Abstract
Purpose
Poly(ADP-ribose) polymerase (PARP) inhibitors are undergoing extensive clinical testing for their single-agent activity in homologous recombination- (HR-) deficient tumors and ability to enhance the action of certain DNA damaging agents. Compared to other PARP inhibitors in development, iniparib (4-iodo-3-nitrobenzamide) is notable for its simple structure and the reported ability of its intracellular metabolite 4-iodo-3-nitrosobenzamide to covalently inhibit PARP1 under cell-free conditions. The present preclinical studies were performed to compare the actions iniparib with the more extensively characterized PARP inhibitors olaparib and veliparib.
Experimental design
The abilities of iniparib, olaparib and veliparib to i) selectively induce apoptosis or inhibit colony formation in HR-deficient cell lines, ii) selectively sensitize HR-proficient cells to topoisomerase I poisons and iii) inhibit formation of poly(ADP-ribose) polymer in intact cells were compared.
Results
Consistent with earlier reports, olaparib and veliparib selectively induced apoptosis and inhibited colony formation in cells lacking BRCA2 or ATM. Moreover, like earlier-generation PARP inhibitors, olaparib and veliparib sensitized cells to the topoisomerase I poisons campto-thecin and topotecan. Finally, olaparib and veliparib inhibited formation of poly(ADP-ribose) polymer in intact cells. In contrast, iniparib exhibited little or no ability to selectively kill HR-deficient cells, sensitize cells to topoisomerase I poisons, or inhibit poly(ADP-ribose) polymer formation in situ. In further experiments, iniparib also failed to sensitize cells to cisplatin, gemcitabine or paclitaxel.
Conclusions
While iniparib kills normal and neoplastic cells at high (>40 µM) concentrations, its effects are unlikely to reflect PARP inhibition and should not be used to guide decisions about other PARP inhibitors.
Keywords: Poly(ADP-ribose) polymerase, enzyme inhibitor, nicotinamide, topoisomerase, homologous recombination
INTRODUCTION
The concept of synthetic lethality centers on targeting two separate molecular pathways that are non-lethal when disrupted individually, but are lethal when inhibited simultaneously. This approach is increasingly being used to guide the development of targeted anticancer therapies (1, 2). For example, based on the observation that poly(ADP-ribose) polymerase (PARP) inhibitors are selectively toxic to cells lacking homologous recombination (HR) proteins such as BRCA1, BRCA2 and ATM (3–5), PARP inhibitors are being tested in tumors harboring BRCA1 or BRCA2 mutations (2, 6, 7). Recent results have shown that the PARP inhibitor olaparib induces partial or complete remissions in 41% of advanced breast and 33% of recurrent ovarian cancers in BRCA1/2 mutation carriers (8, 9). In light these promising results, there has been substantial effort to develop olaparib, veliparib, and a variety of other third-generation PARP inhibitors as antineoplastic agents (reviewed in refs. 2, 6, 7, 10, 11).
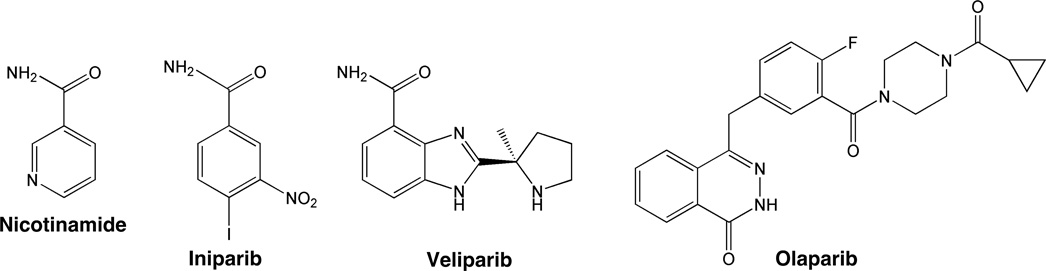
Among the PARP-directed agents currently under development, iniparib is the furthest along in clinical testing (Fig. 1). A simple mimic of nicotinamide, this agent was originally described as the prodrug of 4-iodo-3-nitrosobenzamide, an agent that covalently inhibits PARP1 by binding to its first zinc finger under cell-free conditions (12). Subsequent studies suggested that iniparib exhibits single-agent activity in triple negative breast cancer lines and enhances the cytotoxicity of cisplatin and gemcitabine (13). A phase 2 clinical trial suggested that iniparib not only is well-tolerated when administered with gemcitabine and carboplatin, but also increases the response rate of the gemcitabine/carboplatin regimen in patients with triple-negative breast cancer (14). A subsequent phase 3 trial, however, failed to reproduce these results, raising questions about the future of PARP inhibition as a therapeutic strategy (15).
Figure 1. Structures of nicotinamide and the three drugs used in this study.
Recent studies from our laboratory demonstrated that veliparib and olaparib, which are active site-directed noncovalent PARP inhibitors, selectively kill HR-deficient ovarian and pancreatic cells by causing activation of the error-prone nonhomologous end-joining DNA repair pathway (16). In anticipation of experiments to determine whether covalent modification of PARP by 4-iodo-3-nitrosobenzamide kills in a similar fashion, we examined the cytotoxicity of iniparib in HR-deficient cells. Surprisingly, we observed that iniparib exhibited little selectivity for HR-deficient cells. Further studies failed to demonstrate the ability of iniparib to sensitize cells to topoisomerase (topo) I poisons or inhibit poly(ADP-ribose) (pADPr) polymer synthesis in situ, two other hallmarks of PARP inhibitors. Collectively, these studies argue against the possibility that iniparib is inducing cytotoxicity by inhibiting PARP.
MATERIALS AND METHODS
Materials
Veliparib (ABT-888) was purchased from Enzo Life Sciences (Plymouth Meeting, PA); olaparib (AZD2281), iniparib (BSI-201), and VE-821 were from ChemieTek (Indianapolis, IN); and camptothecin, gemcitabine, paclitaxel, propidium iodide (PI), etoposide, bovine serum albumin, and gelatin from Sigma-Aldrich (St. Louis, MO). Topotecan was provided by the Drug Synthesis Branch of the National Cancer Institute (Bethesda, MD). AZD 7762 was a kind gift from L. Karnitz (Mayo Clinic, Rochester, MN). Polyclonal rabbit 96–10 anti-pADPr antiserum was raised as reported (17).
Cell Culture
Mouse embryo fibroblasts (MEFs) from wildtype or Atm−/− mice (Z. Lou, Mayo Clinic, Rochester, MN) were cultured in DMEM (medium A). GM16666 and GM16667 human fibroblast lines from the Coriell Institute (Camden, NJ) were cultured in DMEM with 100 µg/ml hygromycin. PEO1 and PEO4 cells (18) from F. Couch (Mayo Clinic, Rochester, MN) were cultured in DMEM containing 100 µM nonessential amino acids and 10 µg/ml insulin. SKOV3 cells (V. Shridhar, Mayo Clinic) were cultured in McCoy’s 5A. All media contained 10% heat inactivated fetal bovine serum, 40 units/ml penicillin G, 40 µg/ml streptomycin, and 1 mM glutamine. Lines were genotyped shortly before acquisition and were reinitiated every 2–3 months from stocks that were cryopreserved immediately after receipt from the indicated sources.
Apoptosis Assays
Cells plated at 5 × 104 cells/60 mm dish were allowed to adhere for 24 h, then treated with veliparib, olaparib, or iniparib in 0.1% (v/v) DMSO for 4 days (olaparib) or 6 days (veliparib, iniparib) based on preliminary time course experiments. At the end of treatment, adherent cells were recovered by trypsinization, combined with cells in the supernatant, sedimented at 150 × g for 10 min, washed in ice-cold calcium- and magnesium-free Dulbecco’s phosphate-buffered saline (PBS), and fixed at 4° C by drop-wise addition of ethanol to a final concentration of 50% (v/v). Cells were subsequently rehydrated in cold PBS, sedimented, resuspended in 300 µl 0.1% (w/v) sodium citrate containing 1 mg/ml RNase A, incubated for 15 min at 37 °C, diluted with 300 µl 0.1% sodium citrate containing 100 µg/ml PI, incubated in the dark at 20 °C for 15 min, and analyzed (20,000 events) on a FACSCanto II flow cytometer (Becton Dickinson, Franklin Lakes, NJ) using excitation and emission wavelengths of 488 and 617 nm, respectively. After data were analyzed using Becton Dickinson CellQuest software, normalized apoptosis was calculated as (observed – baseline)/(100 – baseline) to correct for differences in basal apoptosis rates between cell lines. Results are representative of at least 3 independent experiments.
Colony Forming Assays
Wild-type and Atm−/− MEFs were plated at 500 cells/dish in 60 mm dishes containing medium A, allowed to adhere overnight, and treated with the indicated agents continuously. Other cell lines were plated at 1000 cells/plate (PEO1 and PEO4), 750 cells/plate (GM16666 and 16667) or 500 cells/plate (SKOV3) in their respective media, allowed to adhere for up to 14 h, treated with the indicated agents for 48 h (GM16666 and 16667) or continuously, stained with Coomassie Brilliant Blue, and scored for colony formation (≥ 50 cells) manually.
pADPr levels (modified from refs. 19, 20)
Relative pADPr levels were assessed by quantitative fluorescence microscopy. In brief, SKOV3 cells grown on ethanol-washed coverslips were treated with the indicated agents for 4 h prior to addition of 1 mM MMS for 30 min to stimulate polymer formation. Following treatment, cells were fixed in −20 °C methanol:acetone (70:30); incubated for 1 h at 21° C in blocking buffer consisting of 1% (v/v) glycerol, 0.1% (w/v) gelatin from cold-water fish, 0.1% (w/v) bovine serum albumin, 5% (v/v) goat serum and 0.4% (w/v) sodium azide in PBS; exposed to 96-10 anti-pADPr antiserum (1:500 dilution in blocking buffer) overnight at 4 °C; washed; incubated for 1 h with Alexa Fluor 568-conjugated goat anti-rabbit IgG (Invitrogen) in blocking buffer; washed; counterstained with 1 µg/ml Hoechst 33258 in PBS; and examined by confocal microscopy as described (20).
RESULTS
Inability of iniparib to selectively kill HR-deficient cells
Previous studies have identified important cellular effects of PARP inhibitors, including selective toxicity in HR-deficient cells (3–5, 21, 22), synergistic cytotoxicity when combined with topo I poisons (20, 23–27), and ability to inhibit pADPr formation in cells with damaged DNA. In the present study we compared three of the agents currently undergoing clinical testing (Fig. 1) in assays of these effects.
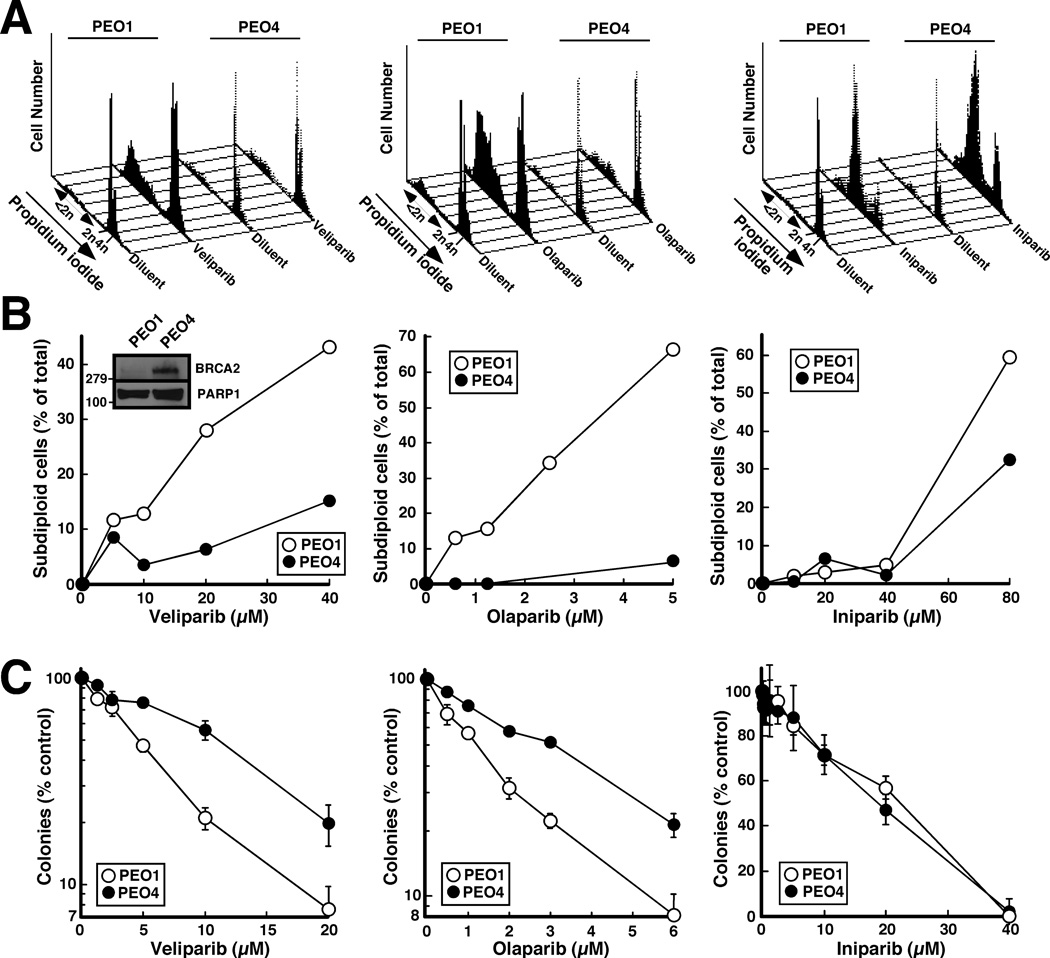
To compare the ability of these agents to selectively induce apoptosis in HR-deficient cells, BRCA2-deficient PEO1 human ovarian cancer cells and their BRCA2-revertant PEO4 counterparts (18, 21) were incubated with olaparib, veliparib or iniparib, then stained with PI and subjected to flow cytometry. As depicted in Fig. 2A and summarized in Fig. 2B, PEO1 cells were much more sensitive to olaparib and veliparib than the PEO4 cells. In contrast, iniparib exhibited much less selectivity (right panels, Fig. 2A and 2B). Similar results were observed when the cells were stained with Hoechst 33258 and examined for apoptotic morphological changes (Fig. S1).
Figure 2. Effects of veliparib, olaparib and iniparib on PEO1 and PEO4 cells.
A, PEO1 and PEO4 treated with diluent, 20 µM veliparib (left), 5 µM olaparib (middle) or 80 µM iniparib (right) were stained with propidium iodide (PI) and subjected to flow microfluorimetry. B, summary of results obtained in panel A and additional concentrations. Inset in B, whole cell lysates were subjected to immunoblotting with antibody to BRCA2 and, as a loading control, PARP1. C, effects of various concentrations of veliparib (left), olaparib (middle) and iniparib (right) on colony formation. Error bars in these and subsequent colony forming assays, ± SD of triplicate aliquots. All results are representative of ≥ 3 independent experiments.
In further experiments, antiproliferative effects of the three agents were compared in colony forming assays. This assay likewise showed that veliparib and olaparib exhibited selectivity for the BRCA2-deficient PEO1 cells (Fig. 2C, left and middle panels), whereas iniparib exhibited no selectivity (Fig. 2C, right panel).
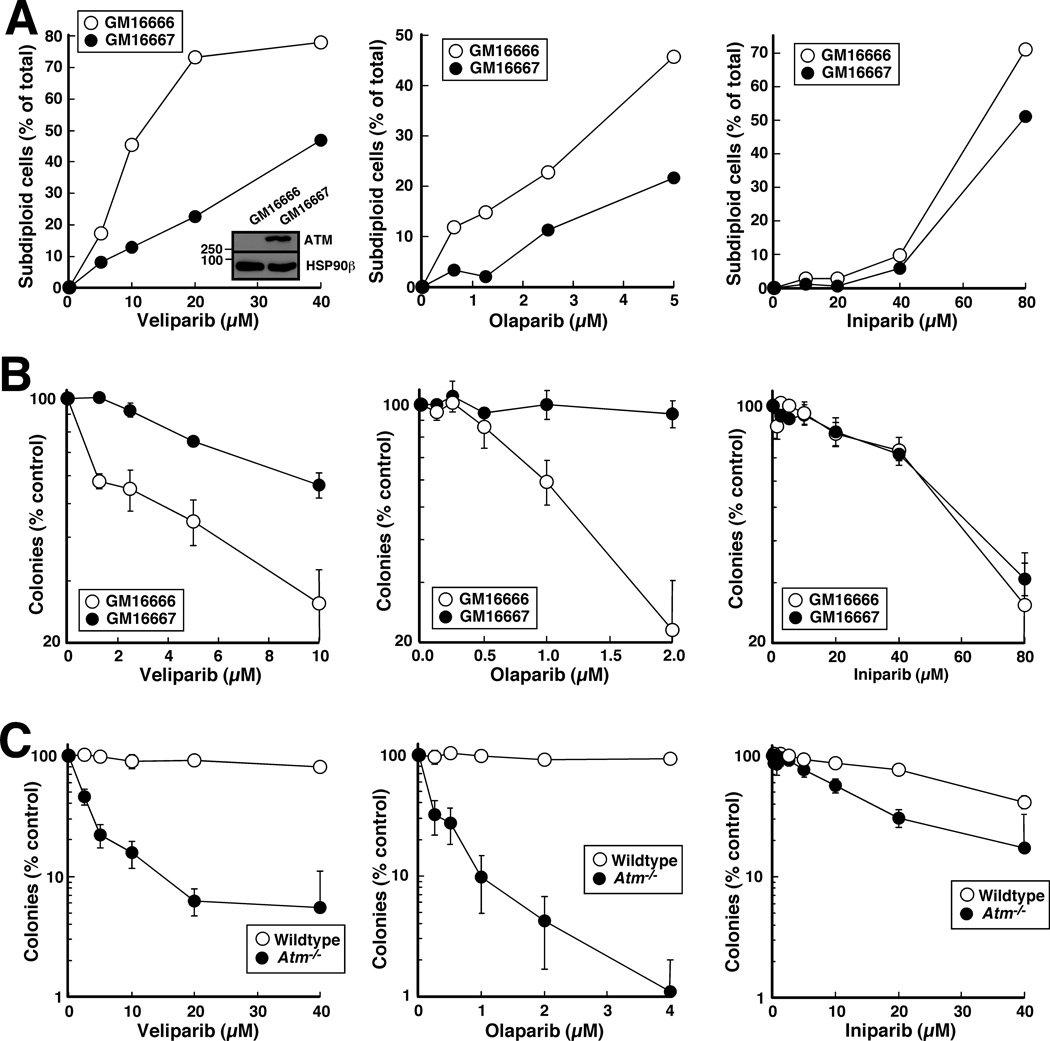
To assure that these observations were not unique to PEO1 and PEO4 cells, we also examined the effects of the three agents in ATM-deficient GM16666 and ATM-restored GM16667 fibroblasts. Once again, veliparib and olaparib exhibited selectivity for the HR-deficient cells (Fig. 3A and B, left and middle panels), whereas iniparib exhibited very little selectivity (Fig. 3A and B, right panels). Similar results were also observed in Atm−/− fibroblasts compared to their wildtype counterparts (Fig. 3C).
Figure 3. Effects of veliparib, olaparib and iniparib on paired GM16666 (ATM-deficient) and GM16667 (ATM restored) human fibroblasts.
A, cells treated with diluent, 20 µM veliparib (left), 5 µM olaparib (middle) or 80 µM iniparib (right) were stained with PI and examined by flow microfluorimetry as in Fig. 2A. B, effects of veliparib (left), olaparib (middle) and iniparib (right) on colony formation. Inset in B, whole cell lysates were subjected to immunoblotting for ATM and, as a loading control, heat shock protein 90β (HSP90β). C, Atm−/− and wildtype MEFs examined by colony forming assays.
Failure of iniparib to synergize with topo I poisons
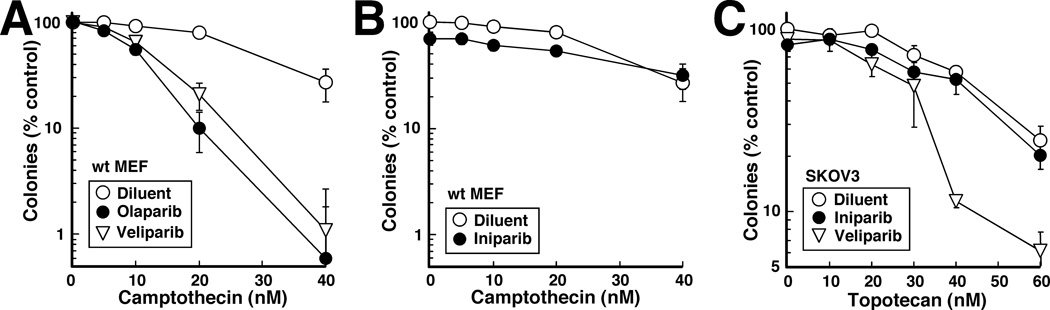
Another hallmark of PARP inhibitors is their ability to synergize with topo I poisons (20, 23–27). To avoid the potential confounding effect of P-glycoprotein, which is constitutively expressed at low levels in rodent cells (28) and has been reported to affect uptake of topotecan (29–31), experiments in MEFs utilized camptothecin. At submicromolar concentrations that were themselves nontoxic, veliparib and olaparib enhanced the sensitivity of wildtype MEFs to camptothecin (Fig. 4A). In contrast, 100-fold higher iniparib concentrations, which were just at the point of inhibiting colony formation by themselves, had no discernible effect on camptothecin sensitivity (Fig. 4B). When topotecan, which is utilized to treat epithelial ovarian cancer (32, 33), was administered to SKOV3 cells, veliparib and olaparib likewise enhanced the cytotoxicity of the topo I poison, whereas iniparib did not (Fig. 4C and data not shown).
Figure 4. Inability of iniparib to sensitize cells to topoisomerase I poisons.
MEFs (A, B) or SKOV3 cells (C) were treated continuously with the indicated concentrations of camptothecin (A, B) or topotecan (C) in the presence of 100 nM veliparib, 100 nM olaparib or 10 µM iniparib, then examined for colony formation.
Further effects of iniparib in combination
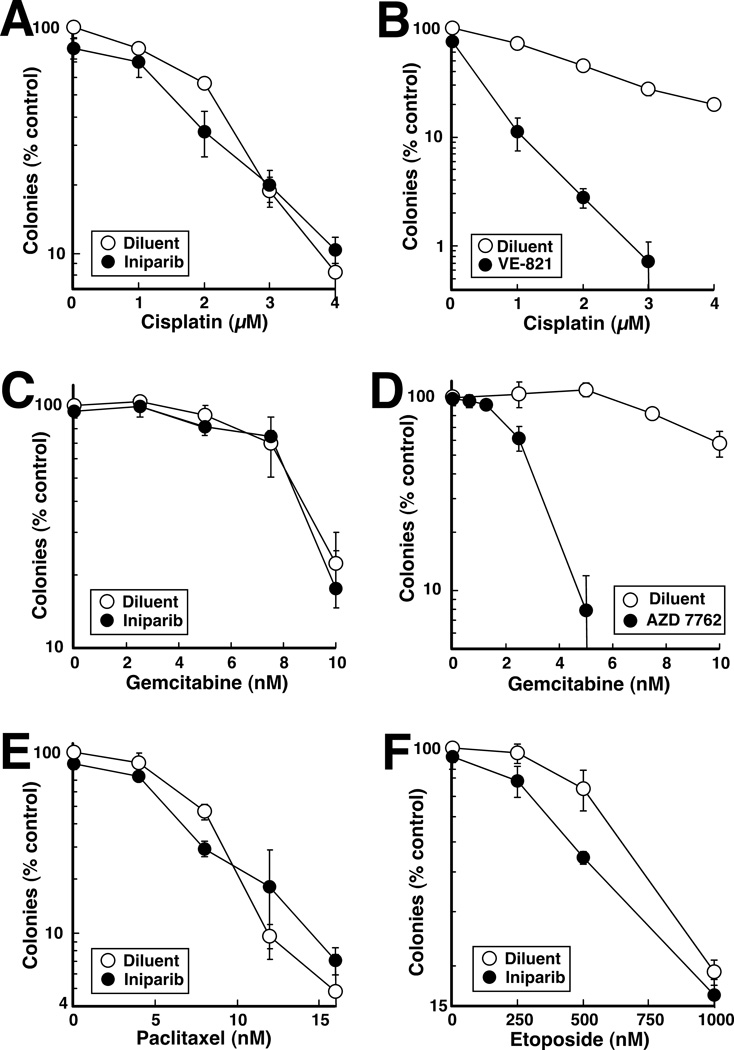
In view of the inability of iniparib to sensitize cells to topo I poisons, we also examined the ability of iniparib to sensitize SKOV3 cells to a number of other classes of agents with which it is being combined in the clinic (www.clinicaltrials.gov). In these experiments, iniparib failed to sensitize cells to cisplatin (Fig. 5A). In contrast, sensitization by the ATR inhibitor VE-821 was readily detected (Fig. 5B) as previously reported (34), indicating that sensitization by iniparib could have been observed if present. Likewise, iniparib failed to sensitize to gemcitabine (Fig. 5C) even though sensitization by the checkpoint kinase inhibitor AZD 7762 (35) was readily demonstrated (Fig. 5D). We also failed to observe sensitization of SKOV3 cells to paclitaxel (Fig. 5E). In contrast, iniparib slightly but reproducibly sensitized SKOV3 cells to etoposide (Fig. 5F).
Figure 5. Effects of iniparib in combination with cisplatin, gemcitabine, paclitaxel or etoposide.
SKOV3 cells were treated continuously with the indicated concentrations of cisplatin (A), gemcitabine (C), paclitaxel (E) or etoposide (F) in the presence of diluent or 10 µM iniparib and examined for colony formation. In panels B and D, the ATR inhibitor VE-821 (1 µM), which is known to sensitize other cells to cisplatin (34), or the Chk1 inhibitor AZD 7762 (100 nM), which is known to sensitize to gemcitabine (35), was substituted for imiparib as a positive control. Error bars in each panel, ± SD of triplicate samples.
Failure of iniparib to inhibit pADPr synthesis
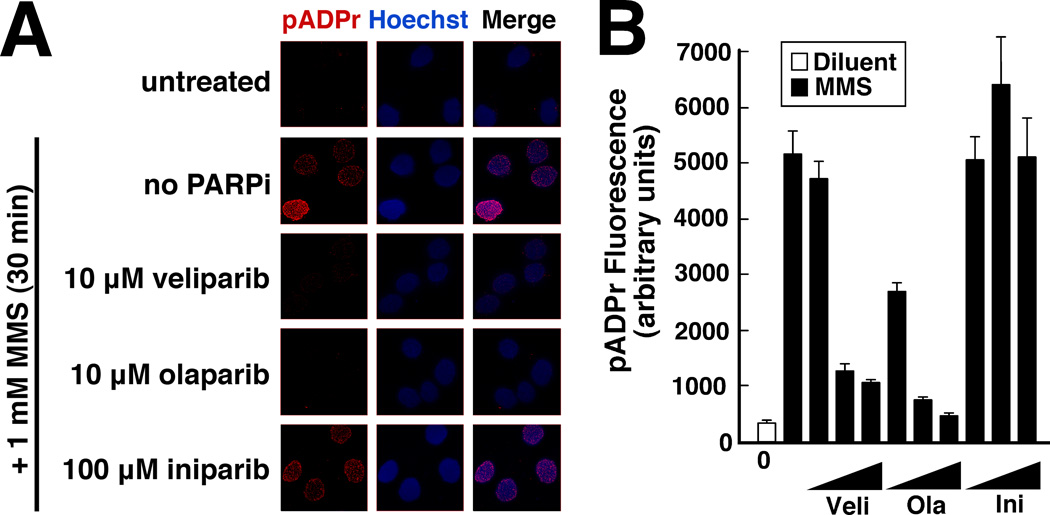
In view of the limited selectively of iniparib for HR-deficient cells (Figs. 2 and 3) and inability of iniparib to sensitize to topo I poisons (Fig. 4), we examined the ability of iniparib to inhibit PARP in situ. For these studies polymer levels were determined by quantitative fluorescence microscopy, a method that is widely used and is more quantifiable than immunoblotting (27, 36). Treatment with veliparib or olaparib caused a dose-dependent decrease in DNA damage-induced pADPr levels (Fig. 6). In contrast, iniparib had no detectable effect on polymer formation in situ at concentrations up to 100 µM.
Figure 6. Effects of veliparib, olaparib and iniparib on pADPr synthesis.
A, following a 4-h treatment with the indicated agent, SKOV3 cells were treated with MMS in the continued presence of the small molecule inhibitor, fixed, stained for pADPr and examined by confocal microscopy. B, mean fluorescence of pADPr in nuclei after treatment of cells with diluent (open bar) or 1 mM MMS (closed bars) in the presence of diluent or the indicated agent at 1, 10 and 100 µM. Error bars, ± SEM for 30 nuclei.
DISCUSSION
Previous studies have identified a number of biological properties of PARP inhibitors, including selective toxicity in HR-deficient cells, ability to synergize with topo I poisons, and, by definition, ability to inhibit pADPr synthesis in intact cells. Iniparib was originally described as a prodrug of a covalent PARP inhibitor (12, 37) and more recently as a small molecule cytotoxic with some PARP inhibitory activity (14). This agent originally displayed promising activity in combination with gemcitabine and cisplatin in phase 2 trials in triple negative breast cancer (14) and ovarian cancer (38, 39) as well as anecdotal activity against BRCA2-deficient pancreatic cancer (40). Interestingly, iniparib has not been previously compared head-to-head with other PARP inhibitors in preclinical studies. Results of the present study, however, indicate that iniparib (in contrast to olaparib and veliparib) fails to exhibit much selectivity for HR-deficient cells (Figs. 2 and 3), fails to sensitize to topo I poisons (Fig. 4) and fails to inhibit pADPr synthesis in intact cells (Fig. 6). These results have important implications for current interpretations of iniparib clinical trials.
Earlier studies demonstrated that PARP knockdown or PARP inhibition is selectively toxic to cells with HR defects (2, 6, 7). More recent studies have suggested that this selective toxicity stems from the ability of active site-directed noncovalent PARP inhibitors to tip the balance toward error-prone nonhomologous end-joining in HR-deficient cells (16). In anticipation of studies that would determine whether the intracellular metabolite 4-iodo-3-nitrosobenzamide, a putative covalent inhibitor of PARP1, acts in a similar fashion, we examined cells lacking BRCA2 (Fig. 2) or ATM (Fig. 3). In both cases, iniparib at concentrations exceeding 40 µM killed cells but failed to show the anticipated selectivity for the HR-deficient cells. This lack of selectivity, which was demonstrated using assays for both apoptosis and colony formation, immediately distinguishes iniparib from other widely studied PARP inhibitors such as veliparib, olaparib and MK-4827 (Figs. 2, 3 and ref. 20).
A wide variety of PARP inhibitors have also been shown to selectively enhance the cytotoxicity of topo I poisons (20, 23–27). This sensitization was readily detected with submicromolar concentrations of olaparib and veliparib (Fig. 4). In contrast, iniparib failed to sensitize multiple cell lines to either camptothecin or topotecan, again indicating a substantial difference in behavior compared to bona fide PARP inhibitors. Conversely, iniparib sensitized cells to etoposide (Fig. 5F), albeit modestly, whereas other PARP inhibitors fail to sensitize cells to this agent (20, 23, 41), again distinguishing iniparib from the PARP inhibitors.
Earlier studies indicated that iniparib can inhibit pADPr synthesis when glutathione levels in cells are depleted with buthionine sulfoximine (42). In contrast, we were unable to detect inhibition of pADPr synthesis in cells containing endogenous levels of glutathione (Fig. 6). This failure to inhibit pADPr synthesis presumably accounts for the inability of iniparib to selectively kill HR-deficient cells or synergize with topo I poisons.
Although iniparib does not appear to be exerting its effects through inhibition of pADPr synthesis, this agent clearly is cytotoxic to a variety of cell lines at concentrations above 40 µM (Figs. 2 and 3). While the mechanism of this cytotoxicity was not explored in the present study, structural similarity of iniparib to nicotinamide (Fig. 1) raises the possibility that the cytotoxic effects of iniparib reflect the collective inhibition of one or more enzymes that bind the nicotinamide derivative NAD+, including possibly GAPDH or sirtuins (42), rather than primary effects on PARP.
In further experiments, the effect of combining iniparib with other chemotherapeutic agents was examined. Current or recently completed trials (www.clinicaltrials.gov) have paired iniparib with paclitaxel (triple-negative breast cancer), carboplatin + gemcitabine (triple-negative breast cancer, ovarian cancer, nonsmall cell lung cancer) or carboplatin + paclitaxel (uterine carcinosarcoma). Accordingly, we specifically focused on the effects of combining iniparib with these classes of agents. Iniparib failed to sensitize cells to cisplatin, gemcitabine or paclitaxel (Fig. 5A, C and E), whereas other enzyme inhibitors caused readily detectable sensitization (Fig. 5B and 5D). Even though carboplatin was not used in these experiments because of its lower solubility and potency in vitro, results with carboplatin would likely be similar to our observations with cisplatin because of the identical mechanism of action of these agents (43). It is possible that different results might be obtained in different cell lines with different genetic and epigenetic changes. Nonetheless, our observation that iniparib has limited impact on sensitivity of cells to platinating agents, taxanes or gemcitabine might be important for interpreting results of recently completed and ongoing clinical trials of iniparib in combination with these agents.
In view of the marked differences between iniparib and other PARP inhibitors described in above, it is important that clinical results obtained with iniparib not be allowed to unduly influence development of bona fide PARP inhibitors. In particular, the recent disclosure that results of the phase 3 iniparib trial in triple negative breast cancer were negative (15) should not be interpreted to mean that bona fide PARP inhibitors will also fail to exhibit activity in this disease, as it is unlikely that iniparib inhibited PARP in this trial. In a similar fashion, if iniparib exhibits clinical activity in a different setting (38–40), these positive findings should also be extrapolated to bona fide PARP inhibitors extremely cautiously, if at all, because of the failure of iniparib to exhibit the properties of a PARP inhibitor as documented in the present study.
TRANSLATIONAL SIGNIFICANCE.
PARP inhibitors are currently undergoing extensive clinical testing. Recent negative results with one putative PARP inhibitor, iniparib, have raised concerns about the usefulness of inhibiting PARP to treat cancer. The present study demonstrates that iniparib fails to exhibit hallmark features of PARP inhibitors, including ability to selectively kill homologous recombination-deficient cells or inhibit synthesis of poly(ADP-ribose) polymer in intact cells. These results raise questions about the classification of iniparib as a PARP inhibitor and encourage caution in making decisions about the utility of PARP inhibitors based on clinical trials of iniparib.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge reagents from Larry Karnitz, Zhenkun Lou, Viji Shridhar and Fergus Couch; secretarial assistance of Deb Strauss; and provocative discussions with Matt Goetz and Lynn Hartmann.
GRANT SUPPORT:
This work was supported in part by P50 CA136393 (SHK and GGP). AGP was supported by T32 GM072474 and a predoctoral fellowship from the Mayo Foundation.
Footnotes
Conflicts of interest: None
AUTHOR CONTRIBUTIONS
AGP and SHK designed the study; AGP, SDL, and KF performed the experiments and interpreted the data; GGP provided critical reagents; and all authors participated in writing and editing the manuscript.
REFERENCES
- 1.Kaelin WG., Jr The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 2.Rehman FL, Lord CJ, Ashworth A. Synthetic lethal approaches to breast cancer therapy. Nat Rev Clin Oncol. 2010;7:718–724. doi: 10.1038/nrclinonc.2010.172. [DOI] [PubMed] [Google Scholar]
- 3.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 4.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 5.McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66:8109–8115. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- 6.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Javle M, Curtin NJ. The role of PARP in DNA repair and its therapeutic exploitation. Br J Cancer. 2011;105:1114–1122. doi: 10.1038/bjc.2011.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Audeh MW, Carmichael J, Penson RT, Friedlander M, Powell B, Bell-McGuinn KM, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376:245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- 9.Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376:235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 10.Penning TD. Small-molecule PARP modulators--current status and future therapeutic potential. Curr Opin Drug Discov Devel. 2010;13:577–586. [PubMed] [Google Scholar]
- 11.Calvert H, Azzariti A. The clinical development of inhibitors of poly(ADP-ribose) polymerase. Ann Oncol. 2011;22 Suppl 1:i53–i59. doi: 10.1093/annonc/mdq667. [DOI] [PubMed] [Google Scholar]
- 12.Mendeleyev J, Kirsten E, Hakam A, Buki KG, Kun E. Potential chemotherapeutic activity of 4-iodo-3-nitrobenzamide. Metabolic reduction to the 3-nitroso derivative and induction of cell death in tumor cells in culture. Biochem Pharmacol. 1995;50:705–714. doi: 10.1016/0006-2952(95)00189-7. [DOI] [PubMed] [Google Scholar]
- 13.Ossovskaya V, Li L, Broude EV. BSI-201 enhances the activity of multiple classes of cytotoxic agaents and irradiation in triple negative breast cancer; American Association for Cancer Research Annual Meeting; 2009. p. 5552. [Google Scholar]
- 14.O'Shaughnessy J, Osborne C, Pippen JE, Yoffe M, Patt D, Rocha C, et al. Iniparib plus chemotherapy in metastatic triple-negative breast cancer. N Engl J Med. 2011;364:205–214. doi: 10.1056/NEJMoa1011418. [DOI] [PubMed] [Google Scholar]
- 15.Guha M. PARP inhibitors stumble in breast cancer. Nat Biotechnol. 2011;29:373–374. doi: 10.1038/nbt0511-373. [DOI] [PubMed] [Google Scholar]
- 16.Patel A, Sarkaria J, Kaufmann SH. Nonhomologous end-joining drives PARP inhibitor synthetic lethality in homologous recombination-deficient cells. Proc Natl Acad Sci U S A. 2011;108:3406–3411. doi: 10.1073/pnas.1013715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Affar EB, Duriez PJ, Shah RG, Sallmann FR, Bourassa S, Kupper JH, et al. Immunodot blot method for the detection of poly(ADP-ribose) synthesized in vitro and in vivo. Anal Biochem. 1998;259:280–283. doi: 10.1006/abio.1998.2664. [DOI] [PubMed] [Google Scholar]
- 18.Langdon SP, Lawrie SS, Hay FG, Hawkes MM, McDonald A, Hayward IP, et al. Characterization and properties of nine human ovarian adenocarcinoma cell lines. Cancer Res. 1988;48:6166–6172. [PubMed] [Google Scholar]
- 19.Haince JF, McDonald D, Rodrigue A, Dery U, Masson JY, Hendzel MJ, et al. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem. 2008;283:1197–1208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- 20.Patel AG, Flatten KS, Schneider PA, Dai NT, McDonald JS, Poirier GG, et al. Enhanced killing of cancer cells by poly(ADP-ribose) polymerase inhibitors and topoisomerase inhibitors reflects poisoning of both enzymes. J Biol Chem. 2012;287 doi: 10.1074/jbc.M111.296475. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakai W, Swisher EM, Karlan BY, Agarwal MK, Higgins J, Friedman C, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–1120. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–1115. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- 23.Bowman KJ, Newell DR, Calvert AH, Curtin NJ. Differential effects of the poly (ADP-ribose) polymerase (PARP) inhibitor NU1025 on topoisomerase I and II inhibitor cytotoxicity in L1210 cells in vitro. Br J Cancer. 2001;84:106–112. doi: 10.1054/bjoc.2000.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calabrese CR, Almassy R, Barton S, Batey MA, Calvert AH, Canan-Koch S, et al. Anticancer chemosensitization and radiosensitization by the novel poly(ADP-ribose) polymerase-1 inhibitor AG14361. J Natl Cancer Inst. 2004;96:56–67. doi: 10.1093/jnci/djh005. [DOI] [PubMed] [Google Scholar]
- 25.Smith LM, Willmore E, Austin CA, Curtin NJ. The Novel Poly(ADP-Ribose) Polymerase Inhibitor, AG14361, Sensitizes Cells to Topoisomerase I Poisons by Increasing the Persistence of DNA Strand Breaks. Clin Cancer Res. 2005;11:8449–8457. doi: 10.1158/1078-0432.CCR-05-1224. [DOI] [PubMed] [Google Scholar]
- 26.Thomas HD, Calabrese CR, Batey MA, Canan S, Hostomsky Z, Kyle S, et al. Preclinical selection of a novel poly(ADP-ribose) polymerase inhibitor for clinical trial. Mol Cancer Ther. 2007;6:945–956. doi: 10.1158/1535-7163.MCT-06-0552. [DOI] [PubMed] [Google Scholar]
- 27.Zhang YW, Regairaz M, Seiler JA, Agama KK, Doroshow JH, Pommier Y. Poly(ADP-ribose) polymerase and XPF-ERCC1 participate in distinct pathways for the repair of topoisomerase I-induced DNA damage in mammalian cells. Nucleic Acids Res. 2011;39:3607–3620. doi: 10.1093/nar/gkq1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kartner N, Evernden-Porelle D, Bradley G, Ling V. Detection of P-glycoprotein in multidrug-resistant cell lines by monoclonal antibodies. Nature. 1985;316:820–823. doi: 10.1038/316820a0. [DOI] [PubMed] [Google Scholar]
- 29.Chen AY, Yu C, Potmesil M, Wall ME, Wani MC, Liu LF. Camptothecin Overcomes mdrl-Mediated Resistance in Human KB Carcinoma Cells. Cancer Res. 1991;51:6039–6044. [PubMed] [Google Scholar]
- 30.Hendricks CB, Rowinsky EK, Grochow LB, Donehower RC, Kaufmann SH. Effect of P-Glycoprotein Expression on the Accumulation and Cytotoxicity of Topotecan (SK&F 104864), a New Camptothecin Analogue. Cancer Res. 1992;52:2268–2278. [PubMed] [Google Scholar]
- 31.Rasheed ZA, Rubin EH. Mechanisms of resistance to topoisomerase I-targeting drugs. Oncogene. 2003;22:7296–7304. doi: 10.1038/sj.onc.1206935. [DOI] [PubMed] [Google Scholar]
- 32.Armstrong DK. Topotecan dosing guidelines in ovarian cancer: reduction and management of hematologic toxicity. Oncologist. 2004;9:33–42. doi: 10.1634/theoncologist.9-1-33. [DOI] [PubMed] [Google Scholar]
- 33.Markman M. Topotecan as second-line therapy for ovarian cancer: dosage versus toxicity. Oncologist. 2005;10:695–697. doi: 10.1634/theoncologist.10-9-695. [DOI] [PubMed] [Google Scholar]
- 34.Reaper PM, Griffiths MR, Long JM, Charrier JD, Maccormick S, Charlton PA, et al. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol. 2011;7:428–430. doi: 10.1038/nchembio.573. [DOI] [PubMed] [Google Scholar]
- 35.Zabludoff SD, Deng C, Grondine MR, Sheehy AM, Ashwell S, Caleb BL, et al. AZD7762, a novel checkpoint kinase inhibitor, drives checkpoint abrogation and potentiates DNA-targeted therapies. Mol Cancer Ther. 2008;7:2955–2966. doi: 10.1158/1535-7163.MCT-08-0492. [DOI] [PubMed] [Google Scholar]
- 36.Haince JF, Poirier GG, Kirkland JB. Nonisotopic methods for determination of poly(ADP-ribose) levels and detection of poly(ADP-ribose) polymerase. Chapter 18. Curr Protoc Cell Biol. 2004 doi: 10.1002/0471143030.cb1807s21. Unit18 7. [DOI] [PubMed] [Google Scholar]
- 37.Rice WG, Hillyer CD, Harten B, Schaeffer CA, Dorminy M, Lackey DA, 3rd, et al. Induction of endonuclease-mediated apoptosis in tumor cells by C-nitroso-substituted ligands of poly(ADP-ribose) polymerase. Proc Natl Acad Sci U S A. 1992;89:7703–7707. doi: 10.1073/pnas.89.16.7703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Birrer MJ, Konstantinopoulos P, Penson RT, Roche M, Ambrosio A, Stallings TE, et al. A phase II trial of iniparib (BSI-201) in combination with gemcitabine/carboplatin (GC) in patients with platinum-resistant recurrent ovarian cancer. J Clin Oncol. 2011;29 suppl doi: 10.1093/oncolo/oyac275. abstr 5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Penson RT, Whalen C, Lasonde B, Krasner CN, Konstantinopoulos P, Stallings TE, et al. A phase II trial of iniparib (BSI-201) in combination with gemcitabine/carboplatin (GC) in patients with platinum-sensitive recurrent ovarian cancer. J Clin Oncol. 2011;29 suppl doi: 10.1093/oncolo/oyac275. abstr 5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fogelman DR, Wolff RA, Kopetz S, Javle M, Bradley C, Mok I, et al. Evidence for the efficacy of Iniparib, a PARP-1 inhibitor, in BRCA2-associated pancreatic cancer. Anticancer Res. 2011;31:1417–1420. [PubMed] [Google Scholar]
- 41.Huehls AM, Wagner JM, Huntoon CJ, Geng L, Erlichman C, Patel AG, et al. Poly(ADP-Ribose) polymerase inhibition synergizes with 5-fluorodeoxyuridine but not 5-fluorouracil in ovarian cancer cells. Cancer Res. 2011;71:4944–4954. doi: 10.1158/0008-5472.CAN-11-0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bauer PI, Mendeleyeva J, Kirsten E, Comstock JA, Hakam A, Buki KG, et al. Anti-cancer action of 4-iodo-3-nitrobenzamide in combination with buthionine sulfoximine: inactivation of poly(ADP-ribose) polymerase and tumor glycolysis and the appearance of a poly(ADP-ribose) polymerase protease. Biochem Pharmacol. 2002;63:455–462. doi: 10.1016/s0006-2952(01)00872-3. [DOI] [PubMed] [Google Scholar]
- 43.Reed E. Cisplatin, carboplatin and oxaliplatin. In: Chabner BA, Longo DL, editors. Cancer Chemotherapy & Biotherapy: Principles and Practice. 4th ed. Philadelphia: Lippincott Williams and Wilkins; 2006. pp. 332–343. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.