Abstract
Studying structural and functional connectivities of human cerebral cortex has drawn significant interest and effort recently. A fundamental and challenging problem arises when attempting to measure the structural and/or functional connectivities of specific cortical networks: how to identify and localize the best possible regions of interests (ROIs) on the cortex? In our view, the major challenges come from uncertainties in ROI boundary definition, the remarkable structural and functional variability across individuals and high nonlinearities within and around ROIs. In this paper, we present a novel ROI prediction framework that localizes ROIs in individual brains based on their learned fiber shape models from multimodal task-based functional magnetic resonance imaging (fMRI) and diffusion tensor imaging (DTI) data. In the training stage, shape models of white matter fibers are learnt from those emanating from the functional ROIs, which are activated brain regions detected from task-based fMRI data. In the prediction stage, functional ROIs are predicted in individual brains based only on DTI data. Our experiment results show that the average ROI prediction error is around 3.94 mm, in comparison with benchmark data provided by working memory and visual task-based fMRI. Our work demonstrated that fiber bundle shape models derived from DTI data are good predictors of functional cortical ROIs.
Keywords: brain network, diffusion tensor imaging, fMRI, ROI prediction, shape analysis
Introduction
Mapping structural and functional connectivities of human cerebral cortex via neuroimaging offers an exciting and unique opportunity to understand cortical architecture and thus has received significant interest (Friston et al. 2003; Goebel et al. 2003; Sporns et al. 2005; Biswal et al. 2010; Hagmann et al. 2010; Van Dijk et al. 2010). A fundamental question in mapping structural and functional connectivities is how to define the best possible regions of interests (ROIs) for the connectivity measurement. Essentially, when mapping human brain connectivities, ROIs provide the structural substrates for measuring connectivities within individual brains and for pooling data across population groups. Thus, identification of reliable, reproducible, and accurate ROIs is critically important for the success of connectivity mapping. However, in our view, this task is challenging for several critical reasons. 1) The boundaries between cortical regions are unclear. It is practically difficult to obtain in vivo cytoarchitectural or chemoarchitectural delineation of ROIs on the cerebral cortex. 2) Individual variability of cortical anatomy, connection, and function is remarkable. Quantitative mapping of the regularity, while accounting for the variability, of cortical structure and function is a challenging task. 3) The properties of ROIs are highly nonlinear. For instance, a slight change of the location of a ROI can dramatically alter its structural connectivity profiles (Li et al. 2010).
Many approaches for identifying cortical ROIs have been developed in the literature. Manual labeling by experts based on their domain knowledge, though widely used, is vulnerable to intersubject and intrasubject variation. Also, it is cumbersome and hardly reproducible. Data-driven methods (Baumgartner et al. 1997; McKeown et al. 1998; Filzmoser et al. 1999; Hansen et al. 1999; Lai and Fang 1999; Ngan and Hu 1999; Calhoun et al. 2001; Duann et al. 2002) via clustering ROIs from brain image itself are typically sensitive to the clustering parameters, and importantly, their neuroscience interpretation might not be clear. Image registration algorithm, for example, FSL FLIRT, HAMMER (Shen and Davatzikos 2002), diffusion tensor image (DTI) registration approaches (Yang et al. 2008), and others as listed in Klein et al. (2009), is another possible approach to identifying ROIs by warping a set of predefined ROIs in the template space into individual subjects. These warping-based ROIs identification methods might be vulnerable to anatomical variability across subjects and the clear functional meaning cannot be guaranteed. For 2 decades, ROI identification via task-based functional magnetic resonance imaging (fMRI) data has been widely used and regarded as the benchmark approach (Haynes et al. 2007; Logothetis 2008; Friston 2009). However, in many applications, there is no task-based fMRI data available. For example, it is challenging to acquire high-quality task-based fMRI data for elderly or children participants (Epstein et al. 2007; Jack et al. 2010). Additionally, the human brain is composed of many functional networks, such as working memory, vision, auditory, language, motor, attention, and emotion systems. Extensive acquisition of task-based fMRI data for all these networks is both time consuming and expensive, which makes it impractical for wide use. Instead, a typical DTI images scan needs less than 10 min, is much less demanding, and is widely available. Therefore, we are strongly motivated to accurately identify and predict functionally meaningful cortical ROIs based only on DTI data.
The close relationship between structural connectivity pattern and brain function has been reported in the literature (Passingham et al. 2002; Honey et al. 2009). An interesting observation from our recent results in Li et al. (2010) is that white matter (WM) fiber connection patterns of the same functional cortical ROI are reasonably consistent across different subjects, suggesting that fiber connection pattern might be a good predictor of functional ROI. This finding is in agreement with the “connectional fingerprint” concept presented in Passingham et al. (2002). Essentially, each brain’s cytoarchitectonic area has a unique set of extrinsic inputs and outputs, called the connectional fingerprint (Passingham et al. 2002), and this is crucial in determining the functions that each brain area performs. Therefore, fiber connectivity pattern may be good predictor of functional cortical ROI.
In this paper, we present a novel computational framework that learns fiber bundle models of functional cortical ROIs based on both task-based fMRI and DTI data in the training stage and applies the predictive models to locate functional ROIs in testing samples based only on DTI data in the prediction stage. Specifically, in the training stage, all subjects were linearly aligned to a standard space first. Then, functionally activated ROIs were detected from task-based fMRI, and the fiber bundles emanating from these ROIs were extracted. Finally, the trained model was learned and it includes 2 major components: the ROI coordinate principal component analysis (PCA) model and the fiber bundle templates. In the prediction stage, DTI data of an individual subject was aligned to the standard space first. Then, starting from the average location of warped ROIs from the training data set, an energy function was designed to iteratively optimize the ROIs’ locations. This energy function consists of internal and external terms. The internal term was measured by ROI coordinate reconstruction error, which corresponds to the ROI coordinate PCA model component; while the external term was defined by the Hausdorff metric of ROIs’ fiber bundles, which corresponds to the fiber bundle template component. Our experimental results demonstrated that average prediction error is approximately 3.9 mm, compared with benchmark ROIs derived from task-based fMRI data.
The major advantages of this computational framework and our contributions lie in the following 2 aspects. 1) In the training stage, the activated brain regions detected from task-based fMRI data provide the benchmark ROI data for learning shape models of WM fibers emanating from these ROIs. Our experimental results from working memory task-based fMRI data (Li et al. 2010) demonstrate that the fiber connection patterns of corresponding functional ROIs are quite consistent, providing direct evidence that WM fiber connection pattern is a good predictor of functional landmark (Honey et al. 2009). This is similar to the connectional fingerprint concept presented in Passingham et al. (2002). 2) In the prediction stage, only DTI data is needed to accurately locate the functional ROIs using our predictive models. Typically, a DTI scan takes less than 10 min and is widely available. Therefore, the general methodology of predicting functional cortical ROIs based on DTI will have wide applications in brain imaging in the future.
Materials and Methods
The flowchart of our ROI prediction framework is illustrated in Figure 1. It consists of 2 major stages: model training and ROI prediction. The model training (purple boxes) is performed on the data set in which each subject has both fMRI and DTI data. As such, we can obtain both the benchmark ROI locations and the fiber bundles emanating from them. In the first step (left purple box), multimodal data of all subjects were linearly aligned into the standard template space; then in the second step (right purple box), a PCA model of ROI locations is learned to characterize statistical relationship among ROI locations. Meanwhile, fiber bundle templates are obtained from the extracted fiber bundles for each ROI. The outputs of the training procedure consist of 4 components and are displayed in the red box. In the prediction stage (the green box in Fig. 1), a new subject with DTI and fMRI data was linearly aligned to the standard space and then the trained model (red box) was applied on the DTI data of the subject by minimizing an energy function starting from the initial ROIs location, thus generating the predicted ROIs. The predicted ROIs are then evaluated by comparing with the benchmark ROIs obtained from task-based fMRI data (black box).
Figure 1.
The flowchart of the model training and ROI prediction framework.
Data Acquisition and Preprocessing
Data set 1
Fifteen university students voluntarily participated in this study under IRB approval, and none of them was reported to have mental or physical disease. Each volunteer performed a modified version of the OSPAN task (3 blocks: OSPAN, Arithmetic, and Baseline) while fMRI data was acquired (Faraco et al. 2011). DTI scans were also obtained for each volunteer. Briefly, both fMRI and DTI scans were acquired on a 3-T GE Signa scanner with acquisition parameters as follows. fMRI: 64 × 64 matrix, 4-mm slice thickness, 220-mm field of view (FOV), 30 slices, time repetition (TR) = 1.5 s, time echo (TE) = 25 ms, ASSET = 2; DTI: 128 × 128 matrix, 2-mm slice thickness, 256-mm FOV, 60 slices, TR = 15 100 ms, TE = variable, ASSET = 2, 3 B0 images, 30 optimized gradient directions, b-value = 1000.
Data set 2
Ten healthy children performed a block-based visual task. Each block has a 30-s rest period and a 20-s task period when pictures were shown as stimuli. For each subject, DTI data were also obtained. Acquisition parameters for the scans were as follows. fMRI: 64 × 64 matrix, 4-mm slice thickness, 220-mm FOV, 30 slices, TR = 2 s, TE = 30 ms; DTI: 256 × 256 matrix, 3-mm slice thickness, 240-mm FOV, 50 slices, TR = 12 000 ms, TE = variable, 1 B0 volume, 15 DTI volumes, b-value = 1000.
The preprocessing of the DTI data is composed of brain skull removal, motion correction, and eddy current correction. Afterward, the diffusion tensor was computed and the fiber was tracked via the MEDINRIA (http://www-sop.inria.fr/asclepios/software/MedINRIA/) toolkit. The fMRI data of the OSPAN task was analyzed using the FSL FEAT (http://fsl.fmrib.ox.ac.uk/fsl/feat5/). Individual activation map reflecting the OSPAN (complex span) contrast was identified. For each subject, the DTI space is used as the standard space, in which the gray matter (GM)/WM surface is generated via an approach similar to that in Liu et al. (2007). The cortical surface was reconstructed from the GM/WM tissue map via the marching cubes algorithm and used as the ROI definition and prediction space. This will significantly reduce the ROI prediction search space from 3D to 2D. Coregistration between DTI and fMRI data is performed using the FSL FLIRT and the resulting global transform matrix is subsequently applied to the activated ROIs in order to map them into the DTI space and the cortical surface. In data set 1, 8 consistently activated working memory ROIs from OSPAN (OSPAN > BASELINE) contrast were used in this paper for ROI modeling and prediction (Table 1). In data set 2, 10 consistently activated cortical ROIs were used (Table 2). Among these subjects in each data set, a randomly selected subject is used as the template, onto which all the others are linearly registered. In this paper, hereafter, standard space denoted as has 2-fold meanings: DTI space for multimodal data of individual subject and the template subject, onto which all subjects are registered.
Table 1.
Eight activated working memory ROIs used in this paper
| ROI 1 | Left occipital pole |
| ROI 2 | Left paracingulate gyrus |
| ROI 3 | Left precuneus |
| ROI 4 | Right dorsolateral prefrontal cortex |
| ROI 5 | Right lateral occipital gyrus |
| ROI 6 | Right paracingulate gyrus |
| ROI 7 | Right precuneus |
| ROI 8 | Right superior frontal gyrus |
Table 2.
Ten activated visual cortex ROIs used in this paper
| ROI 1 | Left occipital pole |
| ROI 2 | Left lateral occipital cortex |
| ROI 3 | Left inferior frontal gyrus |
| ROI 4 | Left dorsolateral prefrontal cortex |
| ROI 5 | Left frontal pole |
| ROI 6 | Right postcentral gyrus |
| ROI 7 | Right occipital pole |
| ROI 8 | Right superior temporal gyrus |
| ROI 9 | Right inferior frontal gyrus |
| ROI 10 | Right frontal pole |
Model Training
Before developing models from the training subjects, we applied the approaches in Li et al. (2010) as a preprocessing step. In Li et al. (2010), 16 most stable and functionally activated working memory ROI locations were extracted individually, and a joint ROI optimization model that considers group-wise anatomical, structural, and functional information was applied to obtain the optimized ROI locations. After this initial optimization, both structural and functional connectivities derived from the ROI locations are more consistent across subjects (Li et al. 2010), which will facilitate the ROI prediction in this paper.
Then, we constructed 2 models for each functional ROI, which are the statistical distribution PCA model for the ROIs locations within the template space and the fiber bundle models of the ROIs. These learned models will be used as the prior information for ROI prediction in the prediction stage. It is noteworthy that a ROI location is defined to be a single vertex on the surface onto which the functional activation peaks from fMRI data are mapped.
PCA Model for ROIs Locations
Despite considerable variation, the spatial distribution patterns of the ROIs have certain degree of consistency (Fig. 2a,b). In this paper, the spatial distribution patterns of ROIs in the training set were captured and modeled by their coordinates PCA model, which is widely used in the biomedical image analysis community. Figure 2a,b illustrates the distribution of the ROIs in the standard space in each data set. We can observe the considerable regularity of the ROI distribution across subjects.
Figure 2.
(a) Visual evaluation of the effectiveness of ROI coordinate PCA model for data set 1. (b) Visual evaluation of the effectiveness of ROI coordinate PCA model for data set 2. (c) Examples of fiber bundles from ROI #8 of different subjects in data set 1. In (a) and (b), the same ROIs are coded with the same color.
Specifically, to train the ROI coordinate PCA model, we first put the coordinates of all ROIs of one subject into a vector as: where n is the number of ROIs. This vector can also be explained as the feature vector of ROI distribution pattern of one subject. Then, the standard PCA method was conducted on feature matrix where m is the number of training subjects. As a result, the 2 components of the ROI coordinate PCA model were obtained: the mean feature and the PCA transformation matrix which consists of p eigen vectors of M corresponding to the largest p eigen values that altogether cover 90% of variance.
Fiber Bundle Models of ROIs
Fiber bundle model is constructed for each ROI separately across the training subjects to embed group-wise fiber shape information. Taking ROI i for example, we define as the local region centered at location of ROI i on subject j’s surface, and fiber bundle penetrating was extracted and denoted as . Then, we construct a fiber bundle template set for ROI i by simply gathering all the fiber bundles from different subjects in it, denoted as where m is the number of training subjects. As an example, Figure 2c shows fiber bundle template set of data set 1, from which it is noted that fiber bundles of the same corresponding ROI across subjects are reasonably consistent in terms of overall shapes and connectivity patterns.
Similarity Measurement
In this section, we will focus on measuring how close a subject to be predicted is to the training ones in terms of the 2 models introduced in Model training. Regarding ROI spatial distribution patterns, the ROI coordinates of the subject to be predicted were projected and reconstructed by the trained PCA model in PCA Model for ROIs Locations, and the reconstruction error was defined as the distance measurement, denoted as . Specifically, the ROI coordinates of the subject to be predicted was extended to be , in the same way as that in PCA Model for ROIs Locations. Then, it was projected and reconstructed from the trained PCA model as follows:
| (1) |
Then, the obtained and were converted into the conventional spatial coordinate format, and the Euclidian distance, represented by , was computed between them, which is the reconstruction error defined as the measurement of how close its distributive pattern is to the training ones.
To measure the similarity between fiber bundles, we adopted the Hausdorff metric, a simple but effective measurement (Zöllei et al. 2010). In our problem, although the overall shape patterns of fiber bundles of the same corresponding ROI are reasonably consistent across subjects, there is still considerable variation in local fiber shapes (Fig. 2c). Therefore, by considering a fiber bundle as a set of individual fiber curves, we measure the supremum of the infimum of the distances between 2 fiber curve sets (2 bundles). Denote 2 fiber bundles as and , and let and be the fiber curve. The Hausdorff metric is defined as follows (Henrikson 1999):
| (2) |
where sup represents the supremum of a set, inf is the infimum of a set, and denotes the distance between 2 fiber curves. In this paper, was defined as the minimum distance between 2 fiber curves.
It should be noted that although we had already applied global alignment between subjects, the distance caused by the ROI location variation across subjects should be eliminated before computing the Hausdorff metric. Currently, we adopted a linear registration method to globally align fiber bundles of corresponding ROIs.
ROI Prediction Framework
As already illustrated in Figure 1, ROI prediction for a subject starts with linearly registering it into the standard space . Then, the entire-brain fibers of the subject are extracted, and the triangulated cortical surface is reconstructed using the same method in Data Acquisition and Preprocessing. The core procedure of the ROI prediction framework is formularized and solved as an energy minimization problem. We defined the energy function as:
| (3) |
where the internal term is expressed by the PCA constraint of ROI coordinates and the external term is formulated by the Hausdorff measurement of fiber bundle similarity of all ROIs, and λ trades off the 2 terms.
Specifically, the external energy term is formulated as:
| (4) |
where n is the number of ROIs, the m is the number of training subjects, is the fiber bundle extracted from the subject to be predicted and is the template fiber bundles introduced in Fiber Bundle Models of ROIs. It is noted that the template fiber bundles in equally contribute to the Hausdorff metric. As to the internal energy term, it is directly formulated as the reconstruction error of the coordinates of all ROIs from the PCA model:
| (5) |
A whole-space search was conducted to minimize the energy function. The search starts from the mean ROIs location of the trained model and ends when the location becomes stable. In order to reduce the computation load, we conducted an iterative global search. In each iteration, the search space for each ROI is adaptable. Considering that potential ROI locations cannot vary too much because of consistent location distribution pattern (Fig. 2a,b), in the initial global search iteration, the search scope is uniformly set as 8-ring neighborhood on the surface. In the following kth iteration, we automatically set the search scope for each individual ROI as follows. Let be the distance that ROI i moves in the kth iteration, and be the search scope of ROI i in the kth iteration on the cortical surface. If is small, will be shrunk. In other word, . Then, forms the search space , on which the kth global search is conducted for all possible combinations of n ROIs. Denote as one combination of ROIs. To further prune the search space , we added a constraint to the energy function in equation (3) by eliminating candidates of ROI i in if the fiber bundle emanating from it satisfies . The constrained search space is denoted as , which induces the final form of equation (3) as:
| (6) |
Finally, the iterative global search stops if all ROI locations are stable and we have . It is worth noting that the 2 measurements and are computed and normalized into [0, 1] before the iterative global search begins.
Experimental Results
Evaluation of ROI Coordinate PCA Model and Hausdorff Metric
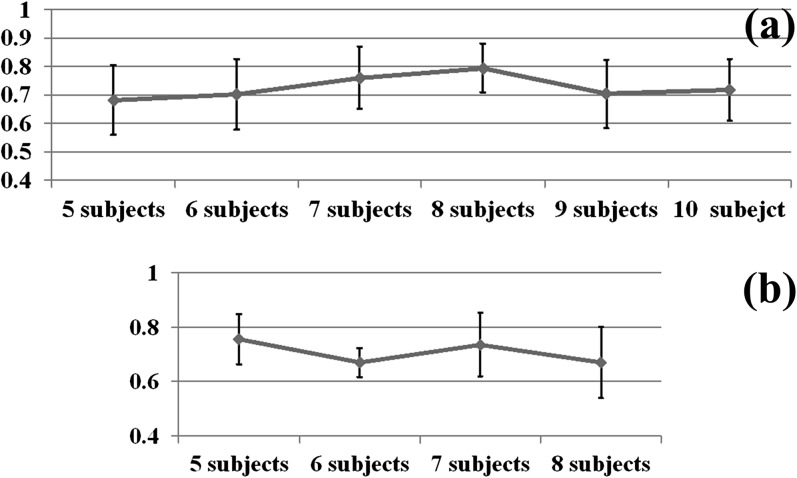
As the internal energy term, the ROI coordinate PCA model was used to constrain the ROI locations based on the assumption that ROI coordinates are roughly following Gaussian distribution. This assumption can be partly verified by checking whether the first principal component of matrix is relatively large, that is, the ratio of the first eigen value to the eigen value sum is expected to be large. Also, we explored if the subject number m will be an influencing factor of the consistency of ROI coordinate distribution. Therefore, we randomly select p subjects from one data set for 50 times and computed the 50 ratios of the first eigen value to the eigen value sum, as well as their means and standard deviations. The results shown in Figure 3 demonstrate 2 points: the ratio is as high as above 0.5; the number of subjects does not significantly affect the consistency of ROI coordinate distribution pattern. This result indicates that the PCA model of ROI locations is valid.
Figure 3.
The ratio of the first eigen value to eigen value sum, against the subject number. (a) Brain network in working memory task and (b) brain network in visual task.
In order to further demonstrate that the PCA model of ROI location coordinates really captures the intrinsic relationship of the common human brain architecture, we designed and performed the following experiment on the working memory data set. This experiment also aims to demonstrate that the reconstruction error defined in PCA Model for ROIs Locations can be used as a meaningful constraint in ROI prediction. Taking one subject from data set 1, for example, we uniformly sampled 100 groups of the 8 ROIs in the neighborhood of each one. The 100 groups of 8 ROIs were then projected and reconstructed by the trained PCA model and thus generated the reconstructed errors. Five subjects from data set 1 were randomly chosen for this experiment. Additionally, the sampling range, that is, sizes of neighborhood, was considered as another parameter. The results in Figure 4 show the mean reconstruction error of each 100-group test. The mean trend line for each subject shows that the minima are always at the original ROI locations for each subject, suggesting that the ROIs identified by task-based fMRI have much less variation in terms of their spatial locations, in comparison to random samplings around their neighborhoods. As the random sampling range was enlarged, grows accordingly. This result demonstrates the validity of for use as the meaningful constraint and similarity measurement in ROI prediction.
Figure 4.
Trend lines of against sampling range. The unit is mm for vertical axis.
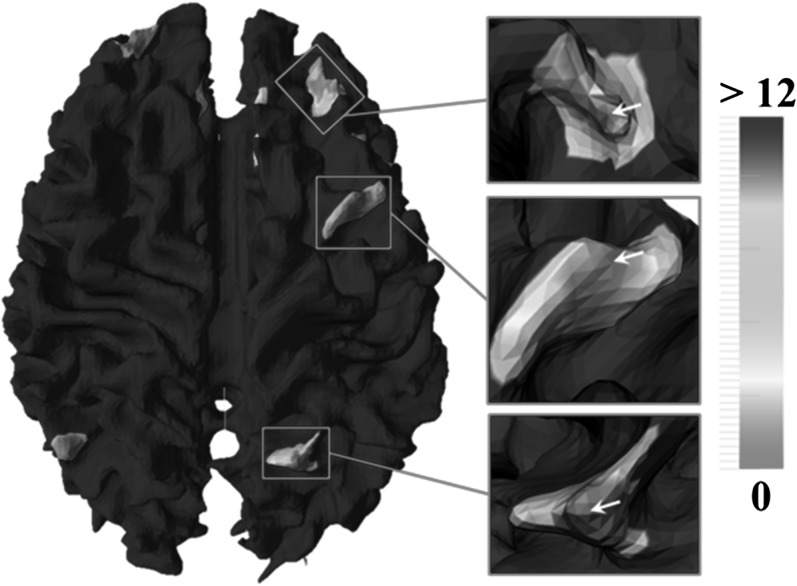
In order to evaluate the effectiveness of Hausdorff metric for fiber bundles, we performed an experiment on the working memory data set to examine the similarity measurements between fiber bundles, as shown in Figure 5. For each benchmark ROI, we extracted fiber bundles for every vertex within its neighborhood. Then, Hausdorff metric were computed between those fiber bundles and the one extracted from the benchmark ROI itself. The local region used to extract fiber bundle is 3-ring neighborhood size here. Afterward, the Hausdorff metrics were color coded and mapped onto the corresponding neighboring vertices. By visual inspection, a rough Gaussian-like distribution centered at the original ROI location (white arrows) can be found, which suggests that the shape pattern of fiber bundle is a distinctive feature and the Hausdorff metric is an effective measurement for fiber bundle similarity. In other words, a ROI’s fiber bundle can distinguish itself from other neighbors. This characteristic of uniqueness is much desired in ROI search and prediction.
Figure 5.
Hausdorff metric measured between fiber bundles extracted from ROI foci and their neighboring vertices. White arrows: original ROI locations. The unit for Hausdorff metric is mm.
ROI Prediction
We used the leave-one-out strategy to evaluate the ROI prediction framework on the 2 data sets individually. In all the experiments, λ was assigned as 0.3 in the global search. In this paper, we did not consider the impact of ROI size. Therefore, the local region centered at location of ROI i on subject j’s surface and from which fiber bundle emanating was extracted, was uniformly assigned to be the size of 3-ring neighborhood.
As the subject to be predicted has task-based fMRI data, the fMRI-derived ROIs are used as the benchmark data to evaluate the ROI prediction results. Table 3 and Table 4 show the Euclidian distances between the predicted ROIs and the benchmark data for ROIs, that is, the prediction errors for all the leave-one-out experiments. Most of the prediction errors are approximately 2–4 mm, which is 1–2 voxels in DTI volumes. On average, the average prediction errors for ROIs are 3.9 ± 2.18 mm and 3.99 ± 2.22 mm in data set 1 and data set 2, respectively. It is interesting that there are ROIs (#2, #3, #6, #8 in data set 1 and #2, #4, #6, #9 in data set 2) that have average prediction errors below 3 mm, which is considered as very accurate.
Table 3.
Distances between the predicted ROIs and the benchmark ROIs (mm) in the working memory task-based fMRI data set
| Subject | ROI 1 | ROI 2 | ROI 3 | ROI 4 | ROI 5 | ROI 6 | ROI 7 | ROI 8 | Subject mean |
| 1 | 9.13 | 2.21 | 1.87 | 4.7 | 3.71 | 1.45 | 2.37 | 2.81 | 3.53 |
| 2 | 5.87 | 2.31 | 2.54 | 5.32 | 2.44 | 3.09 | 8.85 | 2.65 | 4.13 |
| 3 | 9.25 | 2.96 | 2 | 5.1 | 2.99 | 2.36 | 1.59 | 2.78 | 3.63 |
| 4 | 6.17 | 2.1 | 3.11 | 3.93 | 1.95 | 4.85 | 3.78 | 2.15 | 3.51 |
| 5 | 6.03 | 1.11 | 2.81 | 3.35 | 5.04 | 2.19 | 8.06 | 2.2 | 3.85 |
| 6 | 7.5 | 3.07 | 2.67 | 5.89 | 2.61 | 2.93 | 8.18 | 3.19 | 4.51 |
| 7 | 6.44 | 4.05 | 3.19 | 1.64 | 1.53 | 2.67 | 7.54 | 3.71 | 3.85 |
| 8 | 5.07 | 1.45 | 2.87 | 2.79 | 5.33 | 1.25 | 7.72 | 2.07 | 3.57 |
| 9 | 6.08 | 2.93 | 3.35 | 4.71 | 2.11 | 4.78 | 3.77 | 2.48 | 3.78 |
| 10 | 6.92 | 2.89 | 3.84 | 4.75 | 2.31 | 6.3 | 3.3 | 2.39 | 4.09 |
| 11 | 9.6 | 3.65 | 2.93 | 1.53 | 1.73 | 2.89 | 8.12 | 2.96 | 4.18 |
| 12 | 7.98 | 3.5 | 3.47 | 2.18 | 2.07 | 2.59 | 7.59 | 3.03 | 4.05 |
| 13 | 7.98 | 3.14 | 2.79 | 5.87 | 2.84 | 3.57 | 7.99 | 3.52 | 4.71 |
| 14 | 9.59 | 1.98 | 2.02 | 5.14 | 3.5 | 1.09 | 1.54 | 2.37 | 3.4 |
| 15 | 5.72 | 2.17 | 2.7 | 2.52 | 5.52 | 1.92 | 8.21 | 1.62 | 3.8 |
| ROI ± mean | 7.29 ± 1.55 | 2.64 ± 0.82 | 2.81 ± 0.55 | 3.96 ± 1.51 | 3.05 ± 1.31 | 2.93 ± 1.45 | 5.91 ± 2.78 | 2.66 ± 0.57 |
Note: Means across ROIs and subjects are in bold fonts.
Table 4.
Distances between the predicted ROIs and the benchmark ROIs (mm) in the vision task-based fMRI data set
| Subject | ROI 1 | ROI 2 | ROI 3 | ROI 4 | ROI 5 | ROI 6 | ROI 7 | ROI 8 | ROI 9 | ROI 10 | Subject mean |
| 1 | 4.27 | 3.14 | 3.82 | 2.89 | 2.09 | 3.14 | 3.53 | 8.3 | 2.95 | 5.16 | 3.93 |
| 2 | 9.61 | 1.04 | 5.7 | 1.86 | 5.67 | 1.95 | 4.64 | 2.85 | 0.93 | 2.32 | 3.66 |
| 3 | 9.94 | 4.33 | 6.87 | 1.05 | 3.92 | 2.19 | 3.73 | 5.06 | 0 | 4.39 | 4.15 |
| 4 | 9.1 | 3.96 | 2.27 | 2.6 | 5.95 | 2.85 | 1.35 | 7.87 | 1.24 | 6.15 | 4.33 |
| 5 | 7.38 | 1.54 | 4.04 | 4.45 | 5.4 | 2.47 | 5.13 | 3.8 | 1.91 | 5.1 | 4.12 |
| 6 | 6.9 | 2.36 | 1.7 | 4.69 | 2.33 | 3.02 | 3.7 | 6.69 | 2.88 | 5.91 | 4.02 |
| 7 | 7.89 | 2.63 | 2.78 | 1.54 | 2.19 | 3.5 | 4.75 | 5.26 | 3.49 | 5.14 | 3.92 |
| 8 | 8.77 | 2.84 | 4.8 | 1.36 | 5.03 | 2.01 | 3.38 | 6.65 | 2.07 | 5.85 | 4.28 |
| 9 | 8.35 | 3.52 | 2.03 | 3.15 | 4.85 | 3.52 | 1.79 | 6.35 | 1.76 | 6.82 | 4.21 |
| 10 | 7.63 | 1.46 | 1.28 | 1.38 | 2.11 | 2.44 | 3.87 | 5.97 | 1.01 | 5.7 | 3.29 |
| ROI ± mean | 7.98 ± 1.63 | 2.68 ± 1.1 | 3.53 ± 1.84 | 2.5 ± 1.3 | 3.95 ± 1.62 | 2.71 ± 0.58 | 3.59 ± 1.21 | 5.88 ± 1.7 | 1.82 ± 1.07 | 5.25 ± 1.23 |
Note: Means across ROIs and subjects are in bold fonts.
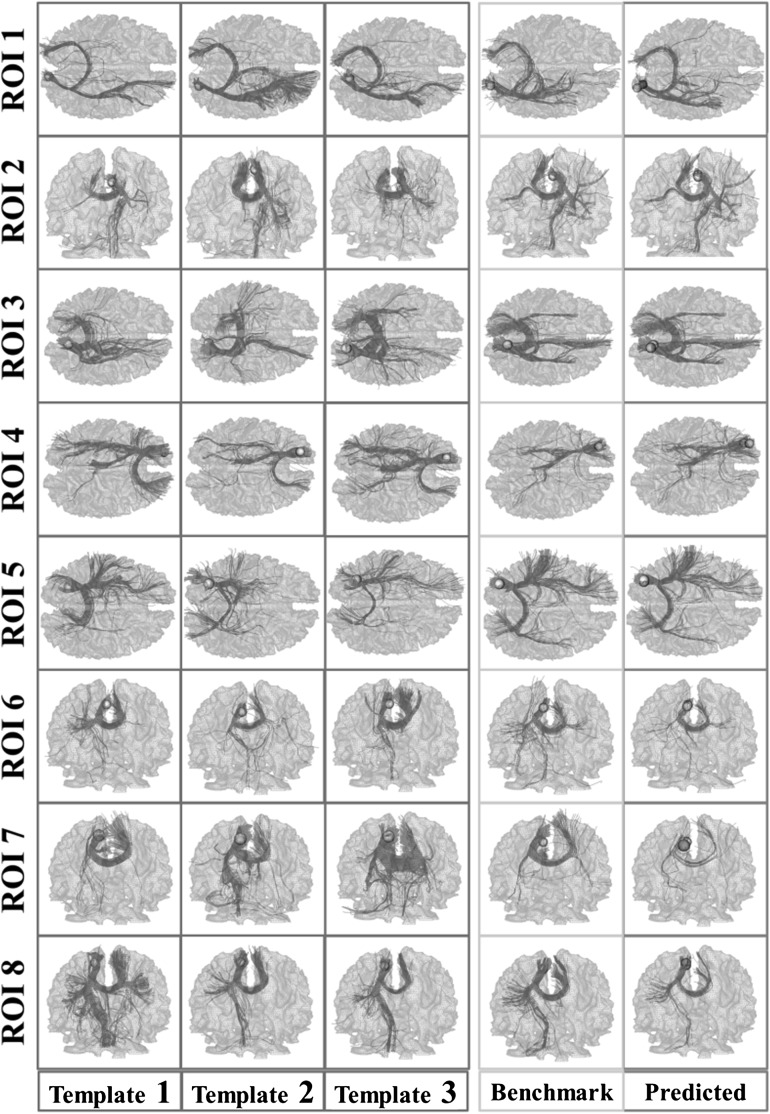
To visualize the consistency of fiber bundles of the predicted ROIs, we showed the fibers emanating from predicted ROIs and fibers emanating from benchmark ROIs for one randomly selected subject from each data set in Figure 6 and Figure 7, respectively. Corresponding fiber bundles of 3 subjects randomly selected from the training subjects are also shown for comparison. It is evident that the fiber bundles of the predicted ROIs (column 5) are quite similar to those of the benchmark ROIs derived from task-based fMRI (column 4), indicating the effectiveness and accuracy of our ROI prediction framework. It is also evident that the fiber bundles of the predicted ROIs and benchmark ROIs are quite similar to those of the corresponding ROIs of the training subjects (column 1–3). These results reveal that there are consistent fiber connection patterns for corresponding functional ROIs in different brains, which provides the neuroscience foundation and technical feasibility for the work in this paper.
Figure 6.
Visualization of fiber bundles of the subjects in data set 1. Yellow frames: fiber bundles emanating from benchmark ROIs (yellow bubbles); Gray frames: the template fiber bundles emanating from corresponding ROIs of 3 training subjects (yellow bubbles); Green frames: fiber bundles emanating from predicted ROIs (green bubbles), and benchmark ROIs in the yellow frame are also displayed for comparison.
Figure 7.
Visualization of fiber bundles of the subject in data set 2. Yellow frames: fiber bundles emanating from benchmark ROIs (yellow bubbles); Gray frames: the template fiber bundles emanating from corresponding ROIs of 3 training subjects (yellow bubbles); Green frames: fiber bundles emanating from predicted ROIs (green bubbles), and benchmark ROIs in the yellow frame are also displayed for comparison.
As an important parameter, the influence of λ in the energy function equation (6) is explored. We tested it on one randomly selected subject in data set 1 by ranging λ from 0.0 to 1.0, and the corresponding mean prediction errors of the 8 ROIs were computed and illustrated in Figure 8. We can observe that the prediction error is the smallest when λ is around 0.3. Therefore, in this paper, λ was assigned as 0.3 for all the experiments.
Figure 8.
The impact of λ on the prediction result. Values of λ are labeled by the horizontal axis, while the prediction errors are represented by the vertical axis with the unit mm.
Notably, some ROIs (e.g., #1, #4, #7 in data set 1 and #1, #5, #8, #10 in data set 2) have relatively large prediction errors, and as an example, the corresponding fiber bundles of those in data set 1 (#1, #4, #7 in data set 1) are shown and examined in Figure 9. We can see that although the distance between the predicted ROI and the benchmark one is relatively large (7.29, 3.96, and 5.91 mm), their fiber bundle similarity is still high, either by the overlapping views (Fig. 9, 1a–3a) or separated views (Fig. 9, 1b–3b and Fig. 9, 1c–3c). This visualization suggests that our energy minimization procedure in ROI Prediction Framework might be trapped in a local minimum, thus resulting in those predicted ROIs converged to a wrong location with similar emanating fiber bundle. In the future, other constraint terms will be considered to further reduce the chance of being trapped in local minima during the energy minimization procedure.
Figure 9.
Fiber bundle illustration for ROI #1, #4, and #7 (sub-fig 1, sub-fig 2, and sub-fig 3). (a) Fiber bundles emanating from predicted ROI and benchmark ROI are overlapped; (b) Fiber bundles emanating from predicted ROI; (c) Fiber bundles emanating from benchmark ROI; (d–f) Template fiber bundles of ROI in the training data set. Yellow bubbles: benchmark ROI locations; Green bubble: predicted ROI locations.
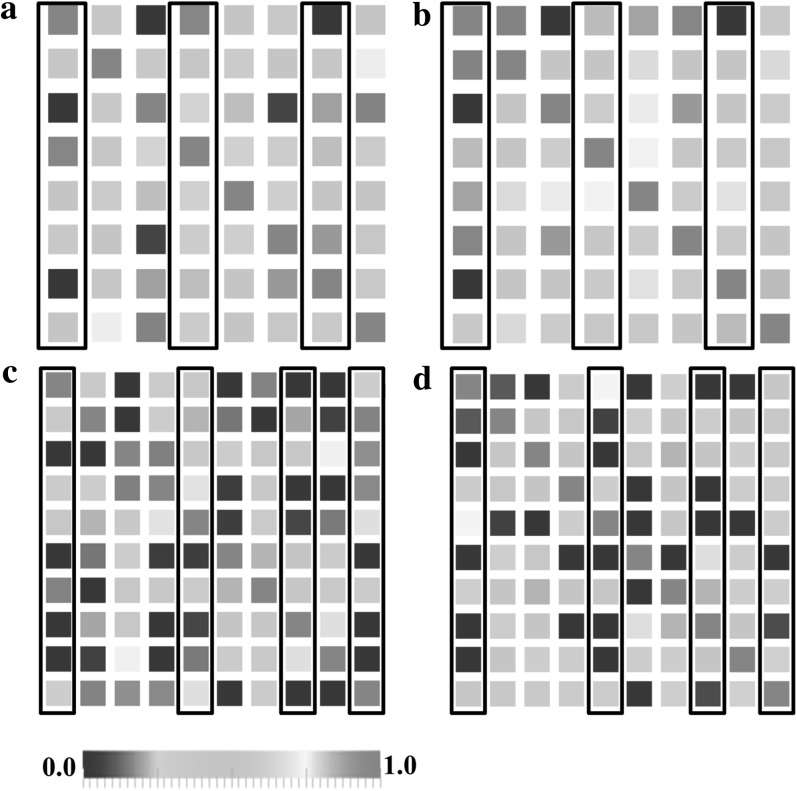
Finally, the functional connectivity matrices between ROIs of one randomly selected subject from each data set were shown in Figure 10. The connectivity matrix was computed as follows: we mapped the ROIs into the fMRI data space, and for each ROI, functional signals from the 27-neighborhood voxels were extracted and averaged. The Pearson correlation was computed between any pair of ROIs. We computed the connectivity matrix for both the benchmark ROIs and the predicted ROIs. In Figure 10, only minor difference between a and b, or c and d, can be observed, demonstrating that the functional connectivity measured by our predicted ROIs is very close to the real connectivity. Importantly, the columns of the ROIs with relatively large prediction errors are highlighted, from which we can find that there is no direct relationship between the prediction error and functional connectivity changes.
Figure 10.
Functional connectivities between ROIs. The color bar is at the bottom. (a) Functional connectivity matrix of the benchmark ROIs of one subject from data set 1. (b) Functional connectivity matrix of the predicted ROIs of one subject from data set 1. (c) Functional connectivity matrix of the benchmark ROIs of one subject from data set 2. (d) Functional connectivity matrix of the predicted ROIs of one subject from data set 2. The columns with relatively large prediction errors (#1, #4, #7 in data set 1 and #1, #5, #8, #10 in data set 2) are highlighted by black frames.
Comparison Studies
For the purpose of comparison, we conducted ROI prediction by linear (via FSL FLIRT), nonlinear registration (via the HAMMER software package [Shen and Davatzikos 2002]), and DTI registration approaches (denoted as DTI Reg) (Yang et al. 2008). The prediction procedure is as follows. Taking the linear registration method for instance, we randomly chose a subject as a template from the training data set and registered its fMRI image and the corresponding ROI location into its DTI space. Then, we registered its fractural anisotropy image obtain from DTI data onto that of a subject from the testing data set via FSL FLIRT. Finally, the predicted ROIs were obtained by applying the registration transform on the ROI location of the template, and the distances between the predicted ROIs and benchmark ones from the testing subject were computed.
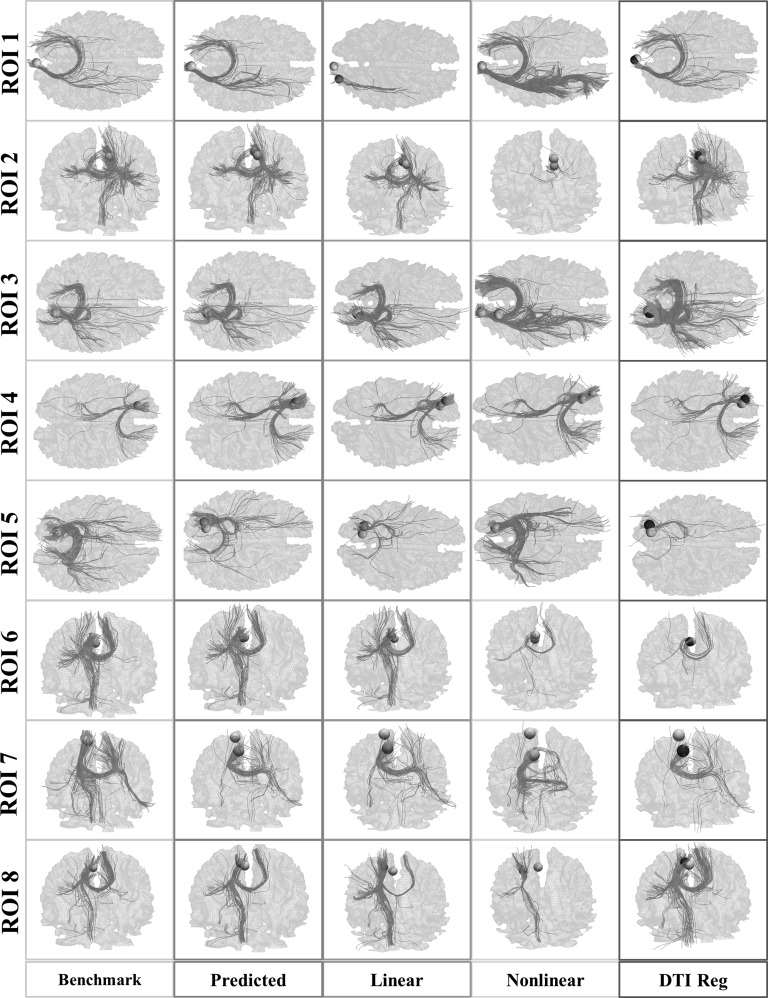
The mean prediction errors for the 3 methods are shown in Table 5. On average, the prediction errors by FSL FLIRT, HAMMER, and DTI Reg are 5.72, 5.53, and 4.94 mm, respectively. As can be seen, our method (3.94 mm) significantly outperforms them. The fiber bundles emanating from predicted ROIs using the 3 methods of one subject were shown in Figure 11. The differences in comparison with the benchmark ones can be easily inspected.
Table 5.
Prediction errors by the FSL FLIRT, HAMMER, and DTI-based registration algorithm (mm)
| ROI 1 | ROI 2 | ROI 3 | ROI 4 | ROI 5 | ROI 6 | ROI 7 | ROI 8 | Mean | |
| FLIRT | 7.48 | 4.95 | 6.24 | 6.18 | 5.41 | 4.51 | 6.46 | 4.54 | 5.72 |
| HAMMER | 6.18 | 4.92 | 6.40 | 7.50 | 5.15 | 3.06 | 6.08 | 4.95 | 5.53 |
| DTI Reg | 6.40 | 4.97 | 4.84 | 7.50 | 2.70 | 5.15 | 3.06 | 4.86 | 4.94 |
Note: Means across ROIs and subjects are in bold fonts.
Figure 11.
Visualization of fiber bundles emanating from predicted ROIs using FSL FLIRT (purple column), HAMMER (light blue column), and DTI Reg (dark blue column) methods. Our prediction (green column) and the benchmark bundles (yellow column), which are the same as columns 4 and 5 in Figure 6, are also shown in the left most columns for comparison. Yellow bubbles: benchmark ROIs; Green bubbles: predicted ROIs via our method; Light blue bubbles: predicted ROIs by HAMMER; purple bubbles: predicted ROIs by FLIRT; dark blue bubbles: predicted ROIs by DTI Reg.
Conclusions
We presented a novel framework for functional brain ROI prediction using the working memory and vision networks as test beds. From the training subjects which include both DTI and fMRI data, we trained the model of ROI location distribution and the model of fiber bundles emanating from functionally cortical ROIs. Predicting ROIs on testing subjects was performed by matching the trained models onto the DTI data of individual subjects. This problem was formulated as an energy minimization one, in which fiber bundle shape and ROI location pattern were, respectively, considered as external and internal terms. Experimental results demonstrated that the ROI prediction framework has promising performance, in comparison to other methods based on linear and nonlinear image registration algorithms.
It should be noted that our ROI prediction framework might be trapped into local minima for some ROIs (as illustrated in Fig. 9), although global search is conducted. This may be caused by the fiber bundle similarity measurement (Hausdorff metric) used in this paper, which should be further investigated. Moreover, variability across subjects in terms of ROI’s structural and anatomical locations and ROI size may be other reasons. Therefore, in the near future, we plan to investigate improved metrics for fiber bundle similarity and model the variability of the ROI's structural and functional profiles with ROI size problem considered. For instance, the group replicator dynamics approach (Ng et al. 2009) could possibly be used to model the variability of the structural and functional connectivities among the ROIs. Notably, our methods focus on the most consistent ROIs at current stage. In the future, we will consider adding other less consistent ROIs into the prediction framework and evaluate the performance of ROI prediction.
Currently, the computational pipeline takes around a few minutes in the training stage and, approximately, 4 h in the prediction stage on a typical PC computing environment. It is worth noting that in the future, optimization strategies like developing robust fiber shape features that require less computation load and automatically pruning the search space could significantly reduce the computation load.
Our work has demonstrated that fiber bundle shape models of functional brain ROIs have remarkable prediction capability, providing direct support to the connectional fingerprint concept (Passingham et al. 2002). In the future, we plan to apply and evaluate this ROI prediction framework in other brain networks, such as executive function, attention, and semantics memory systems and apply and validate this framework on clinical data sets such as the DTI data sets of Alzheimer’s disease and Autism.
Funding
National Institutes of Health Career Award (NIH EB 006878); University of Georgia start-up research funding (to T.L.); Doctorate Foundation of Northwestern Polytechnic University; Chinese Government Scholarship (to T.Z.); The National Natural Science Foundation of China (30830046 to L.L.); The National 973 Program of China (2009CB918303 to L.L.).
Acknowledgments
Conflict of Interest : None declared.
References
- Baumgartner R, Scarth G, Teichtmeister C, Somorjai R, Moser E. Fuzzy clustering of gradient-echo functional MRI in the human visual cortex. Part I: reproducibility. J Magn Reson Imaging. 1997;7:1094–1101. doi: 10.1002/jmri.1880070623. [DOI] [PubMed] [Google Scholar]
- Biswal BB, Mennes M, Zuo XN, Gohel S, Kelly C, Smith SM, Beckmann CF, Adelstein JS, Buckner RL, Colcombe S, et al. Toward discovery science of human brain function. Proc Natl Acad Sci U S A. 2010;107:4734–4739. doi: 10.1073/pnas.0911855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pearlson GD, Pekar JJ. Spatial and temporal independent component analysis of functional MRI data containing a pair of task-related waveforms. Hum Brain Mapp. 2001;13:43–53. doi: 10.1002/hbm.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duann JR, Jung TP, Kuo WJ, Yeh TC, Makeig S, Hsieh JC, Sejnowski JT. Single-trial variability in event-related BOLD signals. Neuroimage. 2002;15:823–835. doi: 10.1006/nimg.2001.1049. [DOI] [PubMed] [Google Scholar]
- Epstein JN, Casey BJ, Tonev ST, Davidson M, Reiss AL, Garrett A, Hinshaw SP, Greenhill LL, Vitolo A, Kotler LA, et al. Assessment and prevention of head motion during imaging of patients with attention deficit hyperactivity disorder. Psychiatry Res. 2007;155:75–82. doi: 10.1016/j.pscychresns.2006.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraco CC, Unsworth N, Lagnely J, Terry D, Li K, Zhang D, Liu Tianming, Miller LS. Complex span tasks and hippocampal recruitment during working memory. Neuroimage. 2011;55(2):773–787. doi: 10.1016/j.neuroimage.2010.12.033. [DOI] [PubMed] [Google Scholar]
- Filzmoser P, Baumgartner R, Moser E. A hierarchical clustering method for analyzing functional MR images. Magn Reson Imaging. 1999;17:817–826. doi: 10.1016/s0730-725x(99)00014-4. [DOI] [PubMed] [Google Scholar]
- Friston K. Modalities, modes, and models in functional neuroimaging. Science. 2009;326:399–403. doi: 10.1126/science.1174521. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Harrison L, Penny W. Dynamic causal modeling. Neuroimage. 2003;19:1273–1302. doi: 10.1016/s1053-8119(03)00202-7. [DOI] [PubMed] [Google Scholar]
- Goebel R, Roebroeck A, Kim DS, Formisano E. Investigating directed cortical interactions in time-resolved fMRI data using vector autoregressive modeling and Granger causality mapping. Magn Reson Imaging. 2003;21:1251–1261. doi: 10.1016/j.mri.2003.08.026. [DOI] [PubMed] [Google Scholar]
- Hagmann P, Cammoun L, Gigandet X, Gerhard S, Ellen Grant P, Wedeen V, Meuli R, Thiran JP, Honey CJ, Sporns O. MR connectomics: principles and challenges. J Neurosci Methods. 2010;194:34–45. doi: 10.1016/j.jneumeth.2010.01.014. [DOI] [PubMed] [Google Scholar]
- Hansen LK, Larsen J, Nielsen FA, Strother SC, Rostrup E, Savoy R, Lange N, Sidtis J, Svarer C, Paulson OB. Generalizable patterns in neuroimaging: how many principal components? Neuroimage. 1999;9:534–544. doi: 10.1006/nimg.1998.0425. [DOI] [PubMed] [Google Scholar]
- Haynes JD, Sakai K, Rees G, Gilbert S, Frith C, Passingham RE. Reading hidden intentions in the human brain. Curr Biol. 2007;17:323–328. doi: 10.1016/j.cub.2006.11.072. [DOI] [PubMed] [Google Scholar]
- Honey CJ, Sporns O, Cammoun L, Gigandet X, Thiran JP, Meuli R, Hagmann P. Predicting human resting-state functional connectivity from structural connectivity. Proc Natl Acad Sci U S A. 2009;106:2035–2040. doi: 10.1073/pnas.0811168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrikson J. Completeness and total boundedness of the Hausdorff metric. MIT Undergrad J Math. 1999;1:69–80. [Google Scholar]
- Jack CR, Jr., Bernstein MA, Borowski BJ, Gunter JL, Fox NC, Thompson PM, Schuff N, Krueger G, Killiany RJ, Decarli CS, et al. Update on the magnetic resonance imaging core of the Alzheimer's disease neuroimaging initiative. Alzheimers Dement. 2010;6:212–220. doi: 10.1016/j.jalz.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang MC, Christensen GE, Collins DL, Gee J, Hellier P, et al. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage. 2009;46:786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai SH, Fang M. A novel local PCA-based method for detecting activation signals in fMRI. Magn Reson Imaging. 1999;17:827–836. doi: 10.1016/s0730-725x(99)00038-7. [DOI] [PubMed] [Google Scholar]
- Li K, Guo L, Faraco C, Zhu D, Deng F, Zhang T, Jiang X, Zhang D, Chen H, Hu X, et al. Individualized ROI optimization via maximization of group-wise consistency of structural and functional profiles. Proceedings of NIPS ′10; 2010 Dec 6–10; In: Lafferty J, Williams CKI, Shawe-Taylor J, Zemel RS, Culotta A, editors. Vancouver, Canada: Neural Information Processing Systems; 2010. http://books.nips.cc/nips23.html. [Google Scholar]
- Liu T, Li H, Wong K, Tarokh A, Guo L, Wong ST. Brain tissue segmentation based on DTI data. Neuroimage. 2007;38:114–123. doi: 10.1016/j.neuroimage.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logothetis NK. What we can do and what we cannot do with fMRI. Nature. 2008;453:869–878. doi: 10.1038/nature06976. [DOI] [PubMed] [Google Scholar]
- McKeown MJ, Makeig S, Brown GG, Jung TP, Kindermann SS, Bell AJ, Sejnowski TJ. Analysis of fMRI data by blind separation into independent spatial components. Hum Brain Mapp. 1998;6:160–188. doi: 10.1002/(SICI)1097-0193(1998)6:3<160::AID-HBM5>3.0.CO;2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng B, Abugharbieh R, McKeown MJ. Discovering sparse functional brain networks using group replicator dynamics (GRD) Inf Process Med Imaging. 2009;21:76–87. doi: 10.1007/978-3-642-02498-6_7. [DOI] [PubMed] [Google Scholar]
- Ngan SC, Hu X. Analysis of functional magnetic resonance imaging data using self-organizing mapping with spatial connectivity. Magn Reson Med. 1999;41:939–946. doi: 10.1002/(sici)1522-2594(199905)41:5<939::aid-mrm13>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Passingham RE, Stephan KE, Kötter R. The anatomical basis of functional localization in the cortex. Nat Rev Neurosci. 2002;3:606–616. doi: 10.1038/nrn893. [DOI] [PubMed] [Google Scholar]
- Shen D, Davatzikos C. HAMMER: hierarchical attribute matching mechanism for elastic registration. IEEE Trans Med Imaging. 2002;21:1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- Sporns O, Tononi G, Kötter R. The human connectome: a structural description of the human brain. PLoS Comput Biol. 2005;1:e42. doi: 10.1371/journal.pcbi.0010042. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk KR, Hedden T, Venkataraman A, Evans KC, Lazar SW, Buckner RL. Intrinsic functional connectivity as a tool for human connectomics: theory, properties, and optimization. J Neurophysiol. 2010;103:297–321. doi: 10.1152/jn.00783.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Shen D, Davatzikos C, Verma R. Diffusion tensor image registration using tensor geometry and orientation features. Med Image Comput Comput Assist Interv. 2008;11:905–913. doi: 10.1007/978-3-540-85990-1_109. [DOI] [PubMed] [Google Scholar]
- Zöllei L, Stevens A, Huber K, Kakunoori S, Fischl B. Improved tractography alignment using combined volumetric and surface registration. Neuroimage. 2010;51:206–213. doi: 10.1016/j.neuroimage.2010.01.101. [DOI] [PMC free article] [PubMed] [Google Scholar]