Abstract
Supervised machine learning methods for clinical natural language processing (NLP) research require a large number of annotated samples, which are very expensive to build because of the involvement of physicians. Active learning, an approach that actively samples from a large pool, provides an alternative solution. Its major goal in classification is to reduce the annotation effort while maintaining the quality of the predictive model. However, few studies have investigated its uses in clinical NLP. This paper reports an application of active learning to a clinical text classification task: to determine the assertion status of clinical concepts. The annotated corpus for the assertion classification task in the 2010 i2b2/VA Clinical NLP Challenge was used in this study. We implemented several existing and newly developed active learning algorithms and assessed their uses. The outcome is reported in the global ALC score, based on the Area under the average Learning Curve of the AUC (Area Under the Curve) score. Results showed that when the same number of annotated samples was used, active learning strategies could generate better classification models (best ALC – 0.7715) than the passive learning method (random sampling) (ALC – 0.7411). Moreover, to achieve the same classification performance, active learning strategies required fewer samples than the random sampling method. For example, to achieve an AUC of 0.79, the random sampling method used 32 samples, while our best active learning algorithm required only 12 samples, a reduction of 62.5% in manual annotation effort.
Keywords: Active learning, Natural language processing, Clinical text classification, Machine learning
1. Introduction
Active learning has been widely studied in the domains where a supervised learning approach is implemented to build a high-performance predictive model, such as text classification [1, 2], information extraction [3], image classification and retrieval [4], gene expression analysis [5], drug discovery [6], etc. It is one of the possible solutions in many supervised learning tasks when labeled training samples are expensive to obtain or otherwise limited. The objective of applying active learning to classification is to build a better classifier with fewer labeled training samples by actively selecting the queries (instances) for labeling rather than choosing them randomly. The active learner is capable of achieving the required performance of the prediction model with reduced annotation costs.
Although researchers have shown that active learning is beneficial in many domains, there is a paucity of published literature on the application of active learning to biomedical text, especially clinical text. With the wide use of electronic health records (EHRs), there are large amounts of textual data available and studies of clinical natural language processing (NLP) technologies have been numerous [7,8,9]. However, statistics-based clinical NLP approaches often depend on physicians or other domain experts for the annotation of textual data, which can be very expensive and time-consuming. Therefore, we believe that pool-based active learning approaches, which can largely reduce annotation effort while retaining high performance for predictive models, will be very useful for clinical NLP research. In this paper, we describe an application of active learning to a clinical text classification task: to determine assertions of clinical concepts, using an annotated corpus from the 2010 i2b2 Clinical NLP Challenge. We implemented and evaluated several active learning algorithms, including some that are newly developed, and our results showed that some active learning strategies outperformed random sampling methods significantly.
2. Background
The pool-based active learning approach to classification [1] is practical for many real-world learning problem domains, including medicine. The learner can access a large quantity of unlabeled data as a pool with low cost and can request the true label from it. An active learning system mainly consists of a classification model and an active sample selection or a querying algorithm. The classification model is built by traditional supervised machine learning algorithms. The model is trained by using the labeled instances (training set) and is then applied to the new unlabeled instances (test set) to predict class labels. The second core component of active learning is the querying method. In general, there are two types of learners: active learner and passive learner. The passive learner just uses a random sampling method, which queries the labels of instances randomly selected from the pool of unlabeled samples, without considering the information about samples in the pool. The active learner, on the other hand, will select the instances that are the most promising in improving the predictive performance of the model.
An active learning protocol is often used for a given dataset and a querying algorithm:
Initialize the labeled training set L = L0, the pool of unlabeled set U =U0, and a test set T.
Train the classification model based on L and predict the probability of class label for each instance in U and T.
Rank the instances in U based on the querying algorithm and assign labels (from human experts) for the top b(i) samples in U, where b(i), the batch size of active learning, is the number of querying samples at iteration i.
Add the b(i) instance(s) with label(s) to L and remove from U.
Iterate steps (2) to (4) until the stop criteria is met.
Finally, the classification performance (AUC score) will be reported for the prediction of T at each iteration i, and the global score based on the learning curve AUC(i) will be computed.
The main issue for the active learner is how to find the good queries from the pool for better classification performance. Many variations of the active learning (querying) algorithms exist, and there are mainly six types: uncertainty sampling [10], query-by-committee (QBC) [11], expected gradient length [12], fisher information [13], estimated error reduction [14] and information density [3]. Detailed information about these algorithms can be found in an active learning literature survey [15]. Some of the algorithms are computationally expensive and not practical, such as expected gradient length, fisher information, and estimated error reduction. QBC is sensitive to the type of classification models selected. This study focused on uncertainty sampling and information density, two widely used methods in active learning, as well as a new framework for a querying algorithm we propose called “model change”. We have worked on active learning tasks in other domains and reported some new uncertainty sampling based querying methods in our previous work [16]. In this paper, we also developed some new querying methods based on uncertainty sampling and model change. Using these novel methods, together with a few well known querying algorithms, we assessed the use of active learning approaches for a clinical text classification task: to determine the assertion status of concepts in clinical text, and comparing the results to the passive learning (random sampling) method.
Many statistics-based text mining and NLP tasks require large numbers of annotated samples, which are very expensive and time-consuming to develop. Therefore, researchers have applied active learning to various NLP tasks [17, 18, 19, 20]. For example, Chen et al. [17] successfully used active learning to reduce the annotation effort while maintaining good performance for a word sense disambiguation task of five English verbs with coarse-grained senses by using two uncertainty sampling based methods. Active learning has also been applied to the biomedical domain. S. Kim et al. [18] presented an active learning strategy that considered both entropy-based uncertainty from classifiers and the diversity of a corpus and used it for the task of biological name entity recognition in MEDLINE abstracts.
In this study, we investigated the application of active learning to clinical text processing, which has not been reported on previously. Specifically, we developed new active learning algorithms and applied them to the assertion classification task for concepts in clinical text. This paper is organized as follows: Section 3 presents datasets and methods that we used in this study, such as cross validation experiments, active learning strategies including classification models and querying algorithms, and evaluation; Section 4 displays the experiment results; Section 5 discusses the significance of our results; and Section 6 summarizes our work and provides a future direction.
3. Methods
3.1 Datasets
We used the manually annotated training set for concept assertion classification in the 2010 i2b2/VA NLP challenge [8], which was organized by i2b2 (the Center of Informatics for Integrating Biology and the Bedside) at Partners Health Care System and Veterans Affairs (VA), Salt Lake City Health Care System. The assertion classification task is to assign one of six labels (“absent”, “associated with someone else”, “conditional”, “hypothetical”, “possible”, and “present”) to medical problems identified from clinical text (discharge summaries and some progress notes collected from three institutions). We participated in the challenge and developed an SVM-based system for the assertion classification task, and we ranked fourth among over 20 participating teams (no statistically significant difference from the top three systems) [21].
For this study, we used the same set of features as described in our previous work and we wanted to assess whether active learning algorithms could reduce sample size while retaining good performance. The feature set includes: (1) window of context, the size of which is optimized; (2) direction with distance in the window of context (e.g., third word on the left); (3) bi-grams identified within the context window; (4) part of speech tags of context words; (5) normalized concepts and semantic types identified by an NLP system (MedLEE) [22], such as certainty, UMLS CUIs, and semantic types; (6) source and section of its clinical note.
The training set from the challenge contained 349 notes, with 11,967 medical problems annotated with one of the six assertion statuses. Given the availability of large annotated data, active learning may not be needed for this specific assertion classification task. However, we utilized this available large data set to evaluate the performance of different active learning algorithms, which should be useful for many other tasks where large annotated data are not available. Moreover, active learning on multi-class classification tasks is more complicated than that on binary classification tasks. Therefore, as an initial study, we focused on the investigation of active learning algorithms for binary classification problems. We converted the multi-class assertion classification task into a binary classification problem, by considering “present” to be the positive class and all others as the negative class. We refer to this dataset as ASSERTION in this study and investigated active learning algorithms for the binary classification of assertion (“present” vs. “non-present”).
In addition, we used NOVA, a dataset of English text from the 2010 active learning challenge [23], as the benchmark for this study. NOVA comes from the 20-Newsgroup dataset [24], which is a popular benchmark dataset for experiments in text applications of machine learning techniques, such as text classification and text clustering. Each text to be classified comes from an email that was posted to one or several newsgroups. The NOVA data are selected from both politics and religion, topics considered as positive and negative class, respectively. The feature set of data is in binary representation using a bag-of-words with a vocabulary of approximately 17,000 words.
Table 1 shows the comparison of the properties of the two datasets. They were both annotated with binary labels. All features for both datasets were binary only. Both datasets were very sparse (sparsity is equal to the ratio between the number of cells with value zero and the total number cells in the data matrix), but the class distribution also was different for two datasets. Additionally, the ASSERTION dataset contained information at the sentence level, while the NOVA dataset was at the document level. The ASSERTION dataset is probably more difficult to classify because it has much higher number of features than NOVA.
Table 1.
Experimental datasets for Active Learning
| Dataset Name | Number of samples | Number of Positive samples | PositiveRate | Number of Features | Feature Type | Sparsity | Class Type |
|---|---|---|---|---|---|---|---|
| ASSERTION | 11,967 | 8,051 | 0.6728 | 71,986 | Binary | 0.9994 | Binary |
| NOVA | 19,466 | 2,769 | 0.2845 | 16,969 | Binary | 0.9967 | Binary |
3.2 Cross Validation on Active Learning
To set up a pool-based active learning framework, a pool of unlabeled samples and an independent test set were initialized. The variability in performance could have been high if many different partitions in the data were created for generating the unlabeled pool and test set. To fully use both datasets and generate reliable results, 3-fold stratified cross validation was performed on active learning. On each of the cross validation iterations, the pool of unlabeled samples was from two folds and the evaluation of performance was based on the remaining fold. The validation results were averaged over three iterations.
3.3 Classification Model
To mainly focus on improving the querying algorithm, the same classifier with the same parameter was used on each run of classification (training and testing). In our preliminary experiments for selecting the best classifier and parameter, the linear Logistic Regression classifier outperformed linear SVM and Naive Bayesian classifiers in 3-fold cross validation for all samples in both the ASSERTION and NOVA datasets. Therefore, the Logistic Regression model implemented in the package “Liblinear” [25] was used. It can output the posterior probability as the prediction value. This output would be used as the input for most querying algorithms.
3.4 Active Learning Strategy
Based on the protocol of active learning described in Section 2, the global performance (learning curve) is influenced by many factors during the active learning process, such as initial performance (the classification performance based on the initial training set), the batch size, the stop criteria, the querying algorithm, etc. However, we designed the active learning experiment so that the querying algorithm would be the most influential factor. We fixed the initial and the final performance points in the learning curve as well as the batch size for each querying algorithm as follows.
We randomly selected three positive samples and three negative samples as the initial training set. In each iteration of the cross validation, all experiments with different querying algorithms would use the same initial training set and, therefore, have the same initial point in the learning curve.
According to the stop criteria, the active learning process stopped when the entire pool of unlabeled samples was queried or U was empty. In each iteration of the cross validation, all experiments with different querying algorithms would have the same final point in the learning curve.
For batch size selection, we used 2i+2 training samples with labels where i is the index of iteration in the active learning process up to the total number of training samples. For example, the size of labeled training set L on each iteration would be 8, 16, 32, 64, 128,…, 4096, …, and the maximum number.
The querying algorithm is the function to assess how informative each instance x is in unlabeled pool U. x* is selected as the most informative sample according to the function x*=argmaxQ(x), where Q(x) is the querying function that outputs the informativeness or querying value (Q value) for data matrix x in U.
3.4.1 Uncertainty Sampling-based Algorithm
Uncertainty sampling queries the sample with the least certainty or on the decision boundary. The simplest uncertainty sampling algorithm is called Least Confidence (LC), which is straightforward for the probabilistic models:
where y* is the most likely label sequence for x. θ is the model that generates the posterior probability P of label y given data matrix x. In the binary classification case, LC is equivalent to querying the instance with the highest Q value (or uncertainty value) that is nearest the 0.5 posterior probability of being in the positive or negative class. In the case of the ASSERTION dataset, if the concept term was classified as “present” with the probability closer to 0.5 versus “non-present,” the term was more likely to be selected for annotation in the next iteration of active learning.
During the active learning process, the class distribution of the training set could become imbalanced (with more positive/negative than negative/positive samples). At this point, we assume that the sample in the minority class is more informative. Moreover, we would like to balance the training set as much as possible in the early iteration of active learning because the classifier would tend to ignore the minority class, resulting in a poor prediction model, especially with a small number of labeled training samples. Therefore, we implemented another uncertainty sampling algorithm called Least Confidence with Bias (LCB) [16], which considers both the uncertainty value from the current prediction model and the proportion of class labels in the training set. LCB is more likely to query the instances around the decision boundary and compensates for class imbalance.
Let pp be the percentage of positive labels in the current training set. We defined Pmax as the posterior probability that gives the highest Q value in LCB function QLCB(x):
Where Pmax = mean(0.5, 1−pp) where Pmax = 0.5 or pp = 0.5, it is equivalent to LC.
Both LC and LCB methods depend on the quality of the prediction model because both algorithms control the sample selection based on the posterior probability output from the model. When the model is poor, it propagates the negative effect to the querying algorithm. LCB could bias the Q value so that the model can converge more quickly to a good one by balancing the training set in the early stage of active learning. However, when the model improves, the bias could increase too much. So we also proposed another modified version of uncertainty sampling called Least Confidence with Dynamic Bias (LCB2). which also considers the size of the current training set. Note that the model is likely to be more reliable when the classification model is trained by a larger set of samples. For the binary classification problem, we have more confidence that the highest Q value is at the point closer to the posterior probability of 0.5 when more labeled training samples are used. QLCB2(x) is the same as QLCB(x) except for Pmax:
where wb is the weight of bias and wu is the weight of uncertainty, and wb = 1−wu, where wu is the ratio of ∣L∣, the size of the current labeled set, and ∣U0∣, the size of initial unlabeled pool: wu = ∣L∣/∣U0∣. When wu = 0, it is equivalent to LCB; when wu = 1, it is equivalent to LC.
3.4.2 Model Change Sampling-based Algorithm
Model change sampling algorithm (MC) is a heuristic method to improve the querying method that relies on the classification model. For example, uncertainty sampling might fail to find the most uncertain samples when given a poor probabilistic model for classification. It is as difficult as finding the true decision boundary by classification model. We implemented the idea of model change for querying on top of model dependent querying methods such as uncertainty sampling. The MC algorithm considers the Q value from not only the current model but also the previous one. It controls the sample selection based on the change of Q values from different models during the active learning process.
We derived the heuristic function based on the following assumption. When the classification model is improving during the active learning process, the posterior predictions for each sample will be closer to either zero or one. In other words, the Q value for each sample, which is the uncertainty value based on LC, LCB or LCB2, becomes smaller and smaller. The heuristic function takes into account the change of Q values over different models. The model change sampling algorithm ranks the unlabeled instances based on the following rule: the instance with the most increasing Q values is the most informative one. If the Q values for all instances are decreasing, the instance with the least decreasing Q value is also considered as the most informative one in the dataset. It also needs to consider the improvement of the model during the active learning process. The Q value for the previous model is discounted because the current model is intuitively better than the previous one.
where i represents the current iteration in the active learning process, i-1 is the index of the previous iteration; wo is the weight of the old model, which is equal to 1/∣L∣ (∣L∣ is the size of the current training set). We applied this formula to uncertainty sampling based querying methods (LC, LCB, and LCB2) so that we had three MC querying algorithms in our study: Least Confidence with Model Change (LCMC), Least Confidence with Bias and Model Change, (LCBMC), and Least Confidence with Dynamic Bias and Model Change (LCB2MC).
3.4.3 Information Density-based Algorithm
The information density (ID) framework proposed by Settles and Craven [3] considers not only the uncertainty of instances but also the data distribution. The most uncertain instance lies on the decision boundary, but it is not necessarily representative of other instances in the distribution. Thus knowing its label is not likely to improve the prediction model.
Here is the ID-based querying function QID(x):
where QUS(x) is the Q value by any uncertainty sampling based method (like LC, LCB, or LCB2); QD(x) is the density function to compute how representative it is for any given instance in the unlabeled set; β is the control factor for the density term. In this study, we implemented an information density approach based on the Euclidean distance to the centers of labeled set L. These centers can represent the dense regions in the input space [26]. In our preliminary study, we only considered one center because it is difficult to determine the appropriate numbers of centers for selecting the most representative sample:
where x̂ is the mean vector for each variable over all samples in the labeled set L; dist(.) is the function for computing the Euclidean distance to this mean vector for each sample in x. We called this method Information Density Based on Distance to Center (IDD). In our experiment, we used method LCB2 in the first term QUS(x) of IDD.
3.5 Evaluation
We applied the same evaluation measures used for the active learning challenge 2010 [27]. The prediction for the performance of active learning was evaluated according to the Area under the Learning Curve (ALC). The learning curve plotted the Area Under the ROC curve score (AUC) computed on all the samples in the test set as a function of the number of labels queried. The global score or ALC score was normalized based on the following function:
where Amax is the area under the best achievable learning curve (1.00 AUC on all points of the learning curve) and Arand is the area under the learning curve obtained by random prediction (0.50 AUC on all points of the learning curve). The learning curve of two neighbor points was interpolated linearly.
In the x-axis of the learning curve, we used log2 scaling. It is consistent with the batch size (2i+2) of active learning, and this scaling actually increases the difficulty of getting a high global score because each additional labeled sample in the early stage of active learning is much more important than the one in the late stage. The performance in the early stages is more significant for the global score, so our target was also to improve the prediction model given a small number of training samples with labels.
Three learning curves were generated in the 3-fold cross validation of active learning for the experiment of each querying algorithm. Then the average learning curve was determined by averaging the AUC scores on each corresponding point from the three learning curves. The final global score of each querying algorithm was the ALC score from the average learning curve.
We ran the active learning experiments for eight querying algorithms and two datasets. The passive learner used the random querying method, while the active learner used other querying approaches. Since the passive learner generated results with high variance from the random factor for sampling, we averaged the learning curves of the random querying method over 50 runs using the same start point, end point, and batch size.
4. Results
Results for the ASSERTION dataset showed that the ALC scores of all active learning methods except IDD outperformed the baseline using the random sampling method. In terms of the global performance, the active learner LCBMC had the best performance on both the ASSERTION and NOVA datasets. Most of the other active learners also performed better than passive leaner. LCB improved the performance by the basic uncertainty sampling method LC, while LCB2 could generate a better learning curve than LCB. The performances of LC, LCB, and LCB2 were consistent for both datasets. The model change-based method improved the uncertainty sampling methods LC and LCB in both datasets, but the performance of LCB2MC was poorer than LCB2. The active learners LC and IDD did not perform well in our experiments on both datasets.
Table 2 shows the cross validation results of ALC scores for both datasets and the different querying algorithms. ALC scores from individual folds, as well as the average of the three folds, were reported
Table 2.
ALC results for Threefold Cross Validation of Active Learning for Two Datasets and Eight Querying Methods.
| Dataset | Querying Method | Fold1 | Fold2 | Fold3 | Average | Standard Deviation | ||
|---|---|---|---|---|---|---|---|---|
| Category | New/Existing | Name | ||||||
| ASSERTION Dataset | Uncertainty Sampling | Existing | LC | 0.7160 | 0.7524 | 0.7586 | 0.7423 | 0.0230 |
| Existing | LCB | 0.7423 | 0.7836 | 0.7560 | 0.7606 | 0.0210 | ||
| New | LCB2 | 0.7536 | 0.7773 | 0.7597 | 0.7635 | 0.0123 | ||
| Model Change | New | LCMC | 0.7171 | 0.7656 | 0.7644 | 0.7490 | 0.0277 | |
| New | LCBMC | 0.7503 | 0.7839 | 0.7803 | 0.7715 | 0.0184 | ||
| New | LCB2MC | 0.7182 | 0.7624 | 0.7615 | 0.7474 | 0.0253 | ||
| Information Density | Existing | IDD | 0.7144 | 0.7268 | 0.6947 | 0.7120 | 0.0162 | |
| Baseline | Existing | Random (50 runs) | 0.7151 | 0.7647 | 0.7434 | 0.7411 | 0.0249 | |
| NOVA Dataset | Uncertainty Sampling | Existing | LC | 0.7643 | 0.6805 | 0.7251 | 0.7233 | 0.0419 |
| Existing | LCB | 0.8524 | 0.8163 | 0.8603 | 0.8430 | 0.0235 | ||
| New | LCB2 | 0.8722 | 0.8344 | 0.8546 | 0.8537 | 0.0189 | ||
| Model Change | New | LCMC | 0.8771 | 0.8144 | 0.8472 | 0.8462 | 0.0314 | |
| New | LCBMC | 0.8702 | 0.8289 | 0.8719 | 0.8570 | 0.0244 | ||
| New | LCB2MC | 0.8768 | 0.8323 | 0.8295 | 0.8462 | 0.0265 | ||
| Information Density | Existing | IDD | 0.7297 | 0.6970 | 0.7161 | 0.7143 | 0.0164 | |
| Baseline | Existing | Random (50 runs) | 0.8151 | 0.7847 | 0.8001 | 0.8000 | 0.0152 | |
Note: LC: Least Confidence; LCB: Least Confidence with Bias; LCB2: Least Confidence with Dynamic Bias; LCMC: Least Confidence with Model Change; LCBMC: Least Confidence with Bias and Model Change; LCB2MC: Least Confidence with Dynamic Bias and Model Change; IDD: Information Density based on Distance to Center.
Figure 1 and Figure 2 show the average learning curves for datasets ASSERTION and NOVA, respectively, for all eight querying methods. In general, LCBMC, which had the highest global score, showed stability with small training sample sizes. On the other hand, the querying methods with low global scores performed poorly or were unstable in the early stage of the active learning process.
Fig. 1.
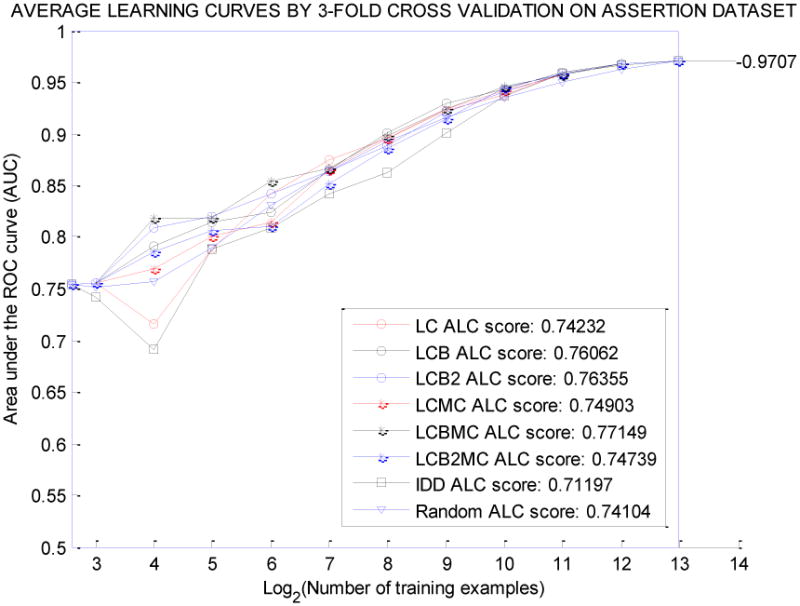
Average Learning Curves for 8 Querying Algorithms on the Assertion Dataset.
Fig. 2.
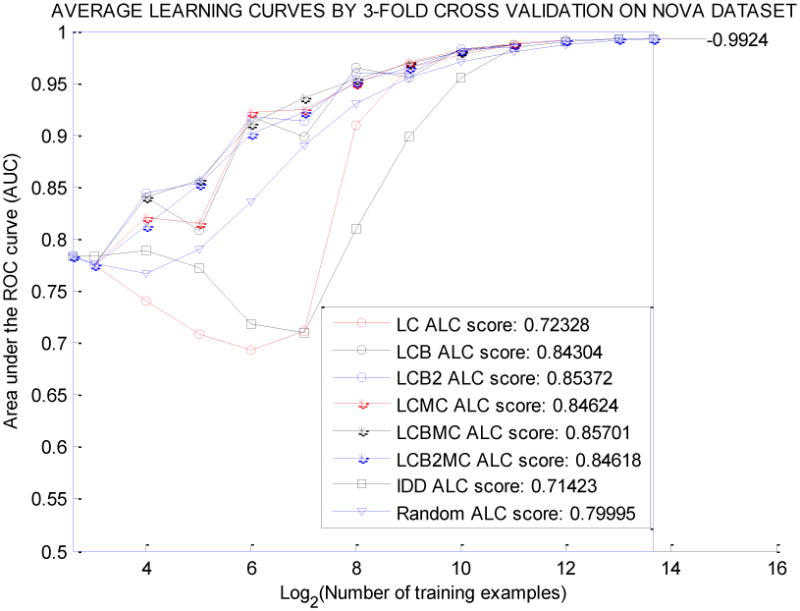
Average Learning Curves for 8 Querying Algorithms on the NOVA Dataset.
We can compare eight querying algorithms on the same figure vertically and horizontally. By reading vertically, we can compare the performance of eight prediction models in AUC at each stage of active learning; by reading horizontally, we can compare the costs of annotation (number of labeled samples used) by eight querying methods for each quality level of the prediction model in AUC.
Table 3 presents the evaluation of prediction models based on the average AUC score and its standard deviation when the size of querying samples was small. This table magnifies the intermediate results in the early stage of the learning curve with 16, 32, and 64 training samples. The average AUC by random querying method was not the worst in the early stage of active learning, but the standard deviation was higher compared to the other methods. The best querying method in our experiments, LCBMC, performed reasonably well with a high average AUC and low standard deviation when only a small number of training samples was used.
Table 3.
Evaluation of the classification model for eight querying algorithms and two datasets on a small training set (with 16, 32, and 64 training samples) based on average AUC score and the standard deviation.
| Dataset | Size of Training Set | LC | LCB | LCB2 | LCMC | LCBMC | LCB2MC | IDD | Random |
|---|---|---|---|---|---|---|---|---|---|
| ASSERTION Dataset | 16 | 71.52% ± 2.55% | 79.16%±3.85% | 80.91% ± 1.31% | 76.87% ±5.89% | 81.92% ± 1.41% | 78.55% ± 4.73% | 69.10% ± 2.70% | 75.65% ±5.83% |
| 32 | 78.85% ± 4.30% | 81.46% ± 2.77% | 82.04% ± 1.38% | 80.11% ± 1.91% | 81.87% ± 2.45% | 80.61% ± 1.31% | 78.77% ± 2.55% | 79.00% ±4.31% | |
| 64 | 84.16% ± 1.42% | 82.33% ± 2.05% | 84.16% ± 0.74% | 81.45% ± 2.34% | 85.42% ± 1.03% | 81.07% ± 1.45% | 80.88% ±3.18% | 83.12% ± 2.25% | |
| NOVA Dataset | 16 | 73.98% ± 6.25% | 83.99% ± 4.93% | 84.42% ±3.66% | 82.01% ± 4.00% | 83.98% ±5.81% | 81.30% ±4.61% | 78.91% ± 4.22% | 76.70% ± 7.06% |
| 32 | 70.88% ± 2.96% | 80.82% ±5.35% | 85.33% ± 1.63% | 81.52% ± 6.97% | 85.69% ±3.05% | 85.25% ±3.15% | 77.27% ± 2.50% | 79.03% ± 6.96% | |
| 64 | 69.38% ±5.05% | 91.79% ± 0.54% | 91.82% ± 0.58% | 92.21% ±0.71% | 91.03% ± 2.80% | 90.16% ± 1.04% | 71.77% ± 2.63% | 83.57% ± 4.88% |
Table 4 presents the evaluation of the prediction model when the training set was large (with 1024, 2048, and 4096 samples). This table magnifies the intermediate results for the late stage of active learning. In this stage, the active learners performed better when compared with the passive learner on the ASSERTION dataset. It is also true for the NOVA dataset with training sample sizes of 2048 or higher.
Table 4.
Evaluation of the classification model for eight querying algorithms and two datasets with a large training set (with 1024, 2048, and 4096 training samples) based on average AUC score and the standard deviation.
| Dataset | Size of Training Set | LC | LCB | LCB2 | LCMC | LCBMC | LCB2MC | IDD | Random |
|---|---|---|---|---|---|---|---|---|---|
| ASSERTION Dataset | 1024 | 93.73% ± 0.48% | 94.34% ± 0.53% | 94.09% ± 0.47% | 94.23% ± 0.57% | 94.64% ± 0.26% | 94.44% ±0.41% | 93.57% ± 0.56% | 93.46% ± 0.46% |
| 2048 | 95.81% ± 0.26% | 95.94% ± 0.24% | 95.80% ±0.41% | 95.67% ± 0.47% | 95.74% ± 0.29% | 95.88% ± 0.54% | 95.77% ±0.11% | 94.99% ± 0.33% | |
| 4096 | 96.67% ± 0.28% | 96.76% ± 0.28% | 96.66% ± 0.36% | 96.76% ± 0.26% | 96.74% ± 0.30% | 96.82% ± 0.32% | 96.70% ± 0.24% | 96.18% ± 0.26% | |
| NOVA Dataset | 1024 | 97.71% ± 0.62% | 98.26% ± 0.36% | 98.29% ± 0.22% | 98.10% ±0.18% | 98.03% ±0.51% | 98.10% ± 0.39% | 95.48% ± 0.46% | 97.05% ± 0.42% |
| 2048 | 98.66% ± 0.23% | 98.69% ± 0.25% | 98.38% ± 0.26% | 98.83% ± 0.22% | 98.65% ± 0.23% | 98.65% ±0.41% | 98.39% ± 0.26% | 98.03% ± 0.28% | |
| 4096 | 99.07% ± 0.20% | 99.10% ±0.19% | 99.02% ± 0.28% | 99.11% ± 0.20% | 99.11% ± 0.25% | 99.08% ± 0.24% | 99.00% ± 0.20% | 98.63% ±0.21% |
In addition, none of the experiments needed much computational time. The querying algorithms could rank or generate Q values for all samples in the unlabeled pool on both datasets (more than 8000 samples) in less than one second. The classifier Logistic Regression in the “Liblinear” package could complete threefold cross validation (for the end point in the learning curve) in less than three seconds for the ASSERTION dataset (with about 12,000 samples) and four seconds for the NOVA dataset (with about 20,000 samples).
To assess whether there are significant differences in terms of mean ALC global scores among different active learners and the passive learner, we conducted a statistical test based on results from bootstrapping. We re-sampled the test set by random sampling with replacement for 200 times and generated 200 bootstrapping data sets. For each bootstrapping data set, we evaluated and reported ALC global scores for different active learners and the passive learner. We used Wilcoxon signed rank test [28], a non-parametric test for paired samples, to assess whether differences between two methods are statistically significant. As there were eight different methods (28 comparisons in total), we applied Bonferroni correction [29] to adjust for multiple comparisons, with family-wise type I error control at alpha = 0.05. Therefore, if the p-value from Wilcoxon signed rank test was less than 0.0018 (0.05/28), we claimed that there was a statistically significant difference between two methods. Table 5 shows the results of the statistical test. Except the ones between Random and LC, Random and IDD, LC and IDD, and LCMC and LCB2MC, all other comparisons showed statistically significant differences.
Table 5.
Results of the statistical test (Wilcoxon signed rank test with Bonferroni correction for multiple testing) among ALC global scores from different active learners and the passive learner (“Y”: Statistically significant; “N”: Not statistically significant)
| LC | LCB | LCB2 | LCMC | LCBMC | LCB2MC | IDD | |
|---|---|---|---|---|---|---|---|
| Random (50 Runs) | N | Y | Y | Y | Y | Y | N |
| LC | Y | Y | Y | Y | Y | N | |
| LCB | Y | Y | Y | Y | Y | ||
| LCB2 | Y | Y | Y | Y | |||
| LCMC | Y | N | Y | ||||
| LCBMC | Y | Y | |||||
| LCB2MC | Y |
5. Discussion
For the concept assertion classification task, active learners generated better prediction models with higher AUC scores, and required less annotation effort than the passive learner (based on the results shown in Tables 3 and Table 4). Using the ASSERTION dataset, the prediction model trained by 32 randomly selected annotated samples had a 0.7900 average AUC score; however, LCBMC could achieve the prediction model with a 0.8192 average AUC by using 16 annotated samples, which saved half of the annotation cost. Overall, the active learning strategy was more efficient in reducing annotation costs and improving prediction models for the clinical dataset ASSERTION. In Figure 1, the best learning curve by LCBMC lay above the average learning curve by random sampling. The result for the general English dataset NOVA was also consistent with the ASSERTION dataset. Such findings show that active learning strategies hold promise in solving similar clinical text classification problems when annotation is expensive and time-consuming.
To further analyze the learning curves for the ASSERTION dataset, we calculated the approximate numbers of training cases at different levels of AUC, for both active learning approaches and random sampling approaches (Table 6). In the early stage of active learning, the random sampling method used 32 samples to achieve an AUC of 0.79, while LCBMC used only 12 samples to achieve the same AUC, a 62.5% of reduction in sample size. In the middle stage of active learning, the random sampling method used 512 labeled cases to train a model with an AUC of 0.92, while LCB used about 369 samples to build the same model. In the late stage, the random sampling method required 4,096 samples to generate a model with an AUC of 0.96, while LCB used only 2,518 samples to reach the same AUC. This analysis demonstrates that active learning methods require fewer training samples than the random sampling method, with similar classification performances.
Table 6.
Approximate numbers of training samples at different levels of AUCs for both active learning algorithms and the random sampling method.
| AUC | 0.79 | 0.83 | 0.86 | 0.89 | 0.92 | 0.93 | 0.95 | 0.96 |
| Random | 32 | 64 | 128 | 256 | 512 | 1024 | 2048 | 4096 |
| LC | 33 | 56 | 103 | 232 | 435 | 903 | 1557 | 2768 |
| LCB | 16 | 73 | 127 | 219 | 369 | 650 | 1354 | 2518 |
| LCB2 | 13 | 46 | 129 | 277 | 462 | 824 | 1471 | 2785 |
| LCMC | 26 | 81 | 127 | 241 | 426 | 770 | 1473 | 2843 |
| LCBMC | 12 | 41 | 118 | 225 | 414 | 713 | 1271 | 2784 |
| LCB2MC | 19 | 91 | 166 | 298 | 524 | 812 | 1330 | 2555 |
| IDD | 35 | 102 | 269 | 443 | 694 | 1002 | 1600 | 2790 |
The basic uncertainty sampling algorithm LC and the information density algorithm IDD did not perform well in active learning on both datasets. LC could not find the most informative samples when the annotated instances were insufficient, because LC relies on the performance of a probabilistic model that was poor in the early stage of active learning. However, LCB and LCB2 could improve the performance for both datasets by also considering the imbalance of class and the quality of the classification model. The model change-based method LCBMC further improved the uncertainty-based method LCB by considering the change of informative values between models. The information density-based method IDD failed to improve the global score, because the density term based on distance to center did not find the most “representative” samples for both datasets. It negatively affected the overall performance, even though the uncertainty term by LCB2 could perform reasonably well by itself on both datasets.
Although LCBMC was the best querying algorithm for both datasets based on the global score in our experiments, its learning curve for the ASSERTION dataset was not flawless. It could generate a classification model with 0.8192 and 0.8187 average AUC scores by using 16 and 32 annotated samples, respectively. Although the difference in AUC did not seem significant, we did not expect that the model would get worse with larger training sets. Further investigation of the querying algorithm is needed to improve the stability of the learning curve. One possible direction worth investigating is to automatically select the batch size as a function of the probabilistic prediction and querying model in the iteration of active learning, instead of presetting this parameter.
Our current experiments were limited to binary classification tasks. In practice, however, more than two class labels are often involved in many classification tasks for clinical text. Active learning for multi-class classification is a more challenging problem, where we need to extend the current querying algorithms so that they can assess the informativeness of samples in multiple classes. In text classification research, a number of studies have applied active learning approaches to multi-class classification problems including word sense disambiguation [17], name entity recognition [18], natural language parsing [19], etc. So far we have only applied and evaluated active learning methods on one set of clinical textual data. To assess the usefulness of active learning in the medical domain, we need to validate it in broader types of text mining applications.
6. Conclusion
This study demonstrated that active learning technologies can be applied to clinical text classification tasks effectively, with improved performance and reduced annotation effort. New querying methods developed here showed good performance on the concept assertion classification task. We plan to extend the active learning formalism to multi-class classification problems, as well as to other applications that are related to clinical text processing.
Highlights.
One of the first attempts to apply active learning to clinical text classification.
Active learning algorithms generally outperformed random sampling method.
Active learning reduced over 50% samples to achieve similar performance.
Acknowledgments
This study was supported in part by NIH grants NLM R01LM010681 and NCI R01CA141307. The datasets used were obtained from 2010 i2b2/VA NLP challenge and we would like to thank the organizers for sharing the dataset.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lewis D, Gale W. Proceedings of the ACM SIGIR Conference on Research and Development in Information Retrieval. ACM/Springer; 1994. A sequential algorithm for training text classifiers; pp. 3–12. [Google Scholar]
- 2.Tong S, Koller D. Support Vector Machine Active Learning with Applications to Text Classification. Journal of Machine Learning Research. 2001:999–1006. [Google Scholar]
- 3.Settles B, Craven M. Proceedings of the Conference on Empirical Methods in Natural Language Processing (EMNLP) ACL Press; 2008. An analysis of active learning strategies for sequence labeling tasks; pp. 1069–1078. [Google Scholar]
- 4.Tong S, Chang E. Proceedings of the ACM International Conference on Multimedia. ACM Press; 2001. Support vector machine active learning for image retrieval; pp. 107–118. [Google Scholar]
- 5.Liu Y. Active learning with support vector machine applied to gene expression data for cancer classification. Journal of Chemical Information and Computer Sciences. 2004;44:1936–1941. doi: 10.1021/ci049810a. [DOI] [PubMed] [Google Scholar]
- 6.Forman G. Incremental machine learning to reduce biochemistry lab costs in the search for drug discovery. BIOKDD02. 2002:33–36. [Google Scholar]
- 7.Meystre SM, Savova GK, Kipper-Schuler KC, Hurdle JE. Extracting information from textual documents in the electronic health record: a review of recent research. Yearbook of Medical Informatics. 2008:128–144. [PubMed] [Google Scholar]
- 8.Uzuner O, South BR, Shen S, Duvall S. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. Journal of the American Medical Informatics Association: JAMIA. 2011 Jun; doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uzuner O, Solti I, Xia F, Cadag E. Community annotation experiment for ground truth generation for the i2b2 medication challenge. Journal of the American Medical Informatics Association (JAMIA) 2010;17:519–523. doi: 10.1136/jamia.2010.004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis D, Catlett J. Proceedings of the Eleventh International Conference on Machine Learning. Morgan Kaufmann; 1994. Heterogeneous uncertainty sampling for supervised learning; pp. 148–156. [Google Scholar]
- 11.Seung HS, Opper M, Sompolinsky H. Query by committee. Proceedings of the ACM Workshop on Computational Learning Theory. 1992:287–294. [Google Scholar]
- 12.Settles B, Craven M, Ray S. Advances in Neural Information Processing Systems (NIPS) Vol. 20. MIT Press; 2008b. Multiple-instance active learning; pp. 1289–1296. [Google Scholar]
- 13.Chaloner K, Verdinelli I. Bayesian experimental design: A review. Statistical Science. 1995;10(3):237–304. [Google Scholar]
- 14.Roy N, McCallum A. Proceedings of the International Conference on Machine Learning (ICML) Morgan Kaufmann; 2001. Toward optimal active learning through sampling estimation of error reduction; pp. 441–448. [Google Scholar]
- 15.Settles B. Computer Sciences Technical Report 1648. University of Wisconsin-Madison; 2009. Active Learning Literature Survey. [Google Scholar]
- 16.Chen Y, Mani S. Study of active learning in the challenge. Proc 2010 International Joint Conference on Neural Networks; Barcelona, Spain. 18-23 July 2010. [Google Scholar]
- 17.Chen J, Schein A, Ungar L, Palmer M. An Empirical Study of the Behavior of Active Learning for Word Sense Disambiguation. Proceedings of the Human Language Technology Conference of the North American Chapter of the ACL; New York. June 2006.pp. 120–127. [Google Scholar]
- 18.Kim S, Song Y, Kim K, Cha J, Lee G. MMR-based active machine learning for bio named entity recognition. Proceedings of the Human Language Technology Conference of the North American Chapter of the ACL; New York. June 2006.pp. 69–72. [Google Scholar]
- 19.Tang M, Luo X, Roukos S. Active Learning for Statistical Natural Language Parsing. Proceedings of the 40th Annual Meeting of the Association for Computational Linguistics (ACL); Philadelphia. July 2002; pp. 120–127. [Google Scholar]
- 20.Gangadharaiah R, Brown R, Carbonell J. Active Learning in Example-Based Machine Translation. The 17th Nordic Conference on Computational Linguistics, (NODALIDA09); Odense, Denmark. May 2009. [Google Scholar]
- 21.Jiang M, Chen Y, Liu M, Rosenbloom S, Mani S, Denny JC, Xu H. A study of Machine Learning based Approaches to Extract Clinical Entities and their Assertions from Discharge Summaries. Journal of the American Medical Informatics Association (JAMIA) 2011 April 20; doi: 10.1136/amiajnl-2011-000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. Journal of the American Medical Informatics Association (JAMIA) 1994;1(2):161–174. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guyon I, Cawley G, Dror G, Lemaire V. Results of the Active Learning Challenge. Workshop on Active Learning and Experimental Design. Journal of Machine Learning Research: Workshop and Conference Proceedings. 2011:19–45. [Google Scholar]
- 24.Joachims T. Computer Science Technical Report. Carnegie Mellon University; 1996. A probabilistic analysis of the Rocchio algorithm with TFIDF for text categorization. CMU-CS-96-118. [Google Scholar]
- 25.Fan R, Chang K, Hsieh C, Wang X, Lin C. LIBLINEAR: A library for large linear classification. Journal of Machine Learning Research. 2008;9:1871–1874. [Google Scholar]
- 26.McCallum A, Nigam K. Proceedings of the International Conference on Machine Learning (ICML) Morgan Kaufmann; 1998. Employing EM in pool-based active learning for text classification; pp. 359–367. [Google Scholar]
- 27.http://www.causality.inf.ethz.ch/activelearning.php?page=evaluation#co
- 28.Wilcoxon F. Individual comparisons by ranking methods. Biometrics Bulletin. 1945 Dec;1(6):80–83. [Google Scholar]
- 29.Hochberg Y, Tamhane AC. Multiple Comparison Procedures. Hoboken, NJ: John Wiley & Sons; 1987. [Google Scholar]


