Abstract
Purpose
Integration of signal transduction inhibitors into chemotherapy regimens generally has generally not led to anticipated increases in response and survival. However, it remains unclear whether this is because of inadequate or inconsistent inhibition of target or other complex biology. The mammalian target of rapamycin (mTOR) signaling pathway is frequently activated in acute myelogenous leukemia (AML) and we previously demonstrated the safety of combining the mTOR inhibitor, sirolimus, with mitoxantrone, etoposide, and cytarabine (MEC) chemotherapy. However, we did not reliably determine the extent of mTOR inhibition on that study. Here we sought to develop an assay that allowed us to serially quantify mTOR kinase’s activation state during therapy.
Experimental design
To provide evidence of mTOR kinase activation and inhibition, we applied a validated whole blood fixation/permeabilization technique for flow cytometry in order to serially monitor S6 ribosomal protein (S6) phosphorylation in immunophenotypically-identified AML blasts.
Results
With this approach, we demonstrate activation of mTOR signaling in 8/10 subjects’ samples (80%) and conclusively show inhibition of mTOR in the majority of subjects’ tumor cell during therapy. Of note, S6 phosphorylation in AML blasts is heterogeneous and, in some cases, intrinsically resistant to rapamycin at clinically achieved concentrations.
Conclusions
The methodology described is rapid and reproducible. We demonstrate the feasibility of real-time, direct pharmacodynamic monitoring by flow cytometry during clinical trials combining intensive chemotherapy and signal transduction inhibitors. This approach greatly clarifies pharmacokinetic/pharmacodynamic relationships and has broad application to pre-clinical and clinical testing of drugs whose direct or downstream effects disrupt PI3K/AKT/mTOR signaling.
Introduction
It has been proposed that molecularly-targeted cancer therapeutics would herald an era of improved clinical response and reduced need for more highly toxic, traditional cytotoxic chemotherapy. However, with few exceptions, neither goal has been met in many, if not most tumor types. There are several possible reasons for this dilemma. Signal transduction inhibitors (STIs) may not adequately inhibit target proteins in vivo.(1, 2) Alternatively, functional redundancy within cancer cells may allow several signaling pathways to share roles in maintaining growth or survival.(3) Finally, it is possible that there is heterogeneity of signaling between different populations of tumor cells so that STI therapy may simply select for a sub-population of cells that do not require the targeted pathway to survive.(4) As none of these effects can be predicted by drug levels alone, direct pharmacodynamic measurements of signal transduction inhibitors during early phase clinical trials may facilitate rational clinical development of molecularly-targeted agents. In developmental therapeutics for most epithelial and soft tissue tumors, repeated tumor biopsy sampling is logistically challenging. However, hematopoietic cancers such as leukemia provide an excellent opportunity to serially sample circulating tumor cells to validate novel agents targeting signal transduction.
Phospho-specific flow cytometry is an emerging technique for single cell evaluation of signal transduction. In hematologic malignancies that are well characterized immunophenotypically, it holds the promise to provide tumor-specific analysis of signal transduction in heterogeneous tissue.(5–7) This includes peripheral blood and/or bone marrow samples containing admixtures of normal and malignant cells. Using antibodies that are highly specific for the phosphorylation state of key residues on signaling proteins, this technology allows the researcher to interpolate a signal transduction pathway’s activation state in immunophenotypically-identified cells. However, the necessary reagents for sample processing potentially alter protein structure and/or antibody binding in ways that can compromise data quality for cell surface epitopes, light scatter, and intracellular phospho-proteins alike.(8) These reagents typically include formaldehyde or paraformaldehyde to fix cells followed by alcohols or detergents to permeabilize their cellular membranes. Although relatively easily applied to highly uniform cell samples, such as cell lines, phospho-specific flow cytometry has thus far resisted routine application to clinical specimens.
Recent advances have identified methodologies that simplify processing and improve data quality from clinical specimens, particularly for evaluation of ras/MAPK and PI3K/AKT/mTOR signaling.(9, 10) Current whole blood and marrow fixation techniques provide excellent preservation of light scatter properties and surface immunophenotype, allowing for evaluation of single or multiple phospho-proteins in specific cell populations.(11, 12) However, few if any clinical trials in hematologic malignancies have used these techniques systematically to evaluate pharmacodynamic effects of drugs with concurrent therapeutic drug monitoring.
Acute myelogenous leukemia (AML) is a clinically and genetically heterogeneous hematopoietic cancer characterized by the accumulation of immature myeloid precursors in the marrow and blood, leading to rapid and inexorable exhaustion of normal hematopoiesis. Clinically, AML has a poor survival due to high relapse rates following initial chemotherapy or de novo chemotherapy resistance, which is seen in half of patients over the age of 65.(13) Investigational approaches in AML have included targeting oncogenic signal transduction, either alone or in combination with chemotherapy. Preclinical data from our group and others highlights a critical role for the activation of phosphotidylinositol 3′ kinase (PI3K) pathway signaling through its downstream effectors AKT.(14, 15) These data suggest that the mammalian target of rapamycin (mTOR) plays a critical role in chemotherapy resistance and inhibition of mTOR may augment chemotherapy response. We therefore designed clinical trials combining rapamycin (sirolimus, Rapamune) with cytotoxic chemotherapy for AML.(16)
The cellular effects of mTOR are largely mediated by its activation of the p70S6 kinase, which itself phosphorylates ribosomal protein S6 (S6). Although p70S6 kinase antibodies are useful readouts by western blot, they have not been optimized for flow cytometric analysis. By contrast, S6 is abundantly expressed in normal and malignant cells and evaluation of S6 phosphorylation is commonly used as a surrogate marker for mTOR activation. Importantly, S6 phosphorylation is rapidly and thoroughly inhibited in the presence of rapamycin, while other functions of mTOR (e.g. phosphorylation of its other major downstream target, 4EBP1) are less clearly modulated by rapamycin treatment.(17)
Here we report the pharmacodynamic results of a pilot study of the rapamycin mTOR inhibitor sirolimus in combination with intensive, AML induction chemotherapy. These data demonstrate the feasibility of serial flow cytometric monitoring of mTOR activation state in immunophenotypically identified tumor cells from fixed whole blood. To measure mTOR activation by flow cytometry, we used S6 phosphorylation as a readout. This approach provides robust data despite low circulating tumor burden, and, in combination with therapeutic drug monitoring and ex vivo dose response modeling, highlights biologic heterogeneity in AML tumor cells that may predict clinical response or resistance to molecularly targeted agents.
Materials and Methods
Clinical trial Subject Selection
Subjects were recruited from the clinical practices of the University of Pennsylvania between February 2008 and November 2008. Eligible subjects were aged >18 with non-M3 AML that either had relapsed following prior chemotherapy or was refractory to induction chemotherapy. Primary refractory leukemia was defined as either persistent disease (>5% marrow blasts) following two induction cycles or recurrent leukemia following a single induction cycle that had resulted in complete tumor clearance from a nadir or recovering marrow biopsy. Patients older than age 60 were eligible with untreated AML provided they had no evidence of SWOG favorable risk karyotype by cytogenetics, FISH, or multiplexed RT-PCR (for AML1-ETO or MYH11-CBFβ). Patients with untreated secondary leukemia--defined as AML arising out of an antecedent hematologic disorder or following chemotherapy or radiation therapy--were also eligible. Pathology review at the Hospital of the University of Pennsylvania was performed to confirm diagnoses in all cases.
All subjects were required to have an ECOG performance status of 0–1 and resolution of prior treatment-related toxicities. Required baseline organ function studies specified an ejection fraction above 45%, creatinine ≤2.0mg/dL, total bilirubin ≤1.5 mg/dL, hepatic transaminases ≤3x upper level of normal, no uncontrolled infections, and no unstable baseline co-morbidities that would jeopardize toxicity assessment. Due to significant drug-drug interactions between sirolimus and systemic imidazole or triazole antifungals, these medications were prohibited for 72 hours prior to study entry. Subjects whose medical regimen required drugs with marked effects upon sirolimus pharmacokinetics (e.g. rifampin, carbemazepine, verapamil, diltiazem, tacrolimus) were ineligible.
The research was approved by the Institutional Review Boards of the University of Pennsylvania and the study was monitored by the Clinical Trials Safety Review and Monitoring Committee of the Abramson Cancer Center of the University of Pennsylvania. Written informed consent was obtained from all subjects according to the Declaration of Helsinki.
Treatment schema
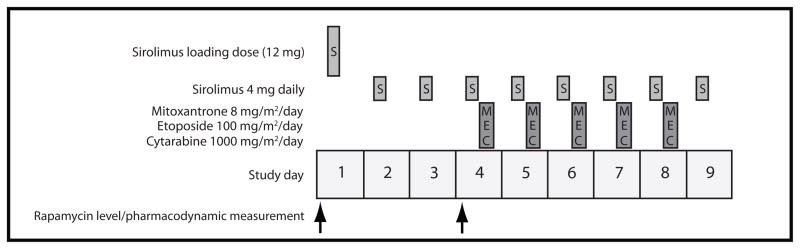
The treatment regimen is shown in Figure 1 and consisted of a loading dose of 12 mg sirolimus by mouth on day 1 followed by 8 daily doses of 4 mg sirolimus administered orally. 72 hours after the sirolimus loading dose, subjects began intravenous MEC chemotherapy (mitoxantrone 8 mg/m2/day on days 1–5, etoposide 100 mg/m2/day on days 1–5, and cytarabine 1000 mg/m2/day on days 1–5) using actual body weight for dose calculations. Each intravenous chemotherapy drug was administered over one hour and actual body weight was used for chemotherapy calculations. Concurrent hydroxyurea was allowed on days 1–3 (i.e. concurrent with sirolimus monotherapy) to control leukocytosis prior to initiation of MEC chemotherapy. Whole blood rapamycin concentration was measured in the clinical laboratory by high pressure liquid chromatography from samples drawn prior to the daily dose on treatment days 4 and 7. Sirolimus doses were not adjusted for weight or altered based upon measured drug concentration. Concurrent with the day 4 trough sirolimus level, peripheral blood was collected and processed as below for flow cytometric pharmacodynamic analysis.
Figure 1. treatment schema.
A 12 mg loading dose of oral sirolimus was administered on day 1, followed by 8 daily doses of 4 mg sirolimus. Mitoxantrone 100 mg/m2/day, etoposide 100 mg/m2/day, and cytarabine 1000 mg/m2/day were each administered over 1 hour for five doses each starting after sirolimus administration on study day 4. Samples were obtained for pharmacodynamic measurement and rapamycin concentration measurement on day 4 prior to sirolimus dosing. A second rapamycin level was obtained prior to sirolimus dosing on day 7.
Supportive care and toxicity assessment
Subjects care guidelines were previously described.(16) Toxicity assessment used the National Cancer Institute’s Common Toxicity Criteria for Adverse Events (CTCAE) version 3.0 (http://ctep.cancer.gov/protocoldevelopment/electronic_applications/docs/ctcaev3.pdf).
Response criteria
Complete remission was defined by a bone marrow aspirate showing >20% cellularity, trilineage hematopoiesis, and <5% blasts as well as the presence of an absolute neutrophil count >1000/μL, an un-transfused platelet count of >100,000/μL, and no circulating or extramedullary leukemic blasts.(18) Complete remission without platelet recovery (CRp) met all criteria for CR except the platelet count failed to exceed 100,000/uL. A partial remission (PR) met all criteria for CR, except the marrow blast percentage was between 5 and 25%. Progressive disease was defined as an increase of at least 25% in the absolute number of leukemic cells in peripheral blood or bone marrow/aspirate, the development of extramedullary disease, or other evidence of increased tumor burden.
Flow cytometric pharmacodynamic analysis
Peripheral blood was collected into heparinized vacutainers and was transported to the laboratory within one hour of collection for samples drawn after sirolimus, and immediately processed upon receipt. Baseline samples were processed within 24 hours of collection. Prior work in our laboratory has shown that this timeframe did not alter measurements of mTOR activation (data not shown) in the absence of signal transduction inhibitor therapy. Whole blood was passed through a 40 micron mesh filter to remove any possible clotted specimen. Multiple 100 μL aliquots were prepared in 15 × 75 mm flow tubes. To these aliquots, sample conditions included unmanipulated aliquots as well as ex vivo activation of S6 phosphorylation by phorbol myristate acetate (PMA, Sigma-Aldrich) 400 nM for 10 minutes, and inhibition by rapamycin (Sigma-Aldrich) 500 or 1000 nM for 30 minutes. These conditions were carried out in a dry bath (humidified incubator at 37C, 21% O2, 5% CO2).
Following incubation in the dry bath, blood samples were fixed, red cells lysed, and remaining nucleated cells permeabilized as previously described.(9, 10) Briefly, cells were fixed for 10 minutes by addition of room temperature, methanol-free formaldehyde (Polysciences) directly to samples to achieve a final concentration of 4%. To permeabilize nucleated cells and lyse red blood cells, fixed samples were then incubated for 30 minutes at room temperature in Triton X-100 (Sigma-Aldrich) diluted in calcium and magnesium-free, phosphate buffered saline (PBS) to a final concentration 0.1%. Triton X-100 was pre-warmed to 37C prior to addition to the fixed blood. Samples were then centrifuged for 3 minutes at 1000g at 4C, washed with 4C 4% bovine serum albumin (BSA) in PBS wash buffer, centrifuged again, and supernatants were discarded. The resulting cell pellets were either resuspended in medium containing 10% weight/volume glycerol (Sigma), 20% fetal calf serum, and 70% PBS and stored at −20C or analyzed immediately. Frozen baseline and day 4 samples were thawed by incubation in a room temperature water bath and then were centrifuged as above, washed once as above, and the cell pellets resuspended in ice cold 50% methanol in 0.9% NaCl for 10 minutes to unmask phospho-epitopes. Samples used for ex vivo drug sensitivity studies were not frozen but were immediately placed in methanol as above and analyzed by flow cytometry on the day of collection. Following methanol incubation, samples were washed twice at 4C and stained with antibodies (30minutes in the dark), then washed once and subjected to flow cytometric analysis.
Antibodies used included murine anti-human CD45 PerCP, CD45 APC-Cy7, CD34 APC, CD33 PE-Texas Red (invitrogen), CD45 PE-Cy5, CD33 PE, CD34 PE, CD14 PE, CD10 APC (all from Becton Dickinson), rabbit monoclonal phospho-S6 Alexa 488 or Alexa 647 (Cell Signaling). A “fluorescence minus one” (FMO) control containing antibodies directed against cell surface markers but not phospho-S6 was included for all subjects to estimate autofluorescence and facilitate definition of a phospho-S6 negative gates. Data were acquired on a FACS-Calibur (Becton Dickinson) using Cell Quest software or LSR-II (Becton Dickinson) using DiVa software and exported as uncompensated FCS3 files. At least 10,000 cell events were collected for all samples. Fluorescence compensation and subsequent analysis was performed using FlowJo software version 8 and 9 (TreeStar, Ashland, OR). Samples with fewer than 1000 cell events in a blast gate defined by CD45 × SSC were considered insufficient for further analysis. Constitutive phosphorylation of S6 required the finding of >5% of blast gate events in the positive phospho-S6 region defined by PMA and/or FMO controls.
Results
10 subjects provided paired pre-treatment and day 4 samples for flow cytometric analysis. The baseline characteristics of subjects who provided paired flow data is shown in table 1. The median day 4 trough rapamycin concentration for these subjects was 8.6 ng/mL (range 3.5–12.8). Day 7 trough rapamycin concentrations were slightly higher (mean 12.6 ng/mL, range 4.8–22.1).
Table 1.
Baseline characteristics of the ten subjects who provided paired samples for flow cytometric pharmacodynamic analysis.
| n=10 | |
|---|---|
| Age (median) | 61 (range 32–75) |
|
| |
| Sex | |
| Male | 9 |
| Female | 1 |
|
| |
| Disease Status | |
| First relapse/primary refractory | 6 |
| Primary refractory/1st CR < 6 mo. | 3 |
| 1st CR 6–12 mo. | 2 |
| 1st CR >12 mo. | 1 |
| > 1st relapse | 2 |
| Secondary AML (antecedent MDS) | 1 |
| Age >60 (without favorable karyotype) | 1 |
|
| |
| Karyotype | |
| Favorable | 2 |
| Intermediate | 8 |
| Unfavorable | 0 |
|
| |
| FLT3-ITD | 3 |
|
| |
| Prior therapy | |
| Anthracycline/cytarabine (7+3) | 8 |
| High dose cytarabine (post-remission) | 3 |
| High dose cytarabine (at relapse) | 2 |
| Myeloablative transplant | 2 |
| no prior AML therapy | 2 |
Note: high dose cytarabine regimens were defined by ara-C doses of at least 1 gm/m2.
Abbreviations: CR = complete remission, FLT3-ITD = Fms-like tyrosine kinase 3 internal tandem duplication, MDS = myelodysplastic syndrome
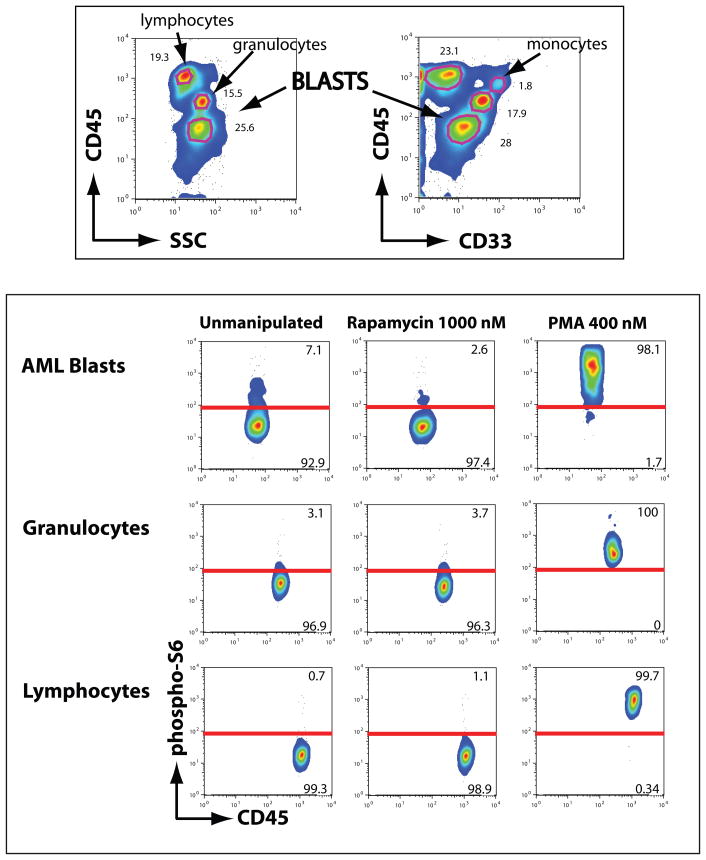
Previous work (reviewed in (19)) has demonstrated that the CD45 by side scatter plot allows cells to be divided into four cell populations: lymphocytes, monocytes, progenitor cells/granulocytes, and blasts. AML cells typically cluster within the blast gate (CD45 dim, side scatter low), although acute myelomoncytic/monoblastic leukemic blast poupulations may extend into or exclusively occupy a CD45 bright region otherwise enriched for monocytes.
Although our cell processing reduced the brightness of cell surface antigen staining to some degree, it nonetheless provided excellent discrimination of light scatter and CD45 expression. This was sufficient to define discrete cell population gates in all subjects (figure 2, top left). Of note, 4/10 subjects on our study had a total white blood cell (WBC) count below 5 × 103/μL and an absolute blast counts (ABC, defined as blast percentage × total white blood cell count) below 1 × 103/μL on at least one pharmacodynamic peripheral blood draw. An additional subject had no visible circulating blasts by peripheral smear and underwent paired marrow aspiration at baseline and day 4 for pharmacodynamic sampling. Morphologic hematopathology review of the marrow aspirate in this subject showed only 15% blasts. Depending upon the subjects’ clinical immunophenotypes, antibodies directed against myeloid and/or immaturity surface antigens (e.g. CD33 and CD34) were used to confirm the blast gate and were informative in all cases. These findings demonstrate the feasibility of obtaining consistent, tumor-specific data on peripheral blood samples even on patients with a low WBC.
Figure 2. Identification of blast gate and establishment of positive gates for phospho-S6.
Top: Color-enhanced contour plot showing CD45 by side scatter or CD33 pattern of this 100 μL sample of peripheral blood obtained on trial. Total white cell count is 5 × 103/μL with 23% blasts by light microscopy. Although not seen in all samples, this subject’s granulocytes show relatively low side scatter, consistent with a history of antecedent myelodysplasia and peripheral blood smear findings of hypogranular neutrophils. Blasts show dim CD33 expression, confirming myeloid lineage. CD34 was not expressed on this sample’s blasts but on other samples helped confirm immaturity (not shown).
Bottom: In the same sample’s blasts, heterogeneity of S6 phosphorylation is noted, unlike granulocyte or lymphocytes. Too few monocytes were present for meaningful data evaluation in this population. PMA stimulated cells (right column) define a positive region for S6 phosphorylated cell events. The majority of blasts at baseline show no constitutive phosphorylation, but a small population has variable, but constitutive activation. Ex vivo rapamycin treatment of the baseline blood sample (middle column) inhibits mTOR and markedly reduces the percentage of blasts with constitutive S6 phosphorylation.
S6 phosphorylation was evaluable in all subjects samples, even in cases where the WBC was as low as 1.1 × 103 with 10% blasts (i.e. ABC= 110/μL). In several subjects with acute monocytic or myelomonocytic leukemias, expansion of both a CD45 dim, SSC low blast gate as well as a CD45, SSC-intermediate monocytic gate was noted. In these cases, each population was evaluated for S6 phosphorylation. Discrimination between monocyte and lymphocyte populations was made based upon CD33 staining and both blast and monocyte population analysis was performed in these cases (figure 2, top right). Thus, we were able to analyze S6 phosphorylation in separate populations of normal and malignant cells in all samples.
Comparing samples drawn at baseline to samples treated ex vivo with PMA or rapamycin, we saw dynamic changes in S6 phosphorylation (figure 2, bottom). In all subjects’ samples, PMA led to marked upregulation of S6 phosphorylation in all CD45+ cell populations, regardless of staining pattern in the unmanipulated sample. This serves as an internal control for cell viability and served as a positive control for the purpose of estimating the percentage of blasts with mTOR activation at baseline. It further shows that the phospho-S6 epitope is fully unmasked by our cell processing method and capable of resolution if phosphorylated. Ex vivo treatment with ≥500 nM rapamycin markedly reduced or abrogated S6 phosphorylation in all samples that showed baseline constitutive S6 phosphorylation. Comparing PMA as a positive control and rapamycin and/or FMO treatment as positive and negative controls, respectively, we created discrimination lines for positive and negative phospho-S6 populations and scored percentage of positive cells for each group of cells.
For baseline samples, the most consistent finding was marked heterogeneity of S6 phosphorylation amongst leukemic blasts. The majority of samples (8/10) showed both a dominant, discrete population that lacked S6 phosphorylation (similar phospho-S6 brightness to either the FMO control or lymphocytes) and a smaller subset of cells with heterogeneous, but constitutive S6 phosphorylation, comparable in brightness to the range of PMA stimulated blasts. In two subjects’ baseline samples, phospho-S6 staining fell uniformly in the negative gate and no obvious positive phospho-S6 events were seen. From these gates, we defined baseline percentages of positive cell events. In cases where there was marked separation between narrowly clustered positive and FMO populations, a midpoint between the two populations was chosen for discrimination, as previously described.(20) These data demonstrate that, consistent with previous results from our lab and others, mTOR is constitutively activated in AML blasts in the majority of primary samples. However, this activation is actually only present in a subset of blasts at any one time.
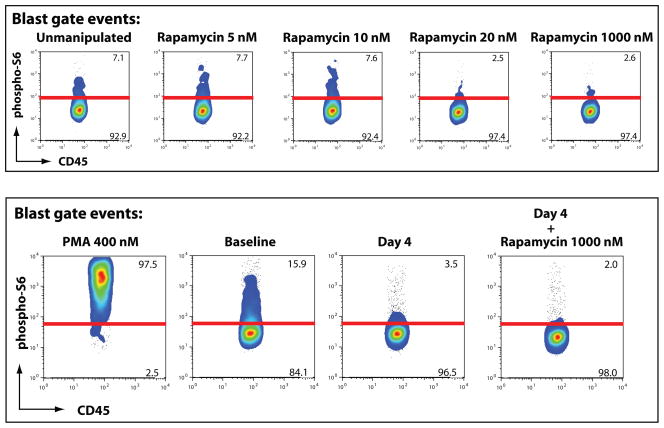
The described intracellular phospho-specific flow cytometry of fixed whole blood allowed for several applications. For example, we used the methodology to determine the sensitivity of AML cells to rapamycin. To do this, we evaluated the effect of a 30 minute ex vivo incubation of the baseline sample in increasing concentrations of rapamycin to estimate a dose-response curve for mTOR inhibition in leukemic blasts. A representative sample showing low nanomolar sensitivity to rapamycin is shown in figure 3 (top). Note that concentrations below 10 nM (corresponding to clinically measured concentrations of 9.1 ng/mL) fail to produce substantial reduction in S6 phosphorylation, but concentrations at or above 20 nM show similar reduction to that of 1000 nM rapamycin. This suggests that near-maximal inhibition occurs between 10–20 nM in this sample, which is similar to clinically achieved concentrations. These studies demonstrate that we can assay in vitro drug sensitivity on AML cells and compare the effects of in vitro inhibition of target with in vivo results. This allows us to discriminate between patients who received inadequate concentrations of drug in vivo compared to samples with endogeneous rapamycin insensitivity.
Figure 3. Ex vivo inhibition of mTOR predicts in vivo effects of oral sirolimus therapy.
Top: Exposing aliquots of a representative baseline sample (see figure 2) to a increasing concentrations of rapamycin ex vivo suggest that target inhibition occurs at a whole blood concentration between 10 and 20 nM (9.1–18.2 ng/mL). Of note, higher concentrations do not further impair S6 phosphorylation, predicting that mTOR inhibition in this sample will occur at clinically achievable concentrations.
Bottom: Serially obtained samples demonstrate abrogation of S6 phosphorylation in vivo during oral sirolimus therapy. Measured rapamycin concentration on day 4 was 4.7 ng/mL (5.1 nM). Exposure of the day 4 sample to ex vivo rapamycin in excess shows no significant additional effect, suggesting that near maximal target inhibition occurred in vivo.
Note: although all samples shown in this figure were obtained from the same patient’s sample, the staining and cytometer data acquisition for the two experiments were not performed simultaneously. This accounts for minor variation in phospho-S6 staining across the ex vivo and in vivo experiments. Within experiments, cell samples were exposed to a single staining antibody cocktail, such that dynamic changes in phospho-signal can directly compared.
Flow cytometry also allowed for analysis of signaling in different subpopulations of blast cells. In one subject with AML arising from underlying chronic myelomonocytic leukemia, the baseline sample demonstrated abnormal expansion of both an immature myeloblast population as well as a more differentiatied monoblast population by CD45 and side scatter (Supplementary figure 1). These two populations were confirmed as discrete populations by CD34, CD33, and CD14 staining (CD34+, CD33 dim+, CD14-myeloblasts vs. CD34-, CD33 bright+, CD14+ monoblasts). Interestingly, the myeloblast population showed no baseline phosphorylation of S6, though constitutive phosphorylation was readily seen in monoblasts. 30 minute ex vivo treatment with rapamycin failed to potently inhibit S6 phosphorylation in monocytes until concentrations above 50 nM were evaluated. Notably, 50 nM exceeds measured blood rapamycin concentrations from the trial. Thus, we are methodologically able to demonstrate variations of signaling within sub-populations of leukemic cells, which may be important for understanding the diverse response of leukemia to signal transduction inhibitors.
Finally, we focused on clinical evaluation of the effect of oral sirolimus therapy upon S6 phosphorylation (figure 3, bottom). Because the majority of the blast events showed phospho-S6 staining in a range comparable to FMO, shifts in the mean fluorescence intensity were less dramatic than changes in the percentage of cells in the positive phospho-S6 gate. To quantify the degree of mTOR inhibition, we calculated an inhibition percentage, defined by 1 minus the fraction of positive phospho-S6 blasts on day 4 divided by the percentage at baseline. Thus, if a subject had 20% phospho-S6 positive blasts at baseline and 5% on day 4, the inhibition percentage was 1- (5/20)]= 75% inhibition. We therefore could use changes in S6 phosphorylation during therapy to group subjects based upon their blasts’ biochemical sensitivity to sirolimus.
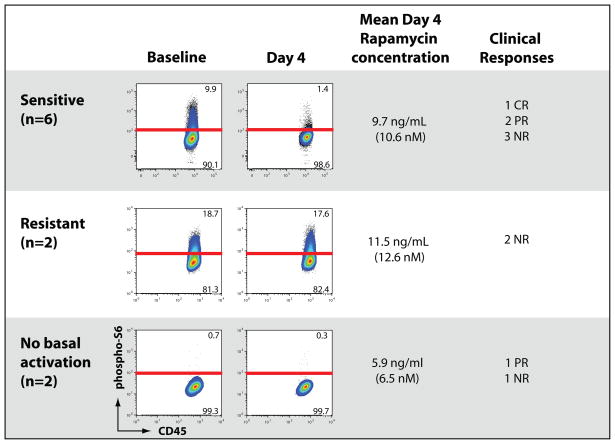
Three patterns of baseline mTOR activation and response to sirolimus emerged from our anaysis. Of the 8 subjects that showed baseline constitutive phosphorylation of S6, 6 showed marked reduction in the percentage of phospho-S6 positive events (Figure 4) Such subjects blasts were considered biochemically “sirolimus-sensitive” and showed a mean inhibition percentage of 75% reduction in the percentage of cells in the positive gate for S6 phosphorylation (range 53–98%) The two subjects without constitutive phosphorylation of S6 showed no appreciable change in their blasts’ S6 phosphorylation on day 4 and were considered uninformative for sirolimus sensitivity. Two subjects showed persistent S6 phosphorylation in their blasts on day 4 and were considered sirolimus-resistant. In one subject, baseline and day 4 percentage positive phospho-S6 events were similar and in the other a marked increase in phospho-S6 events was seen. Of note, this subject’s baseline sample was noted to require relatively higher concentrations of rapamycin in ex vivo testing to demonstrate inhibition of S6 phosphorylation. Interestingly, measured sirolimus troughs were similar in biochemically sirolimus sensitive and resistant leukemia. Overall, 8/10 (80%) of patients demonstrated activation of S6 at baseline. Of these 8 subjects 6/8 (75%) were inhibited in vivo. One of these two patients demonstrated sirolimus resistance both in vivo and in vitro. The other subject’s samples were not evaluated for in vitro sensitivity. 4 subjects had ex vivo testing performed on their baseline samples. 3/3 subjects whose ex vivo testing of their leukemia sample for rapamycin sensitivity demonstrated sensitivity at ≤20 nM had biochemical sensitivity to sirolimus during therapy. One subject baseline showed no inhibition of S6 phosphorylation ≤20 nM, modest reduction at 50 nM, and marked inhibition at 1000 nM. This subject’s day 4 sample showed biochemical sirolimus resistance.
Figure 4.
Patterns of AML blasts’ pharmacodynamic responses to oral sirolimus therapy Subjects’ samples showed either constitutive S6 phosphorylation at baseline (n=8) or no obvious phosphorylation at baseline (n=2). The effects of 72 hours of oral sirolimus therapy are shown as well as the measured rapamycin concentrations at the time of pharmacodynamic monitoring and clinical responses. Reduction in S6 phosphorylation (>50% reduction in percentage of cells in S6 + gate) defined rapamycin sensitivity. Abbreviations: CR= complete remission, PR= partial remission, NR= no response
Although the intent of this pilot study was not to evaluate clinical efficacy, clinical responses to sirolimus and MEC were evaluable in 9 of 10 subjects who provided paired samples for flow analysis. One subject died of bacterial infection prior to hematopoietic recovery from sirolimus MEC and was not evaluable for treatment response. Among the 5 evaluable sirolimus-sensitive subjects, 1 CR and 2 PRs were seen. Neither of the 2 evaluable sirolimus-resistant patients showed a clinical response. One of the two evaluable subjects without baseline S6 phosphorylation had a PR.
Discussion
These data provide the first evidence that serial intracellular flow cytometric monitoring of fixed peripheral blood can be used to evaluate the biochemical efficacy of drugs targeting the PI3K/AKT/mTOR signal transduction pathway in leukemia clinical trials. Additionally, they demonstrate heterogeneity in rapamycin sensitivity and suggest that evaluation for ex vivo drug sensitivity prior to or early in therapy may have a role in predicting chemotherapy response to regimens combining mTOR inhibitors and chemotherapy. This approach may promote more rational enrollment of subjects to clinical trials of novel therapeutics and eventually is hoped to help develop personalized approaches in the future.
Using serial flow cytometric analysis of clinical trial samples, we confirm widespread constitutive activation of mTOR in circulating peripheral blood AML cells. Consistent with prior data, however, we show marked heterogeneity in the degree of this activation among patients and within particular patients’ blast populations. We further show that oral administration of the mTOR inhibitor sirolimus during an intensive combination chemotherapy regimen achieves successful disruption of mTOR enzymatic function in the malignant cells of the majority of treated patients. Finally, we demonstrate that, in a small number of patient samples, constitutive phosphorylation is present but resistant to mTOR inhibition by oral sirolimus. This finding was not predicted by therapeutic drug monitoring, suggesting cell-intrinsic resistance, as confirmed in one case by ex vivo bioassays of rapamycin sensitivity. These results demonstrate that flow cytometric assays highlight biologic heterogeneity that is invisible to techniques such as Western blot.
Drug development of rapamycin analogs for cancer therapy has emphasized parenteral formulations and/or drugs with improved bioavailability in comparison to rapamycin. The goal of these second generation drugs is to produce comparably high rapamycin concentrations. Interestingly, our data show relatively low doses of rapamycin lead to effectively maximal mTOR inhibition in tumor cells. This is particularly true in samples that show sensitivity by ex vivo testing. Thus in many, if not most patients, second generation rapalogs may unnecessarily add expense and/or dose-dependent toxicity compared to sirolimus.
Due to the abundance of tumor cells that are readily accessible during clinical care, leukemia is perhaps an ideal disease in which direct pharmacodynamic monitoring technology can be developed. Despite this apparent ease, pharmacodynamic monitoring in leukemia in the past has not been entirely stratightforward. Studies of signal transduction traditionally used density centrifugation (ficoll separation) and western blot of isolated mononuclear fractions. While marrow aspirates often contain highly purified populations of leukemic cells, the quality of tumor sampling is invariably compromised by peripheral blood “hemodilution” of the specimen during aspiration. Additionally, density centrifugation enriches for, but does not exclusively isolate tumor cells. Since both leukemic and non-malignant cells express target signaling proteins, it is notable that the concentrations necessary to abrogate enzymatic function of several kinases may differ from that of normal tissue and can vary substantially among tumor samples. Finally, cell lysis for Western blot eliminates all ability to resolve the heterogeneity of these samples. Taken together, obtaining pharmacodynamic signaling data in leukemia trials is procedurally simple, but interpretation of these data can prove challenging.
Phospho-specific intracellular flow cytometry thus provides an attractive method to directly measure changes in key signaling proteins in response to clinical therapeutics and has appropriate sensitivity to provide robust readout despite low circulating tumor burden. In spite of these potential advantages, we note several limitations of this report. We recognize that our data were obtained in real time and our expertise in flow cytometry to immunophenotype samples developed during this research effort. While the majority of samples in this study were evaluated using 3 or 4 color cytometry, we currently are using a more sophisticated 6–10 fluorophore panel to provide further specificity for leukemic populations and to evaluate subpopulations among leukemia samples due to observed heterogeneity in signaling activation and inhibition. As only ten subjects were evaluated by these methods, we are currently completing a larger study to better characterize mTOR activation and inhibition to this regimen, as well as the predictive ability for flow cytometric studies for chemotherapy response.
We also recognize that S6 phosphorylation can be upregulated by cytokine stimulation as well as stromal and/or microenvironment marrow factors that are absent or present in low concentrations in peripheral blood. Therefore, another potential limitation is our choice of peripheral blood samples. Direct comparisons of our methods in marrow and blood are ongoing to estimate the magnitude, if any, of these differences. Thus far, we have not seen a consistent pattern to suggest that peripheral blood sampling provides inferior estimation of basal S6 phosphorylation or signal transduction inhibition. However, our measurements of mTOR activation state among circulating blasts should be considered only estimates of the effects of sirolimus therapy upon marrow blasts. Flow cytometric analysis in the context of an in vitro model of the marrow microenvironment (e.g. stroma-supported culture of pretreatment blast samples) might further enhance our ability to predict clinical response to mTOR or other signaling inhibitors.
We hypothesize that mTOR inhibition and chemotherapy are synergistic in their anti-leukemic effect based upon preclinical assays; whether this translates into improved clinical response remains to be determined. Further, if these agents are indeed synergistic, it is unknown whether partial inhibition or abrogation of S6 phosphorylation predicts this interaction. We expect our current studies to clarify whether the degree of S6 signal disruption predicts chemotherapy response to sirolimus MEC and as well whether mTOR inhibition itself is sufficient to reverse chemotherapy resistance. Such larger studies may require multiple treatment sites, which potentially jeopardizes the data quality of our techniques. This is in part because transit time from samples obtained at remote centers might alter the robustness of our data. We therefore are exploring local cell processing such that data quality on multi-center trials is maintained.
In summary, we demonstrate that real time pharmacodynamic evaluation of leukemia cells by serial flow cytometric analysis of S6 phosphorylation is a powerful tool to evaluate in vivo biochemical efficacy of signal transduction inhibitors. Beyond the monitoring of mTOR inhibitors in AML, we are also currently testing whether monitoring peripheral blood S6 phosphorylation by flow can be used to directly confirm the pharmacodynamic effects of other agents that secondarily inhibit PI3K/AKT/mTOR signaling. This includes clinical trials of AKT and FLT3 inhibitors in AML and JAK2 inhibitors in myeloproliferative disorders. We also seek to document whether compensatory changes in upstream or parallel signaling proteins occur during mTOR inhibition with these agents through the use of multiple phospho-specific antibodies. Such data may help elucidate resistance mechanisms to these agents. Finally, we hope that our flow cytometric approach can ultimately be used as screening tools to predict patients likely to benefit from specific clinical therapeutics, thus promoting personalized approaches to molecularly targeted agents.
Supplementary Material
Translational Relevance.
Because activation and inhibition of signaling proteins may be discordant in normal and malignant tissues, pharmacodynamic measurement from surrogate tissue rather than tumor cells sub-optimally estimates target inhibition. Using samples obtained during a clinical trial combining sirolimus and intensive cytotoxic chemotherapy for acute myelogenous leukemia (AML), we used phospho-specific flow cytometry of formaldehyde-fixed whole blood to directly evaluate in vivo mTOR signal inhibition in immunophenotypically identified leukemic blasts. These results were paired with measured rapamycin concentrations to model tumor cells’ biochemical sensitivity or resistance to sirolimus. We demonstrate the feasibility and robustness of this approach, even in clinical samples with minimal circulating tumor burden. Because mTOR is a major downstream effecter of many signaling proteins targeted by experimental therapeutics, these data suggest that this approach may facilitate early phase development of multiple classes of novel drugs for hematologic malignancies.
Acknowledgments
This manuscript is dedicated in memory of Dr. Alan M. Gewirtz, whose intellect, wit, and compassion continue to inspire all who knew him. We wish to thank T. Vincent Shankey, Charles Pletcher, and James E. Thompson for technical assistance and helpful discussions during flow cytometric methods development. We also thank Joy Cannon for assistance with blood sample acquisition and database management. Lastly, we wish to thank the patients and staff of the Hematologic Malignancies unit at the Hospital of the University of Pennsylvania who helped make these studies possible.
AEP was supported by a grant from the National Cancer Institute (K23 CA141054-01) and the University of Pennsylvania’s Institute for Translational Medicine. Additional funding was provided from a grant from the Commonwealth of Pennsylvania Department of Health. The Department specifically disclaims responsibility for any analyses, interpretations or conclusions.
References
- 1.Smith BD, Levis M, Beran M, Giles F, Kantarjian H, Berg K, et al. Single-agent CEP–701, a novel FLT3 inhibitor, shows biologic and clinical activity in patients with relapsed or refractory acute myeloid leukemia. Blood. 2004;103:3669–76. doi: 10.1182/blood-2003-11-3775. [DOI] [PubMed] [Google Scholar]
- 2.Pratz KW, Cortes J, Roboz GJ, Rao N, Arowojolu O, Stine A, et al. A pharmacodynamic study of the FLT3 inhibitor KW–2449 yields insight into the basis for clinical response. Blood. 2009;113:3938–46. doi: 10.1182/blood-2008-09-177030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–7. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corbin AS, Agarwal A, Loriaux M, Cortes J, Deininger MW, Druker BJ. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J Clin Invest. 2011;121:396–409. doi: 10.1172/JCI35721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Irish JM, Hovland R, Krutzik PO, Perez OD, Bruserud O, Gjertsen BT, et al. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004;118:217–28. doi: 10.1016/j.cell.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 6.Han L, Wierenga AT, Rozenveld-Geugien M, van de Lande K, Vellenga E, Schuringa JJ. Single-cell STAT5 signal transduction profiling in normal and leukemic stem and progenitor cell populations reveals highly distinct cytokine responses. PLoS One. 2009;4:e7989. doi: 10.1371/journal.pone.0007989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kotecha N, Flores NJ, Irish JM, Simonds EF, Sakai DS, Archambeault S, et al. Single-cell profiling identifies aberrant STAT5 activation in myeloid malignancies with specific clinical and biologic correlates. Cancer Cell. 2008;14:335–43. doi: 10.1016/j.ccr.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee AW, Sharp ER, O’Mahony A, Rosenberg MG, Israelski DM, Nolan GP, et al. Single-cell, phosphoepitope-specific analysis demonstrates cell type- and pathway-specific dysregulation of Jak/STAT and MAPK signaling associated with in vivo human immunodeficiency virus type 1 infection. J Virol. 2008;82:3702–12. doi: 10.1128/JVI.01582-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chow S, Hedley D, Grom P, Magari R, Jacobberger JW, Shankey TV. Whole blood fixation and permeabilization protocol with red blood cell lysis for flow cytometry of intracellular phosphorylated epitopes in leukocyte subpopulations. Cytometry A. 2005;67:4–17. doi: 10.1002/cyto.a.20167. [DOI] [PubMed] [Google Scholar]
- 10.Chow S, Minden MD, Hedley DW. Constitutive phosphorylation of the S6 ribosomal protein via mTOR and ERK signaling in the peripheral blasts of acute leukemia patients. Exp Hematol. 2006;34:1183–91. doi: 10.1016/j.exphem.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 11.Woost PG, Solchaga LA, Meyerson HJ, Shankey TV, Goolsby CL, Jacobberger JW. High-resolution kinetics of cytokine signaling in human CD34/CD117-positive cells in unfractionated bone marrow. Blood. 2011;117:e131–41. doi: 10.1182/blood-2010-10-316224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marvin J, Swaminathan S, Kraker G, Chadburn A, Jacobberger J, Goolsby C. Normal bone marrow signal-transduction profiles: a requisite for enhanced detection of signaling dysregulations in AML. Blood. 2011;117:e120–30. doi: 10.1182/blood-2010-10-316026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lowenberg B, Ossenkoppele GJ, van Putten W, Schouten HC, Graux C, Ferrant A, et al. High-dose daunorubicin in older patients with acute myeloid leukemia. N Engl J Med. 2009;361:1235–48. doi: 10.1056/NEJMoa0901409. [DOI] [PubMed] [Google Scholar]
- 14.Xu Q, Simpson SE, Scialla TJ, Bagg A, Carroll M. Survival of acute myeloid leukemia cells requires PI3 kinase activation. Blood. 2003;102:972–80. doi: 10.1182/blood-2002-11-3429. [DOI] [PubMed] [Google Scholar]
- 15.Xu Q, Thompson JE, Carroll M. mTOR regulates cell survival after etoposide treatment in primary AML cells. Blood. 2005;106:4261–8. doi: 10.1182/blood-2004-11-4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perl AE, Kasner MT, Tsai DE, Vogl DT, Loren AW, Schuster SJ, et al. A phase I study of the mammalian target of rapamycin inhibitor sirolimus and MEC chemotherapy in relapsed and refractory acute myelogenous leukemia. Clin Cancer Res. 2009;15:6732–9. doi: 10.1158/1078-0432.CCR-09-0842. [DOI] [PubMed] [Google Scholar]
- 17.Tamburini J, Green AS, Bardet V, Chapuis N, Park S, Willems L, et al. Protein synthesis is resistant to rapamycin and constitutes a promising therapeutic target in acute myeloid leukemia. Blood. 2009;114:1618–27. doi: 10.1182/blood-2008-10-184515. [DOI] [PubMed] [Google Scholar]
- 18.Cheson BD, Bennett JM, Kopecky KJ, Buchner T, Willman CL, Estey EH, et al. Revised recommendations of the International Working Group for Diagnosis, Standardization of Response Criteria, Treatment Outcomes, and Reporting Standards for Therapeutic Trials in Acute Myeloid Leukemia. J Clin Oncol. 2003;21:4642–9. doi: 10.1200/JCO.2003.04.036. [DOI] [PubMed] [Google Scholar]
- 19.Jennings CD, Foon KA. Recent advances in flow cytometry: application to the diagnosis of hematologic malignancy. Blood. 1997;90:2863–92. [PubMed] [Google Scholar]
- 20.Hedley DW, Chow S, Goolsby C, Shankey TV. Pharmacodynamic monitoring of molecular-targeted agents in the peripheral blood of leukemia patients using flow cytometry. Toxicol Pathol. 2008;36:133–9. doi: 10.1177/0192623307310952. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.