Abstract
Objective
We sought to identify and characterize two distinct populations of bona fide circulating endothelial cells, including the endothelial colony forming cell (ECFC), by polychromatic flow cytometry (PFC), colony assays, immunomagnetic selection, and electron microscopy.
Methods and Results
Mononuclear cells from human umbilical cord blood and peripheral blood were analyzed utilizing our recently published PFC protocol. A population of cells containing both ECFCs and mature circulating endothelial cells (CEC) were determined by varying expressions of CD34, CD31 and CD146, but not AC133 and CD45. After immunomagnetic separation, these cells failed to form hematopoietic colonies, yet clonogenic endothelial colonies with proliferative potential were obtained, thus verifying their identity as ECFCs. The frequency of ECFCs were increased in cord blood and were extremely rare in the peripheral blood of healthy adults. In addition, we also detected another mature endothelial cell population in the circulation that was apoptotic. Finally, when comparing this new protocol to a prior method we determined that the present protocol identifies circulating endothelial cells while the earlier protocol identified extracellular vesicles.
Conclusions
Two populations of circulating endothelial cells including the functionally characterized ECFC are now identifiable in human cord blood and peripheral blood by PFC.
Keywords: Endothelial Progenitor Cell, Flow Cytometry
Endothelial progenitor cells (EPCs) have been used as biomarkers in cardiovascular disease risk as well as angiogenesis therapies (as reviewed in 1), yet only one defined subset of EPCs has been shown to have clonogenic and proliferative potential, the endothelial colony forming cell (ECFC)2. EPCs were first described after a subset of mononuclear cells were cultured, and cells emerged that displayed some phenotypic and functional characteristics of endothelial cells3. EPCs are thought to circulate in human peripheral blood, home to sites of new blood vessel formation, and facilitate either arteriogenesis or angiogenesis by direct integration into the endothelium or via paracrine stimulation of existing vessel wall derived cells4.
Changes in EPC, ECFC, circulating endothelial progenitor cell (CEP), or circulating endothelial cell (CEC) concentration within peripheral blood have been detected utilizing multiple culture methods, with these changes being coupled to human maladies, including coronary artery disease, diabetes and cancer5, 6. However, these cells were not identifiable through flow cytometry alone, and required laborious and time intensive culture techniques2. By themselves, these culture techniques are not practical for use as a diagnostic tool. This lack of practicality has led to the development of numerous flow cytometry protocols to try to identify these cell types (CEPs and CECs) in circulation7, 8. Until now, the ECFC remained undefined via flow cytometry in either human umbilical cord blood or peripheral blood mononuclear cells, in part because there was not a consensus definition by cell surface antigen expression (CD34, CD45, CD31, CD146 and AC133).
A recent review of the controversies in EPC definition determined that some of the clinical trials published in 2010 claiming to quantify EPCs actually quantified hematopoietic stem cells (HSCs)9. In addition to the lack of clear flow cytometric definition, the necessary functional assays to verify a cell type’s true identity are often lacking, but imperative for full characterization and mechanistic determination of how these cells function in vessel formation in vivo. In order to fully utilize the potential of the ECFCs as a biomarker for angiogenesis and cardiovascular disease risk, an accurate flow cytometry protocol is imperative.
Rare cell flow cytometry analysis, including EPC flow cytometry analysis, have limitations that are only becoming realized through optimizing antibody panels, proper compensation and the appropriate selection of graphic displays10–12. The improvement in gating controls, such as fluorescence minus one controls, provide an accurate estimate of where the positive/negative threshold for a combination of fluorochromes is located13. Utilizing traditional logarithmic dot plots can disguise significant cell populations with low expression and lead to misreporting of actual event frequencies, but re-analysis of these cell populations with contour plot displays has exposed considerable differences in both the identification and frequency analysis of rare circulating cells14, 15. Bi-exponential scaling removes the artifact created by logarithmic scaling which causes a data spreading that is inaccurate since all events are transformed into a positive value and offers a unique ability to visualize all of the events collected11.
Other sources of error in rare event analysis include contamination of cell populations with false positive events and non-specific fluorescent event readings10. Specifically, monocytes, red blood cells, and dead cells auto-fluoresce and non-specifically bind antibodies16–18. A majority of previously published EPC flow cytometry protocols do not address these contaminants by starting with either a forward scatter threshold gate or a mononuclear cell gate8, 19–22. It is now clear that these approaches are insufficient for optimum polychromatic flow cytometry (PFC) analysis23. We attempted to address whether these errors had played a role in the lack of identifying the population of cells containing the ECFC by carefully incorporating all the best practices from the most current literature.
By incorporating PFC, we have found that professed CECs previously identified by conventional flow cytometry approaches7, are not endothelial cells, but mostly endothelial and platelet extracellular vesicles. Further, we also identify two functionally distinct circulating endothelial cell populations in peripheral blood, one a mature endothelial cell without clonogenic potential (CECs) and the other a true endothelial progenitor cell with clonal capability (ECFCs). Without the incorporation of PFC technology and the necessary functional assays verifying their identities, these rare events would not have been discovered. These advances provide a methodological platform for study of these cells in human clinical studies and models of vascular disease and tumor angiogenesis.
METHODS
Isolation of Mononuclear Cells
Peripheral blood samples (16–32mls) were collected from 20 healthy adult donors (10 male and 10 female, age range 20–40 years) and cord blood samples (20–100mls) were collected from 15 full-term newborns. The Institutional Review Board at the Indiana University School of Medicine approved all protocols, informed consent was obtained from adult donors, and cord blood collection was deemed exempt. Granulocyte colony stimulating factor mobilized peripheral blood CD34+ cells were provided through a Program of Excellence in Gene Therapy grant from Shelly Heimfeld at the Fred Hutchinson Cancer Research Centre, Seattle, WA, USA. Mononuclear cells were isolated using the CPT Vacutainer system by centrifuging at 1,600g for 30 minutes at room temperature. The mononuclear cells were removed and washed in phosphate buffered saline (PBS, Invitrogen, Grand Island, NY, USA) with 2% fetal bovine serum (FBS, Hyclone, Logan, UT, USA).
Extracellular Vesicle Enrichment
Peripheral blood collected in CPT Vacutainer tubes was centrifuged at 1,600g for 30 minutes. The serum and mononuclear cells were removed and centrifuged at 13,000g for 2 minutes. The supernatant was transferred to a new tube and centrifuged at 18,000g for 20 minutes to pellet the microvesicles. The microvesicle pellet was re-suspended in PBS with 2% FBS for antibody staining and flow cytometry analysis.
PFC Immunostaining
To assess the surface antigens of the mononuclear cells, we performed flow cytometry analysis as previously described15. The following primary conjugated monoclonal antibodies were used: CD14, CD31, CD34, CD45, AC133, glycophorin A (glyA, CD235a), LIVE/DEAD® (viability/apoptosis marker) and DAPI (nuclear stain). In order to resolve the rare and/or dim populations of interest, specific antigen and fluorochrome conjugate coupling was optimized for the six-antibody plus viability marker panel as previously described10, 15, 24, 25.
Mice
NOD/SCID mice, 6–8 weeks old, were housed according to protocols approved by the Laboratory Animal Research Facility and adhered strictly to National Institutes of Health guidelines and protocols were approved by Indiana University Animal Care and Use Board.
Transmission Electron Microscopy
To confirm extracellular vesicles populations, LIVE/DEAD®−CD14−glyACD31brightCD34+ CD45−AC133− cells obtained via fluorescent activated cell sorting (FACS) were allowed to lay on polycarbonate membranes (Electron Microscopy Sciences, Hatfield, PA, USA) and fixed. After washing the filters in buffer, they were dehydrated and embedded. Thin sections (80nm) were cut and stained with uranyl acetate and lead citrate. Specimens were viewed and photographed in a Philips CM100 transmission electron microscope (FEI Company, Hillsboro, OR, USA).
For immunoelectron microscopy analysis, LIVE/DEAD®−CD14−glyA−CD31+ CD34brightCD45−AC133− cells (i.e. ECFCs) obtained via FACS were spun down and fixed, dehydrated and embedded in Unicryl (Electron Microscopy Sciences) where thin sections (70–90nm) were mounted on formvar/carbon coated nickel grids. The grids were placed into primary polyclonal anti-von Willebrand factor (vWF) antibody (Abcam), after which a secondary antibody with 10nm gold particles was added as previously described26. The grids are viewed with a Tecnai G 12 Bio Twin transmission electron microscope.
Immunomagnetic Selection of Cord Blood CD146+CD45− Cells
Cord blood mononuclear cells were immunomagnetically selected using the human CD45 and CD146 MicroBeads and Magnetic Cell Sorting (MACS) system (Miltenyi Biotec) exactly as directed by the manufacturer. The CD45− fraction was isolated and then the CD146+ fraction was selected. The purity of MACS-separated sub-populations was confirmed by PFC acquisition and analysis. To compare the MACS separated fraction with the current gold standard protocol, a CD146 (Clone P1H12, BD Biosciences) Dynabead (Invitrogen) separation was performed (following the previously published protocol by Woywodt et al.27).
Verification of ECFCs within the CD146+CD45− Cells
To investigate the presence of ECFCs within MACS sub-populations, 50,000 CD45+ cells, CD146−CD45− cells or CD146+CD45− cells were plated into a 24-well collagen coated plate in cEGM-2 and cultured as previously described2. 30×106 cord blood mononuclear cells from the same donor were cultured in parallel as a positive control. ECFCs that arose were expanded and suspended in a collagen gel and implanted into NOD/SCID mice. One month later, animals were sacrificed and grafts excised and analyzed by immunohistochemistry as described previously28. A more detailed methodology can be found in the supporting information section.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism software, version 5.01 for Windows (GraphPad Software, San Diego, CA, USA). Data was tested for normality using the D'Agostino-Pearson normality test (alpha=0.05), and normal data sets were compared using two-tailed Student’s t test or one-way ANOVA.
RESULTS
Masking Cell Populations are Found when Incorporating PFC
To determine if exclusive use of the conventional flow cytometry gating strategies leads to contamination and misidentification of CECs, we co-stained peripheral blood mononuclear cells identified within the forward scatter/side scatter threshold gate with the standard CEC antibody panel (CD34, CD45, CD31, and AC133)7. In addition, we also co-stained mononuclear cells with antibodies to identify monocytes (CD14), red blood cells (glyA), and a viability marker (LIVE/DEAD®) to identify dead/apoptotic cells. By back-gating the glyA+ and LIVE/DEAD®+ cells over the mononuclear cell plot, it becomes clear that there is no way to remove these cells by scatter profile alone as the cells distribute throughout the mononuclear cell gate (Fig 1 and Supplemental Fig I). Therefore, exclusive use of a forward scatter threshold gate on mononuclear cells will not eliminate these contaminants and renders enumeration of purported CECs inaccurate. For more information on the problems associated with the previously published cytometry protocols, refer to reference29. A flow chart is also included (Figure 3) that compares the differences between conventional flow cytometry methods and PFC methods.
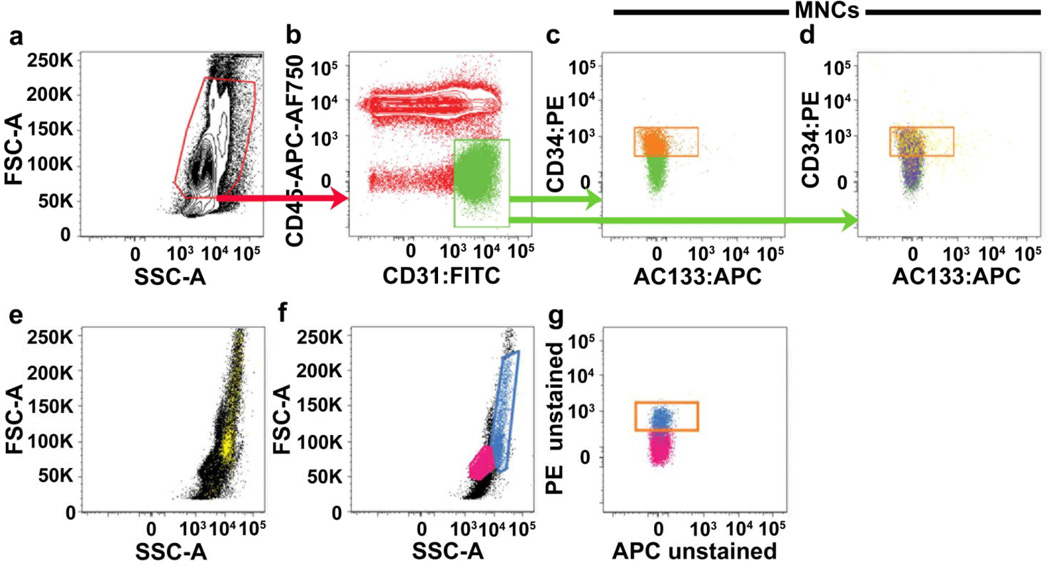
Figure 1. Threshold gating of mononuclear cells based on forward scatter and side scatter does not remove red blood cells, dead cell and monocyte contamination.
Representative PFC analysis of a CPT mononuclear cell preparation of peripheral blood either (a–e) stained with the six-antibody/viability marker panel (as defined in Methods), which includes glyA (red blood cell marker), CD14 (monocyte marker) and LIVE/DEAD® (cell viability marker) or (f–g) completely unstained. Mononuclear cells (red gate in a) are initially gated on a FSC/SSC plot in an attempt to exclude red blood cells, dead cells and debris, and sub-gated onto a bivariant antigen plot (b) for identification of CD31brightCD45− cells (green gate). CD31brightCD45− cells are further sub-gated to identify the CD34+AC133− subset (orange gate in c and d). In (d), glyA+ red blood cells and/or dead/apoptotic cells (purple events) and CD14+ monocytes (yellow events), contained within the initial mononuclear cell gate, are mapped onto the bi-variant plot and can clearly be seen distributed throughout the CD34+AC133− gate (orange gate). Monocytes are difficult to exclude based on their scatter profile. CD14+ monocytes (yellow cells mapped in e) can be seen distributed throughout the mononuclear cells when back-gated onto a forward scatter/side scatter plot (e). In analysis of unstained samples (f–g), cells that have the scatter profile of monocytes (blue gate in f and blue cells mapped onto g), are highly auto-fluorescent and contaminate the orange gate used for frequency analysis of CD34+AC133− cells. Lymphocytes are gated and mapped in pink for reference (f–g). Similar results were seen in 9 other samples from different donors.
Figure 3. Comparison of Conventional Flow Cytometry to Polychromatic Flow Cytometry.
Frequency Analysis and Characterization of CD31brightCD34+CD45−AC133− cells (CECs)
To determine the frequency and cellular identity of CECs, and to estimate the potential experimental error in analyzing this cell type with conventional flow cytometry applications, we compared analysis of CECs utilizing a previously established flow cytometry protocol7 and our recently optimized PFC strategy15. We isolated peripheral blood mononuclear cell samples from 10 healthy, young adults and stained them with the six monoclonal antibodies (CD34, CD45, CD31, AC133, glyA, and CD14) and a viability marker (LIVE/DEAD®) panel with fluorescence minus one or isotype controls15. Stained and fixed samples were acquired on a digital Becton Dickinson (BD) LSRII flow cytometer and assessed for CD31brightCD34+CD45−AC133− events (CECs) using two different analysis paths as shown in Figure 2.
Figure 2. Frequency analysis of CD31brightCD34+CD45−AC133− cells.
Two strategies for frequency analysis of CD31brightCD34+CD45−AC133− cells from peripheral blood stained with the six-antibody/viability marker panel are shown. In the first strategy (a–d), conventional flow cytometry techniques were utilized. In the second strategy (e–i), PFC techniques were utilized, including the removal of monocytes, red blood cells and dead cells that lead to a more uniform population with a tighter expression profile.
Similar to previously published EPC protocols5, 7 peripheral blood mononuclear cells were analyzed in logarithmic dot plots (Fig. 2a–d)7, and CEC identification was determined by placement of population gates, which were based on isotype controls (Fig. 2a–d)7. Mononuclear cells (Fig. 2a) were identified on a forward scatter/side scatter plot and sub-gated onto a bivariant antigen plot to identify CD31brightCD45− cells (Fig. 2b). CD31brightCD45− mononuclear cells were further sub-gated to identify the CD34+ sub-population (Fig. 2c). AC133 expression on the resulting CD31brightCD34+CD45− mononuclear cells was assessed on an AC133 histogram (Fig. 2d). In the first strategy (Fig. 2a–d), compensation was applied manually using singly stained cell controls as described7. Mononuclear cells from the same donor were also analyzed by excluding monocytes, red blood cells, and dead/apoptotic cells before incorporating bi-exponential contour plots to identify CD31brightCD34+CD45−AC133− events (Fig. 2e–i).
Single-color bead controls were used to set automated compensation before analysis. First, CD14− cells (Fig. 2e) were identified. All CD14− cells were then assessed for viability and glyA expression (Fig. 2f). CD14−glyA−LIVE/DEAD®− cells (Fig. 2f) were sub-gated onto a bivariant antigen plot to identify CD14−glyA−LIVE/DEAD®−CD31brightCD45− cells (Fig. 2g). Viable CD14−glyA−CD31brightCD45− cells were further sub-gated to identify the CD34+ sub-population (Fig. 2h). AC133 expression on the resulting viable CD14−glyA−CD31brightCD34+CD45− cells was evaluated on an AC133 histogram (Fig. 2i). To avoid biased gating error (false positives and negatives) which is inherent when utilizing multiple fluorochromes, regional gates were added based on fluorescence minus one gating controls to detect CECs13,15.
Comparison of the two methods revealed stark differences in the percentage of collected events that were CD31brightCD34+CD45−AC133− (Fig. 2h compared to Fig. 2c). When the four-antibody panel and logarithmic dot plots were used (Fig. 2a–d), 0.262±0.986% (mean±s.d., n=10, range 0.140–0.443) of collected events were CD31brightCD34+CD45−AC133−. Surprisingly, in cell preparations from the same donors analyzed in bi-exponential contour plots with the exclusion of monocytes, red blood cells and dead/apoptotic cells before analysis (Fig. 2e–i), CD31brightCD34+CD45−AC133- events constituted 0.150±0.0563% (mean±s.d., n=10, range 0.0844–0.260; logarithmic method vs. bi-exponential method, p=0.0058 by two-tailed, unpaired Student’s t test) of collected events.
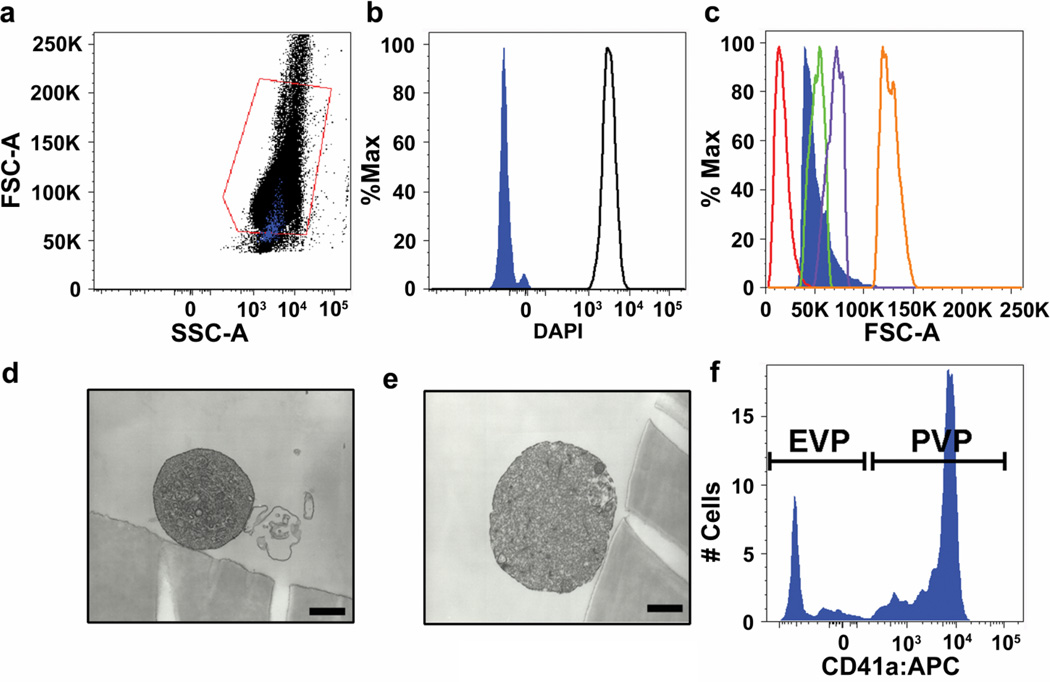
Given this observation, we next determined the cellular identity of CD14−glyA−LIVE/DEAD®−CD31brightCD34+CD45−AC133− events identified by PFC. Interestingly, back-gating of CD14−glyA−LIVE/DEAD®−CD31brightCD34+CD45−AC133− events (as identified in Fig. 2e–i) onto the original forward scatter/side scatter dot plot revealed their location spanning the lower margin of the mononuclear gate in a region that typically harbors small cells and debris and not endothelial cells (Fig. 4a). To determine the percentage of CD14−glyA10−LIVE/DEAD®−CD31brightCD34+CD45−AC133− events that were actual cells, we stained the population with a DAPI nuclear stain. Utilizing lymphocytes as a positive cellular control (open black in Fig. 4b), none of the CD14−glyA−LIVE/DEAD®−CD31brightCD34+CD45−AC133− events contained a nucleus, which confirmed that these events were not endothelial cells (Fig. 4b) as previously reported7. Extracellular vesicles including microvesicles do not contain a nucleus, are small in forward scatter/side scatter dot plots, and are shed from endothelial cells, platelets, lymphocytes, monocytes and other leukocytes30, 31. Since extracellular vesicles are biologically active and correlate with cardiovascular disease risk31–35, we tested whether the CD14−glyA−LIVE/DEAD®−CD31brightCD34+CD45−AC133− events were extracellular vesicles. Extracellular vesicles have also been shown to contaminate early EPC cultures and correlated with the number of colony forming units counted36. As shown in Figure 4c, we initially compared CD14−glyA−LIVE/DEAD®−CD31brightCD34+CD45−AC133− events with beads of standardized size in flow cytometry analysis and determined that CD31brightCD34+CD45−AC133− events were in the range of 1–5µm in size (solid blue in Fig. 4c), which is in the reported size range of extracellular vesicles31, 37.
Figure 4. Characterization of CD31brightCD34+CD45−AC133− cells.
CD14−glyA−LIVE/DEAD®− CD31brightCD34+CD45−AC133− events (as identified in Figure 1e–i) (solid blue in b) stain negatively for the nuclear dye DAPI. (c) CD14−glyA−LIVE/DEAD®−CD31brightCD34+CD45−AC133− events (solid blue in c) are in the range of 1–5µm in size. (d–e) Representative electron microscopy photomicrographs of sorted CD14−glyA−LIVE/DEAD®−CD31brightCD34+CD45−AC133− events. Scale bars represent 500nm. (f) CD41a+ events are platelet-derived microvesicles (PMV) and CD41a− events are endothelial-derived microvesicles (EMV).
After determination of size, we employed electron microscopy of sorted CD14−glyA−LIVE/DEAD®−CD31brightCD34+CD45−AC133− events to further characterize the identity of these events. Analysis of the CD14−glyA−LIVE/DEAD®−CD31brightCD34+CD45−AC133− events by electron microscopy revealed the morphology of microvesicles as previously described (Fig. 4d–e) 38–40. Microvesicles were identified by flow cytometry as leukocyte-, endothelial-, or platelet-shed particles by determining the number of CD45+CD41a− (leukocyte), CD45− CD31+CD34+CD41a− (endothelial), or CD41a+CD31+ (platelets) events41, 42. PFC analysis of the high-density centrifugation isolated microvesicles contained in the CD14−glyA−LIVE/DEAD®−CD31brightCD34+CD45−AC133− gate revealed a heterogeneous population of endothelial- and platelet microvesicles (Fig. 4f). Similar results were seen in 3 independent experiments using cells from different donors. Collectively, this data generated by the application of PFC demonstrates that the previous description of CECs was largely composed of endothelial- and platelet-derived microvesicles that were not detected as such by methods employed.
Prospective Identification and Isolation of Circulating ECFCs by PFC
ECFCs are rare circulating endothelial progenitor cells with clonogenic and in vivo vessel forming capacity, but CECs are thought to have limited proliferative potential2. Given this conundrum, we next sought initially to isolate ECFCs and/or CECs by PFC in human cord blood, since ECFCs are enriched 20 fold in cord blood compared to adult peripheral blood2. Previous studies demonstrate that ECFCs express CD34 and not CD45, and can be enriched by immunomagnetic selection for these defined cells (CD34+CD45−)43. Therefore, we stained mononuclear cells derived from adult peripheral blood and cord blood with antibodies directed against CD34 and CD45 to identify CD34+CD45− cells. Cord blood mononuclear cells harbored a small population of viable CD34+CD45− cells (Fig. 5b), which was not detectable in adult peripheral blood mononuclear cells (Fig. 5a) via PFC. Further, cord blood derived CD34+CD45− cells co-expressed the endothelial cell surface antigens CD31, CD146 and CD105 (Fig. 5c–e respectively), which were previously identified on cultured ECFCs2 and CECs44.
Figure 5. Prospective identification and isolation of circulating ECFCs by PFC and Immunomagnetic Selection.
Representative PFC analysis of (a) adult peripheral blood mononuclear cells and (b) cord blood mononuclear cells. Putative CD34+CD45− ECFCs are gated in green. (c–e) Cells (b gate) were analyzed for expression of (c) CD31, (d) CD146, and (e) CD105. (f–g) PFC analysis of cord blood mononuclear cells (f) prior to and (g) following isolation of CD146+CD45− cells via MACS. (h) Representative photomicrograph of an ECFC colony derived from the CD146+CD45− fraction of cord blood mononuclear cells 6 days after culture in endothelial-specific media. Scale bar represents 200µm. (i) Low (×1,900) and (j) high (×13,000) magnification electron microscopy of sorted cord blood LIVE/DEAD®−CD14−glyACD31brightCD34+CD45− AC133− cells, is consistent with endothelial morphology.
To determine whether this cell population would form endothelial cell colonies, cord blood derived CD146+CD45− cells were initially isolated by a two-step MACS procedure (CD45+ cell depletion followed by CD146+ enrichment of the CD45− fraction) instead of using FACS. Immunomagnetic selection was utilized since multiple experiments clearly demonstrated that ECFCs do not survive traditional FACS isolation methods (data not shown). CD146 was chosen over CD34 for the secondary selection as CD146 was the antigen of choice used in the previously published protocol that is widely viewed as the gold standard technique for counting circulating endothelial cells27. PFC analysis of cord blood mononuclear cells before (Fig. 5f) and after (Fig. 5g) immunomagnetic selection demonstrated that this procedure captured the entire CD34+CD45− cell population. We compared our MACS procedure with that of the previously published gold standard Dynabead CD146 protocol by Woywodt et al.,27 and found that the negative fraction following positive isolation still contained the CD31+CD34brightCD45− cells. We hypothesize that the reason these cells had not been detected by the Woywodt et al., method is that the Miltenyi CD146 uses a proprietary blend of monoclonal antibody clones, whereas we utilized a monoclonal P1H12 to attach to the Dynabead. Importantly, PFC analysis of the MACS cells confirmed that all CD146+CD45− cells also expressed CD34, CD31 and CD105, but not AC133 (data not shown).
We next cultured MACS CD146+CD45− cells in defined endothelial and hematopoietic cell culture conditions for colony formation. CD146+CD45− cells formed multiple ECFC colonies within 3–4 days (Fig. 5h) and never yielded hematopoietic progenitor cell colonies (n=10) (data not shown). These ECFCs were passaged and implanted in collagen gels which subsequently grew human vessels (Figure 6) as previously described45. To further confirm that the CD34+CD45− population identified by PFC prospectively identifies the ECFC at similar frequencies reported via MACS, we identified via FACS, CD34+CD45− cells and analyzed the sorted cells by electron microscopy. Electron microscopy of sorted cord blood LIVE/DEAD®− CD14−glyA−CD31+CD34brightCD45−AC133− cells is consistent with endothelial morphology on low (x1,900) and high (x13,000) magnification (Fig. 5i–j and Supplemental Fig. II). Similar results were seen in 4 cord blood samples from different donors. Thus, PFC identifies CD34+CD146+CD31+CD105+CD45− cells, which contain both highly proliferative ECFCs, and CECs with limited or no clonogenic potential.
Figure 6. Photomicrograph of human endothelial cells expressing CD31 forming blood vessels that contain mouse erythrocytes.
(a) Photomicrograph (original X60) of cellularized gels containing red blood cell perfused blood vessels stained with hematoxylin and eosin. (b) The vast majority of perfused blood vessel are lined by human endothelial cells (derived from the implanted ECFC) that express CD31 (brown stain). The scale bar represents 100µ. Similar results were seen in 4 control and immunomagnetic selected -isolated ECFC cultures.
DISCUSSION
We elucidate circulating endothelial cell populations by a PFC protocol which is able to identify cells based on cell surface antigen expression, as well as the prerequisite colony assays, including electron microscopy, and in vivo function, to definitively identify CECs and ECFCs. Both CECs and ECFCs are rare cells in the peripheral blood of healthy adults, thus by excluding the masking cell populations, specifically red blood cells (glyA), monocytes (CD14+) and dead/apoptotic (LIVE/DEAD®) cells, a more accurate cellular composition can be quantified. As shown by Figures 1 and 2, CD14+, glyA+, and LIVE/DEAD®+ cells cannot be gated out by size and lead to false positive events. In rare cell flow cytometry analysis, there is a statistically significant difference between the conventional and our PFC methodology in the number of positive CEC events. By incorporating a previously published conventional flow cytometry protocol for detection of human CECs7, it has now been clarified to identify both circulating platelet and endothelial extracellular vesicles.
Circulating extracellular vesicles are known modulators of intravascular coagulation, angiogenesis, cancer, metabolism, and immune function, and more recent data demonstrate that extracellular vesicles alter gene expression in cells via delivery of miRNA31, 46–49. As reported herein, the majority of circulating events previously described as CECs are platelet-derived extracellular vesicles, though numerous circulating cells release these elements. The presence of extracellular vesicles expressing the surface antigen profile CD31brightCD34+CD45−AC133− may correlate with various diseases that previously were attributable to CECs. Thus, the phenotypic population of CECs (CD31brightCD34+CD45−AC133−) may correlate with the patient disorder, though the actual cellular elements comprising those events and their mode of action may be different from those proposed in prior reports. Future studies are needed to specifically isolate the various cell-derived extracellular vesicles to define their function in human cardiovascular disease and cancer.
Furthermore, we report both a PFC protocol plus the requisite culture assays, to identify a population of cells containing the rare circulating ECFCs in human cord blood and peripheral blood. ECFCs with robust proliferative potential were identified in cord blood, while few, if any, ECFCs could be quantified in the peripheral blood of normal adults. This is consistent with the cell culture colony assays that have been performed using healthy adult peripheral blood. Proof that the samples contain proliferative cells was obtained when using CD146 beads and MACS to isolate ECFCs from the CD45− and not CD45+ fraction of blood samples. To isolate the population of cells containing the ECFC, CD146 was utilized over CD34 for the secondary selection step as it would correlate best with the previously published standard endothelial cell isolation protocol27. In addition, the population of cells we believed to be ECFCs stained positive for CD146 utilizing PFC (Figure 5d). Thus, ECFCs are circulating in cord blood and can be detected by the present PFC protocol. However, we found FACS isolation methods to be detrimental to the recovery of viable ECFCs (data not shown) and recommend immunomagnetic selection as the preferred mode for ECFC isolation. In contrast to the report of Delorme et al.,50 we verified that cord blood CD146+ cells that give rise to ECFCs are enriched in the CD45− rather than the CD45+ cell fraction. Differences in our data may have resulted from our CD45 depletion studies (removing T cells) prior to the CD146 selection.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Shawn Ahlfeld, MD at Indiana University School of Medicine for critical evaluation of this article. We acknowledge the assistance and state-of-the-art facilities from Sue Rice at the Flow Cytometry Core at the Indiana University Simon Cancer Center and Caroline Miller at the Electron Microscopy Center at Indiana University School of Medicine. We also acknowledge the In Vivo Therapeutics Core of the Indiana University Simon Cancer Center and the nursing staff at St. Vincent Hospital (Indianapolis, IN) for providing human cord blood samples for this study.
SOURCES OF FUNDING
NIH P50 NS052606 (D.A.I.); Department of Defense NF073122 (D.A.I.); Riley Children’s Foundation (D.A.I. and M.C.Y.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURES
All authors declare no conflict of interest.
REFERENCES
- 1.Mund JA, Ingram DA, Yoder MC, Case J. Endothelial progenitor cells and cardiovascular cell-based therapies. Cytotherapy. 2009;11:103–113. doi: 10.1080/14653240802714827. [DOI] [PubMed] [Google Scholar]
- 2.Ingram DA, Mead LE, Tanaka H, Meade V, Fenoglio A, Mortell K, Pollok K, Ferkowicz MJ, Gilley D, Yoder MC. Identification of a novel hierarchy of endothelial progenitor cells utilizing human peripheral and umbilical cord blood. Blood. 2004;104:2752–2760. doi: 10.1182/blood-2004-04-1396. [DOI] [PubMed] [Google Scholar]
- 3.Asahara T, Murohara T, Sullivan A, Silver M, van der Zee R, Li T, Witzenbichler B, Schatteman G, Isner JM. Isolation of putative progenitor endothelial cells for angiogenesis. Science. 1997;275:964–967. doi: 10.1126/science.275.5302.964. [DOI] [PubMed] [Google Scholar]
- 4.Urbich C, Dimmeler S. Endothelial progenitor cells: characterization and role in vascular biology. Circ Res. 2004;95:343–353. doi: 10.1161/01.RES.0000137877.89448.78. [DOI] [PubMed] [Google Scholar]
- 5.Bertolini F, Shaked Y, Mancuso P, Kerbel RS. The multifaceted circulating endothelial cell in cancer: towards marker and target identification. Nat Rev Cancer. 2006;6:835–845. doi: 10.1038/nrc1971. [DOI] [PubMed] [Google Scholar]
- 6.Ingram DA, Lien IZ, Mead LE, Estes M, Prater DN, Derr-Yellin E, DiMeglio LA, Haneline LS. In vitro hyperglycemia or a diabetic intrauterine environment reduces neonatal endothelial colony-forming cell numbers and function. Diabetes. 2008;57:724–731. doi: 10.2337/db07-1507. [DOI] [PubMed] [Google Scholar]
- 7.Duda DG, Cohen KS, Scadden DT, Jain RK. A protocol for phenotypic detection and enumeration of circulating endothelial cells and circulating progenitor cells in human blood. NatProtoc. 2007;2:805–810. doi: 10.1038/nprot.2007.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khan SS, Solomon MA, McCoy JP., Jr Detection of circulating endothelial cells and endothelial progenitor cells by flow cytometry. Cytometry B Clin Cytom. 2005;64:1–8. doi: 10.1002/cyto.b.20040. [DOI] [PubMed] [Google Scholar]
- 9.Mund JA, Case J. The ontogeny of endothelial progenitor cells through flow cytometry. Curr Opin Hematol. 2011 doi: 10.1097/MOH.0b013e328345a16a. [DOI] [PubMed] [Google Scholar]
- 10.Baumgarth N, Roederer M. A practical approach to multicolor flow cytometry for immunophenotyping. J Immunol Methods. 2000;243:77–97. doi: 10.1016/s0022-1759(00)00229-5. [DOI] [PubMed] [Google Scholar]
- 11.Parks DR, Roederer M, Moore WA. A new "Logicle" display method avoids deceptive effects of logarithmic scaling for low signals and compensated data. Cytometry A. 2006;69:541–551. doi: 10.1002/cyto.a.20258. [DOI] [PubMed] [Google Scholar]
- 12.Roederer M. Compensation is not dependent on signal intensity or on number of parameters. Cytometry. 2001;46:357–359. doi: 10.1002/cyto.10008. [DOI] [PubMed] [Google Scholar]
- 13.Roederer M. Spectral compensation for flow cytometry: visualization artifacts, limitations, and caveats. Cytometry. 2001;45:194–205. doi: 10.1002/1097-0320(20011101)45:3<194::aid-cyto1163>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 14.Herzenberg LA, Tung J, Moore WA, Herzenberg LA, Parks DR. Interpreting flow cytometry data: a guide for the perplexed. Nat Immunol. 2006;7:681–685. doi: 10.1038/ni0706-681. [DOI] [PubMed] [Google Scholar]
- 15.Estes ML, Mund JA, Ingram DA, Case J. Chapter 9 Identification of endothelial cells and progenitor cell subsets in human peripheral blood. Current protocols in cytometry / editorial board, J Paul Robinson, managing editor [et al] 2010;33(Unit 9) doi: 10.1002/0471142956.cy0933s52. 31-11. [DOI] [PubMed] [Google Scholar]
- 16.Duda DG, Jain RK, Willett CG. Antiangiogenics: the potential role of integrating this novel treatment modality with chemoradiation for solid cancers. J Clin Oncol. 2007;25:4033–4042. doi: 10.1200/JCO.2007.11.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McFarland DC, Zhang C, Thomas HC, T LR. Confounding effects of platelets on flow cytometric analysis and cell-sorting experiments using blood-derived cells. Cytometry A. 2006;69:86–94. doi: 10.1002/cyto.a.20207. [DOI] [PubMed] [Google Scholar]
- 18.Strijbos MH, Kraan J, den Bakker MA, Lambrecht BN, Sleijfer S, Gratama JW. Cells meeting our immunophenotypic criteria of endothelial cells are large platelets. Cytometry B Clin Cytom. 2007;72:86–93. doi: 10.1002/cyto.b.20156. [DOI] [PubMed] [Google Scholar]
- 19.Bertolini F, Paul S, Mancuso P, Monestiroli S, Gobbi A, Shaked Y, Kerbel RS. Maximum tolerable dose and low-dose metronomic chemotherapy have opposite effects on the mobilization and viability of circulating endothelial progenitor cells. Cancer Res. 2003;63:4342–4346. [PubMed] [Google Scholar]
- 20.Goon PK, Boos CJ, Stonelake PS, Blann AD, Lip GY. Detection and quantification of mature circulating endothelial cells using flow cytometry and immunomagnetic beads: a methodological comparison. Thrombosis and haemostasis. 2006;96:45–52. doi: 10.1160/TH06-04-0185. [DOI] [PubMed] [Google Scholar]
- 21.Mancuso P, Burlini A, Pruneri G, Goldhirsch A, Martinelli G, Bertolini F. Resting and activated endothelial cells are increased in the peripheral blood of cancer patients. Blood. 2001;97:3658–3661. doi: 10.1182/blood.v97.11.3658. [DOI] [PubMed] [Google Scholar]
- 22.Redondo S, Hristov M, Gordillo-Moscoso AA, Ruiz E, Weber C, Tejerina T. High-reproducible flow cytometric endothelial progenitor cell determination in human peripheral blood as CD34+/CD144+/CD3- lymphocyte sub-population. Journal of immunological methods. 2008;335:21–27. doi: 10.1016/j.jim.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 23.Perfetto SP, Chattopadhyay PK, Roederer M. Seventeen-colour flow cytometry: unravelling the immune system. Nature reviews. 2004;4:648–655. doi: 10.1038/nri1416. [DOI] [PubMed] [Google Scholar]
- 24.Mahnke YD, Roederer M. Optimizing a multicolor immunophenotyping assay. Clin Lab Med. 2007;27:469–485. doi: 10.1016/j.cll.2007.05.002. v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tung JW, Heydari K, Tirouvanziam R, Sahaf B, Parks DR, Herzenberg LA, Herzenberg LA. Modern flow cytometry: a practical approach. Clinics in laboratory medicine. 2007;27:453–468. doi: 10.1016/j.cll.2007.05.001. v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fukuchi M, Watanabe J, Kumagai K, Katori Y, Baba S, Fukuda K, Yagi T, Iguchi A, Yokoyama H, Miura M, Kagaya Y, Sato S, Tabayashi K, Shirato K. Increased von Willebrand factor in the endocardium as a local predisposing factor for thrombogenesis in overloaded human atrial appendage. Journal of the American College of Cardiology. 2001;37:1436–1442. doi: 10.1016/s0735-1097(01)01125-1. [DOI] [PubMed] [Google Scholar]
- 27.Woywodt A, Blann AD, Kirsch T, Erdbruegger U, Banzet N, Haubitz M, Dignat-George F. Isolation and enumeration of circulating endothelial cells by immunomagnetic isolation: proposal of a definition and a consensus protocol. J Thromb Haemost. 2006;4:671–677. doi: 10.1111/j.1538-7836.2006.01794.x. [DOI] [PubMed] [Google Scholar]
- 28.Li H, Gerald WL, Benezra R. Utilization of bone marrow-derived endothelial cell precursors in spontaneous prostate tumors varies with tumor grade. Cancer Res. 2004;64:6137–6143. doi: 10.1158/0008-5472.CAN-04-1287. [DOI] [PubMed] [Google Scholar]
- 29.Estes ML, Mund JA, Mead LE, Prater DN, Cai S, Wang H, Pollok KE, Murphy MP, An CS, Srour EF, Ingram DA, Jr, Case J. Application of polychromatic flow cytometry to identify novel subsets of circulating cells with angiogenic potential. Cytometry Part A : the journal of the International Society for Analytical Cytology. 2010;77:831–839. doi: 10.1002/cyto.a.20921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009 doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 31.Gyorgy B, Szabo TG, Pasztoi M, Pal Z, Misjak P, Aradi B, Laszlo V, Pallinger E, Pap E, Kittel A, Nagy G, Falus A, Buzas EI. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci. 2011;68:2667–2688. doi: 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deregibus MC, Cantaluppi V, Calogero R, Lo Iacono M, Tetta C, Biancone L, Bruno S, Bussolati B, Camussi G. Endothelial progenitor cell derived microvesicles activate an angiogenic program in endothelial cells by a horizontal transfer of mRNA. Blood. 2007;110:2440–2448. doi: 10.1182/blood-2007-03-078709. [DOI] [PubMed] [Google Scholar]
- 33.Diamant M, Tushuizen ME, Sturk A, Nieuwland R. Cellular microparticles: new players in the field of vascular disease? European journal of clinical investigation. 2004;34:392–401. doi: 10.1111/j.1365-2362.2004.01355.x. [DOI] [PubMed] [Google Scholar]
- 34.Leroyer AS, Tedgui A, Boulanger CM. Role of microparticles in atherothrombosis. Journal of internal medicine. 2008;263:528–537. doi: 10.1111/j.1365-2796.2008.01957.x. [DOI] [PubMed] [Google Scholar]
- 35.Steppich B, Mattisek C, Sobczyk D, Kastrati A, Schomig A, Ott I. Tissue factor pathway inhibitor on circulating microparticles in acute myocardial infarction. Thrombosis and haemostasis. 2005;93:35–39. doi: 10.1160/TH04-06-0393. [DOI] [PubMed] [Google Scholar]
- 36.Prokopi M, Pula G, Mayr U, Devue C, Gallagher J, Xiao Q, Boulanger CM, Westwood N, Urbich C, Willeit J, Steiner M, Breuss J, Xu Q, Kiechl S, Mayr M. Proteomic analysis reveals presence of platelet microparticles in endothelial progenitor cell cultures. Blood. 2009;114:723–732. doi: 10.1182/blood-2009-02-205930. [DOI] [PubMed] [Google Scholar]
- 37.Zubairov DM, Andrushko IA, Zubairova LD, Svintenok GY. Study of the mechanism of hemostatic effect of desmopressin. Bulletin of experimental biology and medicine. 2007;144:200–202. doi: 10.1007/s10517-007-0288-3. [DOI] [PubMed] [Google Scholar]
- 38.Combes V, Simon AC, Grau GE, Arnoux D, Camoin L, Sabatier F, Mutin M, Sanmarco M, Sampol J, Dignat-George F. In vitro generation of endothelial microparticles and possible prothrombotic activity in patients with lupus anticoagulant. J Clin Invest. 1999;104:93–102. doi: 10.1172/JCI4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hussein MA. Thromboembolism risk reduction in multiple myeloma patients treated with immunomodulatory drug combinations. Thrombosis and haemostasis. 2006;95:924–930. doi: 10.1160/TH06-02-0080. [DOI] [PubMed] [Google Scholar]
- 40.Kushak RI, Nestoridi E, Lambert J, Selig MK, Ingelfinger JR, Grabowski EF. Detached endothelial cells and microparticles as sources of tissue factor activity. Thrombosis research. 2005;116:409–419. doi: 10.1016/j.thromres.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 41.Faure V, Dou L, Sabatier F, Cerini C, Sampol J, Berland Y, Brunet P, Dignat-George F. Elevation of circulating endothelial microparticles in patients with chronic renal failure. J Thromb Haemost. 2006;4:566–573. doi: 10.1111/j.1538-7836.2005.01780.x. [DOI] [PubMed] [Google Scholar]
- 42.Preston RA, Jy W, Jimenez JJ, Mauro LM, Horstman LL, Valle M, Aime G, Ahn YS. Effects of severe hypertension on endothelial and platelet microparticles. Hypertension. 2003;41:211–217. doi: 10.1161/01.hyp.0000049760.15764.2d. [DOI] [PubMed] [Google Scholar]
- 43.Case J, Mead LE, Bessler WK, Prater D, White HA, Saadatzadeh MR, Bhavsar JR, Yoder MC, Haneline LS, Ingram DA. Human CD34+AC133+VEGFR-2+ cells are not endothelial progenitor cells but distinct, primitive hematopoietic progenitors. Exp Hematol. 2007;35:1109–1118. doi: 10.1016/j.exphem.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 44.Widemann A, Sabatier F, Arnaud L, Bonello L, Al-Massarani G, Paganelli F, Poncelet P, Dignat-George F. CD146-based immunomagnetic enrichment followed by multiparameter flow cytometry: a new approach to counting circulating endothelial cells. J Thromb Haemost. 2008;6:869–876. doi: 10.1111/j.1538-7836.2008.02931.x. [DOI] [PubMed] [Google Scholar]
- 45.Schechner JS, Nath AK, Zheng L, Kluger MS, Hughes CC, Sierra-Honigmann MR, Lorber MI, Tellides G, Kashgarian M, Bothwell AL, Pober JS. In vivo formation of complex microvessels lined by human endothelial cells in an immunodeficient mouse. Proc Natl Acad Sci U S A. 2000;97:9191–9196. doi: 10.1073/pnas.150242297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Freyssinet JM. Cellular microparticles: what are they bad or good for? J Thromb Haemost. 2003;1:1655–1662. doi: 10.1046/j.1538-7836.2003.00309.x. [DOI] [PubMed] [Google Scholar]
- 47.Gelderman MP, Simak J. Flow cytometric analysis of cell membrane microparticles. Methods Mol Biol. 2008;484:79–93. doi: 10.1007/978-1-59745-398-1_6. [DOI] [PubMed] [Google Scholar]
- 48.Morel O, Toti F, Hugel B, Freyssinet JM. Cellular microparticles: a disseminated storage pool of bioactive vascular effectors. Curr Opin Hematol. 2004;11:156–164. doi: 10.1097/01.moh.0000131441.10020.87. [DOI] [PubMed] [Google Scholar]
- 49.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, Xiao T, Schafer J, Lee ML, Schmittgen TD, Nana-Sinkam SP, Jarjoura D, Marsh CB. Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Delorme B, Basire A, Gentile C, Sabatier F, Monsonis F, Desouches C, Blot-Chabaud M, Uzan G, Sampol J, Dignat-George F. Presence of endothelial progenitor cells, distinct from mature endothelial cells, within human CD146+ blood cells. Thrombosis and haemostasis. 2005;94:1270–1279. doi: 10.1160/TH05-07-0499. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.