.
Sequence generality for the phosphopeptide substrate. Covalent modification by 8VP1 was examined with DNA-anchored hexapeptide substrate CAAYPAA and several illustrated sequence variants, for which one amino acid adjacent to YP (on either side) was changed to one of F (hydrophobic), E (negatively charged), or K (positively charged). Experiments were performed as in Figure 2. Each peptide was attached to the DNA anchor via the N-terminal cysteine side chain, which enables inclusion of K within the sequence and also allows cleavage of the peptide from the anchor by DTT reduction.
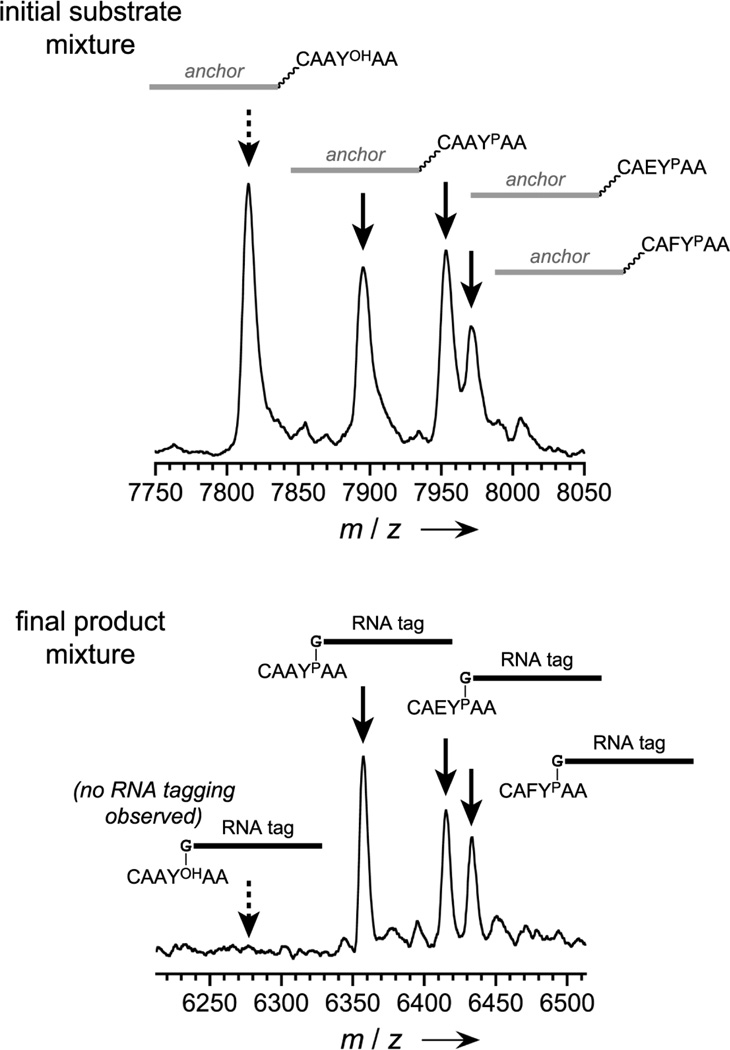
To examine the applicability of deoxyribozymes for analysis of mixtures of phosphorylated and nonphosphorylated peptides, such a mixture (each peptide attached via a disulfide to the DNA anchor) was tagged with RNA by the 8VP1 deoxyribozyme. Analysis of the unpurified product mixture by MALDI mass spectrometry (after DTT cleavage of the DNA anchor) revealed selective RNA tagging of only the phosphopeptides, despite the presence of a large amount of nonphosphorylated peptide (Figure 4).
Figure 4.
Analysis of a peptide mixture by mass spectrometry using the DNA-catalyzed tagging approach. See diagram of this experiment in the Supporting Information. Each of a mixture of nonphosphorylated and phosphorylated peptides (100 pmol nonphosphorylated peptides; 33 pmol each of three phosphorylated peptides) was attached via HEG tether to a common DNA oligonucleotide anchor, and the mixture was tagged with RNA by 8VP1. After PAGE separation of RNA-tagged peptides and removal of the DNA anchors by DTT reduction, analysis by MALDI mass spectrometry revealed that only the phosphorylated peptides were covalently modified with the RNA tag, as desired. No tagging of the nonphosphorylated peptide was observed (lower spectrum, left side; compare with substantial nonphosphorylated peptide signal in the upper spectrum).
In summary, we have demonstrated proof of principle that DNA can catalyze highly selective covalent modification of phosphorylated Tyr or Ser residues in phosphopeptides by attaching an RNA tag at those positions. To our knowledge, this is the first report of any chemical approach for covalent, specific tagging of phosphopeptide side chains. In downstream applications, this RNA tag should be useful to report upon the amount of phosphorylated peptides present in a sample, e.g., by RT-PCR, which may help to avoid issues encountered during mass spectrometric analysis of peptide phosphorylation.[11] A wide range of peptide sequence contexts are accepted by the investigated deoxyribozymes, suggesting that this general approach may be made competitive with more traditional chromatographic separations of phosphopeptides.[2] The phosphopeptide analytical approach outlined here is distinct from methods that depend upon engineering of individual kinases to accept modified ATP substrates.[12] The present findings also expand the repertoire of DNA catalysis to include covalent modification of phosphorylated amino acid side chains. Independently, we have shown that RNA-tagging deoxyribozymes can discriminate against phosphorylated residues in favor of their nonphosphorylated analogues with promising selectivity (>20:1; data not shown). That observation along with the present work suggests the viability of ratiometric analyses in which both phosphorylated and nonphosphorylated peptides are covalently modified with different tags in the same sample.
Several important issues must be addressed in future development of this approach. We will seek DNA catalysts that tag specific sequences of phosphopeptides, rather than accepting a broad range of peptide sequences. The approach also must be developed to work with free, rather than oligonucleotide-anchored, peptide substrates as well as with large phosphorylated proteins, ideally in complex mixtures such as cell lysates. Towards this goal, we have recently demonstrated the first steps towards DNA-catalyzed reactivity of free peptides;[6,13] such efforts must be merged with the present work to establish a useful analytical method.
Experimental Section
General procedures
Hexapeptides were prepared by solid-phase synthesis using Fmoc Rink amide MBHA resin and attached to DNA anchor oligonucleotides as described in the Supporting Information. DNA oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA) or prepared by solid-phase synthesis on an ABI 394 instrument using reagents from Glen Research. 5′-Triphosphorylated RNA oligonucleotides were prepared by in vitro transcription using synthetic DNA templates and T7 RNA polymerase.[14] All oligonucleotides were purified by denaturing PAGE with running buffer 1× TBE (89 mM each Tris and boric acid and 2 mM EDTA, pH 8.3) as described previously.[4,15] The in vitro selection procedure is described in the Supporting Information. Deoxyribozyme sequences (catalytic regions underlined): 8VM1, 5′-CCGTCGCCATCTCTTCGACTGCGGGAGCGGTGAGCGGGTAGGTCTACATGAGGGCTATAGTGAGTCGTATTATCC-3′; 8VP1, 5′-CCGTCGCCATCTCTTCGGACACGATGAGTGACTAAGTGGAATGAGGAAAGCACGAGATAGTGAGTCGTATTATCC-3′.
Kinetic assays
Single-turnover kinetic assays of the 8VM1 and 8VP1 deoxyribozymes with various substrates (Figures 2 and 3) were performed using the following procedure. A 14 µL sample containing 1 pmol of 3′-32P-radiolabeled 5′-triphosphorylated RNA substrate (radiolabel attached with 32P-pCp and T4 RNA ligase), 5 pmol of deoxyribozyme, and 10 pmol of DNA-anchored hexapeptide was annealed in 5 mM HEPES, pH 7.5, 15 mM NaCl, and 0.1 mM EDTA by heating at 95 °C for 3 min and cooling on ice for 5 min. The DNA-catalyzed tagging reaction was initiated by bringing the sample to 20 µL total volume containing 50 mM HEPES, pH 7.5, 40 mM MgCl2, 20 mM MnCl2, 150 mM NaCl, and 2 mM KCl and incubating at 37 °C. At appropriate times, 2 µL aliquots were quenched with 5 µL of stop solution (80% formamide, 1× TBE [89 mM each Tris and boric acid, 2 mM EDTA, pH 8.3], 50 mM EDTA, and 0.025% each bromophenol blue and xylene cyanol). Before PAGE, to each sample was added 50 pmol of a “decoy oligonucleotide”, which was a 60-mer complementary to the deoxyribozyme’s 40 nt enzyme region along with 10 nt of binding arm on each side (added to displace the deoxyribozyme from the substrates and ligation product). Samples were separated by 20% PAGE and quantified with a PhosphorImager. kobs values were obtained by fitting the yield versus time data directly to first-order kinetics.
DNA-catalyzed RNA tagging and mass spectrometry of phosphopeptides
MALDI mass spectrometry was used to verify identities of the products of DNA-catalyzed RNA tagging of DNA-anchored phosphopeptides. Products were prepared using the following procedure. A 15 µL sample containing 300 pmol of DNA-anchored hexapeptide substrate, 330 pmol of deoxyribozyme, and 360 pmol of 5′-triphosphorylated RNA substrate was annealed in 5 mM HEPES, pH 7.5, 15 mM NaCl, and 0.1 mM EDTA by heating at 95 °C for 3 min and cooling on ice for 5 min. The reaction was initiated by bringing the sample to 30 µL total volume containing 50 mM HEPES, pH 7.5, 40 mM MgCl2, 20 mM MnCl2, 150 mM NaCl, and 2 mM KCl and incubating at 37 °C for 14 h. The product was precipitated with ethanol, separated by 20% PAGE, extracted from the polyacrylamide gel in TEN buffer (10 mM Tris, pH 8.0, 300 mM NaCl, and 1 mM EDTA), and precipitated with ethanol. The sample was dissolved in 20 µL of water; 10 µL was desalted by C18 ZipTip and used for mass spectrometry. All observed mass values were in accord with expectations (see Supporting Information for spectra). 8VM1 product with DNA-AAAYPAA substrate: m/z calcd. 12372.1, found 12381.9 (Δ= +0.08%). 8VM1 product with DNA-AAASPAA substrate: m/z calcd. 12296.0, found 12299.3 (Δ = −0.03%). 8VP1 product with DNA-HEG-AAAYPAA substrate: m/z calcd. 12719.4, found 12721.0 (Δ = +0.01%). 8VP1 product with DNA-HEG-AAASPAA substrate: m/z calcd. 12643.3, found 12645.9 (Δ = +0.02%).
To apply deoxyribozymes for RNA tagging of a mixture of phosphorylated and nonphosphorylated peptides (Figure 4; see diagram of experiment in the Supporting Information), the following procedure was used. First, one nonphosphorylated hexapeptide (CAAYOHAA) and three phosphorylated peptides (CAAYPAA, CAEYPAA, and CAFYPAA), each of them HEG-tethered via a disulfide to the same DNA anchor sequence as described above, were mixed in the ratio 100:33:33:33 pmol in 30 µL of water. This sample was desalted using three C18 ZipTips and analyzed by MALDI mass spectrometry, with spectrum shown on the top of Figure 4. All observed mass values were in accord with expectations. DNA-HEG-CAAYOHAA substrate: m/z calcd. 7816.5, found 7815.6 (Δ = −0.01%). DNA-HEG-CAAYPAA substrate: m/z calcd. 7896.5, found 7896.0 (Δ = −0.006%). DNA-HEG-CAEYPAA substrate: m/z calcd. 7954.5, found 7953.8 (Δ = −0.009%). DNA-HEG-CAFYPAA substrate: m/z calcd. 7972.6, found 7971.8 (Δ = −0.01% ). Second, the 8VP1 deoxyribozyme-catalyzed RNA tagging reaction was performed on the same peptide mixture. A 20 µL sample containing 100:33:33:33 pmol of the same four peptides as listed above, 220 pmol of 8VP1 deoxyribozyme, and 240 pmol of 5′-triphosphorylated RNA substrate was annealed in 5 mM HEPES, pH 7.5, 15 mM NaCl, and 0.1 mM EDTA by heating at 95 °C for 3 min and cooling on ice for 5 min. The DNA-catalyzed tagging reaction was initiated by bringing the sample to 40 µL total volume containing 50 mM HEPES, pH 7.5, 40 mM MgCl2, 20 mM MnCl2, 150 mM NaCl, and 2 mM KCl and incubating at 37 °C for 24 h. The sample was purified by 20% denaturing PAGE; the band corresponding to the mixture of RNA-tagged anchored peptides was extracted with TEN and precipitated with ethanol. To remove the DNA anchor by disulfide reduction, the sample was redissolved in 50 µL of 50 mM HEPES, pH 7.5, and 50 mM DTT and incubated at 37 °C for 2 h. The sample was again precipitated with ethanol, redissolved in 30 µL of water, desalted using three C18 ZipTips, and analyzed by MALDI mass spectrometry, with the spectrum shown on the bottom of Figure 4. All observed mass values were in accord with expectations. As expected, no peak was observed for tagging of the nonphosphorylated CAAYOHAA peptide (m/z calcd. 6279.2). CAAYPAA-RNA product: m/z calcd. 6359.2, found 6357.7 (Δ = −0.02%). CAEYPAA-RNA product: m/z calcd. 6417.4, found 6415.5 (Δ = −0.03%). CAFYPAA-RNA product: m/z calcd. 6435.2, found 6433.6 (Δ = −0.02%).
Supplementary Material
Acknowledgements
This research was supported by grants to S.K.S. by the National Institutes of Health (GM065966), the Defense Threat Reduction Agency (BRBAA08-L-2-0001), and the National Science Foundation (0842534). A.S. was partially supported by NIH T32 GM070421.
Footnotes
Supporting information for this article is available on the WWW under http://www.chembiochem.org or from the author.
References
- 1.a) Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. Proteomics. 2004;4:1551–1561. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]; b) Gnad F, Ren S, Cox J, Olsen JV, Macek B, Oroshi M, Mann M. Genome Biol. 2007;8:R250. doi: 10.1186/gb-2007-8-11-r250. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Linding R, Jensen LJ, Pasculescu A, Olhovsky M, Colwill K, Bork P, Yaffe MB, Pawson T. Nucleic Acids Res. 2008;36:D695–D699. doi: 10.1093/nar/gkm902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a) Zhou H, Watts JD, Aebersold R. Nat. Biotechnol. 2001;19:375–378. doi: 10.1038/86777. [DOI] [PubMed] [Google Scholar]; b) Thaler F, Valsasina B, Baldi R, Xie J, Stewart A, Isacchi A, Kalisz HM, Rusconi L. Anal. Bioanal. Chem. 2003;376:366–373. doi: 10.1007/s00216-003-1919-9. [DOI] [PubMed] [Google Scholar]; c) Nühse TS, Stensballe A, Jensen ON, Peck SC. Mol. Cell. Proteomics. 2003;2:1234–1243. doi: 10.1074/mcp.T300006-MCP200. [DOI] [PubMed] [Google Scholar]; d) Thingholm TE, Jorgensen TJ, Jensen ON, Larsen MR. Nat. Protoc. 2006;1:1929–1935. doi: 10.1038/nprot.2006.185. [DOI] [PubMed] [Google Scholar]; e) Li Y, Wu J, Qi D, Xu X, Deng C, Yang P, Zhang X. Chem. Commun. 2008:564–566. doi: 10.1039/b716055k. [DOI] [PubMed] [Google Scholar]; f) Thingholm TE, Jensen ON, Larsen MR. Proteomics. 2009;9:1451–1468. doi: 10.1002/pmic.200800454. [DOI] [PubMed] [Google Scholar]; g) Dunn JD, Reid GE, Bruening ML. Mass Spectrom. Rev. 2010;29:29–54. doi: 10.1002/mas.20219. [DOI] [PubMed] [Google Scholar]; h) Nelson CA, Szczech JR, Dooley CJ, Xu Q, Lawrence MJ, Zhu H, Jin S, Ge Y. Anal. Chem. 2010;82:7193–7201. doi: 10.1021/ac100877a. [DOI] [PMC free article] [PubMed] [Google Scholar]; i) Wang F, Song C, Cheng K, Jiang X, Ye M, Zou H. Anal. Chem. 2011;83:8078–8085. doi: 10.1021/ac201833j. [DOI] [PubMed] [Google Scholar]; j) Xu B, Zhou L, Wang F, Qin H, Zhu J, Zou H. Chem. Commun. 2012;48:1802–1804. doi: 10.1039/c2cc16662c. [DOI] [PubMed] [Google Scholar]
- 3.Brumbaugh K, Johnson W, Liao WC, Lin MS, Houchins JP, Cooper J, Stoesz S, Campos-Gonzalez R. Methods Mol. Biol. 2011;717:3–43. doi: 10.1007/978-1-61779-024-9_1. [DOI] [PubMed] [Google Scholar]
- 4.Flynn-Charlebois A, Wang Y, Prior TK, Rashid I, Hoadley KA, Coppins RL, Wolf AC, Silverman SK. J. Am. Chem. Soc. 2003;125:2444–2454. doi: 10.1021/ja028774y. [DOI] [PubMed] [Google Scholar]
- 5.Sachdeva A, Silverman SK. Chem. Commun. 2010;46:2215–2217. doi: 10.1039/b927317d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong OY, Pradeepkumar PI, Silverman SK. Biochemistry. 2011;50:4741–4749. doi: 10.1021/bi200585n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Breaker RR, Joyce GF. Chem. Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]; b) Silverman SK. Nucleic Acids Res. 2005;33:6151–6163. doi: 10.1093/nar/gki930. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Schlosser K, Li Y. ChemBioChem. 2010;11:866–879. doi: 10.1002/cbic.200900786. [DOI] [PubMed] [Google Scholar]
- 8.a) Silverman SK. Chem. Commun. 2008:3467–3485. doi: 10.1039/b807292m. [DOI] [PubMed] [Google Scholar]; b) Schlosser K, Li Y. Chem. Biol. 2009;16:311–322. doi: 10.1016/j.chembiol.2009.01.008. [DOI] [PubMed] [Google Scholar]; c) Silverman SK. Angew. Chem. Int. Ed. 2010;49:7180–7201. doi: 10.1002/anie.200906345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silverman SK. Acc. Chem. Res. 2009;42:1521–1531. doi: 10.1021/ar900052y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pradeepkumar PI, Höbartner C, Baum DA, Silverman SK. Angew. Chem. Int. Ed. 2008;47:1753–1757. doi: 10.1002/anie.200703676. [DOI] [PubMed] [Google Scholar]
- 11.a) Domon B, Aebersold R. Science. 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]; b) Witze ES, Old WM, Resing KA, Ahn NG. Nat. Methods. 2007;4:798–806. doi: 10.1038/nmeth1100. [DOI] [PubMed] [Google Scholar]; c) Steen H, Jebanathirajah JA, Rush J, Morrice N, Kirschner MW. Mol. Cell. Proteomics. 2006;5:172–181. doi: 10.1074/mcp.M500135-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Blethrow JD, Glavy JS, Morgan DO, Shokat KM. Proc. Natl. Acad. Sci. USA. 2008;105:1442–1447. doi: 10.1073/pnas.0708966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong OY, Mulcrone AE, Silverman SK. Angew. Chem. Int. Ed. 2011;50:11679–11684. doi: 10.1002/anie.201104976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Silverman SK. Biochemistry. 2003;42:15252–15263. doi: 10.1021/bi0355847. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.