Summary
Across the eukaryotic phylogeny, offspring usually inherit their mitochondrial genome from only one of two parents: in animals, the female. While mechanisms that eliminate paternally derived mitochondria from the zygote have been sought, the developmental stage at which paternal transmission of mitochondrial DNA is restricted is unknown in most animals. Here, we show that mature Drosophila sperm lack mitochondrial DNA, and we uncover two processes that eliminate mitochondrial DNA during spermatogenesis. Visualization of mitochondrial DNA nucleoids revealed their abrupt disappearance from developing spermatids in a process requiring the mitochondrial nuclease, Endonuclease G. In Endonuclease G mutants, persisting nucleoids are swept out of spermatids by a cellular remodeling process that trims and shapes spermatid tails. Our results show that mitochondrial DNA is eliminated during spermatogenesis, thereby removing the capacity of sperm to transmit the mitochondrial genome to the next generation.
Introduction
Nearly all eukaryotes examined to date inherit mitochondrial DNA (mtDNA) from only one parent (Birky, 1995). This uniparental inheritance, which has intrigued geneticists since the 1950s, is a keystone principle used in tracing evolutionary lineages and population migrations, and is a fundamental attribute of numerous heritable diseases. Although the purpose of this restricted inheritance pattern is difficult to probe, one proposal suggests that uniparental inheritance exists to foil competition between mtDNAs; which can have outcomes that compromise the fitness of the organism (Hoekstra, 2000; Eberhard, 1981). For example, in S. cerevisiae, in which either mating type can transmit mtDNA, several mtDNA mutations that confer a replicative advantage spread to all progeny in a cross, despite also impairing aerobic respiration in the progeny (Ephrussi et al., 1955; Blanc and Dujon, 1980; de Zamaroczy et al., 1981). Several models have been proposed to explain the molecular basis of uniparental inheritance (Birky, 1995). In one simple model, fusion of asymmetrically sized gametes creates a zygote harboring mostly the mtDNA genotype of the larger gamete. However, uniparental inheritance occurs in the unicellular alga, Chlamydomonas reinhardtii, despite symmetrically sized gametes. Additionally, paternal mtDNA is reliably transmitted in blue mussels of the genus Mytilus, despite the relatively small size of the male gamete (Boynton et al., 1987; Zouros et al., 1994). Furthermore, active mechanisms preventing paternal transmission would eliminate competition more effectively than dilution effects. In metazoans, where mtDNA is usually maternally inherited, several active barriers have been proposed to limit paternal inheritance. In the Japanese medaka fish, Oryzias latipes, paternal mtDNA vanishes within sperm mitochondria after fertilization, and in bovine embryos, paternal mitochondria are eliminated during the first 2 zygotic cell divisions (Nishimura et al., 2006; Sutovsky and Navara, 1996). During the review of this manuscript, two studies in the nematode, Caenorhabditis elegans, identified a mechanism that eliminates the paternal mitochondria after fertilization (Rawi et al., 2011; Sato and Sato, 2011). To further define the molecular barriers to paternal inheritance in animals, we followed the fate of paternal mtDNA in Drosophila melanogaster, an experimentally tractable model organism in which mtDNA is maternally inherited (Rielly and Thomas, 1980). Here, we identify and characterize two developmental barriers to paternal mtDNA transmission that act prior to fertilization. In contrast to previously proposed barriers, these mechanisms, which remove mtDNA from sperm mitochondria during spermatogenesis, suggest that the father primarily enforces maternal-only mtDNA inheritance in D. melanogaster.
Results
Paternal mtDNA is Excluded from Mature Sperm and Fertilized Embryos
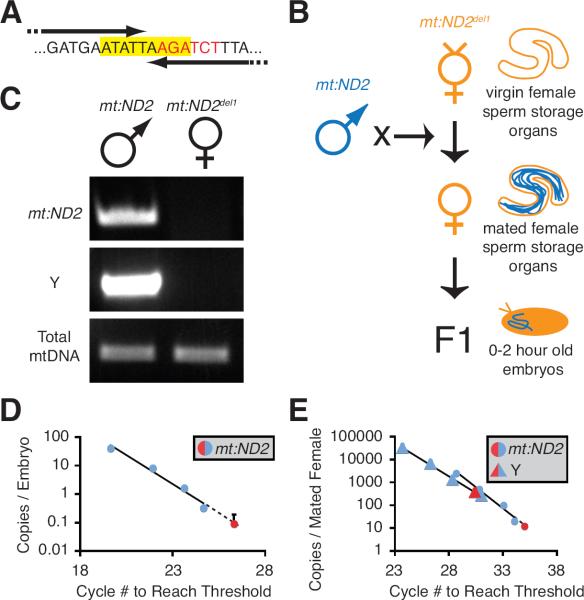
To initially characterize mtDNA inheritance in D. melanogaster, we used PCR to follow paternal mtDNA. Previously, we isolated a healthy fly line homoplasmic for a 9 base-pair deletion that eliminated a BglII site in the mt:ND2 gene of the mtDNA (mt:ND2del1) (Xu et al., 2008). PCR primers ending within the mt:ND2del1 deletion (Fig. 1A) amplified wild type (mt:ND2) but not mt:ND2del1 mtDNA (Fig. 1C). To quantify paternal mtDNA transferred to the egg, we crossed wild type males to mt:ND2del1 females and used quantitative PCR (qPCR) to measure wild type mtDNA in 0–2 hour old embryos (Fig. 1B). However, even though the large sperm mitochondria are transferred to the egg (Perotti, 1973), we did not detect paternal mtDNA above background (signal obtained from embryos in which both parents were mt:ND2del1), and we demonstrated that there is less than one paternal mitochondrial genome per egg (Fig. 1D; Fig. S1A). From this result, we conclude that little or no mtDNA is transferred to the egg, or that it is quickly eliminated after fertilization.
Figure 1. Paternal mtDNA is Excluded from Mature Sperm and Fertilized Embryos.
(A) Sequence of nucleotides deleted in mt:ND2del1 (yellow highlight), and the BgIII restriction site (red font) that was targeted in the generation of mt:ND2del1. Arrows show the 3' portion of PCR primers that anneal to wild type (mt:ND2), but not mutant (mt:ND2del1) mtDNA. (B) Cross scheme and experimental design. (C) PCR genotyping of parental lines that were crossed to follow wild type paternal mtDNA in (D,E). (D) qPCR measuring paternal mtDNA (mt:ND2) transferred to deletion mutant eggs (mt:ND2del1). The mean paternal mtDNA copy number (red dot) in relation to a standard curve (blue dots) is graphed on a “per embryo” basis. Error bar represents standard deviation in three embyro extracts. (E) qPCR measuring the amount of mtDNA (mt:ND2) in sperm stored by mt:ND2del1 females. Y chromosome (red a triangle) and sperm mtDNA (red circle) copy number in representative sperm storage organ extract in relation to standard curves (blue) are graphed on a “per female” basis. In (D,E), standard curves were generated by adding 100 to 12500 copies of mt:ND2 (blue circles) or Y chromosomal DNA (blue triangles) to control extracts, and graphed on a “per embryo” or “per female” basis. See also Fig. S1.
If mtDNA were quickly eliminated in the egg, then it would be still present in mature sperm. Mated females retain hundreds of mature sperm in sperm storage organs. In mt:ND2del1 sperm-storage-organ extracts, we counted the number of sperm with Y chromosome-specific primers and the number of paternal mtDNAs with mt:ND2-specific primers. Our experiments showed far less than one mitochondrial genome per sperm (Fig. 1E, Fig. S1B–D). Therefore, the mitochondrial genome, and hence the capacity of D. melanogaster sperm to contribute to mtDNA inheritance, is eliminated prior to fertilization.
mtDNA Nucleoids Disappear in Late Elongating Spermatids
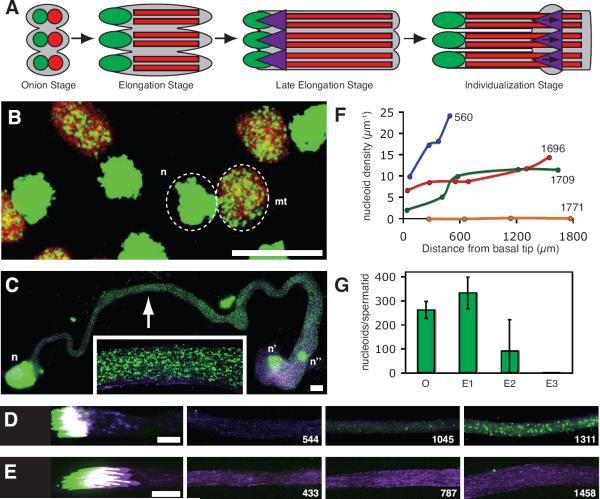
To follow the fate of mtDNA during spermatogenesis (Fig. 2A), we stained testes with PicoGreen (Ashley et al., 2005) (Fig. 2) and other fluorescent DNA binding dyes (Fig. S2). mtDNA appeared as bright puncta, nucleoids. Four mitotic and two meiotic cycles with incomplete cytokinesis produce a cyst of 64 spermatids that undergo extraordinary physical transformations in tight synchrony as they mature (Bate and Arias, 2009). In onion stage spermatids, PicoGreen staining was restricted to the nucleus and the nebenkern, a specialized spherical aggregation of mitochondria (Fig. 2B). An average of 261 nucleoids were counted in each nebenkern (Fig. 2G).
Figure 2. mtDNA Nucleoids Disappear During Spermatogenesis.
(A) Schematic summary of post-meiotic sperm development depicting nuclei (green), mitochondria (red), and investment cones (purple). (B) Onion stage spermatids stained for DNA (PicoGreen: green), and mitochondria (mito-YFP: red). Each nucleus (n) is paired with a large mitochondrial structure, the nebenkern (mt). (C–E) Elongating spermatid bundles stained for DNA (PicoGreen: green) and actin (phalloidin: purple). (C) Elongation R stage bundle, 560 μm long. A cluster of normal spermatid nuclei (n), apically discarded spermatid nuclei (n'), and a somatic cell nucleus belonging to a cyst cell (n”) form large staining foci. mtDNA nucleoids (magnified in inset) form much smaller foci throughout the tail bundle. (D,E) Two late elongation stage bundles, 1729 μm (D) and 1747 μm (E) long. Numbers indicate distance (μm) of image from basal tip of bundle. Scale bars are 10 μm. (F) Nucleoid density (number of nucleoids/μm) along the lengths of 4 representative spermatid bundles (each bundle is a different colored line) of the indicated length (μm). (G) Average number of nucleoids per onion stage spermatid (O), elongating spermatid < 1700 μm (E1), elongating spermatid 1700–1800 μm (E2), and elongating spermatid > 1800 μm (E3). Error bars indicate standard deviation in a least 3 cysts. See also Fig. S2.
As spermatids begin to develop tails, the onion-shaped nebenkern of each spermatid resolves into two mitochondria that elongate next to the extending microtubular axoneme (Tokuyasu, 1975). The 64 axonemes and their associated mitochondria (128 total) elongate in parallel as a bundle. The tails grow enormously long, over 1,800 μm, and the two mitochondria of each spermatid extend the full length of each tail. The bundles elongate at a rate of 0.7 μm/min (Noguchi et al., 2011), and their length provides a measure of developmental progress. As elongation progressed, the nucleoids (average of 332 per spermatid: Fig. 2G) spread along extending mitochondria (Fig. 2C), becoming roughly uniform as tails approached their full length (Fig. 2F). However, bundles extending between 1,700 μm and 1,800 μm showed evidence of nucleoid disappearance. These bundles had few or no nucleoids in the basal (nuclear) end, fewer and fainter nucleoids midway along the tails, and unchanged nucleoid abundance at the apical end (Fig. 2D, F). More dramatically, some bundles longer than 1,750 μm (Fig 2E), and all bundles longer than 1,800 μm (Fig. 2G) lacked nucleoids along their entire length. We interpret this as a wave of mtDNA destruction that begins basally and progresses apically to completion during the final 100 μm of spermatid elongation: a period of about 70 minutes. The time of nucleoid elimination was marked by the recruitment of actin into perinuclear investment cones (Fig. 2D, E) that contribute to the subsequent individualization stage (Tokuyasu et al., 1972a). We conclude that mitochondrial nucleoids are eliminated in coordination with sperm elongation in D. melanogaster.
mtDNA Nucleoid Elimination During Spermatid Elongation Requires EndoG
We next characterized the mechanism that removes mtDNA during spermatogenesis. Since mitochondria are retained throughout sperm elongation (Tokuyasu, 1974), nucleoids are eliminated from within sperm mitochondria. We hypothesized that a nuclease would be involved, and we sought to identify it genetically. An informatics screen for candidate nucleases identified five homologous genes, including the mitochondrial nuclease, Endonuclease G, that have predicted mitochondrial-targeting signals. Four of the EndoG homologues were previously uncharacterized, and we name them Testis EndoG-Like (Tengl) 1–4 due to their testis-specific expression pattern (Fig. S3A). We made mutations that deleted the four Tengl genes (Fig. S3B–D), but even the quadruple mutant still eliminated nucleoids from the elongating spermatids on schedule (Fig. S3E–I). However, a mutation in the ubiquitously-expressed archetype of this group, EndoG (Temme et al., 2009), prevented scheduled nucleoid elimination. EndoGMB07150 is a mutant allele of EndoG created by a minos transposon insertion into protein coding sequences of EndoG (Fig3A). EndoGMB07150 was viable and fertile as a homozygote, or when transheterozygous with a large deletion that removes EndoG (EndoGMB07150/Df). RT-PCR showed that the transposon insertion eliminates full-length EndoG transcripts, but some 5' sequences persist (Fig. 3B). In EndoG mutant spermatid bundles, nucleoids failed to be eliminated from the apical portion of sperm tail bundles during spermatid elongation (Fig. 3C) and persisted throughout spermatid individualization (Fig. 3D), a 10 hour process (Noguchi and Miller, 2003) following sperm elongation. As a control to verify that the EndoGMB07150 transposon insertion caused nucleoids to persist, we precisely excised the transposon and observed that nucleoids disappeared as in wild type (Fig. 3D). In an additional control, we suppressed the EndoGMB07150 mutant phenotype by expressing an EndoG transgene in the germline of the EndoG mutant (Fig. 3D).
Figure 3. mtDNA Nucleoid Elimination During Spermatid Elongation Requires EndoG.
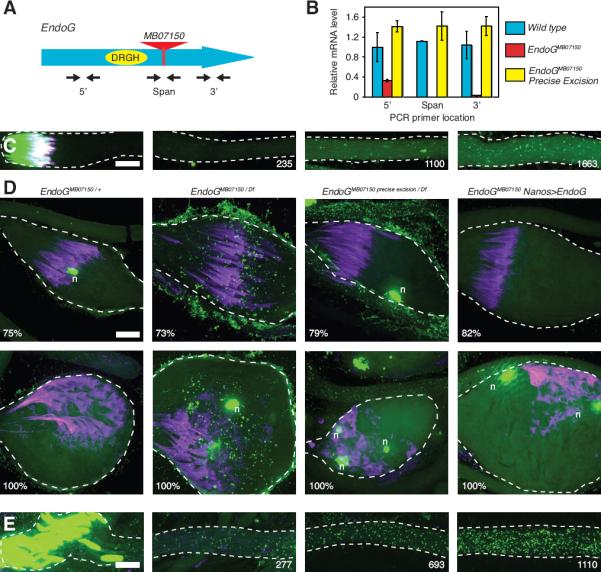
(A) Schematic showing the approximate location of the MB07150 minos transposon insertion (red triangle) in relation to the catalytic residues of EndoG (yellow oval). (B) Quantitative RT-PCR measuring EndoG mRNA abundance in adult male flies. The approximate positions of the 3 primer sets used to measure the abundance of different regions of EndoG mRNA are diagramed in (A). Error bars represent standard deviation in 3 extracts. (C–E) Spermatid bundles stained for DNA (green), and actin (purple). (C) Late elongation stage EndoGMB07150 mutant bundle, 1830 μm long. Numbers indicate distance (μm) of image from basal tip of bundle. (D) Cystic bulge of spermatid bundles of the indicated genotype during spermatid individualization. Numbers indicate % of the length of the bundle that the cystic bulge has traveled. Rare spermatid nuclei (n) culled during the individualization process. (E) Elongation stage EndoGI mutant bundle, 1280 μm long. Numbers indicate distance (μm) of image from basal tip of bundle. Scale bars are 10 μm. See also Fig. S3.
Because EndoG-dependent mtDNA elimination occurs at a precise time in sperm development, we hypothesized that EndoG activation might time mtDNA elimination. One way in which EndoG might be activated is by the removal of an inhibitor. D. melanogaster encodes an EndoG inhibitor (EndoGI), and a loss of function mutant EndoGI has been characterized (Temme et al., 2009). However, we did not observe premature nucleoid elimination elongating EndoGIe00915 mutant spermatids (Fig. 3E), suggesting that EndoGI does not regulate the timing of mtDNA elimination. Thus, the regulation of the developmentally timed EndoG-dependent nucleoid elimination process remains enigmatic.
A Backup Mode of mtDNA Elimination Associated with Spermatid Individualization
If mtDNA destruction during sperm elongation is the only barrier to paternal transmission, then mutation of EndoG might allow paternal transmission. However, qPCR failed to detect paternal mtDNA in eggs fertilized by EndoG mutant fathers or mtDNA in EndoG mutant mature sperm (Fig. S1). We conclude that elimination of mtDNA, while delayed, still occurs during EndoG mutant sperm development.
We next followed mtDNA nucleoids in EndoG mutant sperm to identify a backup mechanism responsible for their elimination. In contrast to wild type spermatids, nucleoids in EndoG mutants persisted beyond spermatid elongation to the individualization stage. During spermatid individualization, actin-containing investment cones, which initially form around each of the 64 spermatid nuclei, move apically in concert. As they travel along the axoneme, the cones invest spermatid tails in membrane, thereby “individualizing” the spermatids, while sweeping the tails clean of extraneous cytoplasm. The cones eliminate structures as small as ribosomes, and trim the mitochondrial mass, producing spherical mitochondrial structures as waste (Fig. 4D) (Tokuyasu et al., 1972a; 1972b). The investment cones, together with the waste they accumulate, traverse the length of the sperm tail in a structure termed the cystic bulge (CB), ultimately producing a waste bag (WB) that is extruded from the apical end of the tails. In the EndoG mutant, we observed mtDNA nucleoids ahead of (Fig. 4B”), but not behind (Fig. 4B) the CB. Furthermore, mtDNA nucleoids accumulated in the CB (Fig. 4B') as the CB traversed regions of the tail with residual mtDNA (Fig. 4C). Given the established capacities of the individualization process, and our observations, we propose a second mechanism for eliminating mtDNA from sperm mitochondria in which the passing individualization complex sweeps up remaining nucleoids and discards them in the waste bag. This second mode of mtDNA nucleoid elimination only became apparent once the early, EndoG-dependent mode of elimination was compromised in the EndoG mutant (Fig. 4E).
Figure 4. Residual mtDNA Nucleoids are Removed from EndoG Mutant Sperm During Spermatid Individualization.
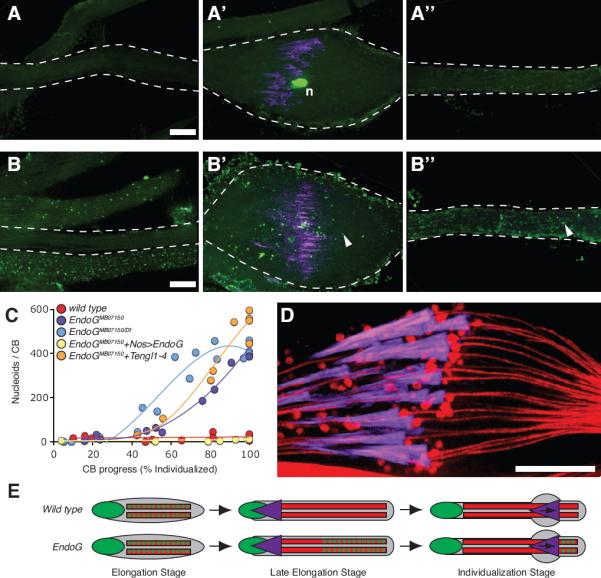
(A–A”,B–B”) Two individualizing sperm-tail bundles stained for DNA (green) and actin (purple). EndoGMB07150/+ (A–A”) and EndoGMB07150/Df (B–B”) bundles imaged ahead of the cystic bulge (A”, B”), at the cystic bulge (A', B') and behind the cystic bulge (A, B). Arrowheads indicate examples of mtDNA nucleoids. Scale bars are 10 μm. (C) Number of nucleoids in cystic bulges at different stages of individualization in bundles from control and EndoG mutants. (D) Focal section through a wild type cystic bulge stained for mitochondria (DJ–GFP: red) and actin (purple). (E) Schematic showing early mtDNA elimination in wild type and late mtDNA elimination in EndoG mutant spermatids; DNA (green), mitochondria (red), and investment cones (purple).
EndoG Mutant Sperm Have Residual Nuclease Activity
As we followed the fate of the mtDNA nucleoids in the EndoG mutant, we realized that the nucleoids are not fully stable. In addition to the disappearance of nucleoids from the basal (nuclear) region of the tail prior to individualization, the number of nucleoids collected by the CB never approached the total number of nucleoids per bundle, even though none were left behind (Fig. 4C). The residual elimination of nucleoids in the EndoG mutant might be due to other nucleases, and/or to residual activity of the EndoGMB07150 protein. To determine whether the four Tengl proteins might provide an alternative activity contributing to mtDNA elimination, we made flies mutant for all five EndoG orthologues and examined the disappearance of mtDNA nucleoids. Quantification of nucleoids showed that multiply mutated males behaved much like the EndoGMB07150 mutant (Fig. 4C). We conclude that the four Tengl proteins, contribution to residual elimination of mtDNA in the EndoG mutant.
To test whether EndoGMB07150 has residual activity, we compared the severity of the phenotype of flies homozygous for EndoGMB07150 (two copies of EndoGMB07150) to the phenotype of flies transheterozygous for EndoGMB07150 and a deficiency for the locus (one copy of EndoGMB07150). The deficiency transheterozygote showed a small increase in the number of nucleoids accumulated in the CB (Fig. 4C). This argues that EndoGMB07150 has residual activity, a portion of which is lost when one copy of the locus is removed. We conclude that residual EndoG activity is responsible for at least part of the residual destruction of mtDNA in our mutant. At present, it is not clear how much of the residual elimination of mtDNA is due to remaining activity of EndoGMB07150 and how much might be due to the action of additional, unidentified nucleases. Nonetheless, EndoG clearly contributes to the normally rapid and complete disappearance of mtDNA nucleoids.
Discussion
Our results demonstrate that in D. melanogaster, the paternal inheritance of mtDNA is restricted by mechanisms that remove mtDNA from developing sperm. In addition to our findings, mtDNA appears to be removed from developing sperm in other species. For example, mtDNA copy number declines during human spermatogenesis (Larsson et al., 1997), and the extremely low abundance of mtDNA (average of 1.4 copies per sperm) in highly purified sperm led to the suggestion that most human sperm lacked mtDNA (May-Panloup et al., 2003). The sperm of mice and the medaka fish,Oryzias latipes, also have reduced mtDNA levels (Nishimura et al., 2006; Shitara et al., 2000; Hecht et al., 1984). While it is unclear if mtDNA is eliminated from all animal sperm as efficiently as it is eliminated in D. melanogaster, it appears that other organisms also exhibit a developmental decline in mtDNA in spermatogenesis.
The molecular mechanisms that remove mtDNA from human, mouse, and medaka fish sperm are currently unknown. In C. reinhardtii a nuclease has been proposed to digest mtDNA to enforce uniparental mtDNA inheritance, but it has yet to be identified (Aoyama et al., 2006). In Arabidopsis thaliana (which lacks an EndoG homologue), a nuclease conserved in angiosperms eliminates mtDNA within mitochondria during pollen development (Matsushima et al., 2011).
Our work implicates the known mitochondrial nuclease, EndoG, in the early elimination of mtDNA during D. melanogaster spermatogenesis. Previous work in C. elegans showed that EndoG contributed to destruction of DNA in apoptotic corpses, and the enzyme was thought to be stockpiled with other pro-apoptotic proteins in the mitochondrial intermembrane space (Li et al., 2001). However, more recent studies questioned the role of EndoG in apoptosis, and argued that EndoG resides in the mitochondrial matrix, where it is proposed to participate in mtDNA replication, mtRNA processing, and mitochondrial biogenesis (David et al., 2006; Côté and Ruiz-Carrillo, 1993; McDermott-Roe et al., 2011). Our findings suggest a function for EndoG in removing mtDNA from within mitochondria, though future work will be necessary to fully understand the generality and regulation of this function.
In addition to EndoG-dependent mtDNA elimination, we found that a cellular remodeling process that trims and shapes the D. melanogaster sperm can remove residual mtDNA. In other species, the cytoplasm is similarly trimmed from developing spermatids. In mammals, this trimming discards at least some mitochondria from each spermatid into a residual body (Breucker et al., 1985; Hecht et al., 1984), suggesting that cellular remodeling contributes to low mtDNA levels in the sperm of other animals.
Most barriers to paternal mtDNA transmission have previously been proposed to act at, or following zygote formation. For example, simple dilution of the small allotment of sperm mtDNA by the large egg contribution is hypothesized to passively limit paternal mtDNA inheritance (Alberts et al., 1994). Furthermore, selective destruction of paternal mitochondria occurs in the early zygotes of several animals (Sutovsky et al., 1999; Sato and Sato, 2011; Rawi et al., 2011). Additionally, unusual cases of paternal mtDNA transmission in interspecies crosses suggest post-fertilization restriction of paternal mtDNA inheritance (Kaneda et al., 1995; Kondo et al., 1990). However, effective mtDNA elimination during D. melanogaster spermatogenesis prevented us from evaluating the contribution of later zygotic processes to paternal mtDNA elimination. Additionally, we could not bypass the pre-zygotic mechanisms. since the individualization process, which removes mtDNA, is also required for sperm production. Importantly, whether or not other barriers to paternal transmission of mtDNA exist, the elimination of mtDNA from developing sperm removes the potential for paternal contribution to mitochondrial inheritance.
In concluding, we would like to consider why mechanisms might have evolved to eliminate mtDNA from sperm. Since biparental inheritance of mtDNA is rare, it presumably carries a disadvantage. Consequently, a sperm that transmits its mtDNA would put the resulting zygote at a disadvantage, while a sperm that eliminated its mtDNA would produce a more successful zygote. The advantage conferred to sperm without mtDNA might provide an evolutionary drive that would act widely to promote the evolution of mechanisms acting in spermatogenesis to limit paternal contributions of mtDNA to progeny.
Materials and Methods
Assaying paternal mtDNA in embryos
To detect paternal mtDNA in the zygote with high sensitivity, we needed many recently fertilized eggs. We collected 300 to 3000 young embryos per experiment, but counting them was prohibitively slow. We used total (maternal) mtDNA copy number in the final DNA preparation to back calculate the number of input embryos. Note that mtDNA copy number remains constant over early embryonic development (Rubenstein et al., 1977)(our unpublished results).
To establish a standard for mtDNA copy number per embryo, 100 mt:ND2del1 embryos were counted and mechanically homogenized with a plastic pestle in 100 μl Homogenization Buffer (HB) 100 mM Tris HCl pH 8.8, 0.5 mM EDTA, 1% SDS. The homogenate was incubated at 65°C for 30 min, treated with 5 μg/ml RNAse A for 30 min at 37°C, and with 0.4 mg/ml Proteinase K for 30 min at 55°C. Proteinase K was inactivated by incubation at 95°C for 2 minutes. Addition of potassium acetate (to 1M) and incubation on ice (30 min) precipitated protein and SDS. DNA was recovered from the supernatant (20,000 × g, 15 min at 4°C) by isopropanol precipitation (0.5 volumes) followed by a wash with 70% ethanol and resuspension in 100 μl water. DNA recovery estimated for an added standard, pUC19, approached 100%. We used different amounts of extract in qPCR experiment to generate a standard curve for qPCR signal/mtDNA copy number versus embryo number.
mt:ND2del1 females were crossed to either wild-type (mt:ND2), EndoG (mt:ND2), or control mt:ND2del1 males. Embryos were collected for 2 hours on standard grape juice agar plates and dechorionated with 50% bleach for 2 min. DNA was prepared as above from 300 to 3000 embryos, except that for this larger scale, we extracted residual protein using phenol:choloroform:isoamyl alcohol extraction followed by 2 chloroform washes before precipitation of DNA. The DNA was precipitated by adding 2 volumes of ethanol on ice and washed with 70% ethanol before resuspension in water. The number of embryos in each sample was determined by qPCR of total mtDNA, and then each sample was diluted to 30 embryo equivalents per μl.
The amount of nucleic acid obtained from 300 embryos binds enough SYBR Green dye to interfere with qPCR quantification. To circumvent this, we used a preliminary PCR without dye. Three hundred embryo equivalents were added to a 50 ul PCR reaction: 400 nM primers, 95° 2 min, 20 cycles of 95°C 30 sec, 48°C 60 sec. The product was then diluted 1:10,000 into a qPCR reaction containing SYBR Green detailed below.
DNA preparation from female reproductive tracts to measure mtDNA copy number in mature sperm
For each cross, 25 two-day-old virgin females were mated to 25 two-day-old virgin males for 15 hours at 25°C. A portion of the female reproductive tract containing the sperm storage organs was dissected in PBS, with care to exclude the ovaries and any embryos in the uterus (which may be fertilized). Pools of 20 female reproductive tracts were then mechanically homogenized in HB, and DNA was prepared as described above.
qPCR parameters
For all qPCR assays, SYBR Green PCR Master Mix (Applied Biosystems) was used in 50 μl reactions with 400 nM of each primer.
To measure the concentration of the DNA standards used to assemble the standard curves, 2-fold serial dilutions of the purified PCR product used to construct each standard curve were run next to 2-fold serial dilutions of a DNA standard of known concentration (100 bp DNA ladder, NEB #N3231S) on an ethidium bromide-contatining agarose gel and the intensities of similarly sized bands were compared.
To measure paternal mtDNA in fertilized embryos or mated females, qPCR of mt:ND2 was performed with the following reaction conditions: 95°C 10 min, 50 cycles of 95°C 30 sec, 48°C 60 sec. Standard curves were constructed by adding 100 to 12500 molecules of DNA standards to the appropriate mt:ND2del1 control extract (either mt:ND2del1 embryos fertilized by mt:ND2del1 males or reproductive tracts from virgin mt:ND2del1 females).
To measure the amount of total mtDNA and pUC19 DNA in embryo extracts, or the number of Y chromosomes in mated female extracts, qPCR was performed with the following reaction conditions: 95°C 10 min, 40 cycles of 95°C 30 sec, 55°C 30 sec, 68°C 60 sec. Standard curves were constructed by adding 100 to 12500 molecules of DNA standards or pUC19 plasmid in the appropriate mt:ND2del1 control extract (either mt:ND2del1 embryos fertilized by mt:ND2del1 males or reproductive tracts from virgin mt:ND2del1 females).
qRT-PCR
5 adult male flies per genotype were homogenized in 500 μl Trizol and total RNA was prepared according to the manufacturer's instructions. cDNA was synthesized from 1 fly equivalent of total RNA using a poly-T primer and Superscript II reverse transcriptase. 30μl qPCR reactions were performed with 0.002 fly equivalents of cDNA, 400 nM of each primer, and SYBR Green PCR Master Mix. Reaction conditions for all primer pairs were 95°C 10 min, 45 cycles of 95°C 30 sec, 55°C 1 min. A standard curve of varying inputs was created for each primer set using 0.02 to 0.00002 wild type fly equivalents of cDNA. The relative abundance of EndoG RNA in each sample was normalized to the abundance of control RpA1 RNA in each sample.
Fixed staining of spermatid cysts
Spermatid staining was performed similarly to (Arama et al., 2003). Testes were dissected from 1–2 day old adult males in ice-cold TB (10mM Tris pH6.8, 183mM KCl, 47mM NaCl, 1mM EDTA) and transferred to a drop of TB on a siliconized coverslip. The base of the testis was severed with a tungsten needle and the contents extruded using a hand-pulled glass capillary tube as a squeegee. The sample was then sandwiched between a poly-lysine treated slide and a coverslip and frozen in liquid nitrogen. The coverslip was popped off with a razor blade and the slide (containing the sample) plunged into ice-cold ethanol. The sample was fixed with 3.7% formaldehyde in PBS for 20 min, washed (PBS, 5 minutes for all washes), permeabilized in PBS + 0.1% Triton×100 for 30 min, washed, treated with 5 μg/ml RNAse A in PBS + 0.1% Tween 20 for 2 h at 37°C, washed, and stained with PicoGreen (1:200) and Rhodamine-Phalloidin (1:200) in PBS for 30 minutes. Samples were washed twice before mounting in Vectashield medium. For antibody staining of mito-YFP flies, rabbit anti-GFP (1:500) (Invitrogen, A11122) was added coincidently with RNAseA, and Alexa546 goat anti-rabbit secondary antibody (1:1000) (Invitrogen, A11010) was added coincidently with PicoGreen.
Image acquisition and processing
Images were acquired on a spinning disc confocal microscope, processed with Volocity 5 software, and presented as a collapsed z-stack. Quantification of mtDNA nucleoid number and spermatid cyst length were performed with Volocity 5 Measurements module. To determine the number of nucleoids in an elongating spermatid bundle, nucleoid density was measured at various points along the length of the bundle and plotted as in Fig. 2F. Then, the area under the curve (which represents nucleoid number per 64 cell bundle) was calculated using Adobe Photoshop.
Supplementary Material
Acknowledgements
This work was supported by the United Mitochondrial Disease Foundation and NIH (GM086854) funding to PHO'F. SZD was supported by a Larry L. Hillblom Foundation award. We gratefully acknowledge the Bloomington Stock Center, The Exelixis Collection at Harvard, and members of the O'Farrell lab, especially Hong Xu, for discussion.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson JD, Hesketh-Moore K. Molecular Biology of the Cell. Third Edition 3rd ed. Garland Science; 1994. [Google Scholar]
- Aoyama H, Hagiwara Y, Misumi O, Kuroiwa T, Nakamura S. Complete elimination of maternal mitochondrial DNA during meiosis resulting in the paternal inheritance of the mitochondrial genome in Chlamydomonas species. Protoplasma. 2006;228:231–242. doi: 10.1007/s00709-006-0155-5. [DOI] [PubMed] [Google Scholar]
- Arama E, Agapite J, Steller H. Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev Cell. 2003;4:687–697. doi: 10.1016/s1534-5807(03)00120-5. [DOI] [PubMed] [Google Scholar]
- Ashley N, Harris D, Poulton J. Detection of mitochondrial DNA depletion in living human cells using PicoGreen staining. Exp Cell Res. 2005;303:432–446. doi: 10.1016/j.yexcr.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Bate M, Arias AM. The development of Drosophila melanogaster. Cold Spring Harbor Laboratory Pr; 2009. [Google Scholar]
- Birky CW. Uniparental inheritance of mitochondrial and chloroplast genes: mechanisms and evolution. Proc Natl Acad Sci USA. 1995;92:11331–11338. doi: 10.1073/pnas.92.25.11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc H, Dujon B. Replicator regions of the yeast mitochondrial DNA responsible for suppressiveness. Proc Natl Acad Sci USA. 1980;77:3942–3946. doi: 10.1073/pnas.77.7.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton JE, Harris EH, Burkhart BD, Lamerson PM, Gillham NW. Transmission of mitochondrial and chloroplast genomes in crosses of Chlamydomonas. Proc Natl Acad Sci USA. 1987;84:2391–2395. doi: 10.1073/pnas.84.8.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breucker H, Schäfer E, Holstein AF. Morphogenesis and fate of the residual body in human spermiogenesis. Cell and Tissue Research. 1985;240:303–309. doi: 10.1007/BF00222339. [DOI] [PubMed] [Google Scholar]
- Côté J, Ruiz-Carrillo A. Primers for mitochondrial DNA replication generated by endonuclease G. Science. 1993;261:765–769. doi: 10.1126/science.7688144. [DOI] [PubMed] [Google Scholar]
- David KK, Sasaki M, Yu S-W, Dawson TM, Dawson VL. EndoG is dispensable in embryogenesis and apoptosis. Cell Death Differ. 2006;13:1147–1155. doi: 10.1038/sj.cdd.4401787. [DOI] [PubMed] [Google Scholar]
- de Zamaroczy M, Marotta R, Faugeron-Fonty G, Goursot R, Mangin M, Baldacci G, Bernardi G. The origins of replication of the yeast mitochondrial genome and the phenomenon of suppressivity. Nature. 1981;292:75–78. doi: 10.1038/292075a0. [DOI] [PubMed] [Google Scholar]
- Eberhard WG. Intraorganism competition involving eukaryotic organelles. Ann N Y Acad Sci. 1981;361:44–52. doi: 10.1111/j.1749-6632.1981.tb46510.x. [DOI] [PubMed] [Google Scholar]
- Ephrussi B, de Margerie-Hottinguer H, Roman H. SUPPRESSIVENESS: A NEW FACTOR IN THE GENETIC DETERMINISM OF THE SYNTHESIS OF RESPIRATORY ENZYMES IN YEAST. Proc Natl Acad Sci USA. 1955;41:1065–1071. doi: 10.1073/pnas.41.12.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht NB, Liem H, Kleene KC, Distel RJ, Ho SM. Maternal inheritance of the mouse mitochondrial genome is not mediated by a loss or gross alteration of the paternal mitochondrial DNA or by methylation of the oocyte mitochondrial DNA. Dev Biol. 1984;102:452–461. doi: 10.1016/0012-1606(84)90210-0. [DOI] [PubMed] [Google Scholar]
- Hoekstra RF. Evolutionary origin and consequences of uniparental mitochondrial inheritance. Hum. Reprod. 2000;15(Suppl 2):102–111. doi: 10.1093/humrep/15.suppl_2.102. [DOI] [PubMed] [Google Scholar]
- Kaneda H, Hayashi J, Takahama S, Taya C, Lindahl KF, Yonekawa H. Elimination of paternal mitochondrial DNA in intraspecific crosses during early mouse embryogenesis. Proc Natl Acad Sci USA. 1995;92:4542–4546. doi: 10.1073/pnas.92.10.4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo R, Satta Y, Matsuura ET, Ishiwa H, Takahata N, Chigusa SI. Incomplete maternal transmission of mitochondrial DNA in Drosophila. Genetics. 1990;126:657–663. doi: 10.1093/genetics/126.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson NG, Oldfors A, Garman JD, Barsh GS, Clayton DA. Down-regulation of mitochondrial transcription factor A during spermatogenesis in humans. Hum Mol Genet. 1997;6:185–191. doi: 10.1093/hmg/6.2.185. [DOI] [PubMed] [Google Scholar]
- Li LY, Luo X, Wang X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature. 2001;412:95–99. doi: 10.1038/35083620. [DOI] [PubMed] [Google Scholar]
- Matsushima R, Tang LY, Zhang L, Yamada H, Twell D, Sakamoto W. A Conserved, Mg2+-Dependent Exonuclease Degrades Organelle DNA during Arabidopsis Pollen Development. Plant Cell. 2011 doi: 10.1105/tpc.111.084012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May-Panloup P, Chrétien M-F, Savagner F, Vasseur C, Jean M, Malthièry Y, Reynier P. Increased sperm mitochondrial DNA content in male infertility. Hum. Reprod. 2003;18:550–556. doi: 10.1093/humrep/deg096. [DOI] [PubMed] [Google Scholar]
- McDermott-Roe C, Ye J, Ahmed R, Sun X-M, Serafín A, Ware J, Bottolo L, Muckett P, Cañas X, Zhang J, et al. Endonuclease G is a novel determinant of cardiac hypertrophy and mitochondrial function. Nature. 2011;478:114–118. doi: 10.1038/nature10490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y, Yoshinari T, Naruse K, Yamada T, Sumi K, Mitani H, Higashiyama T, Kuroiwa T. Active digestion of sperm mitochondrial DNA in single living sperm revealed by optical tweezers. Proc Natl Acad Sci USA. 2006;103:1382–1387. doi: 10.1073/pnas.0506911103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Miller KG. A role for actin dynamics in individualization during spermatogenesis in Drosophila melanogaster. Development. 2003;130:1805–1816. doi: 10.1242/dev.00406. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Koizumi M, Hayashi S. Sustained elongation of sperm tail promoted by local remodeling of giant mitochondria in Drosophila. Curr. Biol. 2011;21:805–814. doi: 10.1016/j.cub.2011.04.016. [DOI] [PubMed] [Google Scholar]
- Perotti ME. The mitochondrial derivative of the spermatozoon of Drosophila before and after fertilization. Journal of Ultrastructure Research. 1973;44:181–198. doi: 10.1016/s0022-5320(73)80055-3. [DOI] [PubMed] [Google Scholar]
- Rawi Al, S., Louvet-Vallée S, Djeddi A, Sachse M, Culetto E, Hajjar C, Boyd L, Legouis R, Galy V. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 2011;334:1144–1147. doi: 10.1126/science.1211878. [DOI] [PubMed] [Google Scholar]
- Rubenstein JL, Brutlag D, Clayton DA. The mitochondrial DNA of Drosophila melanogaster exists in two distinct and stable superhelical forms. Cell. 1977;12:471–482. doi: 10.1016/0092-8674(77)90123-4. [DOI] [PubMed] [Google Scholar]
- Sato M, Sato K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science. 2011;334:1141–1144. doi: 10.1126/science.1210333. [DOI] [PubMed] [Google Scholar]
- Shitara H, Kaneda H, Sato A, Inoue K, Ogura A, Yonekawa H, Hayashi JI. Selective and continuous elimination of mitochondria microinjected into mouse eggs from spermatids, but not from liver cells, occurs throughout embryogenesis. Genetics. 2000;156:1277–1284. doi: 10.1093/genetics/156.3.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutovsky P, Navara C. Fate of the sperm mitochondria, and the incorporation, conversion, and disassembly of the sperm tail structures during bovine fertilization. Biol Reprod. 1996 doi: 10.1095/biolreprod55.6.1195. [DOI] [PubMed] [Google Scholar]
- Sutovsky P, Moreno RD, Ramalho-Santos J, Dominko T, Simerly C, Schatten G. Ubiquitin tag for sperm mitochondria. Nature. 1999;402:371–372. doi: 10.1038/46466. [DOI] [PubMed] [Google Scholar]
- Temme C, Weissbach R, Lilie H, Wilson C, Meinhart A, Meyer S, Golbik R, Schierhorn A, Wahle E. The Drosophila melanogaster Gene cg4930 Encodes a High Affinity Inhibitor for Endonuclease G. J Biol Chem. 2009;284:8337–8348. doi: 10.1074/jbc.M808319200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyasu K. Dynamics of spermxsiogenesis in Drosophila melanogaster* 1:: III. Relation between axoneme and mitochondrial derivatives. Exp Cell Res. 1974 doi: 10.1016/0014-4827(74)90402-9. [DOI] [PubMed] [Google Scholar]
- Tokuyasu K. Dynamics of spermiogenesis in Drosophila melanogaster. VI. Significance of. Journal of Ultrastructure Research. 1975 doi: 10.1016/s0022-5320(75)80089-x. [DOI] [PubMed] [Google Scholar]
- Tokuyasu KT, Peacock WJ, Hardy RW. Dynamics of spermiogenesis in Drosophila melanogaster. I. Individualization process. Z Zellforsch Mikrosk Anat. 1972a;124:479–506. doi: 10.1007/BF00335253. [DOI] [PubMed] [Google Scholar]
- Tokuyasu KT, Peacock WJ, Hardy RW. Dynamics of spermiogenesis in Drosophila melanogaster. II. Coiling process. Z Zellforsch Mikrosk Anat. 1972b;127:492–525. doi: 10.1007/BF00306868. [DOI] [PubMed] [Google Scholar]
- Xu H, DeLuca SZ, O'Farrell PH. Manipulating the metazoan mitochondrial genome with targeted restriction enzymes. Science. 2008;321:575–577. doi: 10.1126/science.1160226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouros E, Oberhauser Ball A, Saavedra C, Freeman KR. An unusual type of mitochondrial DNA inheritance in the blue mussel Mytilus. Proc Natl Acad Sci USA. 1994;91:7463–7467. doi: 10.1073/pnas.91.16.7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.