Abstract
Objectives
Sasang constitutional medicine is a traditional Korean medicine in which an individual is classified into one of four types of constitution: Taeum (TE), Soeum (SE) Soyang (SY), and Taeyang (TY). These constitution types are determined with biologic and physiologic characteristics, so it has been assumed that genetic factors are associated with each constitution type. Identifying the genetic elements underlying each constitution is necessary for the elucidation of the molecular mechanism of Sasang constitutional medicine.
Design
A total of 341,998 genetic loci across the whole genome were genotyped for 1222 subjects of defined constitution type. The genetic loci associated with each constitution type were identified and the functional connectivity of genes within these loci was analyzed using statistical text mining.
Results
From the difference in allele frequencies between constitution types, significant genetic loci associated with each type were identified. Chromosomes 3q27.3 (rs10937331, p=2.71×10−6), 15q22.2 (rs7180547, p=1.58×10−6), and 14q22.3 (rs12431592, p=1.31×10−6) were most significantly associated with TE, SE, and SY constitution types, respectively. From the functional relationship analysis using all loci with a p-value≤10−4, genes associated with each constitution type were identified. Fifteen (15) genes, including GPM6A, SYT4, and GRIK1, were significantly associated with the TE constitution type (p<0.05); 12 genes, including DRGX and AKAP11, were significantly associated with the SE constitution type (p<0.05); and 17 genes, including ZFP42, CDH22, ALDH1A2, OTX2, and EN2, were significantly associated with the SY constitution type (p<0.05).
Conclusions
Genetic loci and genes associated with Sasang constitution types were systematically identified from a genome-wide association study using a large number of subjects.
Introduction
Since the completion of the Human Genome Project, it has been possible to elucidate the genetic elements that underlie the differences between individuals. In spite of rapid progress in recent years, the majority of this research is still conducted at the laboratory level. An alternative and practical approach to laboratory research would be subgrouping the human population according to homogeneous biologic characteristics.
Sasang constitutional medicine is a Korean traditional medicine, in which a person is classified into one of four constitution types—Taeum (TE), Soeum (SE), Soyang (SY), and Taeyang (TY) —based on the nature of his/her physiologic and physical characteristics.1,2 The balance among the physiologic functions of four representative internal organs—lung, spleen, liver, and kidney—is the most important factor for determining the Sasang constitution (SC) type. Previous reports have shown genetic polymorphism associated with SC types on some genes. For example, FTO and MC4R polymorphisms are associated with control of body mass according to SC types.3 Interleukin-1α and -β polymorphisms were also reported to be associated with SC type in obese women.4,5 These results support the idea of genetic involvement in the determination of constitution type.6 However, considering that diverse genetic elements may be involved in the determination of SC type, a single-gene-based approach has limitations for determining complex traits at the genome level. Recently, a genome-wide linkage analysis identified SC-linked genomic regions on chromosome 8 and 11.7 In addition, diverse genetic loci associated with SC types were reported in a genome-wide association (GWA) study of 60 subjects.8 A more expanded population study would be valuable in identifying the genetic elements associated with SC types with a higher degree of statistical certainty.
To address this issue, a GWA study was performed using 1222 subjects to identify the genetic elements associated with SC type. In addition, using a bioinformatics analysis, genes that were significantly related to SC type were identified.
Materials and Methods
Subjects
A total of 1348 subjects who visited Korean Oriental hospitals between 2006 and 2009 were initially included in this study. All experiments were conducted in accordance with the Institutional Guidelines for Human Experimentation set by the ethics committee of the Korea Institute of Oriental Medicine (KIOM). All samples and clinical information were deposited in the KIOM Databank of Sasang Constitutional Medicine. Informed consent was obtained from all subjects for the use of samples and clinicopathological data for research purposes. In order to diagnose SC type, subjects were prescribed a constitution-specific herbal medicine including Panax ginseng, Ephedra herba, and Schisandra chinensis for treatment of their major physical discomfort, as described previously.7 After subjects took the medicine for 30 days or more, improvement of pre-existing symptoms or occurrence of adverse effects was recorded. Constitution types were determined only for subjects showing clear improvements in their chief complaints without any adverse effects.
Genotyping and quality control
Genomic DNA was isolated from the peripheral blood of subjects and was genotyped using an Affymetrix Genome Wide Human SNP array 5.0, as described previously.7 Bayesian Robust Linear Modeling, using a Mahalanobis Distance genotyping algorithm, was used for calling 440,092 genotypes (over 95% call rate) of single nucleotide polymorphisms (SNPs). From a final total of 1222 samples, 12,039 markers were discarded following a Hardy-Weinberg equilibrium test (p<0.001) and 86,324 markers following minor allele frequency <0.01, which left 341,998 SNPs for subsequent analysis.
Population structure
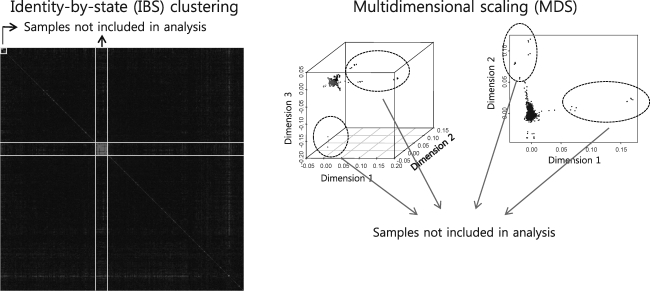
Population structure was measured with identity by state (IBS) and by multidimensional scaling (MDS) of genotypes, calculated with PLINK 1.07,9 using all autosomal markers present in a chip. Calculated IBS values were clustered to estimate the relative distance of the relationship between each pair of individuals. Eighty-three (83) subjects showing IBS similarity over 0.8 were excluded because of their closeness in relationships. From the MDS plot, first- and second-dimension values of 0.05 (arbitrary unit) were used to discard 27 subjects showing heterogeneous genotypes. Sixteen (16) subjects diagnosed with an obscure constitution type were also excluded from the analysis. A final total of 1222 subjects were included in this study. Table 1 shows the clinical implication of constitution type, as measured by analysis of variance in R 2.11.0 (http://cran.r-project.org).
Table 1.
Clinical Implications of Sasang Constitution Types
| |
Male |
|
Female |
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Variables | TE (n=239) | SE (n=103) | TY (n=8) | SY (n=130) | pa | TE (n=273) | SE (n=199) | TY (n=11) | SY (n=259) | pa |
| Age (year) | 46.1±15.4 | 41.5±16.5 | 51.7±14.5 | 48.1±16.0 | 0.0094 | 50.3±14.7 | 43.9±14.8 | 41.7±10.4 | 45.1±14.5 | <0.0001 |
| Height (cm) | 169.9±7.1 | 169.6±7.6 | 169.5±4.4 | 168.6±5.8 | 0.39 | 157.0±5.6 | 158.3±6.0 | 161.7±3.3 | 157.3±6.0 | 0.0093 |
| Weight (kg) | 73.7±11.0 | 61.7±8.7 | 66.6±8.7 | 65.9±8.2 | <0.0001 | 62.1±9.4 | 52.7±6.5 | 56.1±9.6 | 55.1±6.9 | <0.0001 |
| Aspartate aminotransferase (IU/L) | 25.2±8.2 | 24.5±11.9 | 18.6±3.0 | 23.5±8.3 | 0.12 | 23.7±14.6 | 20.7±5.5 | 23.1±10.4 | 21.7±8.5 | 0.022 |
| Alanine aminotransferase (IU/L) | 29.3±14.7 | 23.8±23.2 | 16.4±6.0 | 25.9±15.7 | 0.011 | 22.7±25.8 | 16.8±7.8 | 20.8±16.2 | 19.9±19.0 | 0.018 |
| Alkaline phosphatase (IU/L) | 70.6±43.5 | 75.4±54.1 | 67.1±9.4 | 72.1±41.8 | 0.83 | 62.7±27.6 | 58.4±22.2 | 59.0±19.5 | 58.9±23.8 | 0.21 |
| Total cholesterol (mg/dL) | 192.0±37.2 | 177.9±32.1 | 182.9±38.9 | 184.3±31.5 | 0.0061 | 194.9±37.87 | 181.5±33.3 | 173.5±28.2 | 188.5±33.5 | 0.00042 |
| Triglycerides (mg/dL) | 162.2±115.5 | 113.7±65.0 | 138.8±52.4 | 166.4±111.9 | 0.00043 | 132.7±83.5 | 91.3±46.0 | 106.9±62.1 | 109.3±62.9 | <0.0001 |
| High-density lipoprotein (mg/dL) | 39.8±9.4 | 43.8±9.1 | 36.8±6.5 | 40.6±10.6 | 0.0034 | 44.9±11.3 | 51.0±12.1 | 45.5±9.8 | 48.6±12.0 | <0.0001 |
| Low-density lipoprotein (mg/dL) | 111.9±31.6 | 100.3±26.1 | 109.5±30.4 | 103.1±27.1 | 0.0030 | 112.3±32.4 | 98.7±28.7 | 96.0±21.0 | 105.6±30.5 | <0.0001 |
| Bilirubin (mg/dL) | 0.82±0.29 | 0.86±0.42 | 0.70±0.24 | 0.82±0.37 | 0.51 | 0.70±0.25 | 0.74±0.27 | 0.84±0.31 | 0.78±0.34 | 0.024 |
| Blood urea nitrogen (mg/dL) | 15.4±4.0 | 14.5±3.7 | 16.0±4.9 | 14.9±4.3 | 0.20 | 14.2±4.0 | 13.5±3.8 | 13.3±4.0 | 13.6±3.4 | 0.15 |
Significance was measured by analysis of variance.
TE, Taeum; SE, Soeum; TY, Taeyang; SY, Soyang.
Association analysis
Allele frequencies between each constitution type (case) and other constitution types (control) were compared in a χ2 test using PLINK.9 For multiple adjustments, false discovery rate (FDR) was measured for each allele. To construct quantile–quantile plots, the distribution of observed allelic p-values was plotted against the theoretical distribution of expected p-values. The genomic control inflation factor, λ, was calculated by dividing median χ2 statistics by 0.456. Genomic control was not corrected for, because the inflation factor did not deviate greatly from 1.0 for any of the tests. Subjects with a TY constitution type were excluded due to small sample size (8 males and 11 females). The chromosomal loci were visualized with LocusZoom.10
Text-mining-based network analysis
The degree of functional relationship between genes within loci identified by GWA study was quantified using the bioinformatic approach tool Gene Relationships Across Implicated Loci (GRAIL).11,12 With GRAIL, the functional similarity between genes was measured by applying a text-mining method to a database of PubMed abstracts. SNPs with an association p-value≤10−4 from the GWA study were selected as input for GRAIL. After identifying the candidate genes within a region of an allele, the statistical significance (GRAIL p-value) of the functional relationship between genes was estimated with a null model of the random chance of relationship between genes.
Results
Sasang constitution
From an initial set of 1348 genotyped subjects, 83 subjects were excluded from the study due to close relationships revealed by IBS clustering, 27 subjects were excluded because of heterogeneous genotypes revealed by MDS, and 16 subjects were excluded because of unclear SC type. As a result, 1222 subjects were included in the present study. IBS and MDS patterns are shown in Supplementary Figure 1. Table 1 shows the clinical association of SC types for selected variables. Interestingly, the TE constitution type for both genders showed significant increases in body weight, total cholesterol level, and low-density lipoprotein level (p<0.01).
GWA analysis for SC
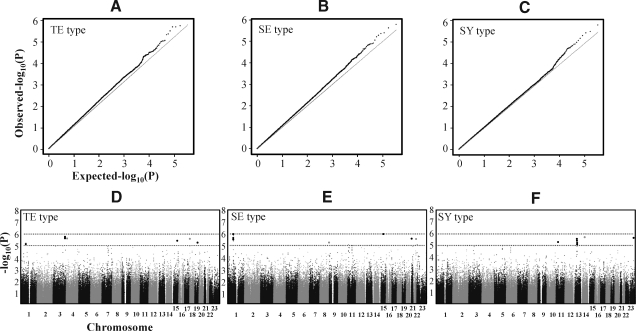
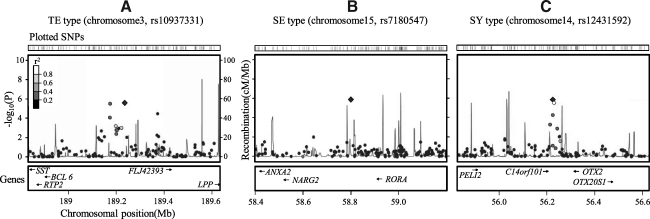
A χ2 analysis for differences in allele frequencies between constitution types was performed. A very small number of subjects (8 males and 11 females) displayed a TY constitution type, so this type was not included in the GWA study. Quantile–quantile plots for each constitution type showed that the distribution of observed p-values deviated from expected p-values in the tail (Figs. 1A–1C). The genome-wide distribution of p-values across all chromosomes for each constitution type is shown in Figs. 1D–1F. Loci with a p-value<10−7 were not detected in all constitution types. Table 2 presents a list of the five most significant SNPs for each constitution type. Although some alleles showed a significance level (p-value) of 10−6, the measured FDR was relatively high. For example, rs10937331 in the TE constitution type showed a p-value of 2.71×10−6 (odds ratio, 1.61), but the FDR value was 0.31. Other constitution types also showed similar levels of statistical significance (i.e., rs7180547, p=1.58×10−6 for SE constitution type and rs12431592, p=1.31×10−6 for SY constitution type). The detailed locus information for the most significant SNPs on chromosomes is shown in Figure 2, which also summarizes information regarding nearby genes, recombination rate, and linkage disequilibrium (LD).
FIG. 1.
Quantile–quantile plots (A–C) and Manhattan plots (D–F) of the p-values of alleles from an association analysis of the constitution types. For quantile–quantile plots, -log10 (observed p-values) were compared with the -log10(expected p-values) under the null distribution for each constitution type ([A] for Taeum (TE) constitution type, [B] for Soeum [SE] constitution type, and [C] for Soyang [SY] constitution type). For Manhattan plots, -log10(p-values) of alleles for each constitution type ([D] for TE constitution type, [E] for SE constitution type, and [F] for SY constitution type) were plotted against chromosomal position. In parts D–F, the upper dotted line indicates p-values of 10−6 and the lower dotted line indicates p-values of 10−5.
Table 2.
Association Between Single Nucleotide Polymorphisms (SNPs) and Sasang Constitution (SC) Types According to Genome-Wide Association Data
| |
|
|
|
|
Allele |
|
|
Minor allele frequency |
|
||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Constitution | SNPa | Locus | Position | Nearby genes | Major | Minor | p | Odds ratiob | Case | Control | FDR |
| TE | rs10937331 | 3q27.3 | 189230771 | LPP | A | G | 2.71×10−6 | 1.61 (1.31,1.96) | 0.24 | 0.16 | 0.31 |
| rs1719953 | 18p11.31 | 5515037 | EPB41L3 | C | T | 3.50×10−6 | 0.58 (0.46, 0.73) | 0.12 | 0.19 | 0.31 | |
| rs4608810 | 4p15.33 | 13708281 | A | G | 3.58×10−6 | 1.55 (1.28, 1.87) | 0.28 | 0.20 | 0.31 | ||
| rs12903518 | 15q25.1 | 77324324 | ANKRD34C, TMED3, KIAA1024 | C | T | 5.43×10−6 | 0.37 (0.24, 0.58) | 0.026 | 0.067 | 0.37 | |
| rs10426235 | 19q13.2 | 46191656 | CYP2B6, CYP2A7P1, CYP2A13, | A | G | 7.39×10−6 | 1.89 (1.42, 2.50) | 0.13 | 0.077 | 0.39 | |
| SE | rs7180547 | 15q22.2 | 58789125 | RORA, NARG2, ANXA2 | T | C | 1.58×10−6 | 2.49 (1.70, 3.67) | 0.081 | 0.034 | 0.35 |
| rs554298 | 1p31.1 | 78377202 | DNAJB4, PTGFR, GIPC2 | T | C | 2.31×10−6 | 1.65 (1.33, 2.03) | 0.29 | 0.20 | 0.35 | |
| rs2830793 | 21q21.3 | 27528341 | GPXP2, ADAMTS5, ADAMTS1, | G | A | 3.92×10−6 | 1.74 (1.37, 2.22) | 0.20 | 0.12 | 0.35 | |
| rs5758072 | 22q13.2 | 39497818 | SLC25A17, ST13, DNAJB7, | T | C | 4.28×10−6 | 1.57 (1.29, 1.91) | 0.38 | 0.28 | 0.35 | |
| rs2735902 | 8q24.3 | 146052174 | ZNF7, COMMD5, ZNF250, | A | T | 8.76×10−6 | 0.61 (0.50, 0.76) | 0.23 | 0.32 | 0.38 | |
| SY | rs12431592 | 14q22.3 | 56213993 | C14orf101, OTX2, PELI2 | C | G | 1.31×10−6 | 1.53 (1.29, 1.83) | 0.43 | 0.33 | 0.36 |
| rs9591534 | 13q14.3 | 53261862 | G | C | 4.23×10−6 | 0.49 (0.36, 0.67) | 0.073 | 0.13 | 0.36 | ||
| rs11033499 | 11p13 | 36193893 | LDLRAD3, COMMD9, PRR5L | G | C | 7.10×10−6 | 2.18 (1.54, 3.08) | 0.086 | 0.041 | 0.37 | |
| rs749539 | 7q36.3 | 154911201 | EN2, CNPY1, BLACE | G | C | 1.27×10−5 | 1.46 (1.23, 1.73) | 0.54 | 0.44 | 0.37 | |
| rs12484954 | 22q13.1 | 37511542 | DNAL4, UNC84B, NPTXR | G | C | 1.35×10−5 | 1.76 (1.36, 2.28) | 0.15 | 0.091 | 0.37 | |
SNPs of linkage disequilibrium static r2>0.5 were regarded as SNPs present in single block.
Numbers in parentheses represent 95% confidence interval.
FDR, false discovery rate; TE, Taeum; SE, Soeum; SY, Soyang.
FIG. 2.
Genomic region of identified loci. The upper panel indicates the location of plotted single nucleotide polymorphisms (SNPs). In the middle panel, -log10 (p-values) are displayed for loci (circles) distributed in a genomic region (0.8 Mb) to either side of the SNPs (diamonds) most strongly associated with each constitution type ([A] for Taeum [TE] constitution type, [B] for Soeum [SE] constitution type, and [C] for Soyang [SY] constitution type). Genomic positions are based on the UCSC browser. Genotyped SNPs were colored based on their correlation with the strongest SNP (white: r2>0.8; gray: 0.4<r2<0.6; black: r2<0.2). Estimated recombination rates (cM per Mb) and linkage disequilibrium information are from HapMap PhaseII (JPT+CHB). The lower panels show the locations of known genes in the region.
Selection of candidate genes associated with SC
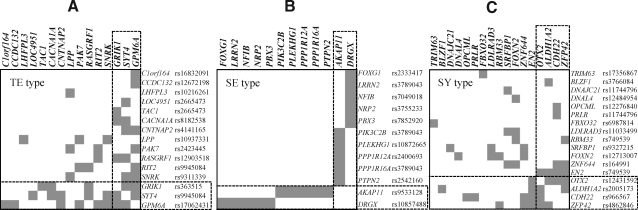
In general, a GWA study focuses on searching for SNPs showing the lowest p-value. As such, the loci of SNPs with p-values around the 10−4 level have not typically been included in genome-wide studies. However, it has been reported that crucial genes can be identified when including these less significant SNPs.11,12 Therefore, text mining was performed based on a network analysis of genes using all loci with p-values≤10−4. For TE, SE, and SY constitution types, 53, 58, and 49 SNPs were included in the analysis, respectively. After selecting the neighboring genes of each locus, relationships among these genes were measured. From a plot of the inter-relatedness of genes (Fig. 3), we found the 15 genes most significantly associated with the genes of the TE constitution type, which included GPM6A, SYT4, and GRIK1 (GRAIL p-value<0.05). In the same way, 12 genes, including DRGX and AKAP11, were significantly associated with the genes of the SE constitution type (p<0.05), and 17 genes, including ZFP42, CDH22, ALDH1A2, OTX2, and EN2, were significantly associated with the genes of the SY constitution type (p<0.05).
FIG. 3.
Interconnectivity among genes in constitution type-associated loci. The literature-based functional connectivity among genes within loci of single nucleotide polymorphisms (SNPs) having p-values less than 10−4 were measured using a Gene Relationships Across Implicated Loci (GRAIL) algorithm and displayed based on the level of interconnectedness. The names of genes and their corresponding SNPs associated with constitution type ([A] for Taeum [TE] constitution type, [B] for Soeum [SE] constitution type, and [C] for Soyang [SY] constitution type) are indicated. The dotted line indicates highly interconnected genes.
Discussion
Classifying human populations based on individual constitution is common in many traditional medicines including Traditional Chinese Medicine and Ayurveda, an ancient system of personalized medicine in India. In all classification methods, the types of constitution are determined by physiologic and physical characteristics of an individual supporting the possible involvement of the genetic elements in constitution determination.6,13–15 Considering the complex nature of the constitution, whole genome approach would be one of the most effective methods to establish genetic basis for constitution, as has been considered in research on Ayurvedic genomics.13 For example, using whole genome expression, diverse genes correlated with the phenotypic class of constitutional types were identified in Ayurveda.16 Recently, GWAs using linkage7 and association methods8 were applied to define SC-associated genetic elements. One of these GWA studies isolated the SNPs associated with SC types in 60 subjects.8 However, the sample size was too small to identify constitution type-associated genetic elements, especially using a high throughput analysis such as a GWA analysis. Therefore, in this study the sample size was greatly increased to 1222 subjects. Although this number still was not sufficient for the precise identification of the genetic loci associated with SC types using a GWA analysis, highly significant genetic loci (with a p-value level of 10−6) associated with each constitution type were obtained (Table 2).
When these results were compared with those of Yin et al.,8 there were no common genetic loci or related genes. Two (2) major factors could contribute to this lack of similarity. In addition to the difference in sample sizes used, the two studies also used different methods for diagnosing an individual's SC type. A determination based on herbal drug response was used. This diagnostic method was originally reviewed1 and, thereafter, has been widely used to determine SC type,2 whereas Yin et al. used a questionnaire (Sasang constitution classification II [QSCCII]) and an interview with a SC specialist to determine an individual's SC type without information about drug response. Although both methods for diagnosing SC type have been designed to prevent subjective diagnosis, there is still the problem of intervention of subjectivity in the diagnosing process such as evaluation by medical doctors. Moreover, the correlation between two diagnosing methods has not been fully investigated. Therefore, a new SC diagnosing tool is now being developed that can operate automatically using only objective physical measurements.
Care is needed when interpreting SC type based only on a couple of genetic elements with high significance levels. Moreover, the high FDR values associated with each SNP show the possible presence of error. The high FDR in this study might result from the heterogeneous nature of phenotypes comprising each SC type. Although we could overcome this phenomenon by greatly increasing the number of test subjects, lower p-values do not always guarantee the identification of SNPs that are actually associated with SC types. Furthermore, biologically significant genetic elements can have less significant p-values. Therefore, as many SNPs as possible, including those with less significant p-values, should be incorporated into the GWA analysis. In the present study, a text-mining-based approach (GRAIL) was applied,11,12 in which the functional similarity among SNPs having p-values<10−4 was estimated. Highly interconnected genes, such as GPM6A, DRGX, and ZFP42 for TE, SE, and SY constitution types, respectively, were selected as the best candidate genes associated with SC type (Fig. 3). To speculate on the main function of these genes from Figure 3, keywords describing functional connections among genes were also measured in GRAIL. Interestingly, neuron-related keywords were mainly enriched in TE types. For example, GPM6A,17 SYT4,18 GRIK1,19 LPP9,20 and CACNA1A21 were reported to be involved in neuronal function. For SE type, cell signaling genes such as AKAP11,22 PTPN2,23 and NRP224 were mainly enriched. For SY type, many genes including ZFP42,25 ALDH1A2,26 and OTX227 are known to be related with the developmental process. The relationship between these molecular functions and physiology of SC types should be further elucidated. Also, the relationships of these SNPs with candidate genes of SC types are now being validated in a more expanded population sample. In the future, the function of candidate genes on the determination of SC type should be studied in detail.
Conclusions
In conclusion, the genetic loci and genes associated with SC type were systematically identified in a large number of test subjects for the first time.
Supplementary Data
SUPPLEMENTARY FIG. 1.
Identity by state (IBS) and multidimensional scaling (MDS) patterns of genotypes. Calculated IBS values were clustered to estimate the relative distance of the relationship between each pair of individuals. Eighty-three (83) subjects showing over 0.8 of similarity of IBS were excluded because of their closeness in relationships. From the MDS plot, first- and second-dimension values of 0.05 (arbitrary unit) were used to discard 27 subjects showing heterogeneous genotypes.
Acknowledgments
This work was supported by National Research Foundation of Korea grant (NRF, No. 20100020617) and a Korea Institute of Oriental Medicine grant (KIOM, No. K10070) funded by the Korea government (MEST).
Disclosure Statement
No competing financial interests exist.
References
- 1.Lee J. Longevity & Life Preservation in Oriental Medicine. Seoul, Republic of Korea: Kyung Hee University Press; 1996. [Google Scholar]
- 2.Shim EB. Lee S. Kim JY. Earm YE. Physiome and sasang constitutional medicine. J Physiol Sci. 2008;58:433–440. doi: 10.2170/physiolsci.RV004208. [DOI] [PubMed] [Google Scholar]
- 3.Cha S. Koo I. Park BL, et al. Genetic Effects of FTO and MC4R polymorphisms on body mass in constitutional types. Evid Based Complement Alternat Med. 2009;(October 11) doi: 10.1093/ecam/nep162. e-pub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee JH. Kwon YD. Hong SH, et al. Interleukin-1 β gene polymorphism and traditional constitution in obese women. Int J Neurosci. 2008;118:793–805. doi: 10.1080/00207450701242883. [DOI] [PubMed] [Google Scholar]
- 5.Song JS. Jeong HJ. Kim SJ, et al. Interleukin-1α polymorphism −889C/T related to obesity in Korean Taeumin women. Am J Chin Med. 2008;36:71–80. doi: 10.1142/S0192415X0800559X. [DOI] [PubMed] [Google Scholar]
- 6.Kim BY. Cha S. Jin HJ. Jeong S. Genetic approach to elucidation of sasang constitutional medicine. Evid Based Complement Alternat Med. 2009;(suppl 1):51–57. doi: 10.1093/ecam/nep058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Won HH. Lee S. Jang E. Kim, et al. A genome-wide scan for the Sasang constitution in a Korean family suggests significant linkage at chromosomes 8q11.22-23 and 11q22.1-3. J Altern Complement Med. 2009;15:765–769. doi: 10.1089/acm.2009.0067. [DOI] [PubMed] [Google Scholar]
- 8.Yin CS. Park HJ. Chung JH, et al. Genome-wide association study of the four-constitution medicine. J Altern Complement Med. 2009;15:1327–1333. doi: 10.1089/acm.2009.0205. [DOI] [PubMed] [Google Scholar]
- 9.Purcell S. Neale B. Todd-Brown K, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pruim RJ. Welch RP. Sanna S, et al. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raychaudhuri S. Plenge RM. Rossin EJ, et al. Identifying relationships among genomic disease regions: Predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raychaudhuri S. Thomson BP. Remmers EF, et al. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat Genet. 2009;41:1313–1318. doi: 10.1038/ng.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patwardhan B. Bodeker G. Ayurvedic genomics: Establishing a genetic basis for mind-body typologies. J Altern Complement Med. 2008;14:571–576. doi: 10.1089/acm.2007.0515. [DOI] [PubMed] [Google Scholar]
- 14.Zwickey H. Schiffke HC. Genetic correlates of Chinese medicine: In search of a common language. J Altern Complement Med. 2007;13:183–184. doi: 10.1089/acm.2007.7026. [DOI] [PubMed] [Google Scholar]
- 15.Chen S. Lv F. Gao J, et al. HLA class II polymorphisms associated with the physiologic characteristics defined by Traditional Chinese Medicine: Linking modern genetics with an ancient medicine. J Altern Complement Med. 2007;13:231–239. doi: 10.1089/acm.2006.6126. [DOI] [PubMed] [Google Scholar]
- 16.Prasher B. Negi S. Aggarwal S, et al. Whole genome expression and biochemical correlates of extreme constitutional types defined in Ayurveda. J Transl Med. 2008;6:48. doi: 10.1186/1479-5876-6-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michibata H. Okuno T. Konishi N, et al. Human GPM6A is associated with differentiation and neuronal migration of neurons derived from human embryonic stem cells. Stem Cells Dev. 2009;18:629–639. doi: 10.1089/scd.2008.0215. [DOI] [PubMed] [Google Scholar]
- 18.Lalonde R. Strazielle C. Brain regions and genes affecting postural control. Prog Neurobiol. 2007;81:45–60. doi: 10.1016/j.pneurobio.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 19.Mozhui K. Karlsson RM. Kash TL, et al. Strain differences in stress responsivity are associated with divergent amygdala gene expression and glutamate-mediated neuronal excitability. J Neurosci. 2010;30:5357–5367. doi: 10.1523/JNEUROSCI.5017-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwindt H. Akasaka T. Zühlke-Jenisch R, et al. Chromosomal translocations fusing the BCL6 gene to different partner loci are recurrent in primary central nervous system lymphoma and may be associated with aberrant somatic hypermutation or defective class switch recombination. J Neuropathol Exp Neurol. 2006;65:776–782. doi: 10.1097/01.jnen.0000229988.48042.ae. [DOI] [PubMed] [Google Scholar]
- 21.Romaniello R. Zucca C. Tonelli A, et al. A wide spectrum of clinical, neurophysiological and neuroradiological abnormalities in a family with a novel CACNA1A mutation. J Neurol Neurosurg Psychiatry. 2010;81:840–843. doi: 10.1136/jnnp.2008.163402. [DOI] [PubMed] [Google Scholar]
- 22.Brannian J. Eyster K. Mueller BA, et al. Differential gene expression in human granulosa cells from recombinant FSH versus human menopausal gonadotropin ovarian stimulation protocols. Reprod Biol Endocrinol. 2010;8:25. doi: 10.1186/1477-7827-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleppe M. Lahortiga I. El Chaar T, et al. Deletion of the protein tyrosine phosphatase gene PTPN2 in T-cell acute lymphoblastic leukemia. Nat Genet. 2010;42:530–535. doi: 10.1038/ng.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neufeld G. Kessler O. Herzog Y. The interaction of Neuropilin-1 and Neuropilin-2 with tyrosine-kinase receptors for VEGF. Adv Exp Med Biol. 2002;515:81–90. doi: 10.1007/978-1-4615-0119-0_7. [DOI] [PubMed] [Google Scholar]
- 25.Galán A. Montaner D. Póo ME, et al. Functional genomics of 5- to 8-cell stage human embryos by blastomere single-cell cDNA analysis. PLoS One. 2010;5:e13615. doi: 10.1371/journal.pone.0013615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma AC. Chung MI. Liang R. Leung AY. A DEAB-sensitive aldehyde dehydrogenase regulates hematopoietic stem and progenitor cells development during primitive hematopoiesis in zebrafish embryos. Leukemia. 2010;24:2090–2099. doi: 10.1038/leu.2010.206. [DOI] [PubMed] [Google Scholar]
- 27.Sakurai Y. Kurokawa D. Kiyonari H, et al. Otx2 and Otx1 protect diencephalon and mesencephalon from caudalization into metencephalon during early brain regionalization. Dev Biol. 2010;347:392–403. doi: 10.1016/j.ydbio.2010.08.028. [DOI] [PubMed] [Google Scholar]