Abstract
Due to the unique redox potential of transition metals, many of these elements serve important roles as cofactors in numerous enzymes. However, the reactive nature of metal becomes an intracellular threat when these ions are present in excess. Therefore, all organisms require mechanisms for sensing small fluctuations in metal levels to maintain a controlled balance of uptake, efflux, and sequestration. The ability to sense metal ion concentration is especially important for the survival of pathogenic bacteria because host organisms can both restrict access to essential metals from invading pathogens and utilize the innate toxicity of certain metals for bacterial killing. Host-induced metal ion fluctuations must be rapidly sensed by pathogenic bacteria so that they can activate metal transport systems, alter their physiology to accommodate differences in metal concentrations, and regulate the expression of virulence factors.
Introduction
Metal starvation signals the up-regulation of acquisition systems [1] while metal excess activates the expression of efflux pumps and other proteins involved in metal resistance [2*]. Additionally, numerous physiological processes in bacteria are controlled in response to fluctuations in ion concentrations, resulting in decreased expression of metal-dependent enzymes upon metal deprivation [3]. This review will highlight recent advances in iron- and copper-responsive metalloregulation in a number of Gram-positive pathogens. The ability to sense fluctuations in these metals is of particular importance to pathogens because host organisms restrict iron availability [4,5] and induce copper toxicity [6**,7**].
Iron metalloregulation: A pathogen’s response to host-induced metal restriction
Vertebrate hosts restrict access to essential nutrients from invading pathogens in a phenomenon known as “nutritional immunity” [4]. Iron sequestration by the host is the archetypal form of nutritional immunity. The mechanisms utilized by the vertebrate host to restrict iron-access to pathogens include a physiological pH that results in low iron solubility, intracellular localization of iron, and storage of iron within iron-binding proteins. The net result of these mechanisms creates an environment virtually devoid of free iron with greater than 90% of iron residing within host cells [1,5].
Highly efficient iron acquisition machinery is essential to the survival of invading pathogens in order to support the function of key iron-dependent enzymatic processes [1]. Many proteins of the respiratory chain possess iron-sulfur clusters, peroxidases and cytochromes utilize the iron-containing molecule heme as a prosthetic group, and other enzymes such as ribonucleotide reductase require iron to catalyze their reactions [8]. While iron is important to the survival of almost all organisms, iron excess produces extreme toxicity due to the ability of this element to catalyze the Fenton reaction, leading to the generation of hydroxyl radicals that damage biological molecules [9]. Therefore, intracellular iron levels are tightly monitored through the action of metalloregulatory proteins.
Numerous families of metalloregulatory proteins have been discovered and characterized over the past few decades. Metalloregulatory families capable of sensing intracellular iron levels include the ferric uptake regulator (Fur) and the diphtheria toxin repressor (DtxR) [2*]. Fur family proteins are typically utilized for iron-sensing in low-G+C Gram-positive microorganisms, and DtxR regulators are more often found in high-G+C Gram-positive bacteria [10]. Some pathogens such as Mycobacterium tuberculosis possess both Fur and DtxR homologues [11]. Both of these metalloregulators bind to DNA operator sites upon association with their cognate metal ligands. Fur and DtxR proteins typically function as repressors, shutting down gene expression upon DNA binding [2*]. However, in a few instances, these metalloregulators have been found to activate the expression of certain genes [12,13].
Fur and DtxR control the expression of numerous gene categories in response to iron starvation including iron acquisition systems [10]. In addition to regulating iron uptake, the DtxR family proteins induce the expression of virulence factors in response to metal deprivation, including the diphtheria toxin of Corynebacterium diphtheriae for which this metalloregulatory protein class is named [14]. While Fur also regulates virulence [15*,16], of particular note is the ability of this protein family to redirect central metabolism from respiration to fermentation in order to alter cellular iron requirements [17,3]. Specific iron-containing respiration enzymes that are repressed in response to iron-starvation include succinate dehydrogenase, aconitate hydratase, and fumarate hydratase. The repression of iron-dependent enzymes upon iron-deprivation has been termed the “iron-sparing response” [3,18].
Because iron-deprivation conditions are directly relevant to the host environment, several recent analyses of transcriptomic and/or proteomic profiles under iron-limiting conditions were performed to identify iron-responsive genes in Staphylococcus aureus [17], Mycobacterium tuberculosis [19], Streptococcus pneumoniae [20], Bacillus anthracis [21], and Listeria monocytogenes [22]. These studies revealed Fur- and/or DtxR-regulated genes as well as iron-responsive genes that may be controlled independently of these known iron-sensing metalloregulators.
An analysis of the S. aureus proteome under conditions of iron deprivation and upon deletion of the fur gene revealed global changes in gene expression and an overall switch from respiration to fermentation [17]. This study demonstrated that this metabolic change not only alters the biological iron requirement of this organism, but also produces an accumulation of acid end products which alter the local pH and allow for iron release from host transferrin.
The transcriptomic analyses of S. pneumoniae, B. anthracis, and L. monocytogenes undergoing iron-starvation reveal similar iron-sparing responses in which iron-dependent proteins are down-regulated [20,21,22]. These studies also uncovered additional, unexpected physiological responses to iron-deprivation. For example, M. tuberculosis undergoes significant changes in lipid content [19] and protein turnover rate [23] in response to fluctuations in iron levels.
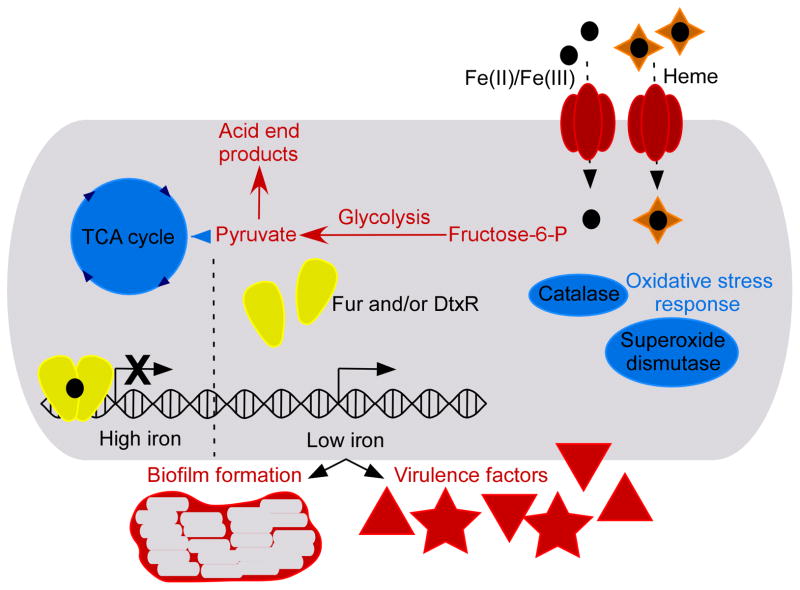
In addition to altering physiologic processes, bacterial pathogens also modulate production of virulence factors upon changes in iron availability. For example, biofilm formation occurs in response to iron limitation through Fur-dependent and Fur-independent mechanisms in S. aureus [24,25]. Similarly, biofilm formation proteins of S. pneumoniae are also up-regulated upon iron-deprivation [20]. The transcriptomic studies of B. anthracis revealed numerous iron-regulated virulence factors including several putative internalin genes that are important during infection [21]. The Fur protein of S. aureus controls the expression of global regulators such as Sae, Agr, and Rot which are known to regulate the expression of virulence factors [16]. Therefore, virulence gene expression in this organism is particularly responsive to iron levels. Deletion of the S. aureus fur gene results in overall reduced virulence in a mouse pneumonia model, underscoring the importance of iron-sensing in the host environment [15*]. Many of the gene categories and physiological processes regulated in response to fluctuations of intracellular iron levels in Gram-positive bacteria are highlighted in Figure 1.
Figure 1. Iron metalloregulation in Gram-positive pathogens.
Fur/DtxR iron-sensors are highlighted in yellow. The figure displays their most common function as iron-responsive repressors; however, these proteins can also act as activators in certain instances. Various physiological processes and proteins known to be regulated in response to iron fluctuations in one or more of the organisms highlighted in this review are depicted. Red coloration indicates that these proteins or processes are up-regulated under iron limitation while blue indicates repression in low iron. Black circles represent iron atoms and orange diamonds are heme.
Copper metalloregulation: A pathogen’s response to host-induced metal toxicity
Like iron, the reversible oxidation of copper contributes to its usefulness as an enzymatic cofactor as well as its toxicity due to the fact that copper generates reactive oxygen species by participating in Fenton-like chemistry [26]. Additionally, copper induces thiol oxidation, leading to sulfhydryl depletion [26,27**]. The final component of copper toxicity can be attributed to the fact that this element is highly competitive for binding sites in metalloproteins [8,2*]. Trace amounts of uncomplexed copper will displace the correct metal cofactors from essential enzymes [28]; therefore, bacteria must maintain an intracellular environment that is virtually devoid of free copper.
Recent studies demonstrated that macrophages have evolved the ability to harness the bactericidal activity of copper [6**]. Macrophages traffic a copper-specific P1 type ATPase, ATP7A, from the Golgi apparatus to phagosomal compartments upon stimulation with proinflammatory agents in order to mediate bacterial killing. New data also suggest that other host environmental niches may be rich in copper. For example, respiratory pathogens such as S. pneumoniae and M. tuberculosis activate a copper stress response in the nasopharynx and the lungs of mice [7**,29]. Therefore, the ability to sense copper fluctuations is critical for pathogenic organisms.
A number of metalloregulatory protein families are capable of sensing fluctuations in intracellular copper levels including ArsR-SmtB, CsoR, and CopY. These metalloregulators control gene expression via de-repression in which metal-binding results in dissociation of the transcriptional regulator from the DNA strand and subsequently allows gene transcription [2*]. In addition to these traditional metalloregulators, two-component systems capable of sensing extracellular copper concentration have been identified in a few bacteria [30,31].
The metalloregulatory mechanisms utilized for copper-sensing within Gram-positive pathogens are diverse. CsoR family copper sensors have been shown to be important in M. tuberculosis [29,32*], S. aureus [33**,34], and L. monocytogenes [35]. Copper-sensing is accomplished through the action of a CopY transcriptional repressor in S. pneumoniae [7**], Enterococcus hirae [36], and Enterococcus faecalis [37]. Finally, Corynebacterium glutamicum, a model organism for studying processes in pathogenic actinomycetes, utilizes a two-component system known as CopRS for sensing extracellular copper levels [31]. CopRS is highly conserved in the pathogenic bacterium C. diphtheriae.
Because various niches within the host environment contain toxic levels of copper, the ability to sequester and export this metal is particularly important in pathogenic bacteria. S. aureus [38,33**], L. monocytogenes [35], S. pneumoniae [7**] and M. tuberculosis [39,40] each encode copper metallochaperones and efflux pumps in order to counter the toxic effects of copper. Deletion of copper detoxification systems results in decreased virulence in S. pneumoniae and M. tuberculosis [7**,39,40]. Interestingly, deletion of an important copper efflux pump in L. monocytogenes does not impact its virulence in a mouse model, indicating that not all host niches are equally copper-rich [35]. Since copper excess is directly relevant to the conditions experienced within the host environment, global transcriptomic responses to copper toxicity have been analyzed for M. tuberculosis [41], S. aureus [27**], E. faecalis [37], and C. glutamicum [31]. All of these analyses reveal up-regulation of efflux and sequestration systems upon copper exposure but other copper-regulated genes vary from organism to organism.
The copper-stress response of M. tuberculosis involves a set of 30 genes, many of which function to protect against oxidative stress. This finding indicates that the major component of copper toxicity in M. tuberculosis is cellular damage caused by the generation of reactive oxygen species. Other copper responsive genes of M. tuberculosis include putative transcriptional regulators of various metalloregulatory families that may represent uncharacterized copper sensors within this organism [41]. Studies have demonstrated that copper sensing is accomplished through at least two CsoR family regulators in M. tuberculosis, underscoring the importance of copper-sensing in bacterial pathogens [32*].
The S. aureus global response to copper is similar to that of M. tuberculosis, including genes involved in copper efflux, protection against oxidative stress, and the misfolded protein response. Interestingly, S. aureus also decreases the expression of global virulence regulators sae and agr upon copper exposure. These virulence regulators control biofilm formation and experimental evidence shows that the presence of copper reduces biofilm development [27**]. This observation provides an explanation for the effectiveness of copper surfaced catheters against the formation of S. aureus biofilms [27**,42].
The copper stimulon of S. pneumoniae differs from that of M. tuberculosis and S. aureus in that expression of genes involved in oxidative stress and misfolded protein responses is not altered. This indicates that the source of copper toxicity is not the same for all Gram-positive pathogens. Instead, the copper stress response of S. pneumoniae includes expression of virulence factors such as StrH [7**]. StrH is an N-acetylglucosaminidase important for the evasion of opsonization and neutrophil-mediated killing [43].
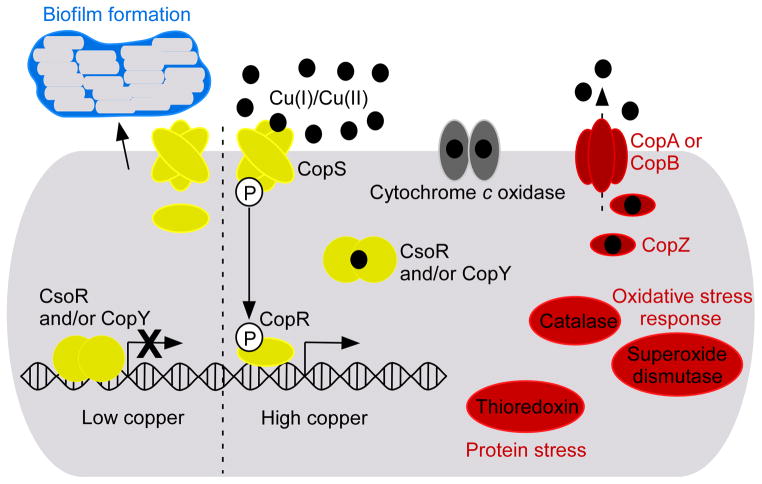
Copper-regulated genes of C. glutamicum are also of particular interest because this organism responds to copper excess by up-regulating its cytochrome c maturation processes [31]. This response is significant because cytochrome c is a copper-dependent enzyme. This observation is proof-of-principal that bacteria not only alter central metabolic pathways in order to accommodate metal deprivation but also regulate metabolic processes to take advantage of metal availability. Many of the major gene categories regulated in response to copper excess in various Gram-positive bacteria are highlighted in Figure 2.
Figure 2. Copper metalloregulation in Gram-positive pathogens.
CsoR, CopY, and CopRS copper-sensors are highlighted in yellow. Various proteins known to be regulated in response to copper excess in one or more of the organisms mentioned in this review are depicted. Red coloration indicates that these proteins are up-regulated by copper excess. Blue coloration indicates repression in high copper. Cytochrome c oxidase has been given a gray coloration because this protein is up-regulated by copper excess in C. glutamicum [31] but it is repressed by copper in M. tuberculosis [41]. Black circles represent copper atoms.
Conclusions
In addition to iron- and copper-metalloregulation, numerous recent advances have been made in studies focused on manganese [44], magnesium [45,46], and zinc [47,48] homeostasis in Gram-positive microorganisms. These studies are of particular importance in the field of pathogenesis because the concentrations of each of these metals are tightly controlled within the vertebrate host [4,49,50,51,52]. Metal ion homeostasis is important for all bacteria. However, the host environment represents a battlefield in which metal ion fluctuations are utilized as a tool for killing invading pathogens. Therefore, maintenance of metal homeostasis represents a particular challenge to pathogenic organisms. Studies focusing on metal ion homeostasis in Gram-positive pathogens are uncovering metal-regulated virulence factors and metabolic processes that are critical for the survival of invading microorganisms and may ultimately yield novel drug targets.
Highlights.
Pathogenic organisms encounter metal stresses in the host environment.
Pathogens alter their physiology to accommodate iron restriction within the host.
Copper excess within the host induces multiple forms of toxicity in bacteria.
Acknowledgments
We would like to thank the members of the Skaar laboratory for critical reading of this manuscript. Work in the Skaar laboratory is supported by grant numbers AI069233, AI073843, and AI091771 from the National Institutes of Health. CAW is supported by grant number T32HL094296 from the National Heart, Lung, and Blood Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, and Blood Institute or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hammer ND, Skaar EP. Molecular mechanisms of Staphylococcus aureus iron acquisition. Annu Rev Microbiol. 2011 doi: 10.1146/annurev-micro-090110-102851. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2*.Osman D, Cavet JS. Bacterial metal-sensing proteins exemplified by ArsR-SmtB family repressors. Nat Prod Rep. 2010;27:668–680. doi: 10.1039/b906682a. This review provides a comprehensive overview of all metalloregulatory classes in addition to a detailed biochemical description of the ArsR-SmtB protein family. [DOI] [PubMed] [Google Scholar]
- 3.Gaballa A, Antelmann H, Aguilar C, Khakh SK, Song K, Smaldone GT, Helmann JD. The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc Natl Acad Sci U S A. 2008;105:11927–11932. doi: 10.1073/pnas.0711752105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kehl-Fie TE, Skaar EP. Nutritional immunity beyond iron: a role for manganese and zinc. Curr Opin Chem Biol. 2010;14:218–224. doi: 10.1016/j.cbpa.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinberg ED. Iron availability and infection. Biochim Biophys Acta. 2009;1790:600–605. doi: 10.1016/j.bbagen.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 6**.White C, Lee J, Kambe T, Fritsche K, Petris MJ. A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J Biol Chem. 2009;284:33949–33956. doi: 10.1074/jbc.M109.070201. The data presented in this article elegantly demonstrate the mechanism employed by macrophages to accumulate bactericidal levels of copper within the phagosome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7**.Shafeeq S, Yesilkaya H, Kloosterman TG, Narayanan G, Andrew PW, Kuipers OP, Morrissey JA. The cop operon is required for copper homeostasis and contributes to virulence in Streptococcus pneumoniae. Mol Microbiol. 2011 doi: 10.1111/j.1365-2958.2011.07758.x. [Epub ahead of print]. This paper not only characterizes the copper stress response of S. pneumoniae but also demonstrates that this response is up-regulated during mouse infection and is required for full virulence. [DOI] [PubMed] [Google Scholar]
- 8.Waldron KJ, Robinson NJ. How do bacterial cells ensure that metalloproteins get the correct metal? Nat Rev. 2009;6:25–35. doi: 10.1038/nrmicro2057. [DOI] [PubMed] [Google Scholar]
- 9.Touati D. Iron and oxidative stress in bacteria. Arch Biochem Biophys. 2000;373:1–6. doi: 10.1006/abbi.1999.1518. [DOI] [PubMed] [Google Scholar]
- 10.Somerville GA, Proctor RA. At the crossroads of bacterial metabolism and virulence factor synthesis in staphylococci. Microbiol Mol Biol Rev. 2009;73:233–248. doi: 10.1128/MMBR.00005-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agranoff D, Krishna S. Metal ion transport and regulation in Mycobacterium tuberculosis. Front Biosci. 2004;9:2996–3006. doi: 10.2741/1454. [DOI] [PubMed] [Google Scholar]
- 12.Delany I, Rappuoli R, Scarlato V. Fur functions as an activator and as a repressor of putative virulence genes in Neisseria meningitidis. Mol Microbiol. 2004;52:1081–1090. doi: 10.1111/j.1365-2958.2004.04030.x. [DOI] [PubMed] [Google Scholar]
- 13.Brune I, Werner H, Hüser AT, Kalinowski J, Pühler A, Tauch A. The DtxR protein acting as dual transcriptional regulator directs a global regulatory network involved in iron metabolism of Corynebacterium glutamicum. BMC Genomics. 2006;9:7–21. doi: 10.1186/1471-2164-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyd J, Oza MN, Murphy JR. Molecular cloning and DNA sequence analysis of a diphtheria tox iron-dependent regulatory element (dtxR) from Corynebacterium diphtheriae. Proc Natl Acad Sci U S A. 1990;87:5968–5972. doi: 10.1073/pnas.87.15.5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15*.Torres VJ, Attia AS, Mason WJ, Hood MI, Corbin BD, Beasley FC, Anderson KL, Stauff DL, McDonald WH, Zimmerman LJ, et al. Staphylococcus aureus Fur regulates the expression of virulence factors that contribute to the pathogenesis of pneumonia. Infect Immun. 2010;78:1618–1628. doi: 10.1128/IAI.01423-09. This paper demonstrates that the ability to respond to iron-sufficient conditions in addition to iron-limited conditions is critical in an S. aureus pneumonia model. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson M, Sengupta M, Purves J, Tarrant E, Williams PH, Cockayne A, Muthaiyan A, Stephenson R, Ledala N, Wilkinson BJ, et al. Fur is required for the activation of virulence gene expression through the induction of the sae regulatory system in Staphylococcus aureus. Int J Med Microbiol. 2011;301:44–52. doi: 10.1016/j.ijmm.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman DB, Stauff DL, Pishchany G, Whitwell CW, Torres VJ, Skaar EP. Staphylococcus aureus redirects central metabolism to increase iron availability. PLoS Pathog. 2006;2:e87. doi: 10.1371/journal.ppat.0020087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janagama HK, Kumar S, Bannantine JP, Kugadas A, Jagtap P, Higgins L, Witthuhn B, Sreevatsan S. Iron-sparing response of Mycobacterium avium subsp. Paratuberculosis is strain dependent. BMC Microbiol. 2010;10:268. doi: 10.1186/1471-2180-10-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bacon J, Dover LG, Hatch KA, Zhang Y, Gomes JM, Kendall S, Wernisch L, Stoker NG, Butcher PD, Besra GS, Marsh PD. Lipid composition and transcriptional response of Mycobacterium tuberculosis grown under iron-limitation in continuous culture: identification of a novel wax ester. Microbiology. 2007;153:1435–1444. doi: 10.1099/mic.0.2006/004317-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nanduri B, Shah P, Ramkumar M, Allen EB, Swiatlo E, Burgess SC, Lawrence ML. Quantitative analysis of Streptococcus pneumoniae TIGR4 response to in vitro iron restriction by 2-D LC ESI MS/MS. Proteomics. 2008;8:2104–2114. doi: 10.1002/pmic.200701048. [DOI] [PubMed] [Google Scholar]
- 21.Carlson PE, Jr, Carr KA, Janes BK, Anderson EC, Hanna PC. Transcriptional profiling of Bacillus anthracis Sterne (34F2) during iron starvation. PLoS One. 2009;4:e6988. doi: 10.1371/journal.pone.0006988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ledala N, Sengupta M, Muthaiyan A, Wilkinson BJ, Jayaswal RK. Transcriptome response of Listeria monocytogenes to iron limitation and fur mutation. Appl Environ Microbiol. 2010;76:406–416. doi: 10.1128/AEM.01389-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao PK, Rodriguez M, Smith I, Li Q. Protein dynamics in iron-starved Mycobacterium tuberculosis revealed by turnover and abundance measurement using Hybrid-Linear Ion Trap-Fourier Transform Mass Spectrometry. Anal Chem. 2008;80:6860–6869. doi: 10.1021/ac800288t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson M, Cockayne A, Williams PH, Morrisey JA. Iron-responsive regulation of biofilm formation in Staphylococcus aureus involves Fur-dependent and Fur-independent mechanisms. J Bacteriol. 2005;187:8211–8215. doi: 10.1128/JB.187.23.8211-8215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson M, Cockayne A, Morrisey JA. Iron-regulated biofilm formation in Staphylococcus aureus Newman requires ica and the secreted protein Emp. Infect Immun. 2008;76:1756–1765. doi: 10.1128/IAI.01635-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Solioz M, Abicht HK, Mermod M, Mancini S. Response of Gram-positive bacteria to copper stress. J Biol Inorg Chem. 2010;15:3–14. doi: 10.1007/s00775-009-0588-3. [DOI] [PubMed] [Google Scholar]
- 27**.Baker J, Sitthisak S, Sengupta M, Johnson M, Jayaswal RK, Morrissey JA. Copper stress induces a global stress response in Staphylococcus aureus and represses sae and agr expression and biofilm formation. Appl Environ Microbiol. 2010;76:150–160. doi: 10.1128/AEM.02268-09. This article uncovers an interesting physiological response to copper stress in S. aureus which provides further explanation for the usefulness of copper as an antimicrobial agent. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macomber L, Imlay JA. The iron-sulfur cluster of dehydratases are primary intracellular targets of copper toxicity. Proc Natl Acad Sci U S A. 2009;106:8344–8349. doi: 10.1073/pnas.0812808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu T, Ramesh A, Ma Z, Ward SK, Zhang L, George GN, Talaat AM, Sacchettini JC, Giedroc DP. CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat Chem Biol. 2007;3:60–68. doi: 10.1038/nchembio844. [DOI] [PubMed] [Google Scholar]
- 30.Munson GP, Lam DL, Outten FW, O’Halloran TV. Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J Bacteriol. 2000;182:5864–5871. doi: 10.1128/jb.182.20.5864-5871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schelder S, Zaade D, Litsanov B, Bott M, Brocker M. The two-component signal transduction system CopRS of Corynebacterium glutamicum is required for adaptation to copper-excess stress. PloS One. 2011;6:e22143. doi: 10.1371/journal.pone.0022143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32*.Festa RA, Jones MB, Butler-Wu S, Sinsimer D, Gerads R, Bishai WR, Peterson SN, Darwin KH. A novel copper-responsive regulon in Mycobacterium tuberculosis. Mol Microbiol. 2011;79:133–148. doi: 10.1111/j.1365-2958.2010.07431.x. The discovery of multiple copper-responsive metalloregulators in a single bacterium demonstrates how crucial copper-sensing is for pathogenic organisms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33**.Grossoehme N, Kehl-Fie TE, Ma Z, Adams KW, Cowart DM, Scott RA, Skaar EP, Giedroc DP. Control of copper resistance and inorganic sulfur metabolism by paralogous regulators in Staphylococcus aureus. J Biol Chem. 2011;286:13522–13531. doi: 10.1074/jbc.M111.220012. The disparate ligand recognition and regulatory networks of two structurally similar CsoR-like family proteins highlight the need for a detailed analysis following identification of putative metalloregulatory proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baker J, Sengupta M, Jayaswal RK, Morrissey JA. The Staphylococcus aureus CsoR regulates both chromosomal and plasmid-encoded copper resistance mechanisms. Environ Microbiol. 2011;13:2495–2507. doi: 10.1111/j.1462-2920.2011.02522.x. [DOI] [PubMed] [Google Scholar]
- 35.Corbett D, Schuler S, Glenn S, Andrew PW, Cavet JS, Roberts IS. The combined actions of the copper-responsive repressor CsoR and copper-metallochaperone CopZ modulate CopA-mediated copper efflux in the intracellular pathogen Listeria monocytogenes. Mol Microbiol. 2011;81:457–472. doi: 10.1111/j.1365-2958.2011.07705.x. [DOI] [PubMed] [Google Scholar]
- 36.Portmann R, Poulsen KR, Wimmer R, Solioz M. CopY-like copper inducible repressors are putative ‘winged helix’ proteins. Biometals. 2006;19:61–70. doi: 10.1007/s10534-005-5381-3. [DOI] [PubMed] [Google Scholar]
- 37.Reyes-Jara A, Latorre M, Lopez G, Bourgogne A, Murray BE, Cambiazo V, Gonzalez M. Genome-wide transcriptome analysis of the adaptive response of Entercoccus faecalis to copper exposure. Biometals. 2010;23:1105–1112. doi: 10.1007/s10534-010-9356-7. [DOI] [PubMed] [Google Scholar]
- 38.Sitthisak S, Knutsson L, Webb JW, Jayaswal RK. Molecular characterization of the copper transport system in Staphylococcus aureus. Microbiology. 2007;153:4274–4283. doi: 10.1099/mic.0.2007/009860-0. [DOI] [PubMed] [Google Scholar]
- 39.Ward SK, Abomoelak B, Hoye EA, Steinberg H, Talaat AM. CtpV: a putative copper exporter required for full virulence of Mycobacterium tuberculosis. Mol Microbiol. 2010;77:1096–1110. doi: 10.1111/j.1365-2958.2010.07273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolschendorf F, Ackart D, Shrestha TB, Hascall-Dove L, Nolan S, Lamichhane G, Wang Y, Bossmann SH, Basaraba RJ, Niederweis M. Copper resistance is essential for virulence of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2011;108:1621–1626. doi: 10.1073/pnas.1009261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ward SK, Hoye EA, Talaat AM. The global responses of Mycobacterium tuberculosis to physiological levels of copper. J Bacteriol. 2008;190:2939–2946. doi: 10.1128/JB.01847-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McLean RJ, Hussain AA, Sayer M, Vincent PJ, Hughes DJ, Smith TJ. Antibacterial activity of multilayer silver-copper surface films on catheter material. Can J Microbiol. 1993;39:895–899. doi: 10.1139/m93-134. [DOI] [PubMed] [Google Scholar]
- 43.Dalia AB, Standish AJ, Weiser JN. Three surface exoglycosidases from Streptococcus pneumoniae, NanA, BgaA, and StrH, promote resistance to opsonophagocytic killing by human neutrophils. Infect Immun. 2010;78:2108–2116. doi: 10.1128/IAI.01125-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosch JW, Gao G, Ridout G, Wang YD, Tuomanen EI. Role of the manganese efflux system mntE for signaling and pathogenesis in Streptococcus pneumoniae. Mol Microbiol. 2009;72:12–25. doi: 10.1111/j.1365-2958.2009.06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dann CE, 3rd, Wakeman CA, Sieling CL, Baker SC, Irnov I, Winkler WC. Structure and mechanism of a metal-sensing regulatory RNA. Cell. 2007;130:878–892. doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- 46.Ramesh A, Winkler WC. Magnesium-sensing riboswitches in bacteria. RNA Biol. 2010;7:77–83. doi: 10.4161/rna.7.1.10490. [DOI] [PubMed] [Google Scholar]
- 47.Smith KF, Bibb LA, Schmitt MP, Oram DM. Regulation and activity of a zinc uptake regulator, Zur, in Corynebacterium diphtheriae. J Bacteriol. 2009;191:1595–1603. doi: 10.1128/JB.01392-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jacobsen FE, Kazmierczak KM, Lisher JP, Winkler ME, Giedroc DP. Interplay between manganese and zinc homeostasis in the human pathogen Streptococcus pneumoniae. Metallomics. 2011;3:38–41. doi: 10.1039/c0mt00050g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buchmeier N, Blanc-Potard A, Ehrt S, Piddington D, Riley L, Groisman EA. A parallel intraphagosomal survival strategy shared by Mycobacterium tuberculosis and Salmonella enterica. Mol Microbiol. 2000;35:1375–1382. doi: 10.1046/j.1365-2958.2000.01797.x. [DOI] [PubMed] [Google Scholar]
- 50.Lucchini S, Liu H, Jin Q, Hinton JC, Yu J. Transcriptional adaptation of Shigella flexneri during infection of macrophages and epithelial cells: insights into the strategies of a cytosolic bacterial pathogen. Infect Immun. 2005;73:88–102. doi: 10.1128/IAI.73.1.88-102.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, Anderson KL, Dattilo BM, Dunman PM, Gerads R, et al. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science. 2008;319:962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- 52.Kehl-Fie TE, Chitayat S, Hood MI, Damo S, Restrepo N, Garcia C, Munro KA, Chazin WJ, Skaar EP. Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell Host Microbe. 2011;10:158–164. doi: 10.1016/j.chom.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]