Abstract
Purpose
EGFR genotyping is now standard in the management of advanced lung adenocarcinoma, as this biomarker predicts marked benefit from treatment with EGFR tyrosine kinase inhibitors (TKIs). EGFR exon 19 insertions are a poorly described family of EGFR mutations, and their association with EGFR TKI-sensitivity in lung adenocarcinoma is uncertain.
Experimental Design
Patients with lung cancers harboring EGFR exon 19 insertions were studied. The predicted effects of the insertions on the structure of the EGFR protein were examined, and EGFR exon 19 insertions were introduced into Ba/F3 cells to assess oncogenicity and in vitro sensitivity to EGFR TKIs. In patients receiving TKI, response magnitude was assessed with serial computed tomography (CT) measurement.
Results
Twelve tumors harboring EGFR exon 19 insertions were identified; patients were predominately female (92%) and never-smokers (75%). The 11 specimens available for full sequencing all demonstrated an 18 bp insertion that resulted in the substitution of a Pro for Leu at residue 747. The mutant EGFR transformed the Ba/F3 cells, which were then sensitive to EGFR TKI. Six patients with measurable disease received TKI and 5 had a response on serial CT.
Conclusions
EGFR exon 19 insertions are a newly appreciated family of EGFR TKI-sensitizing mutations, and patients with tumors harboring these mutations should be treated with EGFR-TKI. While these mutations may be missed through the use of some mutation-specific assays, the addition of PCR product size analysis to multi-gene assays allows sensitive detection of both exon 19 insertion and deletion mutations.
Introduction
EGFR mutation testing has now become the standard of care in the management of non-small cell lung cancer (NSCLC) since identifying that this biomarker can predict which patients will benefit from EGFR tyrosine kinase inhibitors (TKIs) such as erlotinib and gefitinib. Multiple randomized trials have now prospectively demonstrated the unique benefit of TKIs in patients with EGFR-mutant lung cancer (1–3). This has led both the American Society of Clinical Oncology (ASCO) and the National Comprehensive Care Network (NCCN) to recommend EGFR mutation testing to determine which lung cancer patients are likely to benefit from therapy with an EGFR-TKI (4, 5).
Since EGFR mutation testing is now the standard of care, it is important to identify which mutations are associated with benefit from TKIs and how to manage cases with unexpected genotyping results. The most common EGFR mutations are short, in-frame deletions in exon 19 (most often 15 or 18 base pairs (bp)) and the exon 21 point mutation L858R (6), which together are associated with a median progression free survival of 14 months on erlotinib (7). Mutations in exon 20 are also well described and have been associated with TKI resistance (8), the most common being exon 20 in-frame insertions of varying lengths, representing 4–9% of EGFR-mutant lung cancers (6, 9). Exon 20 point mutations like T790M are rarely identified in pretreatment cancers (9, 10), but can be found in more than half of cancers which have acquired resistance to erlotinib or gefitinib (11, 12). In addition, a study of 28 patients with uncommon point mutations in exon 21 (at L861) and exon 18 (at G719) was recently published, finding a 57% response rate to EGFR-TKI (9). While other rare EGFR mutations have been described, none have been clearly characterized as leading to sensitivity to TKI therapy.
In this study, we present a comprehensive evaluation of lung cancers and cell lines harboring insertion mutations in exon 19 of EGFR, a mutation which has been rarely described in the literature and which has an uncertain clinical significance (13–20). Through assessment of clinicopathologic characteristics, in vitro drug sensitivity, and quantitative imaging results, we aimed to determine whether these cancers are clinically and biologically more similar to the TKI-sensitive EGFR exon 19 deletions or to the TKI-insensitive EGFR exon 20 insertions.
Materials and Methods
For an initial prevalence analysis, an institutional database of patients with NSCLC undergoing EGFR mutation testing was queried for tumors harboring exon 19 insertions in the absence of exon 19 deletions (21). The cohort was subsequently expanded for characterization of clinical and pathologic features, at which point additional cases outside of this database were included from two contributing institutions. Patient cases were collected and reviewed through an IRB-approved mechanism. All cases were identified over the course of routine molecular diagnostic testing for EGFR sensitizing mutations at the contributing institution’s diagnostic molecular pathology laboratories.
The initial cohort of exon 19 insertion cases was identified using a PCR-based fragment length analysis previously described (22). Briefly, paraffin-embedded or frozen tissues of tumor samples (biopsy material or cytologic specimens) were submitted to the laboratory where they were macrodissected (if possible) and genomic DNA was extracted. Genomic DNA was amplified by PCR using the forward primer 5’-TGGTAACATCCACCCAGATCA-3’ and reverse primer FAM 5’-AAAAGGTGGGCCTGAGGTTCA-3’; the reverse primer was labeled with the FAM fluorophore. The PCR products were subjected to capillary electrophoresis on an ABI 3730 Genetic Analyzer (Applied Biosystems, Foster City, CA) and compared with the wild type PCR product to determine if differences in length were present, and whether the differences represented a deletion or insertion (Supplementary Figure). All samples were tested in duplicate with positive and negative controls. For the additional cohort, mutations were either identified using the above fragment length analysis or using direct Sanger sequencing. If additional DNA was available, cases with exon 19 insertions were further subjected to PCR-sequencing on the ABI platform mentioned above.
Response to initial EGFR-TKI therapy was assessed by conventional summed measurement of linear tumor diameters on CT scan (23). For patients with advanced disease, best response was defined as the percent change between the smallest measurement while on therapy and the baseline measurement. Patients receiving neoadjuvant TKI had reimaging available after only 3 weeks of therapy, too early to accurately assess partial response (24); for these patients, change in total tumor volume was measured using a previously described semi-automated algorithm (25). Using this algorithm, an operator draws a region of interest (ROI) around the tumor being measured on a single slice, and the computer then automatically delineates the tumor boundaries on all CT slices containing tumor; the radiologist then reviews the resulting tumor boundaries and can correct them if needed. Tumor volume was calculated by summing the tumor areas on all involved CT slices, and change was calculated by subtracting total tumor volume at 3 weeks from that at baseline, as a percent of the baseline tumor volume.
In vitro sensitivity assessment
The full length EGFR gene was cloned into pDNR-Dual (BD Biosciences: San Diego, CA). The exon 19 insertion sequences from two patients were introduced using the Quick Change Site-Directed Mutagenesis kit (Stratagene; La Jolla, CA) with mutant-specific primers according to the manufacturer's instructions and as previously described (26, 27). All the insertions were confirmed by direct sequencing. Ba/F3 cells and NIH-3T3 cells were cultured as previously described (27). Retroviral infection and culture of Ba/F3 and NIH-3T3 cells were performed using previously described methods (26, 27). Ba/F3 and NIH-3T3 cells expressing EGFR E746_A750del (an exon 19 deletion) and Y764_V765insHH (an exon 20 insertion) were used as controls.
For cell proliferation and growth assays, gefitinib and afatinib (BIBW2992) were obtained from commercial sources. Growth inhibition was assessed by MTS assay as described previously (27). Ba/F3 cells were exposed to drugs for 72 hours. All experimental points were set up in six to twelve wells and repeated at least three times. The data was graphically displayed using GraphPad Prism version 5.0 for Windows.
For western blotting, NIH-3T3 cells were lysed in NP-40 buffer and proteins were separated by gel electrophoresis on 4–12% polyacrylamide gels (Invitrogen; Carlsbad, CA). Proteins were transferred to nitrocellulose membranes and detected by immunoblotting performed according to the antibody manufacturer's recommendations. Anti-EGFR antibodies were obtained from Cell Signaling Technology and phospho-EGFR (pY1068) antibodies were purchased from Invitrogen (Carlsbad, CA). The anti-α-tubulin antibody was purchased from Sigma-Aldrich (St. Louis, MO).
Results
Clinical & pathologic characteristics
From 2004 to 2009, 3026 patient specimens at one contributing institution (Memorial Sloan-Kettering Cancer Center) were tested for EGFR exon 19 deletions and L858R point mutations using fragment length analysis (22). Specimens from 347 patients (11.5%) were found to harbor EGFR exon 19 deletions and specimens from 246 (8.1%) harbored L858R point mutations (21). Eight cases (0.26%) tested positive for EGFR exon 19 insertions (Supplementary Figure), thus comprising approximately 2% of exon 19 mutations and approximately 1% of all EGFR mutations.
Four additional patients with tumors harboring EGFR exon 19 insertions were subsequently identified outside of the above analysis, making a total of 12 cases available for study. Median age was 63 (range 41–79). Eleven patients (92%) were female. Ten patients (83%) were white, 2 were African-American, and none were Asian. Nine patients (75%) were never-smokers, two were former smokers with a 5 pack-year smoking history, and one was a former heavy smoker. Four patients (33%) had stage IV disease at diagnosis while 8 patients (67%) were diagnosed with cancer at an earlier stage.
Eleven tumors (92%) had adenocarcinoma histology and one was characterized as adenosquamous carcinoma. All tumors had an exon 19 insertion which was 18 bp in length (Supplementary Table 1). Interestingly, one of the patients had two morphologically different adenocarcinomas in the same lobe; while one tumor harbored an 18 bp insertion in exon 19, the synchronous primary harbored a 15 bp deletion in exon 19.
Protein structure
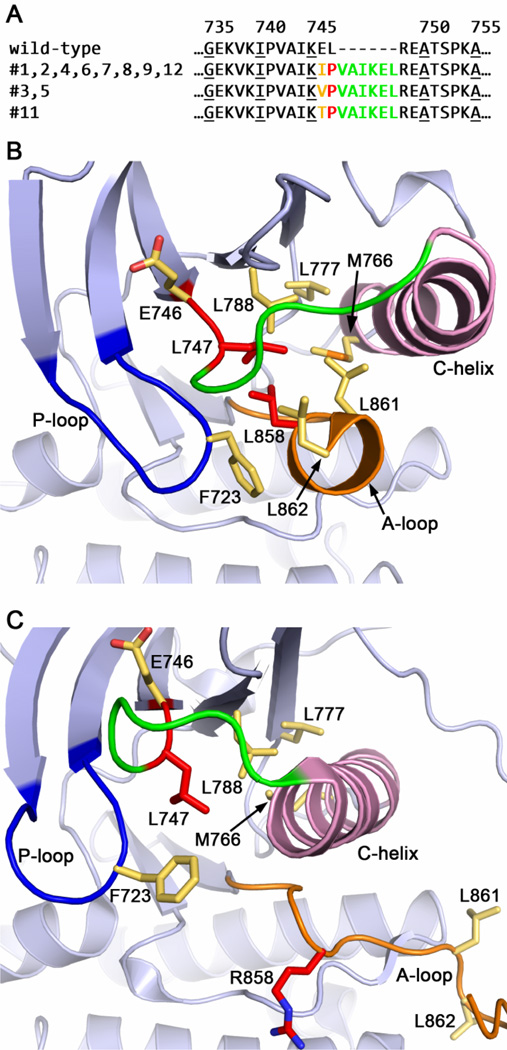
DNA sequencing could be performed on 11 of the 12 cases (Supplementary Table 1). The 11 cases belonged to 5 genotypes, with the insertion beginning at codon 744 in 4 cases and at codon 745 in 7 cases. These five genotypes encode only three distinct amino acid sequences, each conferring a six residue insertion in the protein structure (Figure 1A).
Figure 1.
Structural insights into the mechanism of activation of the EGFR exon 19 insertion mutants. (A) Predicted protein sequence of the 11 exon 19 insertion mutants, aligned with the corresponding region of wild-type EGFR. Structurally, these insertions can be considered as a complex mutation of consisting of E745X (blue), L747P (purple) and an insertion of VAIKEL (red). Note that all variants result in the L747P substitution. (B) In wild-type EGFR, Leu747 contributes to a cluster of hydrophobic residues that is important for stabilizing the inactive state of the kinase. The other participants include L788, L777, M766, L861, L862, L858 and F723. (C) In the exon 19 insertion mutants, the substitution of Leu747 with a Pro can be predicted to disfavor the formation of this hydrophobic core, leading to activation of the mutant EGFR. This mechanism is closely analogous to that proposed for the L858R point mutation. The six residue insertion is expected to lengthen the adjacent loop (green), which leads to the C-helix (pink). Panel B is drawn from the structure of wild-type EGFR in an inactive state (42), Panel C from the structure of the active L858R mutant (28).
Examination of the three-dimensional structure of the EGFR kinase shows that this insertion lies at the end of strand β3 in the N-terminal lobe of the kinase domain. The mutations are expected to have two effects; they will alter the identity of the last two residues of strand β3 (Glu746 and Leu747), and they will result in the addition of the six residue sequence Val-Ala-Ile-Lys-Glu-Leu to the loop connecting this strand with the C-helix. The addition of six residues to this loop is expected to be structurally well-tolerated, as this loop is flexible and typically not well-defined in EGFR crystal structures. Likewise, mutation of Glu746 (to Ile, Val or Thr in the various exon 19 insertion mutants) is likely to have little structural effect, as this residue is exposed on the surface of the kinase. In contrast, Leu747 participates in a key hydrophobic core that stabilizes the inactive form of EGFR (Figure 1B). The non-conservative substitution of Pro for Leu747 (L747P), which occurs in all 11 insertion sequences, can be predicted to disfavor the formation of this hydrophobic core, thereby leading to constitutive activation of the mutant EGFR (Figure 1C). This mechanism is analogous to that proposed for the L858R point mutation (28), which lies immediately adjacent to L747 in this hydrophobic core (Figure 1B).
In vitro sensitivity to TKI
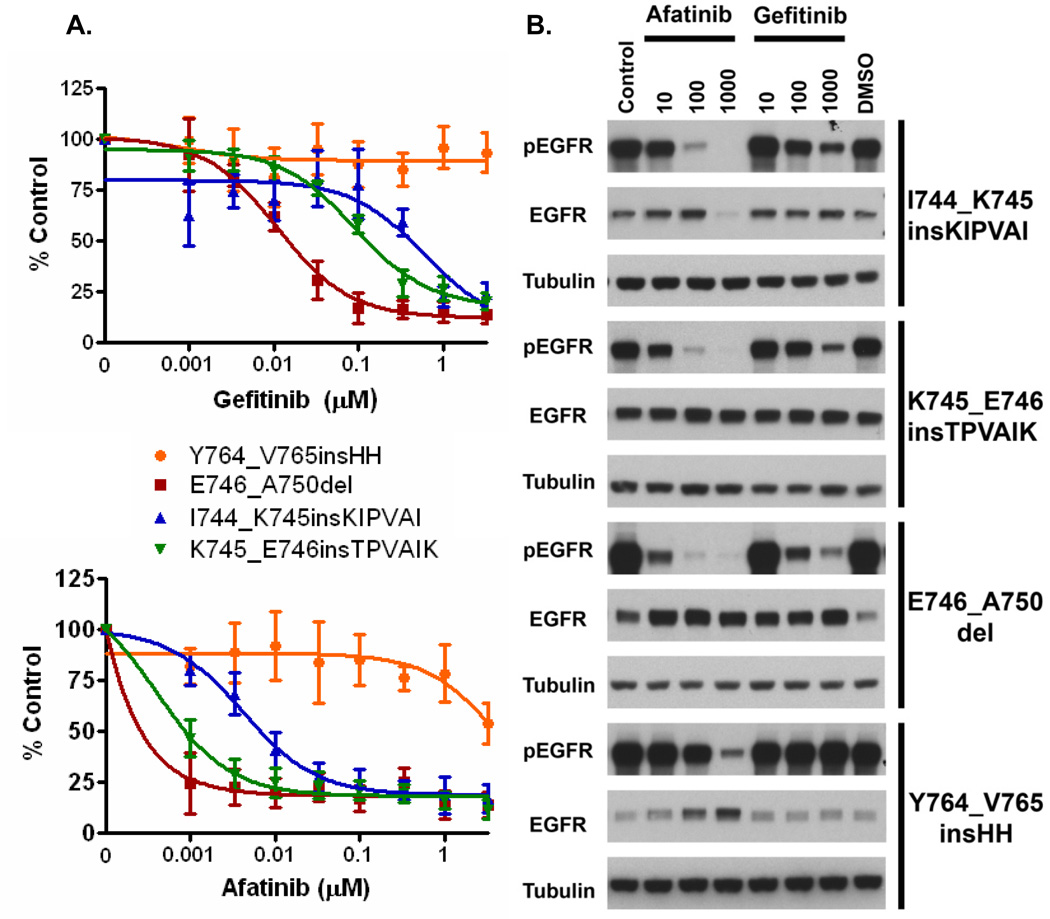
Transfection of Ba/F3 cells with mutant EGFR constructs harboring two different insertions in exon 19 (I744_K745insKIPVAI and K745_E746insTPVAIK) resulted in IL3 independent growth, indicating that these mutations were oncogenic (29). The resulting mutant cell lines were then assessed for sensitivity to TKIs using gefitinib (reversible EGFR-TKI) and afatinib (irreversible EGFR-TKI). The Ba/F3 cells transfected with exon 19 insertions were found to be sensitive to both TKIs, similar to Ba/F3 cells harboring the common EGFR exon 19 deletion (E746_A750del) (Figure 2A). In contrast Ba/F3 cells harboring the exon 20 insertion mutation were resistant to both gefitinib and afatinib. We did note that the sensitivity of cells harboring exon 19 insertions was slightly less than the sensitivity of cells harboring exon 19 deletions for both TKIs.
Figure 2.
In vitro analyses of EGFR exon 19 insertion mutations. (A) Ba/F3 cells harboring different exon 19 insertions are sensitive to gefitinib and afatinib, similar to exon 19 deletion (E746_A750del) Ba/F3 cells. Exon 20 insertion cells (Y764_V765insHH) are resistant to both drugs. (B) NIH-3T3 cells harboring exon 19 insertions demonstrate a marked reduction in phospho-EGFR when treated with either gefitinib or afatinib, consistent with the growth assays (A).
We further analyzed the impact on EGFR phosphorylation using Western blotting (Figure 2B). Consistent with the growth assays, both gefitinib and afatinib inhibited EGFR phosphorylation in NIH-3T3 cells harboring the two EGFR exon 19 insertion mutations and the exon 19 deletion mutation. However, neither drug effectively inhibited EGFR phosphorylation in cells harboring the EGFR exon 20 insertion (Figure 2B).
Clinical sensitivity to TKI
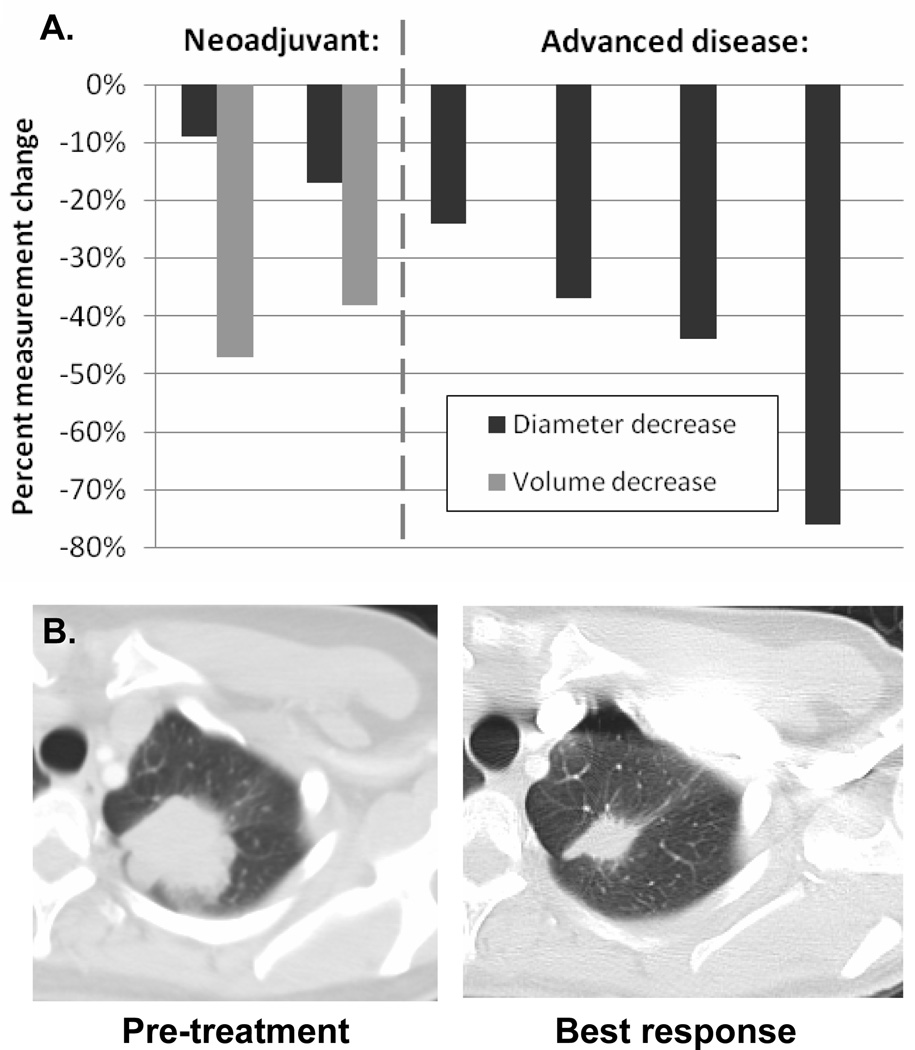
The clinical course of the 12 patients identified is described in Table 1. Eight patients underwent resection of their disease; 4 patients have more than 3 years of follow-up after surgery, and 3 remain disease free. Eight patients received EGFR-TKI therapy: 2 received TKI in the adjuvant setting while they had no evidence of disease, whereas the remaining 6 patients had measurable disease (Figure 3). Two of these patients received single-agent neoadjuvant TKI and were found to have a 3-week tumor diameter decrease of ≥9% and 3-week tumor volume decrease of ≥25%, response characteristics which are closely associated with presence of a sensitizing mutation according to a previous study (24). Of 4 patients who received palliative EGFR-TKI, 3 had a RECIST partial response (23) and a time to progression of greater than 12 months; the fourth patient received a novel EGFR-TKI with uncertain activity (XL647)(30) and had a brief minor response. Re-biopsy material was available from one of the five patients who developed progression on TKI; this specimen did not harbor T790M, but did demonstrate low level MET amplification, which has been found to be associated with acquired resistance to EGFR-TKI (31).
Table 1.
Clinical course of patients with lung cancers harboring EGFR exon 19 insertions
| Patient | Stage | Resected | TKI | Clinical setting | TTR/TTP (months) |
|---|---|---|---|---|---|
| 1 | IA | Yes | None | >1 | |
| 2 | IA | Yes | None | >7 | |
| 3 | IIA | Yes | Erlotinib | Adjuvant | >42 |
| 4 | IIB | Yes | None | >55 | |
| 5 | IIB | Yes | None | >1 | |
| 6 | IIIA | Yes | Erlotinib | Neoadjuvant & adjuvant | >10 |
| 7 | IIIA | Yes | Gefitinib | Neoadjuvant & adjuvant | >48 |
| 8 | IIIB | Yes | Gefitinib | Adjuvant | 38 |
| 9 | IVB | No | Afatinib | Palliative | 14 (PR) |
| 10 | IVB | No | XL647 | Palliative | 4 |
| 11 | IVB | No | Erlotinib | Palliative | 19 (PR) |
| 12 | IVB | No | Erlotinib | Palliative | 50 (PR) |
TKI: Tyrosine kinase inhibitor, TTR: Time to relapse, TTP: Time to progression, NED: No evidence of disease, “>” designates a patient who has not yet relapsed/progressed, PR: partial response
Figure 3.
Response to EGFR-TKI therapy for the 6 patients with measurable disease. (A) Both patients receiving neoadjuvant TKI had a volumetric response (24), while 3 of 4 patients with advanced disease had a RECIST partial response. (B) CT imaging from one patient with advanced disease whose tumor demonstrated a marked response to erlotinib.
Discussion
Through the study of transfected cell lines and lung cancers harboring EGFR exon 19 insertion mutations treated with EGFR-TKIs, we demonstrate that exon 19 insertions are a new family of TKI-sensitizing EGFR mutations. These patients have similar clinical characteristics to other patients with EGFR-mutant lung cancer and can have durable responses to treatment with TKIs. While several case reports have described tumors with EGFR exon 19 insertions as either sensitive or resistant to TKI (Supplementary Table 2) (14, 19, 20), ours is the first large series and strongly suggests a TKI-sensitive phenotype. We recommend that patients with lung cancer harboring an EGFR exon 19 insertion be considered TKI sensitive, and best managed with TKI therapy such as erlotinib.
Our study highlights an important follow-up question raised by the implementation of routine EGFR mutation testing for lung adenocarcinoma, as recommended by multiple recent clinical practice guidelines (4, 5, 32): how should clinicians manage patients whose tumors are found to harbor rare mutations in EGFR? While deletions in exon 19 and point mutations in exons 18 and 21 have been found to be associated with TKI-sensitivity through the study of large patient cohorts (Table 2), no other rare mutations in EGFR have been found to be consistently associated with benefit from TKI (9). In this study, we have shown that additional rare mutations in EGFR can be distinguished as “sensitive” or “insensitive” through the collection of inter-institutional cohorts, rigorous imaging measurement, and accompanying in vitro data. We encourage further inter-institutional collaboration to allow collection and study of patients with other rare EGFR mutations, so their phenotype can be better characterized. Many EGFR mutations that have been reported in NSCLC only appear in the literature a single time (33) – such cases cannot be considered to be clinically relevant until they are comprehensively studied as a series.
Table 2.
Sensitivity phenotype of well-described EGFR mutations in lung cancer
| Phenotype | Genotype |
|---|---|
| TKI sensitive: | Exon 19 deletions (41) Exon 21 L858R (41) Exon 21 L861X* (9) Exon 18 G719X* (9) Exon 19 insertions |
| Baseline resistance: | Exon 20 insertions (8) Exon 20 T790M** (9) |
| Acquired resistance: | Exon 20 T790M (11) |
“X” designates that several amino acid substitutions are possible at this site
When detected using conventional methods
Our structural analysis suggests that the substitution of a Pro for Leu at position 747 underlies the activating effect of the different exon 19 insertions. We note that all of the exon 19 insertions we identified were of the same length (18 bp), all shared an identical 12 bp sequence (TCCCGTCGCTAT), and all resulted in the L747P substitution. Reviewing the literature, there have been 10 other published cases of EGFR exon 19 insertions (Supplementary Table 2) (13–20); amino-acid sequences are available for 9 of these cases, and all are also 18 bp in length and result in an L747P substitution. The tenth case was described as a 15 bp insertion, but the amino acid sequence is not available (19). Interestingly, a review of the common exon 19 deletion mutants reveals that these also result in non-conservative substitutions of L747, often to Pro, Thr or Ser. While it is tempting to speculate that this shared substitution of Leu747 may explain both the activating effect and the inhibitor sensitivity of these otherwise divergent alterations, it must be noted that two cases in a recent series carried L747P point mutations and neither were sensitive to TKI (9). Further study of EGFR point mutations at L747 is needed, and is underway, as separate structural or mechanistic elements may mediate whether a mutation is oncogenic and whether it is TKI sensitive. For example, the EGFR-TKI sensitivity of both exon 19 deletion mutants and the L858R point mutation has been shown to arise in part from the diminished affinity of these mutants for ATP (28, 34), while the secondary T790M mutation has been shown to mediate TKI-resistance by conferring an enhanced affinity for ATP (35). Thus it will be of interest to examine the ATP affinity and other enzyme kinetic properties of the different exon 19 mutants.
As noted above, the exon19 insertion mutants may be structurally and mechanistically similar to the exon 19 deletion mutants. However, they are structurally quite different from the exon 20 insertion mutants, which map to the opposite end of the C-helix and are generally resistant to EGFR-TKIs (8). The underlying reason for the TKI-resistance of exon 20 insertion mutants remains to be elucidated. We do note that exon 20 insertions vary in length (from 3–12 bp) and in their position within the exon (8, 36, 37), in contrast to the similarities between the exon 19 insertions we identified, and this variability may make then biologically more diverse and difficult to treat with a single targeted strategy.
Our finding that 1% of EGFR-mutant lung cancers harbor exon 19 insertions suggests that approximately 250 of such patients are diagnosed annually in the United States. In contrast, a EGFR sequencing of a large cohort of Asian patients with NSCLC identified 627 whose tumors carried EGFR mutation and none were exon 19 insertions (9); however insertions and deletions may be technically difficult to distinguish in Sanger sequencing traces and therefore these data may not be comparable to data based on simple PCR product sizing analysis. Therefore, further data may be needed to assess the possibility of inter-ethnic differences. While these mutations can be detected using conventional Sanger sequencing, they may not be identified when using mutation-specific assays such as the ARMS assay by DxS/Qiagen and the multi-gene mass spectrometry assays being implemented at some academic centers (38). Mutation-specific techniques require development of an individual assay for each specific mutant nucleotide sequence being screened for, which makes these strategies inefficient for detecting insertions/deletions of variable lengths and sequences. However, fragment length analysis (Supplementary Figure) can detect deletion and insertion mutations with high sensitivity despite varying lengths and independent of the specific sequences (22). For this reason, some centers are now using multi-platform assays that include both a mutation-specific multi-gene assay as well as fragment length analysis as part of a comprehensive strategy for detection of driver mutations in lung adenocarcinoma (39, 40).
In conclusion, we have shown that EGFR exon 19 insertions are a new family of sensitizing EGFR mutations in lung adenocarcinoma, and recommend that patients with tumors harboring these mutations be considered for upfront therapy with TKIs like erlotinib or gefitinib. These mutations are associated with durable responses to therapy with tyrosine kinase inhibitors, much like EGFR exon 19 deletions. Though these mutations may be missed by mutation-specific molecular detection techniques due to their varying nucleotide sequences, both exon 19 deletions and insertions can be identified through the incorporation of fragment length analysis into multi-platform genotyping assays.
Statement of Translational Relevance.
EGFR mutation testing is now a standard component of the management of advanced lung adenocarcinoma, creating a need for clinicians to understand how to manage unexpected or rare genotyping results. In this inter-institutional effort, we studied a cohort of patients with EGFR exon 19 insertions, a rare mutation in EGFR, and characterized their response to tyrosine kinase inhibitor (TKI) therapy. Using accompanying in vitro data describing the TKI-sensitivity of cell lines transfected with these mutations, as well as a structural hypothesis supporting the oncogenicity of the resulting mutant protein, we determined that cancers harboring these mutations are sensitive to EGFR-TKI. Our strategy of paired clinical and preclinical functional analyses represents an effective technique for characterizing the biology of other rare tumor genotypes which are too uncommon to be well characterized in patients alone.
Supplementary Material
Acknowledgments
Supported by the National Cancer Institute grants P01-CA129243 (ML & MGK), P50-CA090578 (PJ), and R01-CA114465 (PJ)
References
- 1.Mok TS, Wu Y-L, Thongprasert S, Yang C-H, Chu D-T, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 2.Douillard J-Y, Shepherd FA, Hirsh V, Mok T, Socinski MA, Gervais R, et al. Molecular predictors of outcome with gefitinib and docetaxel in previously treated non-small-cell lung cancer: data from the randomized phase III INTEREST trial. J Clin Oncol. 2009;28:744–752. doi: 10.1200/JCO.2009.24.3030. [DOI] [PubMed] [Google Scholar]
- 3.Janne PA, Wang XF, Socinski MA, Crawford J, Capelletti M, Edelman MJ, et al. Randomized phase II trial of erlotinib (E) alone or in combination with carboplatin/paclitaxel (CP) in never or light former smokers with advanced lung adenocarcinoma: CALGB 30406. J Clin Oncol (Meeting Abstracts) 2010;28:7503. doi: 10.1200/JCO.2011.40.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keedy VL, Temin S, Somerfield MR, Beasley MB, Johnson DH, McShane LM, et al. American Society of Clinical Oncology Provisional Clinical Opinion: Epidermal Growth Factor Receptor (EGFR) Mutation Testing for Patients With Advanced Non-Small-Cell Lung Cancer Considering First-Line EGFR Tyrosine Kinase Inhibitor Therapy. J Clin Oncol. 2011;29:2121–2127. doi: 10.1200/JCO.2010.31.8923. [DOI] [PubMed] [Google Scholar]
- 5.The NCCN Clinical Practice Guidelines in Oncology™. Non-small cell lung cancer (Version 3.2011) National Comprehensive Cancer Network, Inc.; [Accessed May 17, 2011]. p. www.nccn.org. [Google Scholar]
- 6.Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba II, et al. Clinical and Biological Features Associated With Epidermal Growth Factor Receptor Gene Mutations in Lung Cancers. J Natl Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 7.Rosell R, Moran T, Queralt C, Porta R, Cardenal F, Camps C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 8.Wu JY, Wu SG, Yang CH, Gow CH, Chang YL, Yu CJ, et al. Lung cancer with epidermal growth factor receptor exon 20 mutations is associated with poor gefitinib treatment response. Clin Cancer Res. 2008;14:4877–4882. doi: 10.1158/1078-0432.CCR-07-5123. [DOI] [PubMed] [Google Scholar]
- 9.Wu J-Y, Yu C-J, Chang Y-C, Yang JC-H, Shih J-Y, Yang P-C. Effectiveness of tyrosine kinase inhibitors on uncommon epidermal growth factor receptor mutations of unknown clinical significance in non-small cell lung cancer. Clin Cancer Res. 2011;17:3812–3821. doi: 10.1158/1078-0432.CCR-10-3408. [DOI] [PubMed] [Google Scholar]
- 10.Inukai M, Toyooka S, Ito S, Asano H, Ichihara S, Soh J, et al. Presence of epidermal growth factor receptor gene T790M mutation as a minor clone in non-small cell lung cancer. Cancer Res. 2006;66:7854–7858. doi: 10.1158/0008-5472.CAN-06-1951. [DOI] [PubMed] [Google Scholar]
- 11.Arcila ME, Oxnard GR, Nafa K, Riely GJ, Solomon SB, Zakowski M, et al. Rebiopsy of Lung Cancer Patients with Acquired Resistance to EGFR Inhibitors and Enhanced Detection of the T790M Mutation Using a Locked Nucleic Acid-Based Assay. Clin Cancer Res. 2011;17:1169–1180. doi: 10.1158/1078-0432.CCR-10-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and Histological Evolution of Lung Cancers Acquiring Resistance to EGFR Inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res. 2004;64:8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- 14.Mitsudomi T, Kosaka T, Endoh H, Horio Y, Hida T, Mori S, et al. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small-cell lung cancer with postoperative recurrence. J Clin Oncol. 2005;23:2513–2520. doi: 10.1200/JCO.2005.00.992. [DOI] [PubMed] [Google Scholar]
- 15.Yoshida Y, Shibata T, Kokubu A, Tsuta K, Matsuno Y, Kanai Y, et al. Mutations of the epidermal growth factor receptor gene in atypical adenomatous hyperplasia and bronchioloalveolar carcinoma of the lung. Lung Cancer. 2005;50:1–8. doi: 10.1016/j.lungcan.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 16.Okami J, Taniguchi K, Higashiyama M, Maeda J, Oda K, Orita N, et al. Prognostic factors for gefitinib-treated postoperative recurrence in non-small cell lung cancer. Oncology. 2007;72:234–242. doi: 10.1159/000112947. [DOI] [PubMed] [Google Scholar]
- 17.Ilie MI, Hofman V, Bonnetaud C, Havet K, Lespinet-Fabre V, Coelle C, et al. Usefulness of tissue microarrays for assessment of protein expression, gene copy number and mutational status of EGFR in lung adenocarcinoma. Virchows Arch. 2010;457:483–495. doi: 10.1007/s00428-010-0963-z. [DOI] [PubMed] [Google Scholar]
- 18.Job B, Bernheim A, Beau-Faller M, Camilleri-Broet S, Girard P, Hofman P, et al. Genomic aberrations in lung adenocarcinoma in never smokers. PLoS One. 2010;5:e15145. doi: 10.1371/journal.pone.0015145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uruga H, Kishi K, Fujii T, Beika Y, Enomoto T, Takaya H, et al. Efficacy of gefitinib for elderly patients with advanced non-small cell lung cancer harboring epidermal growth factor receptor gene mutations: a retrospective analysis. Intern Med. 2010;49:103–107. doi: 10.2169/internalmedicine.49.2531. [DOI] [PubMed] [Google Scholar]
- 20.De Pas T, Toffalorio F, Manzotti M, Fumagalli C, Spitaleri G, Catania C, et al. Activity of Epidermal Growth Factor Receptor-Tyrosine Kinase Inhibitors in Patients with Non-small Cell Lung Cancer Harboring Rare Epidermal Growth Factor Receptor Mutations. Journal of Thoracic Oncology. 2011;6:1895–901. doi: 10.1097/JTO.0b013e318227e8c6. [DOI] [PubMed] [Google Scholar]
- 21.Dogan S, Ang DC, Brzostowski E, Johnson ML, D'Angelo SP, Paik PK, et al. EGFR and KRAS mutations in 3026 consecutive lung adenocarcinomas: associations with age, sex, and smoking history. J Mol Diagn. 2010;12 Abstracts. [Google Scholar]
- 22.Pan Q, Pao W, Ladanyi M. Rapid polymerase chain reaction-based detection of epidermal growth factor receptor gene mutations in lung adenocarcinomas. J Mol Diagn. 2005;7:396–403. doi: 10.1016/S1525-1578(10)60569-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, et al. New guidelines to evaluate the response to treatment in solid tumors. J Natl Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 24.Zhao B, Oxnard GR, Moskowitz CS, Kris MG, Pao W, Guo P, et al. A pilot study of volume measurement as a method of tumor response evaluation to aid biomarker development. Clin Cancer Res. 2010;16:4647–4653. doi: 10.1158/1078-0432.CCR-10-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao B, Schwartz LH, Moskowitz CS, Ginsberg MS, Rizvi NA, Kris MG. Lung cancer: computerized quantification of tumor response--initial results. Radiology. 2006;241:892–898. doi: 10.1148/radiol.2413051887. [DOI] [PubMed] [Google Scholar]
- 26.Zhou W, Ercan D, Chen L, Yun CH, Li D, Capelletti M, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070–1074. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engelman JA, Zejnullahu K, Gale C-M, Lifshits E, Gonzales AJ, Shimamura T, et al. PF00299804, an Irreversible Pan-ERBB Inhibitor, Is Effective in Lung Cancer Models with EGFR and ERBB2 Mutations that Are Resistant to Gefitinib. Cancer Res. 2007;67:11924–11932. doi: 10.1158/0008-5472.CAN-07-1885. [DOI] [PubMed] [Google Scholar]
- 28.Yun CH, Boggon TJ, Li Y, Woo MS, Greulich H, Meyerson M, et al. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warmuth M, Kim S, Gu XJ, Xia G, Adrian F. Ba/F3 cells and their use in kinase drug discovery. Curr Opin Oncol. 2007;19:55–60. doi: 10.1097/CCO.0b013e328011a25f. [DOI] [PubMed] [Google Scholar]
- 30.Pietanza MC, Lynch TJ, Lara PN, Cho J, Yanagihara RH, Vrindavanam N, et al. XL647, a Multi-Targeted Tyrosine Kinase Inhibitor: Results of a Phase II Study in Subjects with Non-Small Cell Lung Cancer Who Have Progressed after Responding to Treatment with Either Gefitinib or Erlotinib. J Thorac Oncol. 2011 doi: 10.1097/JTO.0b013e31822eebf9. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 31.Oxnard GR, Arcila ME, Chmielecki J, Ladanyi M, Miller VA, Pao W. New Strategies in Overcoming Acquired Resistance to EGFR Tyrosine Kinase Inhibitors in Lung Cancer. Clin Cancer Res. 2011;17:5530–5537. doi: 10.1158/1078-0432.CCR-10-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellis PM, Blais N, Soulieres D, Ionescu DN, Kashyap M, Liu G, et al. A Systematic Review and Canadian Consensus Recommendations on the Use of Biomarkers in the Treatment of Non-small Cell Lung Cancer. Journal of Thoracic Oncology. 2011;6:1379–1391. doi: 10.1097/JTO.0b013e318220cb8e. [DOI] [PubMed] [Google Scholar]
- 33.Murray S, Dahabreh IJ, Linardou H, Manoloukos M, Bafaloukos D, Kosmidis P. Somatic mutations of the tyrosine kinase domain of epidermal growth factor receptor and tyrosine kinase inhibitor response to TKIs in non-small cell lung ancer: an analytical database. J Thorac Oncol. 2008;3:832–839. doi: 10.1097/JTO.0b013e31818071f3. [DOI] [PubMed] [Google Scholar]
- 34.Carey KD, Garton AJ, Romero MS, Kahler J, Thomson S, Ross S, et al. Kinetic analysis of epidermal growth factor receptor somatic mutant proteins shows increased sensitivity to the epidermal growth factor receptor tyrosine kinase inhibitor, erlotinib. Cancer Res. 2006;66:8163–8171. doi: 10.1158/0008-5472.CAN-06-0453. [DOI] [PubMed] [Google Scholar]
- 35.Yun CH, Mengwasser KE, Toms AV, Woo MS, Greulich H, Wong KK, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sasaki H, Endo K, Takada M, Kawahara M, Kitahara N, Tanaka H, et al. EGFR exon 20 insertion mutation in Japanese lung cancer. Lung Cancer. 2007;58:324–328. doi: 10.1016/j.lungcan.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 37.Janne P, Boss DS, Camidge DR, Britten CD, Engelman JA, Garon EB, et al. Phase I dose-escalation study of the pan-HER Inhibitor, PF299804, in patients with advanced malignant solid tumors. Clin Cancer Res. 2011;17:1131–1139. doi: 10.1158/1078-0432.CCR-10-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacConaill LE, Campbell CD, Kehoe SM, Bass AJ, Hatton C, Niu L, et al. Profiling Critical Cancer Gene Mutations in Clinical Tumor Samples. PLoS One. 2009;4:e7887. doi: 10.1371/journal.pone.0007887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kris MG, Lau CY, Ang D, Brzostowski E, Riely GJ, Rusch VW, et al. Initial results of LC-MAP: An institutional program to routinely profile tumor specimens for the presence of mutations in targetable pathways in all patients with lung adenocarcinoma. J Clin Oncol (Meeting Abstracts) 2010;28:7009. [Google Scholar]
- 40.Sequist LV, Heist RS, Shaw AT, Fidias P, Rosovsky R, Temel JS, et al. Implementing multiplexed genotyping of non-small-cell lung cancers into routine clinical practice. Annals of Oncology. 2011 doi: 10.1093/annonc/mdr489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riely GJ, Pao W, Pham D, Li AR, Rizvi N, Venkatraman ES, et al. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin Cancer Res. 2006;12:839–844. doi: 10.1158/1078-0432.CCR-05-1846. [DOI] [PubMed] [Google Scholar]
- 42.Wood ER, Truesdale AT, McDonald OB, Yuan D, Hassell A, Dickerson SH, et al. A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 2004;64:6652–6659. doi: 10.1158/0008-5472.CAN-04-1168. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.